Abstract
Objectives
Host genetic factors contribute to the variable severity of COVID-19. We examined genetic variants from genome-wide association studies and candidate gene association studies in a cohort of patients with COVID-19 and investigated the role of early SARS-CoV-2 strains in COVID-19 severity.
Methods
This case-control study included 123 COVID-19 cases (hospitalized or ambulatory) and healthy controls from the state of Baden-Wuerttemberg, Germany. We genotyped 30 single nucleotide polymorphisms, using a custom-designed panel. Cases were also compared with the 1000 genomes project. Polygenic risk scores were constructed. SARS-CoV-2 genomes from 26 patients with COVID-19 were sequenced and compared between ambulatory and hospitalized cases, and phylogeny was reconstructed.
Results
Eight variants reached nominal significance and two were significantly associated with at least one of the phenotypes “susceptibility to infection”, “hospitalization”, or “severity”: rs73064425 in LZTFL1 (hospitalization and severity, P <0.001) and rs1024611 near CCL2 (susceptibility, including 1000 genomes project, P = 0.001). The polygenic risk score could predict hospitalization. Most (23/26, 89%) of the SARS-CoV-2 genomes were classified as B.1 lineage. No associations of SARS-CoV-2 mutations or lineages with severity were observed.
Conclusion
These host genetic markers provide insights into pathogenesis and enable risk classification. Variants which reached nominal significance should be included in larger studies.
Keywords: SARS-CoV-2, COVID-19, Host genetics, Polygenic risk score, GWAS, Cytokine storm
Introduction
High variability in the clinical presentation of COVID-19 has consistently been described (World Health Organization, 2021). Most patients experience asymptomatic infection or mild disease only, whereas 14% suffer from severe and 5% from critical disease (Hu et al., 2021). The most common symptoms include fever, dry cough, and fatigue; ageusia, anosmia, and gastrointestinal symptoms are also observed (Hu et al., 2021; Velavan and Meyer, 2020a). Severe COVID-19 is characterized by respiratory failure, requiring mechanical ventilation or high-flow oxygen (Table 1 , WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection, 2020). The infection fatality rate has been estimated at 0.36% in Germany (Streeck et al., 2020) and at 0.68% worldwide (Meyerowitz-Katz and Merone, 2020). A cytokine storm is common in severe and critical COVID-19 (Yokota et al., 2021), presenting with an increase in proinflammatory cytokines and inflammatory markers (Velavan and Meyer, 2020b; Velavan et al., 2021a) and causing coagulopathies, oxidative stress, organ damage, and, eventually, death (Fajgenbaum and June, 2020). Although older age, male sex, and concomitant noncommunicable diseases increase the risk of severe COVID-19 courses (Boutin et al., 2021; Phua et al., 2020), severe disease may also occur in younger, healthy individuals, raising the question of what other factors might contribute to disease susceptibility and severity. Distinct ethnic backgrounds have also been found to influence severity, even when controlling for socioeconomic factors (Gu et al., 2020; Mathur et al., 2021). Therefore, human genetic variants might partly determine the course of the disease.
Table 1.
The WHO clinical progression score.
| Patient state | Descriptor | Score |
|---|---|---|
| Uninfected | Uninfected; no viral RNA detected | 0 |
| Ambulatory: mild disease | Asymptomatic; viral RNA detected | 1 |
| Symptomatic; independent | 2 | |
| Symptomatic; assistance needed | 3 | |
| Hospitalized: moderate disease | Hospitalized; no oxygen therapy | 4 |
| Hospitalized; oxygen by mask or nasal prongs | 5 | |
| Hospitalized: severe disease | Hospitalized; oxygen by NIV or high-flow | 6 |
| Intubation and mechanical ventilation, pO2/FiO2 ≥ 150 or | 7 | |
| SpO2/FiO2 ≥ 200 | ||
| Mechanical ventilation pO2/FiO2 <150 (SpO2/FiO2 <200) or vasopressors | 8 | |
| Mechanical ventilation pO2/FiO2 <150 and vasopressors, dialysis, or ECMO | 9 | |
| Dead | Dead | 10 |
NIV = noninvasive ventilation. ECMO = extracorporeal membrane oxygenation. pO2 = partial pressure of oxygen. FiO2 = fraction of inspired oxygen. SpO2 = oxygen saturation. ECMO = extracorporeal membrane oxygenation. Patients hospitalized for isolation only are recorded as ambulatory patients. Adapted from WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection (Lancet Infect dis. 2020).
To understand the role of host genetics on COVID-19 susceptibility, severity, and the requirement of hospitalization, genome-wide association studies (GWASs) as well as candidate gene studies have indicated several loci, which can be linked to COVID-19 pathogenesis (Ovsyannikova et al., 2020; Velavan et al., 2021b; Yildirim et al., 2021). Robust studies were performed by the “COVID-19 Host Genetics Initiative” (COVID-19 Host Genetics Initiative, 2021), the “Genetics Of Mortality In Critical Care” (Pairo-Castineira et al., 2021), and the “COVID Human Genetic Effort” (Zhang et al., 2020a) initiatives and by independent working groups as well as commercial genomics service providers, such as “23andMe” (Shelton et al., 2021) and “AncestryDNA” (Roberts et al., 2020). Meta-analyses of GWAS datasets have yielded high-confidence loci, such as 3p21.31, which are associated with susceptibility to and severity of COVID-19, which are increasingly corroborated by mechanistic studies (Velavan et al., 2021b).
GWAS results have enabled the construction of polygenic risk scores (PRSs) for COVID-19 (Dite et al., 2021; Horowitz et al., 2021; Huang et al., 2022; Powell et al., 2021). A PRS estimates a trait or disease risk on the basis of individual genetic profiles. Summing up the effect sizes of distinct variants can explain a larger portion of the individual risk (Velavan et al., 2021b). Identification of host genetic variants and risk scores, together with demographic and clinical parameters, could allow to predict a patient's course of disease, choose treatment strategies, and support the design of drug candidates or repurposing of available medicines.
In this case-control study, we described factors relevant in the early phase of the pandemic in southern Germany. We aimed to replicate GWAS and candidate gene variants previously found associated with COVID-19, SARS, other respiratory infections, and pneumonia using a custom-designed single nucleotide polymorphism (SNP) panel. Variants identified were used to develop a PRS and phylogeny of the viral genomes was assessed.
Methods
Study population
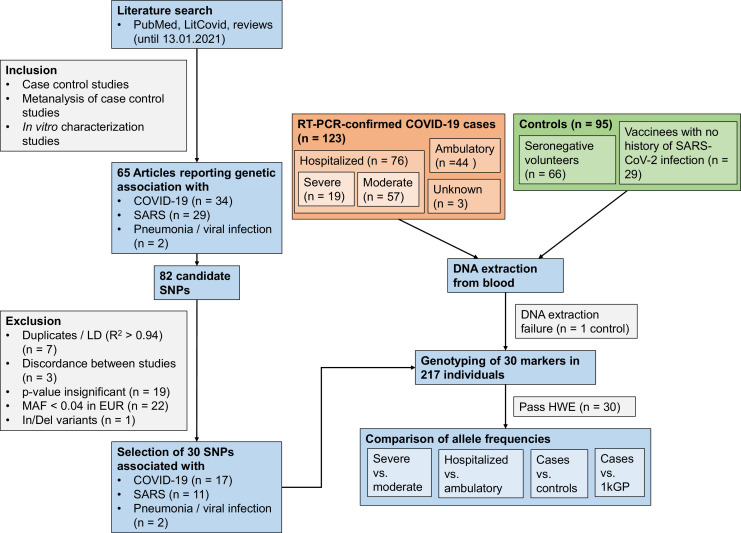
The study population included 123 COVID-19 cases and 94 healthy controls. The 123 patients came from the district of Tübingen, Baden-Wuerttemberg, Germany. Of them, 76 were hospitalized and 44 were ambulatory patients. For three patients, no information on hospitalization was available; these patients were included only in susceptibility analyses. On the basis of the need for mechanical ventilation or the WHO severity score (Table 1; WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection, 2020), hospitalized patients were classified as severe (WHO score ≥6 or mechanical ventilation) and a mild disease subset (WHO score 4-5 or no mechanical ventilation), providing subgroups of 19 and 57 patients, respectively. The SARS-CoV-2 PCR was performed in patients (March 23, 2020 to May 20, 2020). Of the 94 healthy controls, 65 participants were from Tübingen, Germany and were seronegative for SARS-CoV-2 at the time of sample collection (May 26, 2020 to March 16, 2021) (Becker et al., 2021). An additional 29 participants from Heidelberg, Germany, were included, who had no history of SARS-CoV-2 infection. An additional susceptibility analysis was carried out using the European cohort of the 1000 genomes project (1kGP) as control data (Fig. 1 ).
Fig. 1.
Flowchart illustrating the study design.
SNP = single nucleotide polymorphism. LD = linkage disequilibrium. MAF = minor allele frequency. In/Del = Insertion/Deletion. HWE = Hardy-Weinberg equilibrium. 1kGP = 1000 genomes project.
Prioritization of SNPs
To prioritize distinct SNP variants, a PubMed and LitCovid search was conducted until Januray 13, 2021 (Supplementary methods, Supplementary table 1, Fig. 1).
DNA isolation and genotyping
DNA was isolated from blood (QIAamp Blood DNA Mini Kit or FlexiGene DNA Kit; Qiagen, Hilden, Germany). Quantity and quality of DNA were assessed (NanodropTM and QubitTM 4 fluorometer; Thermo Fisher Scientific, Darmstadt, Germany) and the DNA concentration was verified by double measurement. Genotyping was performed with TaqMan® SNP Genotyping Assays (Pallerla et al., 2021) using 5 ng of genomic DNA per assay. Data were loaded into the TaqMan® Genotyper Software v1.5.0 (Thermo Fisher Scientific) to retrieve genotyping calls automatically.
SARS-CoV-2 screening, sequencing; phylogenetic analysis
RNA was extracted from oro-/nasopharyngeal swabs (QIAamp Viral RNA Mini Kit; Qiagen, Hilden, Germany) and SARS-CoV-2 infection was screened by reverse transcription–PCR (Ntoumi et al., 2021). All positive samples with Ct values <30 were subjected to next-generation sequencing using Illumina and Oxford Nanopore Sequencing platforms, as described previously (Ntoumi et al., 2021). All genomes were deposited in GISAID (https://www.gisaid.org/; Supplementary table 2). The Nextclade tool (https://clades.nextstrain.org) was used to identify nucleotide, and amino acid substitutions and lineages were obtained using the Pangolin online tool (https://cov-lineages.org/resources/pangolin.html) (Rambaut et al., 2020).
The maximum likelihood phylogenetic tree was reconstructed using 26 SARS-CoV-2 genomes from this study by applying 1000 bootstrapping iterations using MEGA X (Kumar et al., 2018). From the same period, SARS-CoV-2 genomes from the federal state of Baden-Wuerttemberg, Germany (seven sequences representative per week for the region or all sequences if less than seven in a week, n = 69) and from Germany and neighboring European countries from the “nextregions” dataset (n = 67) retrieved from GISAID were included, as was the Wuhan reference sequence (NC_045512.2). The tree was displayed with the Interactive Tree of Life tool (Letunic and Bork, 2019).
Statistical analysis
Statistical analyses were performed in R (version 4.0.5, Supplementary methods). For the construction of a PRS, data were split into a training and a testing set (70:30 ratio). Four exploratory versions of the PRS were calculated for susceptibility and hospitalization phenotypes, using the P-value of the association in the log-additive model as selection criterion for SNPs to include in the score (see Supplementary methods).
Results
A total of 123 cases and 94 healthy controls were included for genotyping (Table 2 ). The mean age and the proportion of men were higher in patients than in controls. The sex distribution was similar between controls and ambulatory patients. Age was higher in hospitalized than in ambulatory patients. All SNPs passed Hardy-Weinberg equilibrium (P >0.05, Supplementary table 3) and were included in the analyses. First, we looked at the phenotypes “susceptibility to SARS-CoV-2 infection”, “hospitalization”, and “severity of COVID-19”.
Table 2.
Demographic characteristics of the patients with COVID-19 and controls.
| Patients with COVID-19 (n = 123) |
||||||
|---|---|---|---|---|---|---|
| Ambulatory (n = 44) | Moderate (n = 57) | Severe (n = 19) | All (n = 123) | Controls (n = 94) | All (n = 217) | |
| Sex | ||||||
| Female (%) | 27 (61%) | 25 (44%) | 7 (37%) | 59 (48%) | 60 (64%) | 119 (55%) |
| Male (%) | 17 (39%) | 30 (53%) | 12 (63%) | 62 (50%) | 34 (36%) | 96 (44%) |
| Missing (%) | NA | 2 (3%) | NA | 2 (2%) | NA | 2 (1%) |
| Age | ||||||
| Mean (SD) | 47 (14) | 65 (14) | 69 (17) | 59 (17) | 33 (12) | 47 (20) |
| Median [Min-Max] | 48 [18 - 71] | 65 [28 - 91] | 76 [32 - 88] | 59 [18 - 91] | 29 [17 - 67] | 45 [17 - 91] |
| SARS-CoV-2 genome sequenced (%) | 3 (7%) | 16* (28%) | 7* (35%) | 26 (21%) | NA | NA |
SD = standard deviation. * Including one patient who was not SNP-genotyped because no blood sample was available
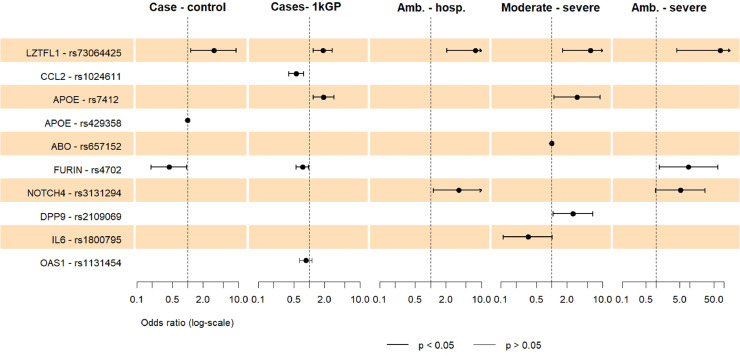
In these comparisons of patients with healthy controls, associations of two variants could be confirmed after Bonferroni correction. rs73064425 in the LZTFL1 gene reached significance in the hospitalization analysis and when comparing ambulatory and severe hospitalized patients. It was also nominally associated with susceptibility and severity (Table 3 , Fig. 2 ). The T allele was a risk allele for hospitalization and severity. For susceptibility to infection, the overdominant model of inheritance was significant, indicating C/T as risk genotype. However, the dominant model also reached nominal significance, where the T allele was associated with a greater risk for SARS-CoV-2 infection (OR = 2.19, 95% CI = 1.08-4.46, P = 0.026).
Table 3.
Association of human genetic variants with susceptibility to COVID-19 and phenotype severity.
| Gene | SNP ID | Susceptibility |
Susceptibility (1kGP) |
Hospitalization |
Severity |
Ambulatory vs. severe disease |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic model | ORadj (95% CI) | padj | Genetic model | OR (95% CI) | p | Genetic model | ORadj (95% CI) | padj | Genetic model | ORadj (95% CI) | padj | Genetic model | ORadj (95% CI) | padj | ||
| LZTFL1 | rs73064425 C/T | Overdominant (CT) | 3.24 (1.16 - 8.99) | 0.020 | Allelic (T) | 1.92 (1.20 - 3.07) | 0.006 | Log-additive (T) | 7.75 (2.1 - 28.55) | 0.0004 | Log-additive (T) | 5.78 (1.66 -20.14) | 0.003 | Log-additive (T) | 78.36 (4.16 – Inf) | 2.26 × 10−5 |
| CCL2 | rs1024611 A/G | Allelic (G) | 0.56 (0.39 - 0.78) | 0.001 | ||||||||||||
| APOE | rs7412 C/T | Allelic (T) | 4 (1.3 - 12.33) | 0.006 | Log-additive (T) | 3.16 (1.1 - 9.04) | 0.030 | |||||||||
| APOE | rs429358 C/T | Recessive (CC) | 1 (1-1) | 0.042 | ||||||||||||
| ABO | rs657152 A/C | Recessive (AA) | 1 (1-1) | 0.020 | ||||||||||||
| FURIN | rs4702 A/G | Dominant (AG – GG) | 0.43 (0.19 - 0.95) | 0.036 | Allelic (G) | 0.74 (0.56 - 0.99) | 0.042 | Recessive (GG) | 9.10 (1.25 - 66.18) | 0.022 | ||||||
| NOTCH4 | rs3131294 A/G | Log-additive (A) | 3.6 (1.14 - 11.4) | 0.016 | Log-additive (A) | 5.23 (0.97 - 28.16) | 0.034 | |||||||||
| DPP9 | rs2109069 G/A | Log-additive (A) | 2.61 (1.07 - 6.37) | 0.030 | ||||||||||||
| IL6 | rs1800795 C/G | Overdominant (CG) | 0.34 (0.11 - 1.01) | 0.049 | ||||||||||||
| OAS1 | rs1131454 A/G | Codominant (AG / GG) | AG: 0.57 (0.36 - 0.89)GG: 0.89 (0.52 - 1.52) | 0.035 | ||||||||||||
OR = odds ratio. 95% CI = 95% confidence interval. adj = P-value, OR and 95% CI after adjustment for age and sex in the logistic regression model. P-values which remained significant after Bonferroni correction are indicated in bold. 1kGP = 1000 genomes project. LZTFL1 = leucine zipper transcription factor like 1. APOE = Apoprotein E. ABO = ABO blood group (Alpha 1-3-N-Acetylgalactosaminyltransferase and Alpha 1-3-Galactosyltransferase). FURIN = Furin (Paired Basic Amino Acid Cleaving Enzyme). NOTCH4 = Notch Receptor 4. CCL2 = C-C Motif Chemokine Ligand 2. DPP9 = Dipeptidyl-Peptidase 9. IL6 = Interleukin-6. OAS1 = 2′-5′-Oligoadenylate Synthetase 1.
Fig. 2.
Loci associated with susceptibility, hospitalization, severity
Forest plot showing the odds ratios with 95% confidence intervals of SNPs which reached nominal significance (P <0.05) in any comparison and any genetic model. The case-control, ambulatory-hospitalized, moderate-severe and ambulatory-severe analyses were adjusted for age and sex. For the susceptibility analysis including the 1000 genomes project (1kGP), the association results of the allelic model are displayed, the codominant model is not shown. Amb. = Ambulatory. Hosp. = Hospitalized.
The second variant reaching significance after correction was rs1024611 G, near the CCL2 gene (OR = 0.56, 95% CI = 0.39-0.78], P = 0.001). This variant was associated with reduced susceptibility in the codominant and allelic model in the analysis, including the European 1kGP cohort (Table 3, Fig. 2).
The results of all four comparisons (susceptibility, hospitalization, severity, susceptibility using 1kGP) can be found in Supplementary Tables 9 – 13.
Susceptibility to SARS-CoV-2 infection
Two more SNPs reached P <0.05 for nominal significance in the susceptibility analysis: rs4702 (FURIN) and rs429358 (APOE) (Table 3, Fig. 2). The rs429358 C/C genotype along with rs7412 C/C, constituting the APOE e4e4 genotype were only identified in four controls. This suggests a protective effect of the APOE e4e4 C/C genotype. For rs73064425 in the LZTFL1 gene, the overdominant model of inheritance was most significant, suggesting C/T as risk genotype. The dominant model also reached significance, where the T allele was associated with a greater risk of SARS-CoV-2 infection (OR = 2.19, 95% CI = 1.08-4.46, P = 0.026).
Hospitalization
rs7412 (APOE) and rs3131294 (NOTCH4) showed nominally significant associations with hospitalization (Table 3, Fig. 2). For rs7412 in the APOE gene, the T allele was associated with severity in the log-additive model. To further investigate the influence of the APOE genotype, consisting of rs429358 C and rs7412 C, it was included in a logistic regression model (age- and sex-adjusted). The APOE e4e4 or e3e4 genotypes were not associated with infection or hospitalization.
Severe COVID-19
Patients with moderate and severe COVID-19 were compared in a severity analysis. Five SNPs reached nominal significance in the adjusted analysis: rs73064425 (LZTFL1), rs7412 (APOE), rs657152 (ABO), rs1800795 (IL6), and rs2109069 (DPP9) (Table 3, Fig. 2). For ABO rs657152, the recessive model fitted best, but no odds ratio could be calculated because the A/A genotype was not observed among patients with severe COVID-19. However, relative protection of the A/A genotype may be assumed because it occurred in 16% of patients with moderate disease.
In a second severity analysis comparing ambulatory with hospitalized patients with severe disease, two SNPs reached nominal significance in the adjusted analysis, rs4702 (FURIN) and rs3131294 (NOTCH4) (Table 3, Fig. 2).
Susceptibility analysis using data from the 1000 genomes project
To increase the sample size for the susceptibility analysis, allele and genotype counts were compared between COVID-19 cases and data retrieved from the 1kGP for European ancestry (Table 3, Fig. 2). Apart from the highly significant CCL2 variant, four more nominal associations were observed. In the comparison of genotype counts (codominant model), a new association was found for rs1131454 in the OAS1 gene. A/G and G/G genotypes were less frequent among patients. For the LZTFL1, APOE, and FURIN variants, the results replicated the findings indicated previously.
Polygenic risk score
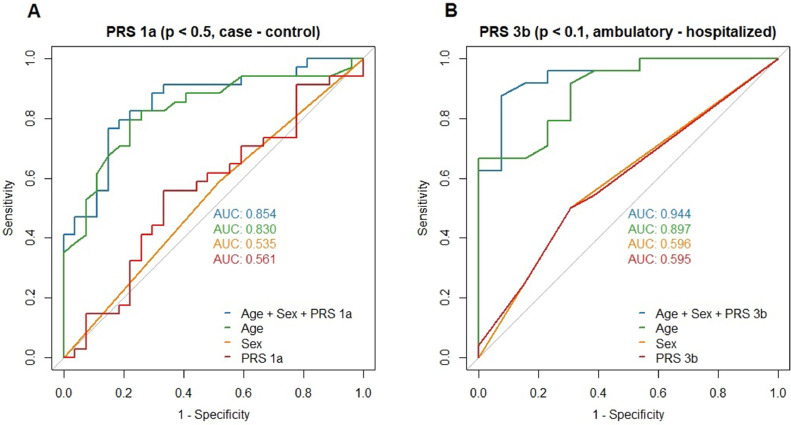
To capture the summed genetic effects of individual variants, different PRS models were developed in a training and a testing dataset. For the susceptibility analysis in the training set (86 cases, 66 controls; Analysis A, Supplementary table 4), all PRS versions succeeded in predicting a patient's case-control status in a logistic regression model, with an area under the ROC (AUC) ranging from 0.73 (PRS 1a, including SNPs reaching P <0.5 in the association analysis) to 0.59 (PRS 4a, including SNPs with P <0.05; Supplementary Fig. 1). In the full model, including age and sex, no PRS version was a significant predictor of the case-control status in the training set. In the testing data set (37 cases, 28 controls), no PRS version was a predictor of susceptibility, which did not change after adjustment for age and sex. The AUC ranged between 0.56 (PRS 1a, including SNPs with P <0.5) and 0.47 (PRS 3a, including SNPs with P <0.1). PRS 1a performed best in the testing set (b = 0.169, P = 0.556, AUC = 0.561, without adjustment, Fig. 3 ).
Fig. 3.
Receiver-operator curves (ROC) for the best-performing versions of the susceptibility and hospitalization PRS.
Area under the ROC (AUC) are shown for logistic regression models including age, sex, and PRS, as well as models with only one of the predictors in the testing dataset. A) Performance of susceptibility PRS including SNPs associated with SARS-CoV-2 infection at P <0.5 (PRS1a) in testing dataset of 37 cases and 28 controls. B) Performance of severity PRS including SNPs associated with hospitalization at P <0.1 (PRS3b) in testing dataset of 24 hospitalized and 13 ambulatory cases.
In the hospitalization analysis (Analysis B, Supplementary table 5), each PRS version could predict hospitalization and remained significant after adjustment for age and sex in the training set (51 hospitalized, 31 ambulatory patients). AUCs of the models containing only PRS ranged from 0.658 (PRS 4b, including SNPs with P <0.05) to 0.824 (PRS 1b, including SNPs with P <0.5, Supplementary Fig. 2). In the testing set (24 hospitalized, 13 ambulatory patients), only PRS 3b (including SNPs with P <0.1) reached significance after age- and sex-adjustment (b = 1.542, P = 0.048). The AUC of PRS 3b was 0.595 (Fig. 3), whereas the full model containing PRS 3b, age, and sex had the best discriminatory ability of all models with an AUC of 0.9439 (McFadden pseudo-R2 = 0.57, P = 5 × 10−6, Fig. 3).
SARS-CoV-2 variants and phylogenetic analysis
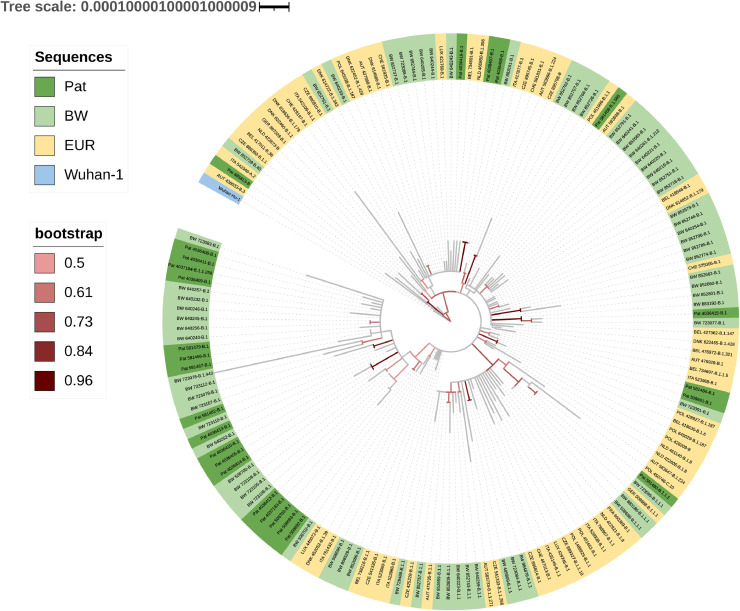
Most sequences were classified as B.1 lineage by the Pangolin software (23/26, 89%). A median of nine nucleotide exchanges (range zero to 12) and six amino acid substitutions (range zero to nine) per sequence was observed. A total of 43 different amino acid substitutions were present in the 26 samples (Supplementary table 6, Supplementary Fig. 3). A total of 25 of 26 sequences carried the spike protein D614G mutation and the ORF1b P314L substitution. The number of total and missense mutations per sequence did not differ between ambulatory and hospitalized patients. Neither lineages nor amino acid substitutions were associated with hospitalization. However, as SARS-CoV-2 sequences were available only for three ambulatory patients, the analysis was underpowered. Phylogenetic analysis showed that 17 sequences formed a distinct cluster with sequences from Baden-Wuerttemberg, Germany (Fig. 4 ). Two-thirds of the reference sequences from Baden-Wuerttemberg clustering with the genomes of this study also originated from Tübingen.
Fig. 4.
Maximum likelihood phylogenetic tree of SARS-CoV-2.
The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Pat = patient-derived sequences (n = 26). BW = Baden-Wuerttemberg, Germany (n = 69). EUR = Germany and it's neighboring countries (n = 67). Wuhan-1: reference root sequence. 1000 bootstrapping iterations were used. Branches with bootstrapping values above 50% are colored.
Discussion
Of the 30 genetic markers in our SNP panel, 10 were associated with at least one of the phenotypes “susceptibility to infection”, “hospitalization”, or “disease severity” after adjustment for age and sex; although for four single nucleotide polymorphisms, the direction of the effect was inconsistent with the original report in at least one comparison.
The strongest Bonferroni-corrigible association was found for rs73064425 in the LZTFL1 gene, where the T allele was consistently identified as a risk factor for severe COVID-19 and increased susceptibility to the infection. This 3p21.31 variant was described as a risk variant in all GWASs of severe COVID-19 (Velavan et al., 2021b). Meta-analyses performed by the HGI found two distinct signals at this locus, one associated with susceptibility (rs2271616), the other with severity (rs10490770) (COVID-19 Host Genetics Initiative, 2021). The LZTFL1 variant rs73064425 is in LD, with the severity lead variant rs10490770 (r 2 = 0.99), which aligns with the stronger association with severity than susceptibility in our cohort. Similar findings were recently reported in a Latvian study (Rescenko et al., 2021). A large analysis using HGI data could also show that the LZTFL1 risk variant had a similar or higher association with severe disease and death from COVID-19 than established clinical risk factors, especially in patients aged under 60 years (Nakanishi et al., 2021).
Uncertainties remain regarding the causal gene(s) at the 3p21.31 locus. Recently, a study combined multiomics and machine learning and determined rs17713054G>A in LZTFL1, which is also in linkage disequilibrium with our rs73064425 (r2 = 0.99), as the probable causative variant for the severity locus. This variant associates with upregulation of LZTFL1 and epithelial-mesenchymal transition in pulmonary epithelial cells, which is an antiviral response pathway and might be a mechanism behind the increased risk of severe COVID-19 in patients with the 3p21.31 variant (Downes et al., 2021).
The other marker reaching significance after Bonferroni correction was rs1024611, a susceptibility marker localized upstream of CCL2 (C-C Motif Chemokine Ligand 2, monocyte chemoattractant protein-1), a chemokine regulating tissue infiltration of monocytes, memory T-cells, and natural killer cells (Deshmane et al., 2009). Although the rs1024611 G allele was previously defined as a risk-increasing variant for SARS, our analysis indicates an opposite effect for COVID-19; the G allele and the G/G genotype showed a protective effect. The G allele confers increased CCL2 production in vitro and in vivo and enhanced leukocyte migration (Tu et al., 2015). It was also shown that SARS-CoV-2 induces CCL2 expression in the host (Chu et al., 2020), and other CCL isoforms characterize a proinflammatory macrophage subpopulation (Paludan and Mogensen, 2022) and promote inflammatory heart injury (Zhang et al., 2022). However, we did not observe enrichment of the high-producing rs1024611 G allele in severe cases. Whether CCL2 might play a different and protective role in early antiviral defense, thereby reducing susceptibility to SARS-CoV-2 infection requires further research.
Among nominally significant associations in this study, the directions of effects were consistent with original reports for rs2109069 (DPP9) (Pairo-Castineira et al., 2021) and rs1131454 (OAS1) (Banday et al., 2021). For rs4702 (FURIN), the A/G + G/G genotype was associated with susceptibility, which was concordant with the original report, where this genotype was shown to reduce FURIN expression and decrease SARS-CoV-2 infection of alveolar and neural organoids in vitro (Dobrindt et al., 2021). However, in the ambulatory versus severe disease comparison, GG was associated with risk for severe disease. Notably, the genotype-tissue expression database (GTEx Consortium, 2015) lists rs4702 GG both as an expression-increasing (e.g., in whole blood or lung) and an expression-decreasing (e.g., in the frontal cortex or tibial artery) eQTL. A study linked increased soluble furin to severity of COVID-19 (Kocyigit et al., 2021). Therefore, the relevance of changed FURIN expression in various tissues during infection and in severe disease requires further clarification. As previously reported (Kuo et al., 2020), the APOE e4e4 genotype was not associated with severity in our study. This may be because the e4e4 genotype was observed only in four individuals, all belonging to the controls. Therefore, the genotype could not be examined in patients with COVID-19. Furthermore, the direction of effect was inconsistent for the rs3131294 (NOTCH4) variant (Pairo-Castineira et al., 2021). rs3131294 lies in the HLA region on chromosome 6 and acts as an eQTL of several MHC II genes, with over 400 reports in the genotype-tissue expression database. Given the highly diverse effects of the rs3131294 polymorphisms on MHC II genes and potential links to immune-mediated diseases, detailed analyses of this locus should follow. Lastly, for rs657152 (ABO) (COVID-19 Host Genetics Initiative, 2021, Severe COVID-19 GWAS Group et al., 2020; Shelton et al., 2021) and rs1800795 (IL6) (Ulhaq and Soraya, 2020), an effect allele could not be determined because the overdominant model was most significant. Therefore, they cannot easily be compared with original studies.
We constructed PRSs to predict susceptibility to SARS-CoV-2 infection and hospitalization due to COVID-19 from our panel SNPs. The summed effect of several variants with primary low-confidence associations can improve a predictive model containing only age and sex because the hospitalization PRS reached significance in the full model. Regarding discrimination between cases and controls or hospitalized and ambulatory patients, the AUCs of the best-performing PRSs were similar to that of the variable sex, whereas age was the most meaningful predictor, suggesting a similar impact of sex and genetic background in our cohort. Interestingly, for susceptibility, the PRS model with the highest P-value threshold for inclusion of SNPs into the score, which was P <0.5, was best suited to predict infection risk; however, for hospitalization, the PRS model with a threshold of P <0.1 outperformed the other PRSs. This highlights the benefit of including variants into predictive models which otherwise would be excluded in stringent frequentist analyses. However, our sample sizes were small and the PRS should be evaluated further in larger cohorts. For this purpose, we provided effect sizes for all SNPs included in the panel, derived from association analysis in the whole cohort in Supplementary Tables 7 and 8. Developing a PRS with acceptable discriminatory capacity to identify individuals at risk for severe disease could aid therapeutic decisions, especially as more therapies are developed, which are administered during early infection to prevent progression to severe stages. These could be given to patients with genetic and clinical risk factors. For the implementation of a PRS in clinical practice, it is favorable if such scores consist only of a limited number of SNPs to reduce genotyping costs and efforts.
In addition to host genetics, we also investigated SARS-CoV-2 genetics. Most SARS-CoV-2 genomes belonged to the B.1 lineage, characterized by the ORF1b P314L and spike protein D614G mutations. SARS-CoV-2 isolates carrying D614G became dominant in Europe from February to March 2020 and were reported to have increased fitness but not increased disease severity (Korber et al., 2020; Volz et al., 2021). In this study, no lineages or individual mutations were associated with disease severity, concordant with studies from the United States and China (Esper et al., 2021; Zhang et al., 2020b). The phylogenetic analysis of the genomes suggests local community transmission. However, the high similarity of viral genomes in our study complicates the construction of a high-confidence phylogenetic tree (Morel et al., 2021), as reflected by low bootstrapping values in our tree.
This study has limitations. First, our cohort size was too small to detect small effects of common genetic variants, although we included variants with a higher a priori probability of association. Second, due to different sampling strategies for cases and controls, the age and sex distribution differed between groups, although women and men are affected by SARS-CoV-2 infection almost equally (Ortolan et al., 2020). This might reflect a selection bias. Third, we did not include uninfected controls with known SARS-CoV-2 exposure history. To balance this effect, controls were enrolled from the same federal state as cases, suggesting comparable exposure. Furthermore, population-reflecting controls from the 1kGP were included. Fourth, individuals included in our study all reside in Germany and are predominantly of European ancestry, contributing to the unequal representation of people of other descent. Fifth, our analysis of SARS-CoV-2 variants remains descriptive and association testing remain exploratory owing to the limited availability of viral samples.
Conclusion
We have replicated several genetic markers, including the 3p21.31 locus, reported for susceptibility to SARS-CoV-2 infection and COVID-19 severity. Furthermore, we have calculated a PRS, which may be used to predict the hospitalization risk. Because of the design of our candidate gene panel, additional genetic data have been made available. Therefore, new variants need to be incorporated, and the panel's utility for predicting disease should be validated and refined in independent cohorts of diverse ethnicities. A credible set of host genetic markers, combined with demographic factors and medical history, could enable risk stratification and targeted protective measures for those at highest risk for severe COVID-19.
Acknowledgments
Acknowledgments
The authors acknowledge the support of field workers and participants who consented to be part of this study. We also acknowledge the authors, originating and submitting laboratories of the genetic sequences, and metadata made available through the GISAID. We acknowledge Andrea Kreidenweiss, Jana Held, and Aline Sähr for their help in patient and healthy control recruitment. The author TPV is a member of the Pan African Network for Rapid Research, Response, and Preparedness for Infectious Diseases Epidemics consortium (PANDORA-ID-NET) and PAN-ASEAN Coalition for Epidemic and Outbreak Preparedness (PACE-UP; DAAD Project ID: 57592343).
Conflict of interest
All authors have no competing interests to declare.
Funding
This work was supported by the Federal Ministry of Education and Research (BMBF) (BMBF-01KI2052) and the Federal Ministry of Health (BMG) (BMG-ZMVI1-1520COR801).
Ethical approval
Ethics approval was obtained from the ethics commission of the Medical Faculty of the Eberhard Karls University, the University Hospital of Tübingen (286/2020BO1, 190/2020AMG1, 225/2020AMG1, 256/2020BO2), and the Heidelberg University Hospital (S-103/2019). All participants provided written informed consent to use their samples. All methods were performed in accordance with applying national and international regulations.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.06.030.
Appendix. Supplementary materials
References
- Banday AR, Stanifer ML, Florez-Vargas O, Onabajo OO, Zahoor MA, Papenberg BW, et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with risk of severe COVID-19. medRxiv Preprint posted online July 13, 2021. doi:10.1101/2021.07.09.21260221 [DOI] [PMC free article] [PubMed]
- Becker M, Dulovic A, Junker D, Ruetalo N, Kaiser PD, Pinilla YT, et al. Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat Commun. 2021;12:3109. doi: 10.1038/s41467-021-23473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S, Hildebrand D, Boulant S, Kreuter M, Rüter J, Pallerla SR, et al. Host factors facilitating SARS-CoV-2 virus infection and replication in the lungs. Cell Mol Life Sci. 2021;78:5953–5976. doi: 10.1007/s00018-021-03889-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Chan JF-W, Wang Y, Yuen TT-T, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dite GS, Murphy NM, Allman R. Development and validation of a clinical and genetic model for predicting risk of severe COVID-19. Epidemiol Infect. 2021;149:e162. doi: 10.1017/S095026882100145X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt K, Hoagland DA, Seah C, Kassim B, O'Shea CP, Murphy A, et al. Common genetic variation in humans impacts in vitro susceptibility to SARS-CoV-2 infection. Stem Cell Rep. 2021;16:505–518. doi: 10.1016/j.stemcr.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes DJ, Cross AR, Hua P, Roberts N, Schwessinger R, Cutler AJ, et al. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat Genet. 2021;53:1606–1615. doi: 10.1038/s41588-021-00955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper FP, Cheng YW, Adhikari TM, Tu ZJ, Li D, Li EA, et al. Genomic epidemiology of SARS-CoV-2 infection during the initial pandemic wave and association with disease severity. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Mack JA, Salvatore M, Prabhu Sankar S, Valley TS, Singh K, et al. Characteristics associated with racial/ethnic disparities in COVID-19 outcomes in an academic health care system. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz JE, Kosmicki JA, Damask A, Sharma D, Roberts GHL, Justice AE, et al. Genome-wide analysis in 756,646 individuals provides first genetic evidence that ACE2 expression influences COVID-19 risk and yields genetic risk scores predictive of severe disease. medRxiv Preprint posted online June 10, 2021. doi: 10.1101/2020.12.14.20248176.
- Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QM, Zhang PD, Li ZH, Zhou JM, Liu D, Zhang XR, et al. Genetic risk and chronic obstructive pulmonary disease independently predict the risk of incident severe COVID-19. Ann Am Thorac Soc. 2022;19:58–65. doi: 10.1513/AnnalsATS.202102-171OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocyigit A, Sogut O, Durmus E, Kanimdan E, Guler EM, Kaplan O, et al. Circulating furin, IL-6, and presepsin levels and disease severity in SARS-CoV-2–infected patients. Sci Prog. 2021;104(2) doi: 10.1177/00368504211026119. 00368504211026119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA, et al. ApoE e4e4 genotype and mortality with COVID-19 in UK Biobank. J Gerontol A Biol Sci Med Sci. 2020;75:1801–1803. doi: 10.1093/gerona/glaa169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Rentsch CT, Morton CE, Hulme WJ, Schultze A, MacKenna B, et al. Ethnic differences in SARS-CoV-2 infection and COVID-19-related hospitalisation, intensive care unit admission, and death in 17 million adults in England: an observational cohort study using the OpenSAFELY platform. Lancet. 2021;397:1711–1724. doi: 10.1016/S0140-6736(21)00634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz-Katz G, Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection fatality rates. Int J Infect Dis. 2020;101:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Barbera P, Czech L, Bettisworth B, Hübner L, Lutteropp S, et al. Phylogenetic analysis of SARS-CoV-2 data is difficult. Mol Biol Evol. 2021;38:1777–1791. doi: 10.1093/molbev/msaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Pigazzini S, Degenhardt F, Cordioli M, Butler-Laporte G, Maya-Miles D, et al. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J Clin Invest. 2021:131. doi: 10.1172/JCI152386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoumi F, Mfoutou Mapanguy CC, Tomazatos A, Pallerla SR, Linh LTK, Casadei N, et al. Genomic surveillance of SARS-CoV-2 in the Republic of Congo. Int J Infect Dis. 2021;105:735–738. doi: 10.1016/j.ijid.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan A, Lorenzin M, Felicetti M, Doria A, Ramonda R. Does gender influence clinical expression and disease outcomes in COVID-19? A systematic review and meta-analysis. Int J Infect Dis. 2020;99:496–504. doi: 10.1016/j.ijid.2020.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev. 2020;296:205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pallerla SR, Elion Assiana DO, Linh LTK, Cho FN, Meyer CG, Fagbemi KA, et al. Pharmacogenetic considerations in the treatment of co-infections with HIV/AIDS, tuberculosis and malaria in Congolese populations of Central Africa. Int J Infect Dis. 2021;104:207–213. doi: 10.1016/j.ijid.2020.12.009. [DOI] [PubMed] [Google Scholar]
- Paludan SR, Mogensen TH. Innate immunological pathways in COVID-19 pathogenesis. Sci Immunol. 2022;7 doi: 10.1126/sciimmunol.abm5505. eabm5505. [DOI] [PubMed] [Google Scholar]
- Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV, et al. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir Med. 2020;8:506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell TR, Hotopf M, Hatch SL, Breen G, Duarte RRR, Nixon DF. Genetic risk for severe COVID-19 correlates with lower inflammatory marker levels in a SARS-CoV-2-negative cohort. Clin Transl Immunology. 2021;10:e1292. doi: 10.1002/cti2.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Holmes EC, Á O'Toole, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescenko R, Peculis R, Briviba M, Ansone L, Terentjeva A, Litvina HD, et al. Replication of LZTFL1 gene region as a susceptibility locus for COVID-19 in Latvian population. Virol Sin. 2021;36:1241–1244. doi: 10.1007/s12250-021-00448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GHL, Park DS, Coignet MV, McCurdy SR, Knight SC, Partha R, et al. AncestryDNA COVID-19 host genetic study identifies three novel loci. medRxiv Preprint posted online October 9, 2020. doi: 10.1101/2020.10.06.20205864.
- Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Severe COVID-19 GWAS Group Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Sonmez T, Coker D, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- Streeck H, Schulte B, Kümmerer BM, Richter E, Höller T, Fuhrmann C, et al. Infection fatality rate of SARS-CoV2 in a super-spreading event in Germany. Nat Commun. 2020;11:5829. doi: 10.1038/s41467-020-19509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Chong WP, Zhai Y, Zhang H, Zhang F, Wang S, et al. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect. 2015;71:101–109. doi: 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulhaq ZS, Soraya GV. Anti-IL-6 receptor antibody treatment for severe COVID-19 and the potential implication of IL-6 gene polymorphisms in novel coronavirus pneumonia. Med Clin (Barc) 2020;155:548–556. doi: 10.1016/j.medcle.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan TP, Kieu Linh LT, Kreidenweiss A, Gabor J, Krishna S, Kremsner PG. Longitudinal monitoring of lactate in hospitalized and ambulatory COVID-19 patients. Am J Trop Med Hyg. 2021;104:1041–1044. doi: 10.4269/ajtmh.20-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan TP, Meyer CG. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan TP, Pallerla SR, Rüter J, Augustin Y, Kremsner PG, Krishna S, et al. Host genetic factors determining COVID-19 susceptibility and severity. EBiomedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, Á O'Toole, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. COVID-19 Clinical management: living guidance. https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-2,2021 (accessed 22 December 2021 ).
- WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis 2020;20:e192–7. [DOI] [PMC free article] [PubMed]
- Yildirim Z, Sahin OS, Yazar S. Bozok Cetintas V. Genetic and epigenetic factors associated with increased severity of Covid-19. Cell Biol Int. 2021;45:1158–1174. doi: 10.1002/cbin.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S, Miyamae T, Kuroiwa Y, Nishioka K. Novel coronavirus disease 2019 (COVID-19) and cytokine storms for more effective treatments from an inflammatory pathophysiology. J Clin Med. 2021;10:801. doi: 10.3390/jcm10040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen X, Zuo W, Ji Z, Qu Y, Su Y, et al. Inflammatory activation and immune cell infiltration are main biological characteristics of SARS-CoV-2 infected myocardium. Bioengineered. 2022;13:2486–2497. doi: 10.1080/21655979.2021.2014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.