Abstract
Prostate cancer (PCa) incidence and mortality vary across territories and populations. This can be explained by the genetic factor of this disease. This article aims to correlate the epidemiological data, worldwide incidence, and mortality of PCa with single-nucleotide polymorphisms (SNPs) associated with the susceptibility and severity of this neoplasm in different populations. Eighty-four genetic variants associated with prostate cancer susceptibility were selected from the literature through genome association studies (GWAS). Allele frequencies were obtained from the 1000 Genomes Project, and epidemiological data were obtained from Surveillance, Epidemiology, and End Results (SEER). The PCa incidence, mortality rates, and allele frequencies of variants were evaluated by Pearson’s correlation. Our study demonstrated that 12 SNPs (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs6983267, rs11649743, rs2075110, rs114798100, rs855723, and rs2075109) were correlated with epidemiological data in different ethnic groups. Ten SNPs (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs11649743, rs2075110, rs114798100, and rs2075109) were positively correlated with the mortality rate. Seven SNPs (rs1048169, rs2961144, rs7000448, rs4430796, rs2066827, rs12500426, and rs114798100) were positively correlated with incidence. Positive correlations of incidence and mortality rates were more frequent in the African population. The genetic variants investigated here are likely to predispose to PCa and could play a role in its progression and aggressiveness. This genetic study demonstrated here is promising for implementing personalized strategies to screen for prostate cancer in diverse populations.
Keywords: prostate cancer, genetic risk variant, susceptibility, severity, epidemiology, population
1. Introduction
Prostate cancer (PCa) is the second most common cancer and the fifth leading cause of cancer death among men. In 2020, it was estimated that approximately 1.4 million new cases and 375,000 deaths from prostate cancer occurred worldwide. The etiology of prostate cancer is multifactorial, with some risk factors associated with its development, including advanced age, positive family history, and African ancestry [1].
PCa incidence varies across ethnic groups and geographic variations. There are higher incidence and mortality rates in people of Black African descent worldwide [2]. In the United States, there is variability among different ethnic groups, with the highest incidence and mortality in Black men of African descent. The difference in incidence and mortality can be attributed to the prevalence of multiple prostate cancer genetic risk loci across ethnic groups [3,4].
Recent studies of epidemiology suggest that PCa is a highly heritable disease and suggests a strong causal association between genetic factors and the development of prostate cancer. A twin study revealed that 57% of prostate cancer individuals had a family link to prostate cancer [5]. Recent studies indicate that more than 100 well-recognized SNPs have been found to be associated with prostate cancer and constitute a major risk factor in the development of prostate cancer [6].
GWAS have changed the field of cancer genetics in the last decade. GWAS have improved the understanding of the key genes and gene variants involved in molecular and biological mechanisms that involve PCa etiology. Although PCa incidence and mortality rates are higher in African descent, most large-scale GWAS have been conducted in the European population [7].
This article aims to correlate epidemiological data on the incidence and mortality of prostate cancer worldwide with frequencies of important SNPs in studies of GWAS associated with the susceptibility and severity of this neoplasm in different populations.
2. Materials and Methods
2.1. SNP Determination
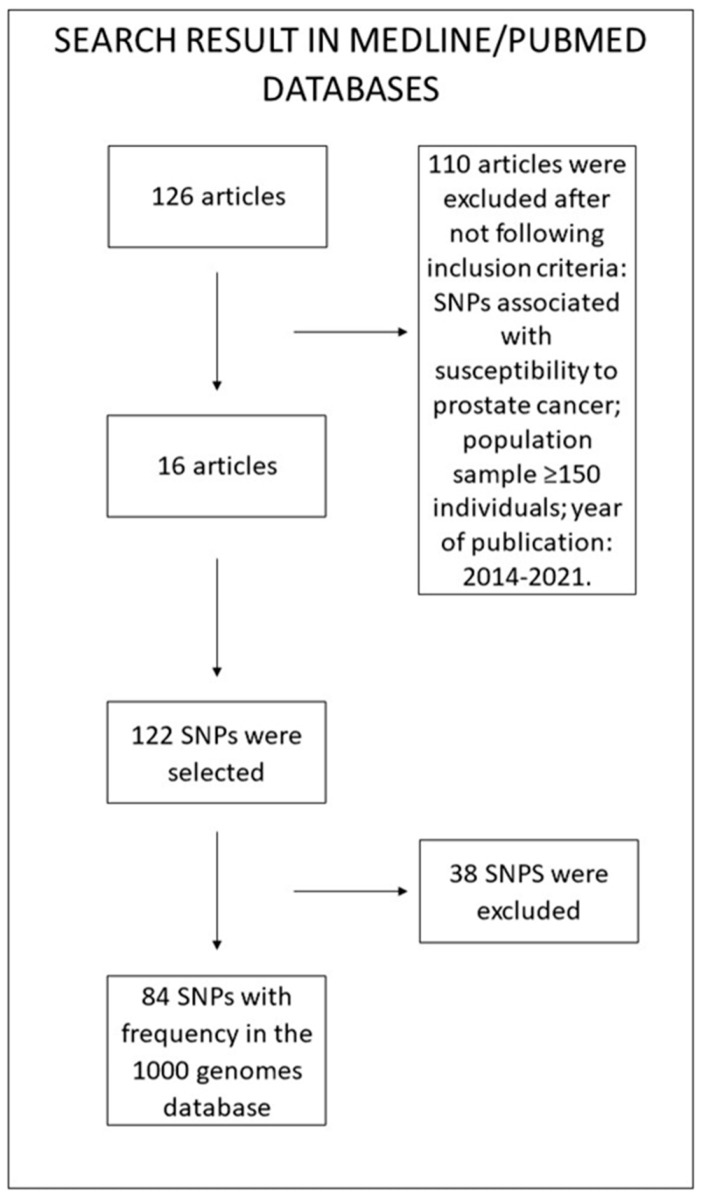
We searched online Medline/PubMed databases for articles published in English using several key terms relating to prostate cancer and susceptibility. The key terms were prostate cancer, GWAS, risk, susceptibility, and population, resulting in 126 articles. There were 16 articles after using the following inclusion criteria: studies on SNPs associated with susceptibility to prostate cancer, with a population sample greater than 150 individuals, and in the period from 2014 to 2021. The result was 122 polymorphisms related to prostate cancer susceptibility. Among these, we selected 84 SNPs that had their frequency detected in the 1000 Genomes database, and 38 were excluded from the study because they were not detected in this database (Figure 1).
Figure 1.
SNP selection flowchart.
2.2. Epidemiological and Genetic Data
The incidence and mortality rates of PCa were obtained from Surveillance, Epidemiology, and End Results (SEER)-Medicare [3] in the United States. In this database, the ethnicities reported were White non-Hispanic, White Hispanic, Black, Asian/Pacific Islander, Amerindian/Alaska Native, and Hispanic. In the 1000 Genomes Project, the ethnicities reported were from: Europe (EUR), Africa (AFR), East Asia (EAS), South Asia (SAS), and the Americas (AMR). In our study, we classified the populations from SEER into 6 groups. The EUR group was composed of White non-Hispanic and White Hispanic populations. The AFR group was composed of the Black population. The EAS and SAS groups were averages of the Asian/Pacific Islander population. The AMR group was composed of Amerindian/Alaska Native and Hispanic groups. Gene allele frequencies were assessed from phase 3 data from the 1000 Genomes Project for Africa, East Asia, Europe, South Asia, and the Americas.
2.3. Statistical Analysis
The prostate incidence, mortality rates, and allele frequencies of the variants were evaluated by Pearson’s correlation. The data were evaluated with previously described groups, using the “cor. test” function of the “stats” package of the R programming language. After this procedure, the values of r, r2, p-value, and 95% CI were obtained. All plots were created using the “ggplot2” graphics package. A p-value less than 0.05 (p ≤ 0.05) was statistically significant.
3. Results
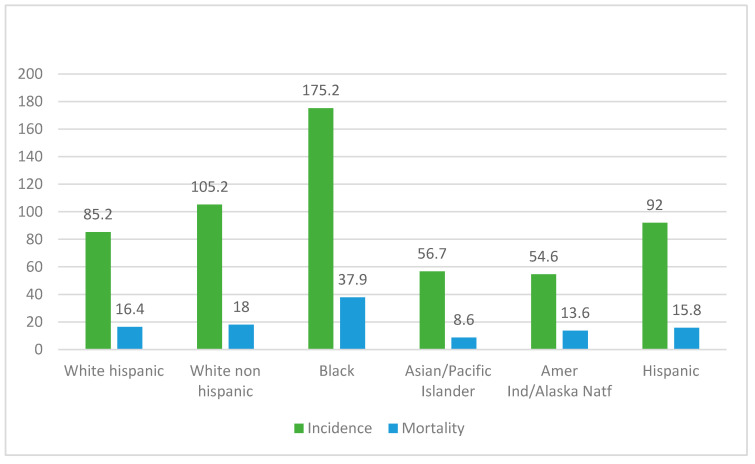
The incidence rate of prostate cancer was 175.2 per 100.000 men in the Black population, followed by White non-Hispanic (105.2), Hispanic (92), White Hispanic (85.2), Asian/Pacific Islander (56.7), and Amerindian/Alaska Native populations (54.6). The mortality rate of prostate cancer was also higher in the Black population at 37.9 per 1000.000, followed by White non-Hispanic (18), White Hispanic (16.4), Hispanic (15.8), Amerindian/Alaska Native (13.6), and Asian/Pacific Islander (8.6) (Figure 2).
Figure 2.
Incidence and mortality rates per 100.000 habitants from SEER prostate cancer registries from 2013 to 2017 in different populations.
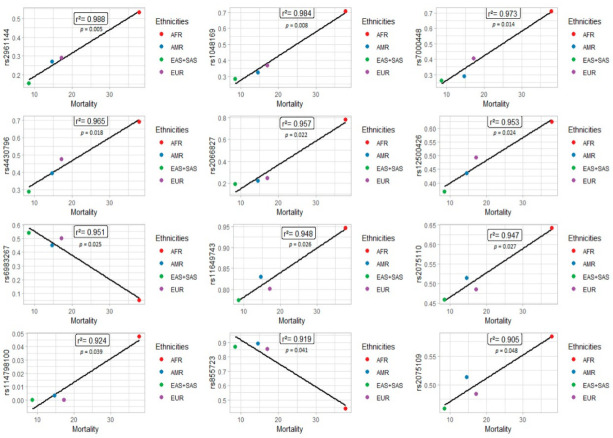
A total of 84 SNPS (Supplementary Tables S1–S3) were selected from the literature, as described in Figure 1. Among those polymorphisms, twelve SNPs (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs6983267, rs11649743, rs2075110, rs114798100, rs855723, and rs2075109) were significant in Pearson’s correlation analysis (Table 1 and Table 2). Among these 12, 10 (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs11649743, rs2075110, rs114798100, and rs2075109) were positively correlated with the mortality rate (Figure 3).
Table 1.
SNPs significantly correlated with PCa mortality in different populations.
| Gene | SNP ID | Gene Consequence | Alleles | Ancestral | Placement | Clinical Impact | MAF | Frequency from the 1000 Genomes Project | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | EAS | SAS | EUR | AMR | ||||||||
| OR2A5 | rs2961144 | Missense Variant | A/G | A | GRCh38.p13 chr 7 | Not Reported | 0.30 | 0.5318 | 0.0079 | 0.298 | 0.2903 | 0.269 |
| HAUS6 | rs1048169 | 3’ UTR Variant | T/C/G | C | GRCh38.p13 chr 9 | Not Reported | 0.42 | 0.7073 | 0.2073 | 0.36 | 0.3668 | 0.323 |
| CASC8 | rs7000448 | Intron Variant | C/T | T | GRCh38.p13 chr 8 | Not Reported | 0.41 | 0.711 | 0.2659 | 0.258 | 0.4046 | 0.29 |
| HNF1B | rs4430796 | Intron Variant | A/C/G/T | G | GRCh38.p13 chr 17 | Not Reported | 0.49 | 0.6906 | 0.2768 | 0.297 | 0.4751 | 0.395 |
| CDKN1B | rs2066827 | Missense Variant | T/A/C/G | T | GRCh38.p13 chr 12 | Benign | 0.36 | 0.7837 | 0.0556 | 0.319 | 0.2425 | 0.218 |
| PDL1M5 | rs12500426 | Intron Variant | A/C | C | GRCh38.p13 chr 4 | Not Reported | 0.48 | 0.6225 | 0.4931 | 0.241 | 0.4931 | 0.435 |
| CCAT2 | rs6983267 | Non-Coding Transcript Variant | G/T | G | GRCh38.p13 chr 8 | Not Reported | 0.39 | 0.041 | 0.0477 | 0.6121 | 0.468 | 0.5010 |
| HNF1B | rs11649743 | Intron Variant | A/G | G | GRCh38.p13 chr 17 | Not Reported | 0.37 | 0.9470 | 0.6617 | 0.885 | 0.8012 | 0.829 |
| EGFR | rs2075110 | Intron Variant | C/G/T | T | GRCh38.p13 chr 7 | Not Reported | 0.48 | 0.643 | 0.3522 | 0.565 | 0.565 | 0.514 |
| PCAT2 | rs114798100 | Intron Variant | A/G | A | GRCh38.p13 chr 8 | Not Reported | 0.01 | 0.0477 | 0 | 0 | 0 | 0.003 |
| WNT1 | rs855723 | Regulatory Region Variant | G/A/C/T | G | GRCh38.p13 chr 12 | Not Reported | 0.25 | 0.4365 | 0.996 | 0.737 | 0.8549 | 0.889 |
| EGFR | rs2075109 | Intron Variant | T/C | C | GRCh38.p13 chr 7 | Not Reported | 0.50 | 0.584 | 0.3522 | 0.564 | 0.4841 | 0.513 |
Table 2.
SNPs significantly correlated with PCa incidence in different populations.
| Gene | SNP ID | Gene consequence | Alleles | Ancestral | Placement | Clinical Impact | MAF | Frequency from the 1000 Genomes Project | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | EAS | SAS | EUR | AMR | ||||||||
| CASC8 | rs7000448 | Intron Variant | C/T | T | GRCh38.p13 chr 8 | Not Reported | 0.41 | 0.711 | 0.2659 | 0.258 | 0.4046 | 0.29 |
| HAUS6 | rs1048169 | 3 Prime UTR Variant | T/C/G | C | GRCh38.p13 chr 9 | Not Reported | 0.42 | 0.7073 | 0.2073 | 0.36 | 0.3668 | 0.323 |
| NF1B | rs4430796 | Intron Variant | A/C/G/T | G | GRCh38.p13 chr 17 | Not Reported | 0.49 | 0.6906 | 0.2768 | 0.297 | 0.4751 | 0.395 |
| OR2A5 | rs2961144 | Missense Variant | A/G | A | GRCh38.p13 chr 7 | Not Reported | 0.30 | 0.5318 | 0.0079 | 0.298 | 0.903 | 0.269 |
| PDL1M5 | rs12500426 | Intron Variant | A/C | C | GRCh38.p13 chr 4 | Not Reported | 0.48 | 0.6225 | 0.4931 | 0.241 | 0.4931 | 0.435 |
| CDKN1B | rs2066827 | Missense Variant | T/A/C/G | T | GRCh38.p13 chr 12 | Benign | 0.36 | 0.7837 | 0.0556 | 0.319 | 0.2425 | 0.218 |
| WNT1 | rs855723 | Regulatory Region Variant | G/A/C/T | G | GRCh38.p13 chr 12 | Not Reported | 0.25 | 0.4365 | 0.996 | 0.737 | 0.8549 | 0.889 |
| CCAT2 | rs6983267 | Non-Coding Transcript Exon Variant | G/T | G | GRCh38.p13 chr 8 | Not Reported | 0.39 | 0.0477 | 0.6121 | 0.468 | 0.5010 | 0.450 |
| PCAT2 | rs114798100 | Intron Variant | A/G | A | GRCh38.p13 chr 8 | Not Reported | 0.01 | 0.0477 | 0 | 0 | 0 | 0.003 |
Figure 3.
SNPs associated with prostate cancer mortality in different populations.
The higher the frequency of the variant allele of these polymorphisms in populations, the higher the estimated mortality rates. All ten variants (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs11649743, rs2075110, rs114798100, and rs2075109) were more frequent in the African population, which demonstrates the greater aggressiveness of prostate cancer in the African population.
Nevertheless, two polymorphisms (rs6983267 and rs855723) were inversely correlated with the mortality rate. Consequently, the higher the frequency of the variant allele, the lower the estimated mortality rate. Both variants were more frequent in the East Asian population, possibly because East Asian ancestry may be a protective factor against prostate cancer.
Only twelve genetic variants were significantly correlated with worldwide prostate cancer mortality data. More information about the location, impact, relative risk, and allele frequencies of the variants are presented in Table 1.
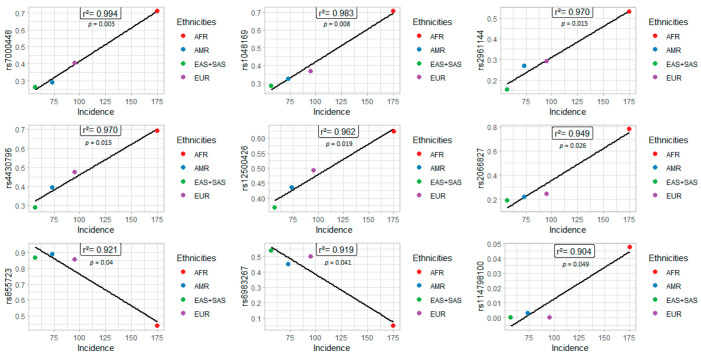
Nine variants (rs7000448, rs1048169, rs2961144, rs4430796, rs12500426, rs2066827, rs6983267, rs855723, and rs114798100) had a statistically significant correlation with the incidence rate (Figure 4). Among nine SNPs, seven variants (rs7000448, rs1048169, rs2961144, rs4430796, rs12500426, rs2066827, and rs114798100) were positively correlated with the incidence rate. The higher the frequency of the variant, the greater the number of new cases of prostate cancer. Seven polymorphisms (rs1048169, rs2961144, rs7000448, rs4430796, rs2066827, rs12500426, and rs114798100) were more frequent in the African population, which indicates high susceptibility to prostate cancer in Africans.
Figure 4.
SNPs associated with prostate cancer incidence in different populations.
However, two polymorphisms (rs6983267 and rs855723) were inversely correlated with the incidence rate, so the higher the frequency of the variant allele, the lower the incidence rate. Both variants also had a lower mortality rate, which reinforces the protective action of the allele against prostate cancer in the East Asian population.
For nine variants, there were significant correlations between variations in allelic frequencies and the incidence of prostate cancer in different populations (Table 2). More information about the genes and their respective IDs, the location of these variants, clinical impact, and the relative risk are presented in Table 2.
4. Discussion
Prostate cancer incidence and mortality rates vary significantly by ethnicity, showing a wide fluctuation in epidemiological rates and evidencing a higher incidence and worse prognosis in men of African descent [8]. Our results showed that populations of African origin had higher incidence and mortality rates of the disease, and, contrary to populations of Asian origin, they obtained better results related to susceptibility and mortality.
This study correlated epidemiological data on the incidence and mortality of prostate cancer worldwide with frequencies of important SNPs in studies of GWA associated with the susceptibility and severity of this neoplasm in different populations. Our results indicated correlations for 12 genetic variants (rs2961144, rs7000448, rs4430796, rs2066827, rs12500426, rs6983267, rs1649743, rs1649743, rs20751010, rs2075109, rs114798100, rs114798100, and rs855723) in 10 genes (HNF1B, EGFR, CCAT2, WNT, PCAT2, PDLIM5, CDKN1B, CASC8, HAUS6i, and OR2A5) with incidence and mortality rates. Our results show that the variants related to high incidence and mortality are more frequent in the African population and less frequent in the Asian population.
Hepatocyte Nuclear Factor 1 β (HNF1B) is a transcription factor-2 that regulates metabolic pathways and genes important for human embryonic development [9]. The gene is essential for the embryonic formation of the genital tract, pancreas, liver, biliary tract, and gastrointestinal system [10]. In our studies, SNPs rs4430796 and rs11649743 were concomitantly associated with a higher incidence and mortality of prostate cancer. Both variants have been linked to an increased risk of prostate cancer in several studies [11,12].
The Epidermal Growth Factor Receptor (EGFR) gene acts in the regulation of proliferation, differentiation, division, survival, and cancer development [13]. In 2004, Shuch et al. reported increased gene expression in African Americans with prostate cancer. In our study, the SNPs in EGFR (rs2075110 and rs2075109) were correlated with higher mortality and susceptibility to prostate cancer. Both variants were more frequent in Africans.
The Colon cancer-associated transcript 2 (CCAT2) gene was first associated with colon cancer; however, it is currently associated with several types of cancer, including prostate cancer [14]. Peng He et al., when analyzing 18 patients with prostate cancer, found a high expression of CCAT2, indicating a role of this gene in the pathogenesis and progression of the disease [15]. In our study, the rs6983267 SNP of the CCAT2 gene was inversely related to mortality and incidence rates. The higher the frequency of this marker, the lower the mortality and incidence rates. This SNP was more frequent in the Asian population and less frequent in Africans.
The Wingless-type (WNT) proteins are involved in bone metabolism, affecting several diseases [16]. The WNT gene is related to PCa bone metastasis and is found at high levels in the advanced stages of this disease [17]. In our study, the variant rs855723 allele was inversely correlated with incidence and mortality. Wang et al. found that rs855723 was associated with lower expression of WNT1, and this corroborates our finding on mortality.
The PDLIM5 gene is a protein that binds to several proteins, such as PKC, PKA, PKD, and AMPK, and is important in signal modulation pathways [18]. In our study, rs12500426 was associated with higher mortality and incidence, being more frequent in Africans.
The CDKN1B gene is responsible for encoding a protein that inhibits a cyclin-dependent kinase that plays an important role in the cell cycle [19]. Other studies, such as that by Farashi et al. (2018), have demonstrated the possible association of rs2066827 with the risk of developing PCa, resulting from a missense mutation (V109G) in the CDKN1B gene that contributes to prostate cancer tumorigenesis through the deregulation of cell cycle checkpoints [20]; these data corroborate the findings in our study, which demonstrate a positive correlation between the increase in the frequency of this SNP and the increase in the incidence of cases.
The CASC8 gene is responsible for expressing a lncRNA (long non-coding RNA) that plays an important role in the regulation of MYC, which is related to susceptibility to various cancers, such as breast, colorectal, and prostate cancers [21,22]. Our findings demonstrate a directly proportional correlation of rs7000448 in this gene with the mortality and incidence of prostate cancer, especially in the African population. These findings are in agreement with international studies, which demonstrated that CASC8 and rs7000448 are mainly related to PCa, such as in the meta-analysis carried out by Tong (2020). This polymorphism was related to the increase in cases in Caucasians and Africans [23,24,25].
HAUS6 is part of the group of augmin complex subunits, which play an important role in the recruitment and amplification of microtubule molecules during cell division [26]. Our analysis demonstrated a significant correlation of rs1048169 in this gene with incidence and mortality, which were higher in the African population. There are few studies correlating this polymorphism with the risk of developing PCa; however, Schumacher and colleagues pointed out a possible role of this variant in the risk of developing the neoplasm [19].
OR2A5 is a gene responsible for expressing hormone receptors and neurotransmitters. Lin and colleagues found results in agreement with our study, where OR2A5 was also related to the risk of developing prostate cancer [27]. In our findings, rs2961144 showed a positive correlation with the mortality rate mainly in African populations.
The SNP rs114798100 in the PCAT2 gene is described as an important risk marker for prostate cancer development. It is considered a rare variant and found only in African American populations [28]. In our study, this polymorphism was also related to the risk of developing prostate cancer.
This study had limitations regarding ethnic group information, such as the Amerindians. The study also had a lack of homogeneity in the socioeconomic levels in studies. Despite these limitations, this study contributes important findings to the literature, and it may help to choose genetic markers that are more globally homogeneous in relation to the prognosis and predisposition to prostate cancer.
These genetic variants likely predispose to PCa and may play a role in PCa progression and aggressiveness. The SNPs studied, which were positively correlated with incidence and mortality rates, were more frequent in the African population. This genetic study holds promise for implementing personalized strategies for prostate cancer screening in diverse populations.
5. Conclusions
Our study demonstrated that 12 SNPs (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs6983267, rs11649743, rs2075110, rs114798100, rs855723, and rs2075109) were correlated with epidemiological data in different ethnic groups. Ten SNPs (rs2961144, rs1048169, rs7000448, rs4430796, rs2066827, rs12500426, rs11649743, rs2075110, rs114798100, and rs2075109) were positively correlated with the mortality rate. Seven polymorphisms (rs1048169, rs2961144, rs7000448, rs4430796, rs2066827, rs12500426, and rs114798100) were positively correlated with incidence. In contrast, two polymorphisms (rs6983267 and rs855723) were negatively correlated with mortality and incidence rates.
Acknowledgments
We acknowledge funding from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FAPESPA (Fundação Amazônia de Amparo a Estudos e Pesquisa), UFPA (Universidade Federal do Pará) and PROPESP (Pró-reitoria de Pesquisa e Pós-graduação) from UFPA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13061039/s1. Table S1: SNPs correlated with PCa mortality in different populations; Table S2: SNPs correlated with PCa incidence in different populations. Table S3: SNPs correlated with PCa mortality and incidence in different populations.
Author Contributions
L.P.A.G. and G.M.V. designed the study, processed the data, and wrote the article. D.F.d.V.B.L., L.W.M.S.V. and V.T.A. contributed to the writing of the article. L.F.P. contributed to the writing of the article and data analysis. M.R.F., R.M.R.B., P.P.d.A., S.E.B.d.S. and N.P.C.d.S. were project coordinators. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable for this study.
Informed Consent Statement
Not applicable for this study. All data used were obtained from secondary sources available on the 1000 Genomes Project platform/database and GBD Compare website.
Data Availability Statement
All relevant data will be shared as Supporting Information files if the manuscript is accepted for publication.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FAPESPA (Fundação Amazônica de Amparo a Estudos e Pesquisas), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and UFPA (Universidade Federal do Pará). These funding agencies played no role in the study design, data collection and analysis, or the decision to publish, or the preparation of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Conti D.V., Darst B.F., Moss L.C., Saunders E.J., Sheng X., Chou A., Schumacher F.R., Al Olama A.A., Benlloch S., Dadaev T., et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Cancer Institute, Surveillance, Epidemiology, and End Results Program (SEER) Cancer Stat Facts: Prostate Cancer. [(accessed on 30 March 2021)]; Available online: https://seer.cancer.gov/statfacts/html/prost.html.
- 4.Saad M., Mokrab Y., Halabi N., Shan J., Razali R., Kunji K., Syed N., Temanni R., Subramanian M., Ceccarelli M., et al. Genetic predisposition to cancer across people of different ancestries in Qatar: A population-based, cohort study. Lancet Oncol. 2022;23:341–352. doi: 10.1016/S1470-2045(21)00752-X. [DOI] [PubMed] [Google Scholar]
- 5.Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Möller S., Unger R.H., et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA J. Am. Med. Assoc. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allemailem K.S., Almatroudi A., Alrumaihi F., Almansour N.M., Aldakheel F.M., Rather R.A., Afroze D., Rah B. Single nucleotide polymorphisms (SNPs) in prostate cancer: Its implications in diagnostics and therapeutics. Am. J. Transl. Res. 2021;13:3868–3889. [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson J.R., Woods-Burnham L., Hooker S.E.J., Batai K., Kittles R.A. Genetic contributions to prostate cancer disparities in men of West African descent. Front. Oncol. 2021;11:4327. doi: 10.3389/fonc.2021.770500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebbeck T.R. Prostate cancer genetics: Variation by race, ethnicity, and geography. Semin. Radiat. Oncol. 2016;27:3–10. doi: 10.1016/j.semradonc.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu W., Sun J., Zhou H., Wang F., Zhao C., Li K., Fan C., Ding G., Wang J. HNF1B inhibits cell proliferation via repression of SMAD6 expression in prostate cancer. J. Cell. Mol. Med. 2020;24:14539–14548. doi: 10.1111/jcmm.16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dundr P., Bártů M., Hojný J., Michálková R., Hájková N., Stružinská I., Krkavcová E., Hadravský L., Kleissnerová L., Kopejsková J., et al. HNF1B, EZH2 and ECI2 in prostate carcinoma. Molecular, immunohistochemical and clinico-pathological study. Sci. Rep. 2020;10:14365. doi: 10.1038/s41598-020-71427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng S.L., Sun J., Wiklund F., Smith S., Stattin P., Li G., Adami H.-O., Hsu F.-C., Zhu Y., Bälter K., et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 12.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Purcell L.D., Gao Z., Hsu F.-C., Kim S.-T., Liu W., Zhu Y., et al. Evidence for two independent prostate cancer risk–associated loci in the HNF1B gene at 17q12. Nat. Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsudomi T., Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2009;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 14.Xin Y., Li Z., Zheng H., Chan M.T., Wu W.K.K. CCAT2: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:e12342. doi: 10.1111/cpr.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He P., Xiong G., Guo W., Jiang G., Li Y., Li H. Long non-coding RNA CCAT2 promotes prostate cancer cell proliferation and invasion by regulating the Wnt/β-catenin signaling pathway. Oncol. Lett. 2020;20:97. doi: 10.3892/ol.2020.11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall C.L., Keller E.T. The role of Wnts in bone metastases. Cancer Metastasis Rev. 2006;25:551–558. doi: 10.1007/s10555-006-9022-2. [DOI] [PubMed] [Google Scholar]
- 17.Thiele S., Rauner M., Goettsch C., Rachner T.D., Benad P., Fuessel S., Erdmann K., Hamann C., Baretton G.B., Wirth M.P., et al. Expression profile of WNT molecules in prostate cancer and its regulation by aminobisphosphonates. J. Cell. Biochem. 2011;112:1593–1600. doi: 10.1002/jcb.23070. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y., Wang X., Xu Z., He Y., Guo C., He L., Huan C., Cai C., Huang J., Zhang J., et al. PDLIM5 inhibits STUB1-mediated degradation of SMAD3 and promotes the migration and invasion of lung cancer cells. J. Biol. Chem. 2020;295:13798–13811. doi: 10.1074/jbc.RA120.014976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farashi S., Kryza T., Clements J., Batra J. Post-GWAS in prostate cancer: From genetic association to biological contribution. Nat. Cancer. 2018;19:46–59. doi: 10.1038/s41568-018-0087-3. [DOI] [PubMed] [Google Scholar]
- 21.Beuten J., Gelfond J.A.L., Martinez-Fierro M.L., Weldon K.S., Crandall A.C., Rojas-Martinez A., Thompson I.M., Leach R. Association of chromosome 8q variants with prostate cancer risk in Caucasian and Hispanic men. Carcinogenesis. 2009;30:1372–1379. doi: 10.1093/carcin/bgp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan C., Hu W., Huang Y., Zhou J., Zheng S. Risk of eighteen genome-wide association study-identified genetic variants for colorectal cancer and colorectal adenoma in Han Chinese. Oncotarget. 2016;7:77651–77663. doi: 10.18632/oncotarget.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Z., Gao M., Yin Z., Yan L., Cui L. Association between lncRNA CASC8 polymorphisms and the risk of cancer: A meta-analysis. Cancer Manag. Res. 2018;10:3141–3148. doi: 10.2147/CMAR.S170783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy A.B., Ukoli F., Freeman V., Bennett F., Aiken W., Tulloch T., Coard K., Angwafo F., Kittles R.A. 8q24 risk alleles in West African and Caribbean men. Prostate. 2012;72:1366–1373. doi: 10.1002/pros.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong Y., Tang Y., Li S., Zhao F., Ying J., Qu Y., Niu X., Mu D. Cumulative evidence of relationships between multiple variants in 8q24 region and cancer incidence. Medicine. 2020;99:e20716. doi: 10.1097/MD.0000000000020716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen A., Liu L., Huang Y., Shen Z., Wu M., Chen X., Wu X., Lin X., Chen Y., Li L., et al. Targeting Spindle-Associated Protein HAUS6 Prevents Cell Growth of Colorectal Cancer Cells by Activating the P53/P21 Pathway. Res. Square. 2020 preprint . [Google Scholar]
- 27.Lin X., Chen Z., Gao P., Gao Z., Chen H., Qi J., Liu F., Ye D., Jiang H., Na R., et al. TEX15: A DNA repair gene associated with prostate cancer risk in Han Chinese. Prostate. 2017;77:1271–1278. doi: 10.1002/pros.23387. [DOI] [PubMed] [Google Scholar]
- 28.Han Y., Rand K.A., Hazelett D., Ingles S.A., Kittles R.A., Strom S.S., Rybicki B.A., Nemesure B., Isaacs W.B., Stanford J.L., et al. Prostate cancer susceptibility in men of African ancestry at 8q24. JNCI J. Natl. Cancer Inst. 2016;108:djv431. doi: 10.1093/jnci/djv431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data will be shared as Supporting Information files if the manuscript is accepted for publication.