Abstract
An unusually thick (∼1 cm) slime developed on a slump of finely disseminated pyrite ore within an extreme acid mine drainage site at Iron Mountain, near Redding, Calif. The slime was studied over the period of 1 year. The subaerial form of the slime distinguished it from more typical submerged streamers. Phylogenetic analysis of 16S rRNA genes revealed a diversity of sequences that were mostly novel. Nearest relatives to the majority of sequences came from iron-oxidizing acidophiles, and it appears that iron oxidation is the predominant metabolic characteristic of the organisms in the slime. The most abundant of the 16S rRNA genes detected were from organisms related to Leptospirillum species. The dominant sequence (71% of clones) may represent a new genus. Sequences within the Archaea of the Thermoplasmales lineage were detected. Most of these were only distantly related to known microorganisms. Also, sequences affiliating with Acidimicrobium were detected. Some of these were closely related to “Ferromicrobium acidophilus,” and others were affiliated with a lineage only represented by environmental clones. Unexpectedly, sequences that affiliated within the delta subdivision of the Proteobacteria were detected. The predominant metabolic feature of bacteria of this subdivision is anaerobic sulfate or metal reduction. Thus, microenvironments of low redox potential possibly exist in the predominantly oxidizing environments of the slime. These results expand our knowledge of the biodiversity of acid mine drainage environments and extend our understanding of the ecology of extremely acidic systems.
Dissolution of sulfide ores exposed to oxygen, water, and microorganisms results in acid production and environmentally detrimental acid mine drainage (AMD) (35). For the aqueous dissolution of sulfide ores dominated by pyrite (FeS2) at low pH, ferric ion is the predominant oxidant. The overall reaction is written: FeS2 + 14Fe3+ + 8H2O→15Fe2+ + 2SO42− + 16 H+. The reaction is limited by the availability of ferric ion. At pH values of less than ∼3.0, the inorganic rate of ferrous oxidation is slow, and acidophilic organisms can mediate production of ferric iron and conserve energy from this. Thus, it is not surprising that the oxidation of pyrite is greatly increased in the presence of iron-oxidizing species such as Thiobacillus ferrooxidans over the abiotic rate; see Nordstrom and Southham (36) for a discussion. Presently the understanding of biological enhanced pyrite oxidation is incomplete, but it is clear that microbial iron oxidation would replenish ferric ions for the above reaction.
The best-studied organism with respect to microbial enhancement of AMD is T. ferrooxidans. Models have been proposed for its energetic characteristics (28) and role in pyrite dissolution (41), and several investigations have studied iron-oxidizing and respiratory enzymes (7, 11, 17). However, numerous microorganisms are known for their acidiphily and iron-oxidizing capabilities, and it is apparent that different microorganisms have different mechanisms and perhaps different abilities for iron oxidation (7). It has therefore been the endeavor of some investigations to increase the understanding of the microbial ecology of AMD systems (29, 43). Such investigations are essential to understanding important microbial processes that underpin the ecology of these environments.
Our research is directed towards improving the understanding of AMD microbial ecology and changes in microbial community structure as a function of geochemical conditions at major sites of acid generation. This information is relevant to constrain laboratory studies and for the modeling of AMD generation. Understanding of the different iron-oxidizing mechanisms is critical for the future development of assays for iron oxidation activity based on probes to conserved features of metabolic pathways.
Our field site at Iron Mountain, Redding, Calif., comprises an AMD system associated with a pyrite-dominated ore body (up to 95% pyrite). Typically the drainage water pH measures between 0.5 and 1.0, metal ion concentrations are in the decagrams per liter range (35), and temperatures range from 30 to 50°C over the course of a year. Presently, mine waters are treated to remove metals and raise pH. Ultimately, an improved understanding of the mine subsurface microbiology, geochemistry, and resultant dissolution may provide information useful for the development of strategies aimed at reducing acid generation rates.
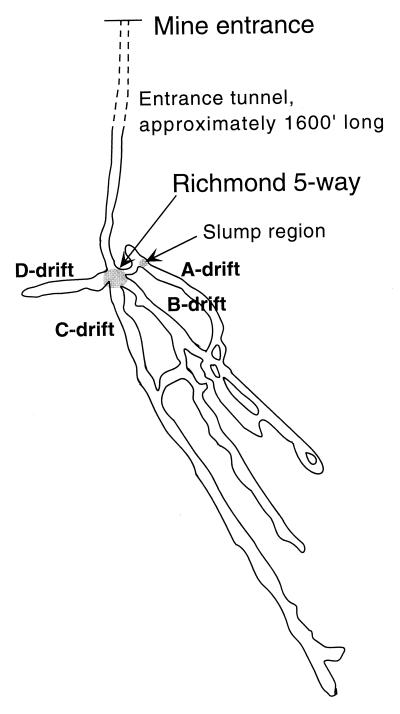
Investigations of microbial communities have focused on samples taken from submerged sediment within the mine at the Richmond 5-way and from the various tunnels extending into the ore body (Fig. 1) (15, 43). Within the mine there are obvious macroscopic differences in the forms of microbial biomass occurring in different localities. Submerged slimes and streamers predominate in flowing water in the mine tunnels. In addition to visible slimes, the surfaces of pyrite grains are often covered by microbial cells. T. ferrooxidans, the organism often associated with AMD, has been detected at only low levels in samples collected within the ore body (43). Cells detected with a Leptospirillum ferrooxidans-specific probe were distributed throughout these samples but made up small proportions of the cells attached to sediment particles (43). Analysis of 16S rRNA gene sequences had previously detected Leptospirillum sequences throughout the mine (40). More recently, extensive analysis of samples collected throughout 1997 indicated substantial fluctuations in geochemical conditions and microbial community compositions and confirmed the scarcity of T. ferrooxidans (15). In the high-ionic-strength conditions, archaea dominated microbial communities. Subsequently, an iron-oxidizing archaeon predominating in some microenvironments within the mine was isolated by us and tentatively named “Ferroplasma acidarmanus” (14). Many of these organisms are inferred to contribute significantly to pyrite dissolution. Peripheral to the ore body, the pH values are higher (those measured in the entrance tunnel are around pH 2.5), and distinct microbial communities exist. Assemblages include T. ferrooxidans, but these are probably irrelevant to pyrite dissolution (43).
FIG. 1.
Cartoon of mine tunnels within the mountain, showing various drifts and position of the slump in the A drift.
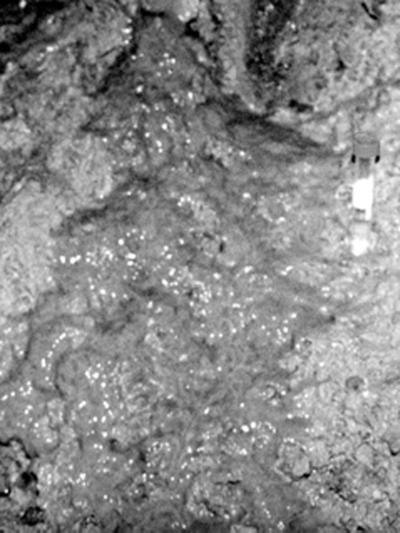
In a region of the A drift, collapse of an overhead stope had resulted in the formation of a slump on the tunnel floor. Hot air streams down from the stope, and water carrying biofilm drips down onto the slump. The dripping slimes are referred to as snottites, a term coined to describe pendulous slime accumulations from other subsurface environments (26). The slump was typically covered in a subaerial slime up to ∼1 cm thick (Fig. 2). Both the slime and the snottite materials differed in appearance from other microbial growths in the mine. This paper describes the results of a molecular phylogenetic analysis of the slump slime and snottite materials. While the majority of the sequences obtained were related to known acidophiles, most of them were novel. These results considerably extend the known diversity of microorganisms in AMD environments.
FIG. 2.
Thick subaerial slime covering part of the slump in the A drift. A 15-ml Falcon centrifuge tube is positioned for scale.
MATERIALS AND METHODS
Sample collection.
Samples were collected from various locations within the mine in November 1998 and in February and May 1999. Specimens for DNA extraction were collected directly into sterile 15-ml Falcon tubes (Becton Dickinson, Paramus, N.J.) and kept on ice or at 4°C until processed (within 3 days). Samples for fluorescence in situ hybridization (FISH) were fixed in freshly prepared 3% paraformaldehyde–phosphate-buffered saline (PBS; 130 mM sodium chloride, 10 mM sodium phosphate buffer [pH 7.2]) solution. The fixed samples were transported to the laboratory on ice, and within 30 h of fixation they were washed in PBS, resuspended in a PBS–96% ethanol solution (1:1, vol/vol), and stored at −20°C prior to hybridization (2).
Microscopy.
For microscopic observations, sample smears were prepared on glass slides. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) at 1.0 μg/ml at room temperature for 5 min (25). Smears were then mounted in Vectorsheild (Vector Laboratories, Burlingame, Calif.) and viewed using a Leica DMRX microscope fitted with UV epifluorescence (Chromatech filter set 31000) and transmitted differential interference contrast.
DNA extraction and purification.
Two methods were used for extraction of nucleic acids from mine samples. Both were based on a bead-beating protocol described by Barns et al. (6) and included initial wash steps to exclude iron and to raise the pH prior to cell lysis. This treatment was necessary as the low pH may cause hydrolysis of DNA and a high iron concentration would have contaminated the extracted DNA, having a deleterious effect on subsequent PCR. Extractions were performed on 0.5-ml samples that were pelleted by centrifugation, washed once with PBS (pH 1.2), washed with one part 2× buffer A (200 mM Tris [pH 8.0], 50 mM EDTA, 200 mM NaCl, 2 mM sodium citrate, 10 mM CaCl2) and one part 50% glycerol, and then resuspended in 0.5 ml of 2× buffer A in 2-ml screw-cap tubes. Pyrophosphate (0.2%), polyadenylic acid (200 μg/ml), and lysosyme (3 mg/ml) were added to the suspension, and this was incubated for 40 min at 37°C. Proteinase K (2 mg/ml) was added to the mixture, and this was incubated for 30 min at 50°C. Samples were treated by bead beating or by freeze-thawing for cell lysis. For bead beating, samples were supplemented with 50% (vol/vol) phenol-chloroform-isoamyl alcohol (24:24:1), 5% (wt/vol) sodium dodecyl sulfate (SDS), and approximately 0.3 g of acid-washed zirconia-silica beads (0.1-mm diameter). The mixture was agitated on a Mini-Beadbeater (Biospec, Bartlesville, Okla.) at low speed for 2 min. For freeze-thawing, the cells were amended with 5% (wt/vol) SDS, subjected to three cycles of freezing in a dry ice-ethanol bath and heating for 3 min at 65°C, and mixed with 50% (vol/vol) phenol-chloroform-isoamyl alcohol (24:24:1). All cell lysates (bead beating and freeze-thawing) were extracted with 50% (vol/vol) phenol-chloroform-isoamyl alcohol (24:24:1), and nucleic acids were precipitated in 50% (vol/vol) isopropanol and 10% (vol/vol) 3 M sodium acetate (pH 5.2) on ice for 20 min. Nucleic acids were pelleted by centrifugation, washed with 70% ethanol, and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). DNA was purified by passage through a ChromaSpin+TE1000 column (Clonetech, Palo Alto, Calif.) and quantified by ethidium bromide-UV detection on an agarose gel. Multiple DNA extracts from a sample were combined prior to PCR amplification.
PCR and fractionation of 16S rRNA genes.
Community 16S rRNA genes were amplified by PCR in mixtures containing approximately 40 ng of purified DNA per ml, 1× PCR buffer (Perkin Elmer, Norwalk, Conn.), a 200 μM concentration of each of the four deoxynucleoside triphosphates, 2.5 mM MgCl2, 350 mM (each) forward and reverse primers, and 0.025 U of AmpliTaq Gold (Perkin Elmer) per μl. In reactions, the reverse primer was the universal 1492R (5′-GGTTACCTTGTTACGACTT-3′) (31), and the forward primer was either Bacteria-specific 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) (31), Archaea-specific 21Fa (5′-TTCCGGTTGATCCYGCCGGA-3′) (12), or the universal 533F (5′-GTGCCAGCMGCCGCGGTAA-3′) (27). A Gene Amp 2400 (Perkin Elmer) was used to incubate reactions through an initial denaturation at 94°C for 12 min, followed by 30 cycles of 94°C for 1 min, 45°C for 45 s, and 72°C for 1.5 min, and completed with an extension period of 20 min at 72°C. For all primer sets, products from four separate reactions were pooled and purified using QIAquick PCR purification columns (Qiagen, Valencia, Calif.). These were quantified on an agarose gel (as above), ligated into the vector pGEM-T (Promega, Madison, Wis.), and used to infect competent host cells following the manufacturer's instructions. When possible for the ligation, we used an insert-vector ratio of 3:1.
RFLP and sequencing of inserted 16S rRNA genes.
For restriction fragment length polymorphism (RFLP) and sequencing, the inserted fragment was amplified by PCR as described above except that DNA was provided by contact of a small pipette tip with a colony of cloned host cells which was immersed in the PCR mixture. Primers for the PCR were the vector-specific T7 and SP6. Restriction enzyme digestion of the unpurified PCR products was carried out, and the restricted fragments were observed, as previously described (27). RFLP patterns were grouped, and representative cloned fragments were sequenced.
Purified PCR products (QIAquick column; see above) were sequenced using the Prism Big Dye terminator sequencing kit (Applied Biosystems, Foster City, Calif.) with 50 to 100 ng of template DNA, according to the manufacturer's instructions. For initial analysis, partial sequences were obtained using the universal primer 533F. Extended sequences were obtained using additional primers 27F and 1492R in separate sequencing reactions. The extension products were purified using Auto Seq G-50 columns (Amersham Pharmacia Biotech, Piscataway, N.J.) and DNA sequences were determined on an automated sequencer (ABI 377XL) at the University of Wisconsin Biotechnology Center.
Phylogenetic analysis.
Phylogenetic affiliations of the partial sequences were initially estimated using the program BLAST (basic local alignment search tool) (1) and available nucleotide databases. Single primer sequences were aligned using the GDE (Genetic Data Environment) multiple sequence editor against close relatives in ARB (a software environment for sequence data) (45). Similarity of partial sequences was determined using ARB, and those with more than 98% similarity were grouped (Table 1). Extended sequences of representatives from the groups were compiled on SeqEd (Applied Biosystems), checked for chimeras on the program CHECK_CHIMERA (34), and used in the phylogenetic analyses. Sequences for analysis were managed in ARB and reduced to unambiguously alignable positions. For bacterial sequences, the Lane mask (31) was used. Evolutionary analyses of alignments were performed by distance methods using ARB and PAUP (47), parsimony in PAUP, and maximum likelihood using fastDNAml (34) essentially as described previously (5, 6). The program MODELTEST was used to determine parameter settings for analyses using PAUP (38).
TABLE 1.
Inventory of the slime (BA) and snottite (SC) 16S rDNA cloned fragments arranged into groups according to RFLP patterns and sequence similarity
| Representative sequencea | No. of clonesb | Microbial group affiliation | Postulated metabolism |
|---|---|---|---|
| BA18 | 1 | δ Proteobacteria | Sulfate/iron reduction |
| BA71 | 4 | δ Proteobacteria | Sulfate/iron reduction |
| BA29 | 69 | Nitrospira group | Iron oxidation/autotrophic |
| BA24 | 4 | Nitrospira group | Iron oxidation/autotrophic |
| BA46 | 13 | Acidimicrobium group | Iron oxidation/heterotrophic |
| BA48 | 1 | Chimera | |
| BA50 | 1 | Chimera | |
| BA84 | 2 | Acidimicrobium group | |
| SC02 | 13 | Nitrospira group | Iron oxidation/autotrophic |
| SC07 | 10 | Nitrospira group | Iron oxidation/autotrophic |
| SC17 | 3 | Nitrospira group | Iron oxidation/autotrophic |
| SC28 | 3 | Nitrospira group | Iron oxidation/autotrophic |
| SC12 | 4 | Thermoplasmales group | |
| SC36 | 2 | Thermoplasmales group | |
| SC38 | 3 | Thermoplasmales group | |
| SC42 | 1 | Thermoplasmales group | Iron oxidation/autotrophic |
Representative clone fragment sequenced for phylogenetic analysis.
Grouped by RFLP pattern and by having ≥98% sequence similarity to the representative sequence within that particular clone library.
Nucleotide sequence accession numbers.
Sequences (excluding potential chimeras) have been submitted to GenBank with accession numbers from AF225446 to AF225459.
RESULTS
The slump slime and snottite materials were first noticed during a sampling trip made to the Iron Mt. mine in November 1998. Samples of the slump slime, snottite, and sediment on the surface of the slump were taken for analyses on this occasion and on subsequent trips. Temperature and pH in water associated with the slime measured in the ranges from 31.5 to 36.8°C and pH 0.77 to 1.21, respectively, throughout the course of sampling. Similar biofilms have been observed in mine tunnels D and C (Fig. 1).
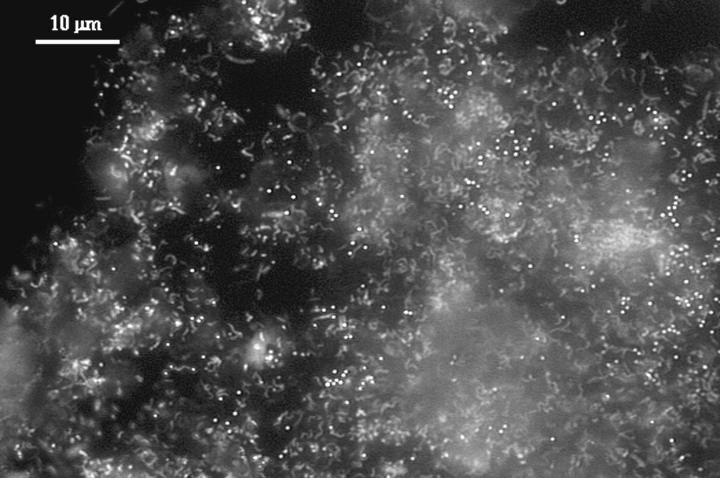
DAPI-stained smears indicated that the slime and snottite were predominantly biological, as opposed to mineralogical (Fig. 3). The biofilms were made up mostly of an extracellular polymeric substance infused with spirillum-shaped cells (approximately 70% of the slime cells and 50% of the snottite cells) and small cocci (approximately 1 μm in diameter). Sediment particles sampled from the base of the slime layer were covered in similar cells. Preliminary analysis of the microbial communities by FISH using domain-specific oligonucleotide probes indicated that the biofilms were mostly bacterial (results not shown). Although the majority of cells were spirillum shaped, these were not detected by FISH with the previously used Leptospirillum group probe LF581 (43) (results not shown). To further analyze the community structures of slime and snottite biofilms, clone libraries of PCR-amplified 16S rRNA genes were prepared.
FIG. 3.
DAPI stain of the slump slime biofilm occurring in the A drift. Curved and straight rods and cocci are evident.
Obtaining 16S rRNA sequences from snottites.
During extraction from AMD samples, we have noticed that the freeze-thaw method produces a greater quantity and less sheared DNA than is produced by bead beating. Thus, the freeze-thaw extraction method was used.
DAPI staining indicated that eukaryotic cells are not abundant in the slime or snottite samples. PCR amplification of the slime 16S rRNA genes was performed with the Archaea- and Bacteria-specific primer sets. From the slime, no PCR amplificates using the archaeon-specific primer pair were observed on an agarose gel. Nevertheless, the archaeal primer amplificate products were concentrated 10-fold for ligation into the cloning vector. However, no ligated inserts could be detected. Therefore, it appeared there were very few or no archaea in the slime. PCR product from the Bacteria-specific primer pair was observed, and this was ligated into the cloning vector. To obtain 16S rRNA gene sequence data, 96 clones were chosen from the bacterial primer clone library (BA library). Extremely effective cloning occurred, as 95 of 96 clones chosen had inserted fragments of the correct size. These clones were given the prefix BA.
Evidence from other investigations indicated that while the Archaea-specific primers failed to amplify 16S rRNA genes from archaea present in the mine, the universal primer set did work (14). Consequently, the universal primer set was used to obtain 16S rRNA gene sequences from the snottite, and 40 clones were analyzed from the snottite library. Of these, 39 had fragments of the correct size that could be sequenced and were given the prefix SC.
Clones were grouped within each of the libraries according to RFLP patterns and sequence similarity. Cloned inserts were amplified by PCR and digested with restriction enzymes (see Materials and Methods). RFLP banding patterns were clearly distinguishable. The 95 BA clones consolidated into 18 groups, and the 39 SC clones reduced to 14 groups. Single-primer sequencing was performed on representatives from the RFLP groups, and these ranged in length from 446 to 721 nucleotides. Sequence similarity then reduced the RFLP groups to eight representative BA sequences and eight representative SC sequences (Table 1). Efficacy of the grouping was confirmed by partial sequencing of multiple representatives from the larger RFLP groups. Partial sequences from the same groups were always greater than 99% similar. Only two clones were found to be strongly chimeric, and highest similarity scores of fragments occurred against sequences from this clone library. BA50 appeared to be a hybrid of clone types BA29 and BA46, and clone BA48 was possibly chimeric to fragments originating from sequence types BA29 and BA24. These possible chimeras were excluded from further analyses.
Phylogenetic relationship of slime and snottite sequence types.
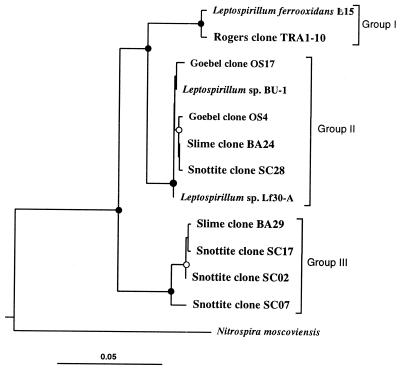
The most abundant sequence type, representing some 69 of the BA clones, was BA29. This sequence was positioned firmly in the Nitrospira group and formed a monophyletic group (group III) with snottite clones SC17, SC02, and SC07 (Fig. 4). These SC clones also collectively represented the majority of the SC library. The group III clones clustered with Leptospirillum sequences from cultured organisms, and this topology was well supported by the phylogenetic analyses (Fig. 4). BA24 and SC28, representing four and three clones of their respective libraries, affiliated strongly with existing Leptospirillum sequences, having >98% similarity to group II types. Clone sequences falling within the Nitrospira group made up 76% of the total clone sequences. This result correlates somewhat with the observed abundance of spirillum-shaped cells (Fig. 3). Considering problems associated with quantification of microbial communities by PCR amplification of 16S rRNA genes (46), this correlation could be fortuitous. However, the dominance of these clones likely reflects the abundance of Leptospirillum in the sample.
FIG. 4.
Evolutionary distance dendogram of slime and snottite clones with “Nitrospira” group sequences based on 781 nucleotides of 16S rDNA. Magnetobacterium bavaricum was used as the outgroup (not shown). Branch points supported by distance, maximum-likelihood, and parsimony estimations (bootstrap values, ≥75%) are indicated by solid circles. Marginally supported branch points (supported by most phylogenetic analyses with bootstrap values of 50 to 74%) are indicated by open circles. Branch points without circles are not supported by the majority of analyses. Evolutionary distances are indicated by the sum of horizontal branch lengths. The scale bar represents changes per nucleotide. Database sequence accession numbers are as follows: Leptospirillum ferrooxidans L15, X86776; Rogers clone TRA1-10, AF047641; Goebel clone OS17, X86772; Leptospirillum sp. BU-1, M79383; Goebel clone OS4, X86770; Leptospirillum sp. Lf30-A, X72852; and Nitrospira moscoviensis, X82558.
Previously, phylogenetic analysis of Leptospirillum sequences defined two groups, I and II (16), that have approximately 93% 16S rDNA sequence similarity (Fig. 4). Results presented here significantly extend the diversity of the Leptospirillum cluster (Fig. 4). Sequences represented by group III clones have only 89 to 93% similarity to sequences from both existing Leptospirillum groups I and II. These likely represent a new group of organisms that are yet to be isolated in culture.
Considerable differences between the group III clone sequences and other Leptospirillum sequences are apparent. A shorter stem in the V5 region is predicted by covariation analysis (23). Also, in the V3 region, from Escherichia coli base numbers 450 to 480, a string of base differences is apparent on both sides of the suggested stem structure. This includes nine consecutive tranversions from the Leptospirillum group II sequence Lf30-A. Coincidentally, this sequence segment (differing V3 region) has been observed previously in a clone derived from an acidic bioleaching environment (21). While that sequence was chimeric, it possibly indicates that Leptospirillum group III types are not restricted to the Iron Mt. mine.
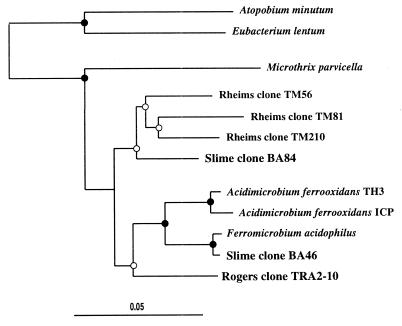
After the Nitrospira-affiliated sequences, clone types clustering with the Acidimicrobium group of the Actinomycete line of descent were the next most abundant of the BA clones (Table 1). BA46, representing 13 clones, was well positioned in the group and clustered with “Ferromicrobium acidophilus,” having 99% similarity to this (Fig. 5). Sequence BA84 was closely related to the Acidimicrobium group and affiliated with clone sequences originating from a peat bog (Fig. 5) (39), having 92 to 93% similarity to these. This lineage contains only environmental clone sequences from a number of different sites (39). As yet no cultured organism of this group has been obtained. Sequence related to the Acidimicrobium group has been detected previously from Iron Mt. mine samples (16). However, that sequence, TRA2-10, was not particularly closely related to those obtained here (Fig. 5).
FIG. 5.
Evolutionary distance dendogram of slime clones with Acidimicrobium group sequences based on 1,107 nucleotides of 16S rDNA. Indications of branch point support and evolutionary distance are as described for Fig. 4. Database sequence accession numbers are as follows: Atopobium minutum, M59059; Eubacterium lentum, AB011817; Microthrix parvicella, X89560; Rheims clone TM56, X92695; Rheims clone TM81, X92697; Rheims clone TM210, X92704; Acidimicrobium ferrooxidans TH3, M79433; Acidimicrobium ferrooxidans ICP, U75647; Ferromicrobium acidophilus, unpublished data; and Rogers clone TRA2-10, AF047642.
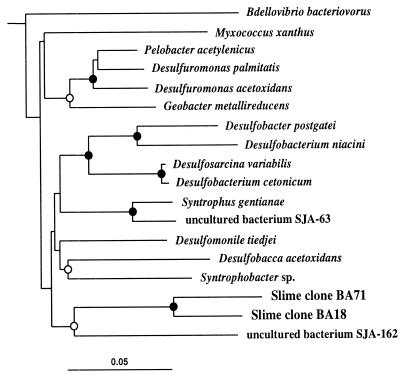
Sequences representing five slime clones were positioned within the delta subdivision of the Proteobacteria (Fig. 6). Sequences BA18 and BA71 were most similar to each other (91%) and ranged from 82 to 78% similarity with other representative delta Proteobacteria sequences. After the phylogenetic analyses (Fig. 6), it was not possible to affiliate the clone sequences with any particular representatives of the subdivision. Nonetheless, to our knowledge, this is the first occasion that microorganisms from the delta subdivision have been detected in such acidic environments.
FIG. 6.
Evolutionary distance dendogram of slime clones within the delta subdivision of the Proteobacteria based on 1,121 nucleotides of 16S rDNA sequences. Desulfovibrio desulfuricans (not shown) was used as the outgroup. Indications of branch point support and evolutionary distance are as described for Fig. 4. Database sequence accession numbers are as follows: Bdellovibrio bacteriovorus, M59297; Myxococcus xanthus, M34114; Pelobacter acetylenicus, X70955; Desulfuromonas palmitatis, U28172; Desulfuromonas acetoxidans, M26634; Geobacter metallireducens, L07834; Desulfobacter postgatei, M26633; Desulfobacterium niacini, U51845; Desulfosarcina variabilis, M34407; Desulfobacterium cetonicum, AJ237603; Syntrophus gentianae, X85132; uncultured bacterium SJA-63, AJ009471; Desulfomonile tiedjei, M26635; Desulfobacca acetoxidans, AF002671; Syntrophobacter sp. X94911; and uncultured bacterium SJA-162, AJ009498.
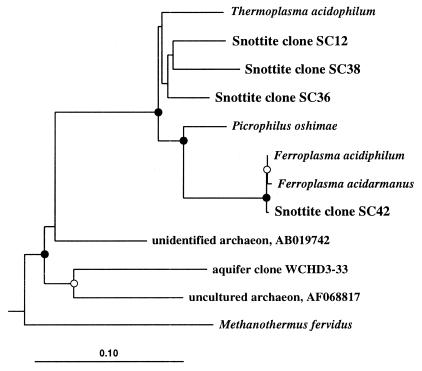
Other clone types detected only in the snottite library were those affiliating within the Thermoplasmales of the domain Archaea (Table 1 and Fig. 7). One clone, SC42, had high sequence similarity with “F. acidarmanus.” This archaeon has been isolated from and detected in other localities within the Iron Mt. mine (14). Other SC clones grouped firmly within the Thermoplasmales and constituted a considerable proportion of the snottite clones (23%). These clones are only distantly related (<93% sequence similarity) to other sequences within the Thermoplasmales group, and the phylogenetic position of these within the group could not be resolved further (Fig. 7).
FIG. 7.
Evolutionary distance dendogram of snottite clones with representative sequences of the Thermoplasmales and relatives based on 870 nucleotides of 16S rDNA sequences. Sulfolobus solfataricus (not shown) was used as the outgroup. Indications of branch point support and evolutionary distance are as described for Fig. 4. Database sequence accession numbers are as follows: Thermoplasma acidophilum, M20822; Picrophilus oshimae, X84901; Ferroplasma acidiphilum, AJ224936; Ferroplasma acidarmanus, AF145441; unidentified archaeon, AB019742; uncultured archaeon, AF068817; aquifer clone WCHD3-33, AF050619; and Methanothermus fervidus, M32222.
DISCUSSION
Microbial diversity and community variability in low-pH biofilms.
Previous culture-based analyses and some molecular analyses have suggested that microbial populations from highly acidic environments tend to have low diversity and be dominated by culturable organisms (20, 48). Consequently, organisms implicated in AMD generation are typically the chemolithoautotrophs T. ferrooxidans and L. ferrooxidans and a variety of acidophilic heterotrophs or facultative heterotrophs (29). Most of the clone sequences from this study affiliate with recognized, cultured acidophiles. This was the expected result and indicates that contamination was not a problem during construction of the clone libraries. However, only clones from Acidimicrobium and Leptospirillum, comprising 10% of the total clone libraries, could be considered closely related to cultured organisms. Most of the sequences detected here represent organisms that constitute new taxa.
New organisms have been detected at this site previously. An archaeon, “F. acidarmanus,” is the dominant prokaryote in some submerged slime streamers. It also colonizes pyrite surfaces associated with more concentrated solutions (14). In combination, this prior study (14) and our current results indicate that microorganisms not previously reported from AMD sites probably contribute significantly to pyrite dissolution and acid production. The detection of novel microorganisms could relate to the characteristics of the study site. Other investigations have sampled AMD from regions peripheral to the sulfide ore bodies, where the pH is higher and culturable bacteria such as T. ferrooxidans thrive (see the report by Schrenk et al. [43]). Furthermore, the novel microorganisms detected in this study are not readily isolated and thus may have been overlooked previously.
Based on the proximity and general similarity in biogeochemical conditions of the slime and snottite, similar microbial communities were expected. Group III type Leptospirillum sequences predominate in both libraries. However, Thermoplasmales-related clones were only detected in the snottite. It is important to note that the primer sets used to construct libraries from the slime would amplify the 16S rRNA genes from this group poorly. Thus, it is probable that Thermoplasmales-related archaea are present in the slime as well. Also, a greater diversity of bacterial clones was detected in the slime library, suggesting a more complex community there (however, this result may be somewhat biased by the size of the clone library analyzed). Although the snottites and slimes are exposed to essentially the same solutions, some differences in chemical conditions could occur and result in variation in the microbial population compositions. The snottites are exposed to a continuous flow of solutions in contact with air, whereas slimes form a more coherent biofilm and associated fluids are probably less oxygenated. Thus, there is the possibility that microenvironments exist within the slime for the support of anaerobes and microaerophilic bacteria.
Annual changes in microbial population compositions have been observed at specific sites within the mine (43), and these variations show a strong correspondence with ambient physical and chemical conditions. Thus, it appears that microorganisms are finely adapted to suit specific temperature-, pH-, or solution chemistry-defined niches. The clone library reported in this study is distinct from those in cloning studies of other, much less extreme environments (8, 18) in that the vast majority of these acidic biofilm clones affiliate with only a handful of species. In situ hybridization studies (7a; P. L. Bond, G. K. Druschel, and J. F. Banfield, submitted for publication) support the conclusion that, at the scale of sampling, the subaerial biofilms studied have a relatively simple microbial composition. This is probably a consequence of a limited supply of electron donors and acceptors and the multiple challenges presented by the very high hydrogen ion activity, high metal activities, and elevated temperature.
Metabolic inferences and microbial ecology.
Although microbial diversity in individual samples is low, microbial growth is prolific. Water entering the mine workings is provided by precipitation on a steep, moderately vegetated hillslope a few hundred feet above the ore body. The levels of organic carbon in mine solutions are low and are attributed to degradation of lithotrophic cells and not to externally derived organic carbon. Consequently, carbon fixation in this ecosystem is based on chemolithoautotrophic oxidation of iron and sulfur compounds.
Although microorganisms of the biofilms have not been isolated, it may be possible to predict their metabolic capabilities based on those of phylogenetically related organisms. We consider this valid because the organisms tend to fall within clusters that utilize the same substrate(s) and have distinct, shared physiological characteristics. Consequently, we use phylogeny-based metabolic and physiological inferences to correlate geochemical with microbial characteristics so as to develop an initial understanding of the ecology of these environments.
Leptospirillum species are acidophilic, gram-negative bacteria that are morphologically helical curved rods (37). Different types of Leptospirillum have been detected in different acidic environments. Leptospirillum has been formally recognized as a coherent bacterial genus (25a), and its members form a phylogenetically distinct cluster within the taxon Nitrospira (32). Genomic diversity, as shown by DNA hybridization analyses, has been demonstrated for some isolates (24). Based on phylogeny, two possibly genus-level groups were suggested initially (16). These are referred to here as group I (the original isolate) and group II. Groups I and II are autotrophic (4), oxidize iron for energy (7, 37), and have optimum growth temperatures of 26 to 30°C (group I) and 30 to 40°C (group II) (19). Group III sequences (this paper) have low similarity with the existing sequences and probably represent a new genus within the Leptospirillum. Because Leptospirillum group III clusters with groups I and II, we suggest that group III members are also autotrophic iron oxidizers. An additional species, referred to as Leptospirillum ferrooxidans, has a higher (∼45 to 50°C) temperature optimum (22). However, the phylogenetic placement of L. thermoferrooxidans, and thus its relationship to group III organisms, is not known. The predominance of the Leptospirillum clones (78%) likely reflects the fact that the prevalent metabolism within the biofilms is iron oxidation. Phenotypic variability of Leptospirillum groups requires further consideration.
It was suggested that group I Leptospirillum organisms are the less abundant type (21). Our initial work at Iron Mt. mine detected only group I type Leptospirillum associated with sediments and pore fluids (16). However, in the biofilms, only sequences from group II and group III types were detected. We speculate that group III is distinct in that it is optimized for different conditions. Confirmation of the factors controlling distribution of Leptospirillum species awaits in situ characterization of their habitats in a variety of acidic environments.
Sequence data reported here have important implications for the development of oligonucleotide probes for characterization of microbe distributions in environmental samples. Prior studies used a Leptospirillum group probe (LF581) developed on the group I and II sequences available at the time (43). However, two mismatches occur between the target region for LF581 and group III Leptospirillum. Under the hybridization conditions used here, group III Leptospirillum cells would not be detected with LF581. Comparison of the sequence data indicates that probes could be developed for each group, as well a probe for all Leptospirillum species. This would allow resolution of the geochemical habitats for group I versus group II versus group III Leptospirillum.
Clones that positioned in the Archaea were placed in the Thermoplasmales and, except for SC42, were only distantly related to known organisms. Thermoplasmales lack a cell wall and are hyperacidophilic (42, 44). Most Thermoplasmales are aerobic heterotrophs. However, T. acidophilum can respire anaerobically using sulfur compounds other than sulfate (44). “F. acidarmanus” is an iron oxidizer that is almost certainly autotrophic as well as heterotrophic (14). The detection of new Thermoplasmales taxa extends our understanding of archaeal biodiversity in acid microbial communities and emphasizes the probable importance of these organisms in AMD production.
Both Acidimicrobium and “Ferromicrobium” species are iron-oxidizing, heterotrophic, acidophilic bacteria. In this environment their source of organic carbon is almost certainly the slime-associated biomass. Acidimicrobium ferrooxidans is capable of autotrophic growth (10). However, “Ferromicrobium acidophilus” requires organic carbon (3). Strains of A. ferrooxidans and “F. acidophilus” also possess iron-reducing capabilities, using ferric iron as an electron acceptor during low redox potential (3, 9). From its phylogenetic placement, BA46 may represent iron-oxidizing, heterotrophic bacteria. It has been suggested that heterotrophic growth of “F. acidophilus” removes dissolved organic carbon inhibitory to coexisting autotrophs (3). Acidimicrobium and “Ferromicrobium” may play a similar synergistic role in the biofilm microbial communities described here.
Clones falling within the delta Proteobacteria could represent anaerobic sulfate-reducing heterotrophs, as sulfate reduction is the dominant physiological trait of this group (13). They could also be iron-reducing bacteria, as similar anaerobic respiration involving metal (iron) reduction has also been detected in this group (30, 33). As well, the clones may represent aerobic bacteria of the Myxobacteria and Bdellovibrio genera. These genera also fall into the delta Proteobacteria. Although sulfate and ferric iron concentrations are high in solutions, sulfate and/or iron reducers have not been previously observed in such acidic environments. It is possible that local concentrations of bioavailable carbon overlap with oxygen-limited microenvironments and that iron- or sulfate-reducing microbes can operate in more highly acidic environments than has been previously recognized. Within the microbial consortium, sulfate- or iron-reducing heterotrophs could have the same synergistic effect as mentioned for Acidimicrobium and “Ferromicrobium.”
Implications for future studies.
Presently there is some understanding of metabolic and physiological characteristics of acidophiles pertinent to AMD production (29). However, the results of this study emphasize that understanding of microbial diversity and proportions comprising communities where AMD production occurs is incomplete, if not inaccurate. Ultimately, we aim to present a more complete picture of microbially mediated AMD production. At this point it is not appropriate to use the sequence data to quantify relative proportions of taxa within these samples. Variation in the ability of the primers to amplify target genes during PCR will bias the pool of sequences obtained. An extreme case of this was observed here when archaeal genes were not amplified with the Archaea-specific primers, but these were obtained using the universal primers. More importantly, the sequence data obtained here will be used to design specific oligonucleotide probes for in situ quantification of the microbial communities. Results of this work will be reported separately (7a; Bond et al., submitted).
Understanding of the ecology of highly acidic environments requires investigation of metabolic pathways and understanding of microbial survival strategies. In the long term, an assay for the level of iron oxidation in the environment is needed. In order to develop such an assay, understanding of the diversity of iron-oxidizing organisms and the biochemistry of their metabolic pathways is critical. Thus, culturing, isolation, and characterization of newly detected species are essential. Results of this study provide information about species habitats that can be used to direct future isolation and culturing strategies.
ACKNOWLEDGMENTS
We thank Norman Pace (UC Berkeley) and Michael Tanner for assistance with the ARB software and for helpful discussion. We also thank Brett Goebel and Katrina Edwards for ongoing discussion of acidophile diversity and many helpful suggestions. Thomas Gihring and Greg Druschel are thanked for their help with sample collection.
This project received NSF support via a supplement to grant CHE 9521731, as well as support from NSF CHE 9807598. Assistance from Bob Hamers is also gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacelar-Nicolau P, Johnson D B. Leaching of pyrite by acidophilic heterotrophic iron-oxidizing bacteria in pure and mixed cultures. Appl Environ Microbiol. 1999;65:585–590. doi: 10.1128/aem.65.2.585-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balashova V V, Vedenina I Y, Markosyan G E, Zavarzin G A. The auxotrophic growth of Leptospirillum ferrooxidans. Microbiology. 1974;43:491–494. [PubMed] [Google Scholar]
- 5.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily, and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot-spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake R C, Shute E A, Greenwood M M, Spencer G H, Ingledew W J. Enzymes of aerobic respiration on iron. FEMS Microbiol Rev. 1993;11:9–18. doi: 10.1111/j.1574-6976.1993.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 7a.Bond, P. L., and J. F. Banfield. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol., in press. [DOI] [PubMed]
- 8.Borneman J, Skroch P W, Osullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridge T A M, Johnson D B. Reduction of soluble iron and reductive dissolution of ferric iron-containing minerals by moderately thermophilic iron-oxidizing bacteria. Appl Environ Microbiol. 1998;64:2181–2186. doi: 10.1128/aem.64.6.2181-2186.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark D A, Norris P R. Acidimicrobium ferrooxidans gen nov, sp nov—mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology. 1996;142:785–790. doi: 10.1099/00221287-142-4-785. [DOI] [PubMed] [Google Scholar]
- 11.Cox J C, Boxer D H. The role of rusticyanin, a blue copper protein, in the electron transport chain of Thiobacillus ferrooxidans grown on iron or thiosulfate. Biotechnol Appl Biochem. 1986;8:269–275. [Google Scholar]
- 12.Delong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereux R, Delaney M, Widdel F, Stahl D A. Natural relationships among sulfate-reducing eubacteria. J Bacteriol. 1989;171:6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards K J, Bond P L, Gihring T M, Banfield J F. A new iron-oxidizing, extremely acidophilic archaea is implicated in acid mine drainage generation. Science. 2000;287:1796–1799. doi: 10.1126/science.287.5459.1796. [DOI] [PubMed] [Google Scholar]
- 15.Edwards K J, Gihring T M, Banfield J F. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol. 1999;65:3627–3632. doi: 10.1128/aem.65.8.3627-3632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards K J, Goebel B M, Rodgers T M, Schrenk M O, Gihring T M, Cardona M M, Hu B, McGuire M M, Hamers R J, Pace N R, Banfield J F. Geomicrobiology of pyrite (FeS2) dissolution: case study at Iron Mountain, California. Geomicrobiol J. 1999;16:155–179. [Google Scholar]
- 17.Elbehti A, Nitschke W, Tron P, Michel C, Lemesle-Meunier D. Redox components of cytochrome bc-type enzymes in acidophilic prokaryotes. I. Characterization of the cytochrome bc(1)-type complex of the acidophilic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J Biol Chem. 1999;274:16760–16765. doi: 10.1074/jbc.274.24.16760. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goebel B M, Stackebrandt E. The biotechnological importance of molecular biodiversity studies for metal bioleaching. In: Priest F G, Ramos-Cormenzana A, Tindall B J, editors. Bacterial diversity and systematics. VIII. New York, N.Y: Plenum Press; 1994. pp. 259–273. [Google Scholar]
- 20.Goebel B M, Stackebrandt E. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol. 1994;60:1614–1621. doi: 10.1128/aem.60.5.1614-1621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goebel B M, Stackebrandt E. Molecular analysis of the microbial biodiversity in a natural acidic environment. In: Jerez C A, Vargas T, Toledo H, Wiertz J V, editors. Biohydrometallurgical processing. II. Santiago, Chile: University of Chile; 1995. pp. 43–52. [Google Scholar]
- 22.Golovacheva R S, Golyshina O V, Karavaiko G I, Dorofeev A G, Pivovarova T A, Chernykh N A. A new iron-oxidizing bacterium, Leptospirillum thermoferrooxidans sp. nov. Microbiology. 1992;61:744–750. [Google Scholar]
- 23.Gutell R R. Collection of small-subunit (16S and 16S-like) ribosomal RNA structures—1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison A P, Jr, Norris P R. Leptospirillum ferrooxidans and similar bacteria: some characteristics and genomic diversity. FEMS Microbiol Lett. 1995;30:99–102. [Google Scholar]
- 25.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Hippe H. Leptospirillum gen. nov. (ex Markosyan 1972), nom. rev., including Leptospirillum ferrooxidans sp. nov. (ex Markosyan 1972), nom. rev. and Leptospirillum thermoferrooxidans sp. nov. (Golovacheva et al. 1992) Int J Syst Evol Microbiol. 2000;50:501–503. doi: 10.1099/00207713-50-2-501. [DOI] [PubMed] [Google Scholar]
- 26.Hose L D, Pisarowcz J A. Cueva de Villa Luz, Tabasco, Mexico: reconnaissance study of an active sulfur spring cave and ecosystem. J Cave Karst Studies. 1999;61:13–21. [Google Scholar]
- 27.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingledew W J, Houston A. The organization of the respiratory chain of Thibacillus ferrooxidans. Biotechnol Appl Biochem. 1986;8:242–248. [Google Scholar]
- 29.Johnson D B. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol. 1998;27:307–317. [Google Scholar]
- 30.Jones J G, Davison W, Gardener S. Iron reduction by bacteria: range of organisms involved and metals reduced. FEMS Microbiol Lett. 1984;21:133–136. [Google Scholar]
- 31.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 32.Lane D J, Harrison A P, Stahl D, Pace B, Giovannoni S J, Olsen G J, Pace N R. Evolutionary relationships among sulfur-oxidizing and iron-oxidizing eubacteria. J Bacteriol. 1992;174:269–278. doi: 10.1128/jb.174.1.269-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovley D R, Giovannoni S J, White D C, Champine J E, Phillips E J P, Gorby Y A, Goodwin S. Geobacter metallireducens gen nov sp nov, a microorganism capable of coupling the complete oxidation of organic-compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 34.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordstrom D K, Alpers C N. Negative pH, efflorescent mineralogy, and consequences for environmental restoration at the Iron Mountain Superfund site, California. Proc Natl Acad Sci USA. 1999;96:3455–3462. doi: 10.1073/pnas.96.7.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordstrom D K, Southam G. Geomicrobiology of sulfide mineral oxidation. In: Banfield J F, Nealson K H, editors. Geomicrobiology: interactions between microbes and minerals. Vol. 35. Washington, D.C.: Mineralogical Society of America; 1997. pp. 361–390. [Google Scholar]
- 37.Pivovarova T A, Markosyan G E, Karavaiko G I. The auxotrophic growth of Leptospirillum ferrooxidans. Microbiology. 1981;50:339–344. [Google Scholar]
- 38.Posada D, Crandall K A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 39.Rheims H, Sproer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers T M. Bacterial diversity in acid mine drainage from Iron Mt., Shasta County, California: a 16S ribosomal RNA approach. Masters thesis. University of Wisconsin, Madison; 1996. [Google Scholar]
- 41.Sand W, Gerke T, Hallmann R, Schippers A. Sulfur chemistry, biofilm, and the (in)direct attack mechanism—a critical evaluation of bacterial leaching. Appl Microbiol Biotechnol. 1995;43:961–966. [Google Scholar]
- 42.Schleper C, Puehler G, Holz I, Gambacorta A, Janekovic D, Santarius U, Klenk H P, Zillig W. Picrophilus gen. nov., fam. nov.—a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J Bacteriol. 1995;177:7050–7059. doi: 10.1128/jb.177.24.7050-7059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrenk M O, Edwards K J, Goodman R M, Hamers R J, Banfield J F. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans—implications for generation of acid mine drainage. Science. 1998;279:1519–1522. doi: 10.1126/science.279.5356.1519. [DOI] [PubMed] [Google Scholar]
- 44.Segerer A H, Stetter K O. The genus Thermoplasma. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 712–718. [Google Scholar]
- 45.Strunk O, Ludwig W. ARB—a software environment for sequence data. Munich, Germany: Department of Microbiology, Technical University of Munich; 1995. [Google Scholar]
- 46.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swofford D L. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b2 ed. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 48.Walton K C, Johnson D B. Microbiological and chemical characteristics of an acidic stream draining a disused copper mine. Environ Pollut. 1992;76:169–175. doi: 10.1016/0269-7491(92)90105-j. [DOI] [PubMed] [Google Scholar]