Abstract
Oxytetracycline-resistant (OTr) mesophilic aeromonads were recovered from untreated hospital effluent (72 isolates) and from fish farm hatchery tanks (91 isolates) at sites within the English Lake District, Cumbria, England. The transfer of OTr plasmids from these isolates was investigated. Using Escherichia coli J53-1 as a recipient, 11 isolates from the hospital site and 6 isolates from the fish farm site transferred OTr plasmids (designated pFBAOT1 to 17). Original isolates were identified using fatty acid methyl ester and fluorescent amplified fragment length polymorphism comparisons as either Aeromonas hydrophila HG3 (eight isolates), A. veronii b.v. sobria HG8 (six isolates), and A. caviae HGB5 (one isolate). One isolate remained unidentified, and one could not be assigned a taxonomic designation beyond the genus level. Plasmids pFBAOT1 to -17 were screened for the presence of the tetracycline resistance determinants Tet A to E and Tet G. Only determinant Tet A (10 plasmids) was detected in these plasmids, with 7 tet gene determinants remaining unclassified. In all cases, Tet A was located on a 5.5-kb EcoRI restriction fragment. Hybridization with inc-rep probes N, P, Q, W, and U showed pFBAOT3, -4, -5, -6, -7, -9, and -11, from the hospital environment, to be IncU plasmids. Further, restriction fragment length polymorphism (RFLP) analyses and DNA probing demonstrated that pFBAOT plasmids were closely related to IncU OTr plasmids pASOT, pASOT2, pASOT3, pRAS1 (originally isolated from A. salmonicida strains from fish farms in Scotland and Norway, respectively), and pIE420 (isolated from a German hospital E. coli strain). In addition, DNA analyses demonstrated that plasmids pRAS1 and pIE420 had identical RFLP profiles and that all fragments hybridized to each other. The presence of tetracycline resistance transposon Tn1721 in its entirety or in a truncated form in these plasmids was demonstrated. These results provided direct evidence that related tetracycline resistance-encoding plasmids have disseminated between different Aeromonas species and E. coli and between the human and aquaculture environments in distinct geographical locations. Collectively, these findings provide evidence to support the hypothesis that the aquaculture and human compartments of the environment behave as a single interactive compartment.
Members of the genus Aeromonas are ubiquitous in most aquatic environments (19). Several species have been implicated in fish disease (e.g., Aeromonas hydrophila, A. sobria, A. allosaccharophila, A. salmonicida, and A. veronii; 6), and pathogenicity in humans has been demonstrated by A. hydrophila, A. veronii, A. jandaei, A. trota, and A. schubertii (23). Treatment or prevention of disease in both humans and fish has been undertaken extensively using antimicrobial agents. It has been estimated that the total amount of antibiotics used in aquaculture and agricultural practices is approximately equal to that employed in the therapy of human disease (46), as typified by Norway in 1989 (14). This has resulted in an increase in the frequency of bacteria resistant to these agents to the extent that they may affect the treatment of human and fish diseases, in addition to impacting the aquaculture environment directly (40). It was concluded that there was inadequate information to allow a quantitative or even a qualitative assessment of the risk to human health posed by antimicrobial agent usage (28, 40). Smith et al. (40) suggested that the greatest potential risk was presented by the transfer of plasmid-encoded resistance genes between the aquatic compartment and the human compartment of the environment. Furthermore, they identified our lack of understanding of the survival of bacterium-plasmid complexes in the environment as the major limitation in our knowledge (40).
Tetracyclines (TCs) have been used exclusively in aquaculture, particularly to control furunculosis in salmonids (40). Inevitably, resistance to oxytetracycline (OT) and TC emerged and was found to be plasmid encoded (1, 37, 38). The frequency of resistance to TCs in A. salmonicida has increased with greater usage from 4% of isolates from 1979 to 1981 (4) to greater than 50% of isolates examined from Scottish fish farms in the early 1990s (34). Assessing the extent of antibiotic resistance in bacterial isolates presents many difficulties (25, 26). Adams and coworkers (1) recognized that there was a need to assess the true nature of OTr at the molecular level. Their strain set contained 66% of isolates with transferable OTr which was subsequently shown to be encoded by large plasmids (the pASOT group) that carried a 5.4-kb EcoRI restriction fragment containing the Tet A determinant (1). The detection of OTr bacteria in the aquaculture industry showed that this environment, like hospitals, was facing a threat from the use of antibiotics (1). Their study also highlighted the need to extend the investigation to isolates from other geographical locations and to other fish pathogens (1). Furthermore, they suggested that the ability of plasmids to spread among the bacterial population present in, and beyond, fish farm environments should be investigated (1).
In this study, we have assessed whether there is sufficient evidence to suggest that the aquaculture and human environments exist as two separate entities with distinct transfer events or whether they comprise a single interactive compartment of the environment where free exchange of genetic information occurs. To test these hypotheses, we investigated the dissemination of conjugative OTr plasmids between isolates in the human and aquaculture compartments of the environments using OTr mesophilic aeromonads recovered from untreated hospital effluent and from a fish farm facility as model organisms and environments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and sampling.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains and Pseudomonas putida PaW340 (a derivative of P. putida nt-2) were maintained on Iso-Sensitest agar (ISA; Oxoid, Basingstoke, United Kingdom). All OTr control strains were maintained on ISA supplemented with OT-HCl (25 μg ml−1) obtained from Vetrapharm (Hampshire, United Kingdom).
TABLE 1.
Strains and plasmids used in this study
| Bacterium | Plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|---|
| E. coli J53-1 | Rifrpro met | NUIa | |
| P. putida PaW340 | Smrtrp | 47 | |
| E. coli J53 | pIE420 | Tcr Sur Smr Tmr | 44 |
| A. salmonicida MT0903 | pASOT | Tcr Smr | 1 |
| A. salmonicida MT1431 | pASOT2 | Tcr | 1 |
| A. salmonicida MT0336 | pASOT3 | Tcr Smr | 1 |
| A. salmonicida 718 | pRAS1 | Tcr Sur Smr Tmr | 37 |
| E. coli | pMT1286 | Apr Tcr; pBR322::Tn1722 | 45 |
| E. coli | pULB2429 | Tcr; source of IncU probe | 9 |
| E. coli | pULB2432 | Apr Tcr; source of IncN probe | 9 |
| E. coli | pULB2420 | Kmr; source of IncP probe | 9 |
| E. coli | pULB2426 | Tcr; source of IncW probe | 9 |
| E. coli | pULB2424 | Apr Tcr; source of IncQ probe | 9 |
| E. coli JM83 | pUC18::750-bp SmaI fragment of pSL18 | Source of Tet A probe | 3 |
| E. coli HB101 | pRT11 | Source of Tet B probe | 3 |
| E. coli DO-7 | pBR322 | Source of Tet C probe | 3 |
| E. coli C600 | pACYC177::3.05-kb HindIII-PstI region of pSL106 | Source of Tet D probe | 3 |
| E. coli HB101 | pACYC177::2.5-kb ClaI-PvuI region of pSL1504 | Source of Tet E probe | 3 |
| E. coli | pMOSBlue::2.6-kb region of pJA8122 | Source of Tet G probe | 48 |
NUI, National University of Ireland, Galway.
Mesophilic aeromonads were recovered from two sample sites. Hospital effluent isolates were recovered from a sewer at Westmorland General Hospital (Kendal, Cumbria, United Kingdom) from a point where domestic (i.e., from lavatories) and pathological effluents converge. Samples were taken on dry days in order to prevent mixture of effluent with storm water runoff from surrounding areas. Samples were collected on four separate occasions between September and October 1997 and were obtained by lowering an ethanol-washed plastic container into the effluent flow. From this master sample, 25 individual sterile McCartney bottles were immediately filled at the site and returned to the laboratory and processed within 1 h. Fish farm samples were taken from a hatchery facility located within the grounds of the Freshwater Biological Association (Cumbria, United Kingdom). The facility was sampled on four separate occasions between October and November 1997 by dislodging independent areas of biofilm from the bottom of runoff channels into 25 individual sterile McCartney bottles. Samples were placed on ice and returned to the laboratory for processing within 30 min.
Environmental mesophilic aeromonads from each individual McCartney bottle were recovered by dilution plating onto m-Aeromonas selective agar (BIOLIFE; ampicillin (AP)-dextrin agar; 16) supplemented with OT (25 μg ml−1) and incubated at 28 ± 0.5°C for 24 h. OTr was confirmed by streaking five randomly selected presumptive aeromonads from each bottle onto ISA supplemented with OT (25 μg ml−1) and incubation at 28°C for 24 h. One of the five confirmed isolates was then maintained for further study.
Antibiotic susceptibility testing.
Susceptibility of aeromonad field isolates and transconjugants to the antibiotics AP (25 μg), OT (30 μg), TC (30 μg), streptomycin (SM; 10 and 30 μg), nalidixic acid (NA; 30 μg), rifampin (RD; 25 μg), kanamycin (KM; 30 μg), and trimethoprim (TM; 5 μg) was assessed on ISA in accordance with the established Kirby-Bauer procedure (7) using disks supplied by Oxoid.
Identification of isolates.
All isolates were initially confirmed as Aeromonas spp. at the genus level by PCR amplification of the aroA gene as described previously (42). Further characterization was carried out by gas-liquid chromatographic analysis of cellular fatty acid methyl esters using the Microbial Identification System (Microbial ID Inc.) as previously described (22). The resulting fatty acid methyl ester patterns were identified automatically and compared with the database AER48C (20) for the identification of mesophilic aeromonads. Isolates that could not be unambiguously classified into one of the known Aeromonas DNA hybridization groups (HGs) or genomic species by comparison with the AER48C database were subjected to whole-genome analysis with the fluorescent amplified fragment length polymorphism analysis technique as described previously (21). Digitized fluorescent amplified fragment length polymorphism fingerprints were further processed using the GelCompar software package (Applied Maths) and compared for identification with the laboratory database AEROLIB (21).
Conjugation experiments.
Filter matings were performed by separately resuspending a loopful of freshly cultured donor and recipient cells in 300 μl of 1× phosphate-buffered saline (pH 7.4) (36), followed by overlaying 10 μl of each suspension onto a 0.22-μm-pore-size membrane filter (Supor-200; Gelman) and then incubation at 28 ± 0.5°C for 24 h. Controls (unmixed donors and recipient cells) were treated in the same manner. After incubation, cells and controls were resuspended in 450 μl of phosphate-buffered saline and transconjugants were selected by spreading onto either ISA supplemented with OT (25 μg ml−1) and RD (50 μg ml−1) at 37°C for 24 h (when E. coli J53-1 was used as the recipient) or ISA supplemented with OT (25 μg ml−1) and SM (500 μg ml−1) for 24 h at 30°C (when P. putida was the recipient). E. coli J53-1 transconjugants were confirmed by demonstration of OT and RD resistance phenotypes and auxotrophy for proline and methionine. P. putida transconjugants were confirmed by OT and SM resistance and auxotrophy for tryptophan.
DNA manipulations.
Restriction endonuclease digestion and agarose gel electrophoresis were carried out using established techniques (36). Plasmid DNA was extracted from control strains and E. coli J53-1 transconjugants after growth in Luria-Bertani medium supplemented with OT (25 μg ml−1) at 37°C with shaking at 150 rpm for 18 h using Qiagen mini and midi columns or using the sucrose gradient plasmid extraction technique (47).
Construction and labeling of DNA probes.
DNA probes for the determinants Tet B and Tet D were constructed as previously described (31) and were purified using GenElute agarose gel purification spin columns (Supelco). Probes for the determinants Tet A, Tet C, Tet E, and Tet G were created via PCR using primers specific to these determinants in accordance with an established protocol (15). PCR probes were purified using the Nucleon QC PCR/oligo clean up kit (Amersham-Pharmacia Biotech). Probes specific to Tn1722 and the methyl-accepting chemotaxis protein (MCP)-encoding gene of Tn1721 were constructed from pMT1286 (45). The entire Tn1722 region was produced by digesting pMT1286 with EcoRI and excision and purification of the 5.6-kb fragment. The MCP probe was prepared by digestion of pMT1286 with EcoRI and HindIII to produce a fragment of 1.2 kb. Excised DNA was purified using the Qiaex II DNA purification kit (Qiagen). DNA probes were labeled with [α-32P]dCTP as specified in the random-primed hexanucleotide labeling kit (Roche-Molecular Biochemicals).
DNA blotting and hybridizations.
In all cases, DNA was blotted onto positively charged nylon membrane (Roche-Molecular Biology). Southern transfers (43) were carried out in accordance with standard procedures (36) using 0.4 M NaOH as the transfer, denaturation, and fixation buffer. Dot blots were performed with QIAGEN plasmid DNA (denatured at 95°C for 10 min), and the DNA was fixed with UV light (302 nm) for 3 min. Prehybridization was carried out in prewarmed (68°C) 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 5× Denhardt's solution, 0.5% (wt/vol) sodium dodecyl sulfate (SDS), and 0.25% (wt/vol) N-lauryl sarcosine for 5 to 6 h. Hybridization was performed in fresh prewarmed solution (prehybridization solution without the addition of Denhardt's solution) at 68°C for 18 to 20 h. Unbound radioactive probe DNA was removed by washing membranes twice for 10 min (each time) in 2× SSPE–0.1% (wt/vol) SDS at room temperature (20 to 25°C), followed by 15 min at 68°C in 1× SSPE–0.1% (wt/vol) SDS and two washes of 15 min (each) in 0.1× SSPE–0.1% (wt/vol) SDS at 68°C. The membranes were then wrapped in cling film and exposed to X-ray film (Hyperfilm-MP; Amersham-Pharmacia) at −70°C for up to 3 days.
Cloning.
Plasmid DNA was cloned into pUC18 (Amersham-Pharmacia-Biotech) in accordance with standard procedures (36). Transformation was carried out using E. coli DH5α competent cells in accordance with the methodology of the supplier (Clontech).
DNA sequencing and analyses.
Cloned DNA was partially sequenced using the universal forward and reverse primers homologous with pUC18 using an ABI Prism 373 sequencer. FASTA and BLAST DNA homology searches were performed using the programs within the DNA Data Bank of Japan (DDBJ) at the following internet address: http://www.ddbj.nig.ac.jp/E-mail/homology.html.
RESULTS AND DISCUSSION
Isolation of OTr aeromonads and conjugation experiments.
A total of 163 presumptive OTr mesophilic aeromonads were isolated from the hospital (72 isolates) and fish farm (91 isolates) sample sites. All isolates were assessed for susceptibility to the antibiotics TC, OT, RD, SM, NA, and KM using sensitivity test disks. All isolates were confirmed as resistant to TC and OT. The most common resistance observed in both environmental groups was to NA (94% of isolates from the hospital and 52% of isolates from the fish farm), while 14% of hospital isolates and 40% of the fish farm isolates were resistant to SM. Resistance to both NA and SM was observed in isolates from both environments. A total of 90% of those isolates that were SM resistant, and 13% of the NA-resistant isolates from the hospital were resistant to both antibiotics, while 75% of SM-resistant and 57% of NA-resistant isolates from the fish farm were resistant to both. No isolates were RD or KM resistant.
The susceptibility of all of the isolates to RD permitted the use of E. coli J53-1 (plasmid free, RD-resistant, and OTs) as a recipient strain in conjugation experiments. Conjugation experiments involving all of the isolates and E. coli J53-1 were carried out, and 17 transconjugants demonstrating the correct phenotype (11 from the hospital and 6 from the fish farm) were obtained. Agarose gel electrophoresis showed that for each transconjugant, only one plasmid was transferred (data not shown). The plasmids were designated pFBAOT1 to -11 (hospital donors) and pFBAOT12 to -17 (fish farm donors).
The cotransfer of other antibiotic resistances was assessed by screening E. coli J53-1 transconjugants for resistance to the antibiotics AP, minocycline, and TM in addition to those mentioned previously. Transconjugants containing plasmids pFBAOT6 (from the hospital) and pFBAOT13 (from the fish farm) were found, in addition to being OT resistant to be resistant to SM at 10 μg/ml but not at 25 μg/ml. The original isolates were also resistant to these antibiotics. Therefore, SM resistance was assumed to be plasmid encoded. Resistance to AP, minocycline, and TM was not observed in any of the transconjugants.
Identification of host strains.
The identification of host strains that carried the 17 transferable OTr plasmids is shown in Table 2. Eight isolates were identified as members of A. hydrophila DNA HG3 and contained plasmids pFBAOT3 to -5, -7, -9 to -11 and -15. Six others were identified as members of A. veronii bv. sobria HG8 (plasmids pFBAOT1, -8, -12, -13, -14, and -17). The isolate that originally carried pFBAOT16 could not be identified beyond the genus level (Table 2).
TABLE 2.
Characterization of OTr pFBAOT plasmids
| Plasmid | Original host | Resistance phenotype | Tet determinant | Incompatibility group | Host rangea |
|---|---|---|---|---|---|
| pFBAOT1 | A. veronii bv. sobria HG8 | Sms Tms | A | Unknown | E. coli |
| pFBAOT2 | Unknownb | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT3 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT4 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT5 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT6 | A. caviae HGB5 | Smr Tms | A | U | E. coli, P. putidac |
| pFBAOT7 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT8 | A. veronii bv. sobria HG8 | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT9 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT10 | A. hydrophila HG3 | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT11 | A. hydrophila HG3 | Sms Tms | A | U | E. coli, P. putidac |
| pFBAOT12 | A. veronii bv. sobria HG8 | Sms Tms | A | Unknown | E. coli |
| pFBAOT13 | A. veronii bv. sobria HG8 | Smr Tms | A | Unknown | E. coli |
| pFBAOT14 | A. veronii bv. sobria HG8 | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT15 | A. hydrophila HG3 | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT16 | Aeromonas sp. | Sms Tms | Unknown | Unknown | E. coli |
| pFBAOT17 | A. veronii bv. sobria HG8 | Sms Tms | Unknown | Unknown | E. coli |
| pASOT | A. salmonicidad | Smr Tms | A | U | E. coli, P. putidad |
| pASOT2 | A. salmonicidad | Sms Tms | A | U | E. coli, P. putidad |
| pASOT3 | A. salmonicidad | Smr Tms | A | U | E. coli, P. putidad |
| pRAS1 | A. salmonicidae | Smr Tmr | A | U | E. coli, P. putidae |
| pIE420 | E. colif | Smr Tmr | A | U | NDg |
Host range assessments.
In an attempt to determine whether the pFBAOT plasmids are capable of transfer to recipients other than E. coli, conjugal transfer was carried out between each of the 17 original isolates and P. putida PaW340. Transfer of OTr to P. putida was demonstrated for plasmids pFBAOT3 to -7, -9, and -11. It was also shown that these plasmids could be retransferred from P. putida transconjugants back into OTs plasmid-free E. coli J53-1 (Table 2). These results suggested that pFBAOT3 to -7, -9, and -11 were potentially broad-host-range plasmids.
Molecular characterization of pFBAOT plasmids.
We assessed which tet determinants and replicons were carried by the pFBAOT plasmids. Minipreparations of plasmids pFBAOT1 to -17 were screened by dot hybridization using the Tet A to E and Tet G probes and those specific to broad-host-range replicons N, P, Q, W, and U. It was shown that the Tet A determinant was carried by 10 plasmids (pFBAOT1, -3 to -7, -9, and -11 to -13) (Table 2), and no other characterized determinants were detected. Therefore, the remaining seven plasmids (pFBAOT2, -8, -10, -14, -15, -16, and -17) may possess previously undescribed tet determinants. Seven plasmids hybridized with the IncU probe (Table 2), and these were the same plasmids that displayed broad-host-range characteristics (plasmids pFBAOT3 to -7, -9, and -11). In addition, all of these plasmids carried Tet A (Table 2). The remaining 10 plasmids did not hybridize with the inc-rep probe set.
The dominance of determinant Tet A, as demonstrated in this study, is not uncommon (1, 24, 30), although the relatively small sample size of the present investigation may have precluded the detection of other determinants. The occurrence of Tet A alone in 19 Scottish A. salmonicida isolates recovered over a period of 11 years has been reported (1). In a study of 68 OTr plasmids from a diverse group of bacteria within the National Collection of Type Cultures, it was shown that Tet A and Tet B were carried most frequently (32.4 and 50% of the isolates, respectively), followed by Tet C (8.8%) and Tet D (5.9%), while 2.9% of the isolates that did not possess any of the determinants Tet A to E (24). Furthermore, Tet B was shown to be the most common determinant in bacteria isolated from pigs (30) while Tet E was dominant in marine sediments (3), as it was with Tet A and Tet D in A. hydrophila from cultured catfish (11, 12). Tet D has also been associated with other fish pathogens, including Pasteurella piscicida (now called Photobacterium damselae subsp. piscicida) and Edwardsiella tarda (5). Tet G has not been detected in any bacteria other than Vibrio anguillarum (35). It is therefore unsurprising that it was not detected in this study.
Analysis of Tet A-containing plasmids.
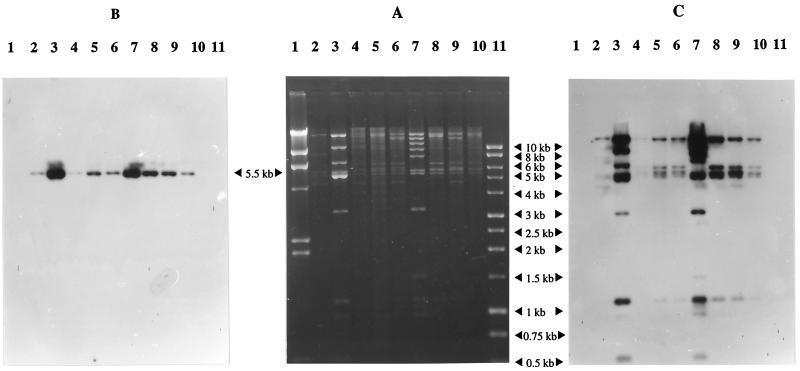
The restriction fragment length polymorphism (RFLP) patterns of plasmid DNA from Tet A-carrying pFBAOT plasmids were compared after digestion with EcoRI. Plasmids from hospital isolates were shown to be closely related to each other, with pFBAOT7 and pFBAOT11 being identical (Fig. 1A). Plasmid extractions from fish farm E. coli transconjugants were repeatedly of poor quality, and so RFLP patterns for plasmids pFBAOT12 to -17 were difficult to interpret. For this reason, the profiles of these plasmids are absent from Fig. 1. However, it was clear that they did not share close homology and were larger than the hospital-derived plasmids (pFBAOT1 to -11) (data not shown). Two EcoRI restriction fragments of approximately 5.2 and 5.5 kb were common to seven of the eight Tet A-carrying hospital-derived plasmids (pFBAOT3 to -7, -9, and -11). pFBAOT1, which was not IncU (and therefore was not included in Fig. 1) and was originally found in A. veronii bv. sobria HG8, carried only the 5.5-kb fragment. In addition, common fragments of approximately 5.7 and 6.5 kb were observed in pFBAOT3 to -5, -7, -9, and -11 (all originally found in A. hydrophila HG3) but were absent from pFBAOT1 and -6 (from A. caviae HG5A) (Fig. 1A; see Fig. 4A). It was difficult to discern from RFLP data alone whether any of the fish farm-derived plasmids carried these common fragments.
FIG. 1.
(A) RFLP profiles (generated by EcoRI digestion) of IncU OTr plasmids from aeromonads and corresponding Southern hybridizations with determinant Tet A and pASOT plasmid. Lanes: 1 and 11, λ DNA markers digested with HindIII and a 1-kb ladder, respectively; 2, pIE420; 3, pASOT; 4, pFBAOT3; 5, pFBAOT4; 6, pFBAOT5; 7, pFBAOT6; 8, pFBAOT7; 9, pFBAOT9; 10, pFBAOT11. (B) Hybridization of the Tet A determinant probe to plasmids. All plasmids harbored Tet A on a 5.5-kb fragment. (C) Hybridization of plasmid pASOT to IncU plasmids.
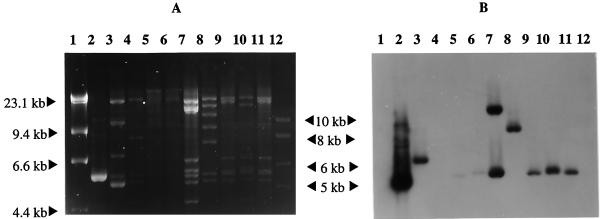
FIG. 4.
(A) EcoRI-generated RFLP patterns for IncU OTr plasmids. The gel was electrophoresed for an extended period in order to separate the fragments in the 5- to 7-kb range. Lanes: 1 and 12, λ DNA markers digested with HindIII and a 1-kb ladder, respectively; 2, pMT1286; 3, pASOT; 4, pIE420; 5, pFBAOT3; 6, pFBAOT4; 7, pFBAOT5; 8, pFBAOT6; 9, pFBAOT7; 10, pFBAOT9; 11, pFBAOT11; 12, 1-kb marker DNA. (B) Southern hybridization of the MCP probe to the DNA in panel A.
Previously, Adams and coworkers (1) reported the localization of Tet A on an EcoRI restriction fragment approximately 5.4 kb in size in OTr plasmids (pASOT plasmids) from Scottish isolates of A. salmonicida (recovered after furunculosis outbreaks). These workers also showed that OTr-encoding plasmid pRAS1 (originally discovered in A. salmonicida isolated from Atlantic salmon with furunculosis in a fish farm near Bergen, Norway; R.-A. Sandaa, personal communication) carried Tet A on the same fragment. Plasmid pRAS1 has also been shown to possess the IncU replicon (R.-A. Sandaa, A. G. Eide, K. Y. Mazengia, B. K. Thorsen, and Ø. Enger, First Symp. EU-Concerted Action Mobile Elements' Contrib. Bacterial Adaptability Diversity (MECBAD), p. 84, 1998) and to be closely related to the pASOT plasmids (based upon RFLP assessments, antibiotic resistances, and transfer frequencies; 1). It was not established whether the pASOT plasmids also contained the IncU replicon. However, it was suggested that the Tet A fragment had “established a foothold among Scottish isolates of A. salmonicida and had persisted for at least 11 years” (1). The discovery of the same Tet A fragment in pRAS1 from Norwegian A. salmonicida was evidence for the foothold extending to other geographical regions. This study, in light of these findings, assessed whether the pFBAOT plasmids are closely related to the pASOT plasmids and pRAS1 and whether they too carry Tet A on the 5.4- to 5.5-kb fragment. We also included pIE420, an OTr IncU plasmid originally isolated from E. coli (44), in our study. This plasmid produced an EcoRI digestion fragment of approximately 5.4 kb, although the determinant conferring its OTr had not been characterized (44).
Southern blots of EcoRI-digested DNA from the IncU pFBAOT plasmids, the pASOT plasmids, and pIE420 were probed with the Tet A probe. The determinant was shown to be carried on a 5.4- to 5.5-kb fragment by all of the IncU plasmids (Fig. 1B). Further study showed that the non-IncU plasmids pFBAOT1 (hospital) and pFBAOT12 and -13 (fish farm) also carried Tet A on a 5.5-kb EcoRI fragment. This was also confirmed using pASOT as a probe, despite these plasmids showing no overall homology with the IncU pFBAOT plasmids (data not presented). These plasmids originated in A. veronii bv. sobria HG8. It was clear from RFLP patterns that the IncU pFBAOT plasmids, the pASOT group, pRAS1, and pIE420 are closely related to each other. Plasmids pRAS1 and pIE420 have identical EcoRI-derived RFLP patterns (data not shown). In addition, both plasmids are resistant to sulfonamides, SM, TC, and TM (8, 38). We hybridized pIE420 to pRAS1 and showed that they produced identical autoradiograph patterns (Fig. 2). These results indicated that pIE420 and pRAS1 are probably identical plasmids although only DNA sequencing can demonstrate this beyond doubt. The relatedness of the pFBAOT plasmids with pASOT and pIE420 or pRAS1 was also demonstrated by hybridization of the entire pASOT plasmid to a duplicate Southern blot of EcoRI-digested DNA shown in Fig. 1A (Fig. 1C). Six of the seven IncU pFBAOT plasmids shared a core of six to seven common fragments, while plasmid pFBAOT6 possessed a different core but shared more common fragments with pASOT than with the other pFBAOT plasmids. Both pFBAOT6 and pASOT possess an EcoRI fragment of approximately 3.1 kb that has been implicated with conferring SM resistance in the latter (1). Our findings supported this hypothesis, as pFBAOT6 demonstrated resistance to SM whereas the other IncU plasmids devoid of this EcoRI fragment were sensitive (Table 2). The relatedness of these plasmids suggested that the pASOT group contained the U replicon. This was confirmed by hybridization of the IncU probe to pASOT, pASOT2, and pASOT3 (Table 2). The carriage of the U replicon by these plasmids is consistent with the findings of other researchers, as it is commonly associated with aeromonads (18). Plasmid RA3, the original IncU plasmid from which the IncU probe is derived, was originally isolated from A. hydrophila in Japan (8, 17). However, this plasmid does not confer TC resistance upon its host, although other A. hydrophila isolates have been shown to carry TC-conferring resistant IncU plasmids (e.g., plasmids pUG1001 and R1463 recovered from A. hydrophila isolated in Ireland and France, respectively; 18). IncU plasmids have also been shown to occur outside the genus Aeromonas. For instance, plasmid pIE420 was originally isolated from E. coli from a hospital patient with pyelonephritis in Osterweick, Germany (44).
FIG. 2.
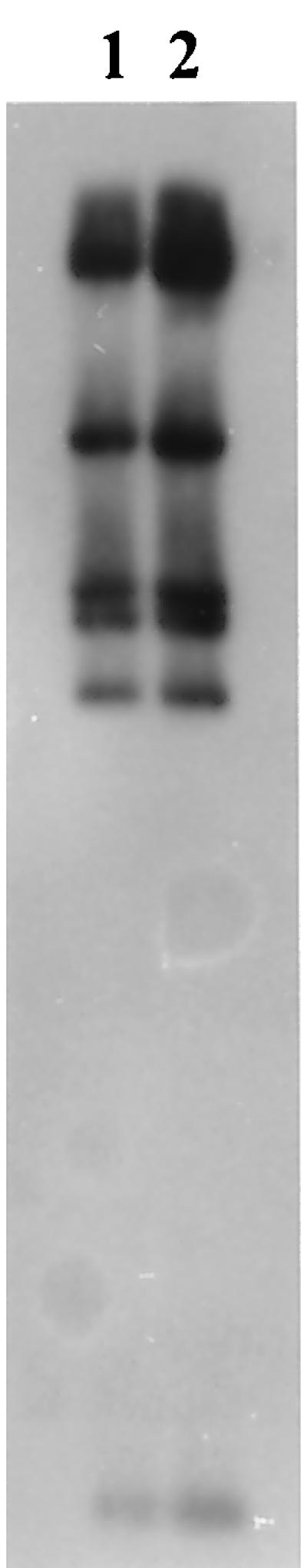
Comparison of plasmids pRAS1 and pIE420 by hybridization of radiolabeled plasmid pRAS1 to both plasmids. Lanes: 1, pRAS1; 2, pIE420.
The lack of DNA homology between pFBAOT plasmids and probes specific to IncN, -P, -Q, and -W is consistent with the findings of other workers (10, 29, 41). It is increasingly clear that the Inc probes described by Couturier et al. (9) are not generally suitable for analyzing the replicons of environmental plasmids (10, 29, 41). We have not assessed whether the non-IncU pFBAOT plasmids belong to one of the other Inc groups outside our original screening strategy. Future work would include screening for IncC plasmids, as they have been found to be associated with TC resistance-encoding plasmids in Aeromonas sp. (18). These included plasmids in A. hydrophila from Japan (plasmid RA1) and France (where the plasmids were not given a designation; 33) and in A. salmonicida from the United Kingdom (plasmid R1491).
Jones and coworkers (24) assessed the relationship between enterobacterial TC resistance and plasmid incompatibility. Their findings suggested that a limited correlation exists between the class of TC resistance group and the plasmid incompatibility group. Tet A, for example, was associated with IncP and IncM plasmids, while Tet B was associated mostly with the IncF and IncH complexes and the IncC group. However, Mendez and coworkers (32) found no such correlation although only 24 different plasmids were analyzed in their study, compared to 68 in that of Jones' group (24).
Cloning and partial sequencing of the 5.5-kb EcoRI Tet A-carrying fragment.
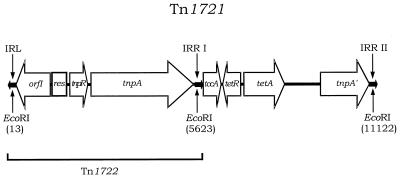
The results presented here show that the 5.5-kb EcoRI Tet A-carrying fragment is more widespread than has been previously suggested (1) and that it is not exclusively associated with IncU plasmids. This fragment was shotgun cloned from plasmid pFBAOT6 into pUC18, and sequencing was carried out from each end of the cloned insert such that partial sequences of 647 and 620 bp were obtained. The first 647 bp of one end of the insert shared 95% homology with Tn1721 (encompassing the first right inverted repeat (IRR-I), the tccA gene (27), and part of the tetR gene, respectively). The first 620 bp of the opposite end of the insert shared 95% homology with IRR-II and part of the tnpA′ region of Tn1721 (data not shown). This demonstrated that the 5.5-kb EcoRI fragment from pFBAOT6 was highly homologous to the 5.5-kb EcoRI fragment found in Tn1721. In order to assess whether the complete transposon Tn1721 was present on these plasmids, a DNA probe was constructed from plasmid pMT1286 (45) that was specific to the gene (orfI) which encodes MCP, situated at the left-hand end of Tn1721 (2) (Fig. 3). This probe was hybridized to plasmid pASOT (pASOT2 and pASOT3 were not assessed), pIE420, and the IncU pFBAOT plasmids. The results showed that all of the IncU pFBAOT plasmids and pASOT carry this gene, unlike plasmid pIE420 (Fig. 4C). Plasmids pFBAOT3, -4, -5, -7, -9, and -11 carry the MCP-encoding gene on a 5.6- to 5.8-kb EcoRI fragment (slightly larger than the region carrying the Tet A determinant). Plasmid pFBAOT5 also carried a second copy of the gene on a fragment of approximately 12 to 13 kb, suggesting that this plasmid contains two copies of a Tn1721-like element. Plasmid pASOT carries the region on a 7-kb EcoRI restriction fragment, while a 9-kb fragment was detected in pFBAOT6 (Fig. 4). The presence of the MCP region on a 5.6- to 5.8-kb EcoRI fragment, in addition to a 5.5-kb Tet A-carrying EcoRI fragment, is indicative of possession of the complete Tn1721 transposon (Fig. 3). Therefore, plasmids pFBAOT3 to -5, -7, -9, and -11 appear to possess complete transposon sequences (indicating that the MCP-encoding gene-containing region is 5,610 bp in length; Fig. 3). The differences between this group and pASOT and pFBAOT6 might be explained in light of recent findings by Schnabel and Jones (39). They demonstrated that the Tet A-mediated TC resistance observed in phylloplane Pseudomonas sp. was due to a truncated form of Tn1721 (designated Tn1720). Tn1720 lacks the MCP-encoding region (orfI) and has minor variations in the sequence of the three inverted repeats on Tn1721. This variation within the left inverted repeat (IRL) resulted in an alteration at the EcoRI site (39). Plasmids pASOT and pFBAOT6 possessed the MCP-encoding region and therefore did not harbor Tn1720. However, the location of the MCP-encoding region on fragments larger than 5.6 kb along with a 5.5-kb Tet A-carrying EcoRI fragment is indicative of the loss of the EcoRI site within the IRL. The absence of the MCP-encoding region from pIE420 suggested that it may contain a Tn1720-like transposon. If this were the case, then the transposition genes tnpR and tnpA would still be present on the plasmid. To test this hypothesis, the plasmid was probed with the complete Tn1722 transposon and no hybridization signal was obtained (data not shown). This result suggested that the 5.5-kb Tet A-carrying EcoRI fragment was deposited by Tn1721 but with subsequent loss of minitransposon Tn1722 (Fig. 3). This scenario might also be applicable to the unrelated plasmids pFBAOT1, -12, and -13, which also carry Tet A on the 5.5-kb EcoRI fragment.
FIG. 3.
Physical and genetic structure of transposon Tn1721 (length, 11,139 bp). The 38-bp inverted repeats IRL, IRR-I, and IRR-II are highlighted. The designation, position, and direction of transcription of genes are shown by the arrowed boxes. The region that comprises the minor transposon Tn1722 is highlighted, along with the location of EcoRI restriction sites. This diagram was adapted from Schnabel and Jones (39) and Tsuda et al. (45).
The most closely related pFBAOT plasmids (pFBAOT3, -4, -5, -7, -9, and -11), isolated from A. hydrophila HGB3, each contain regions specific to the whole of Tn1721, while plasmids pFBAOT1, pFBAOT12, and pFBAOT13 were isolated in A. veronii bv. sobria HG8 and carried only the 5.5-kb Tet A region of Tn1721. Plasmid pFBAOT6, which originated from A. caviae HG5A, harbored a Tn1721-like element that appeared to have an altered IRL region. The differences observed between these plasmids in the carriage of Tn1721 and Tn1721-like regions may be indicative of differences in the nature or rate of host-related transposition events. Coupled to the plasmids themselves and Tn1721, the host strain could represent an important factor in the manner by which dissemination of these mobile elements is governed.
Conclusions.
Adams et al. (1) highlighted the need for research on OT usage in aquaculture and the resistance associated with relevant bacteria to extend to isolates from other geographical locations and to other fish pathogens. In addition, these authors highlighted the need to determine whether these resistance plasmids extend beyond fish farm environments (1). The main objectives of this study were to carry out these assessments and to investigate whether the aquaculture and human compartments of the environment should be considered separate entities with distinct transfer events or a single interactive compartment of the environment. In the course of these analyses, we showed that closely related IncU R plasmids previously associated only with fish farm environments were common to those impacting humans (i.e., pASOT plasmids and pRAS1 from fish farms and pFBAOT plasmids and pIE420 from hospital sewage and a German hospital patient, respectively). In addition, this dissemination occurred among at least four separate countries (Norway, Scotland, England, and Germany). Furthermore, the occurrence of pFBAOT plasmids in A. hydrophila and A. caviae demonstrated that they could disseminate to other related bacteria under natural conditions. This study also showed for the first time that plasmids pRAS1 and pIE420 are probably identical. The original carriage of pIE420 in E. coli (44) and pRAS1 in A. salmonicida (37) demonstrated interspecies transfer from (or to) a human commensal (and potentially pathogenic) organism under natural conditions. Collectively, these findings provide evidence that suggests that we should consider the two environments (fish farm and hospital) one interactive compartment and are therefore contrary to the hypothesis of Smith and coworkers (40). The involvement of Tn1721 and Tn1721-like elements in the dissemination of the Tet A determinant, as demonstrated here and by other workers (13, 39), is extremely relevant to this interaction and, globally, to the potential dissemination of these and similar plasmids.
ACKNOWLEDGMENTS
The support of the European Community (FAIR CT96 1703) is gratefully acknowledged.
We thank Louisa Faulkner for technical assistance and Ruth-Anne Sandaa, Dougie McIntosh, Joyce Petri, Erhard Tietze, and Masataka Tsuda for provision of plasmid-containing control strains.
REFERENCES
- 1.Adams C A, Austin B, Meaden P G, McIntosh D. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl Environ Microbiol. 1998;64:4194–4201. doi: 10.1128/aem.64.11.4194-4201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organisation and a novel gene product with features of a chemotaxis protein. Gene. 1992;111:11–20. doi: 10.1016/0378-1119(92)90597-i. [DOI] [PubMed] [Google Scholar]
- 3.Andersen S R, Sandaa R-A. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl Environ Microbiol. 1994;60:908–912. doi: 10.1128/aem.60.3.908-912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki T, Kitao T, Iemura N, Mitoma Y, Nomura T. The susceptibility of Aeromonas salmonicida strains isolated in cultured and wild salmonids to various chemotherapeutants. Bull Jpn Soc Sci Fish. 1983;49:17–22. [Google Scholar]
- 5.Aoki T, Takahashi A. Class D tetracycline resistance determinants of R plasmids from the fish pathogens Aeromonas hydrophila, Edwardsiella tarda, and Pasteurella piscicida. Antimicrob Agents Chemother. 1987;31:1278–1280. doi: 10.1128/aac.31.8.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin B, Adams C. Fish pathogens. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. New York, N.Y: John Wiley & Sons Publishers; 1996. pp. 197–229. [Google Scholar]
- 7.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45:93–496. [PubMed] [Google Scholar]
- 8.Bradley D E, Aoki T, Kitao T, Arai T, Tschäpe H. Specification of characteristics for the classification of plasmids in incompatibility group U. Plasmid. 1982;8:89–93. doi: 10.1016/0147-619x(82)90045-2. [DOI] [PubMed] [Google Scholar]
- 9.Couturier M, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlberg C, Linberg L, Torsvik V L, Hermansson M. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePaola A, Flynn P A, McPhearson R M, Levy S B. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl Environ Microbiol. 1988;54:1861–1863. doi: 10.1128/aem.54.7.1861-1863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePaola A, Roberts M C. Class D and E tetracycline determinants in gram-negative bacteria from catfish ponds. Mol Cell Probes. 1995;9:311–313. doi: 10.1016/s0890-8508(95)91572-9. [DOI] [PubMed] [Google Scholar]
- 13.Frech G, Schwartz S. Plasmid-encoded tetracycline resistance in Salmonella enterica subsp. enterica serovars choleraesuis and typhimurium: identification of complete and truncated Tn1721 elements. FEMS Microbiol Lett. 1999;176:97–103. doi: 10.1111/j.1574-6968.1999.tb13648.x. [DOI] [PubMed] [Google Scholar]
- 14.Grave K, Engelstad M, Søli N E, Håstein T. Utilisation of antibacterial drugs in salmonid farming in Norway during 1980–1988. Aquaculture. 1990;86:347–358. [Google Scholar]
- 15.Hansen L M, Blanchard P C, Hirsh D C. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob Agents Chemother. 1996;40:1558–1560. doi: 10.1128/aac.40.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havelaar A H, During M, Versteegh J F. Ampicillin-dextrin agar medium for the enumeration of Aeromonas species in water by membrane filtration. J Appl Bacteriol. 1987;62:279–287. doi: 10.1111/j.1365-2672.1987.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 17.Hedges R W, Datta N. fi− R factors giving chloramphenicol resistance. Nature. 1971;234:220–221. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- 18.Hedges R W, Smith P, Brazil G. Resistance plasmids of aeromonads. J Gen Microbiol. 1985;131:2091–2095. [Google Scholar]
- 19.Holmes P, Niccolls L M, Sartory D P. The ecology of mesophilic Aeromonas in the aquatic environment. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. New York, N.Y: John Wiley & Sons Publishers; 1996. pp. 127–150. [Google Scholar]
- 20.Huys G, Kersters I, Vancanneyt M, Coopman R, Janssen P, Kersters K. Diversity of Aeromonas sp. in Flemish drinking water production plants as determined by gas-liquid chromatographic analysis of cellular fatty acid methyl esters (FAMEs) J Appl Bacteriol. 1995;78:445–455. doi: 10.1111/j.1365-2672.1995.tb03432.x. [DOI] [PubMed] [Google Scholar]
- 21.Huys G, Swings J. Evaluation of a fluorescent amplified fragment length polymorphism (FAFLP) methodology for the genotypic discrimination of Aeromonas taxa. FEMS Microbiol Lett. 1999;177:83–92. [Google Scholar]
- 22.Huys G, Vancanneyt M, Coopman R, Janssen P, Falsen E, Altwegg M, Kersters K. Cellular fatty acid composition as a chemotaxonomic marker for the differentiation of phenospecies and hybridization groups in the genus Aeromonas. Int J Syst Bacteriol. 1994;44:651–658. [Google Scholar]
- 23.Janda J M, Abbott S L. Human pathogens. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. New York, N.Y: John Wiley & Sons Publishers; 1996. pp. 151–173. [Google Scholar]
- 24.Jones C S, Osbourne D J, Stanley J. Enterobacterial tetracycline resistance in relation to plasmid incompatibility. Mol Cell Probes. 1992;6:313–317. doi: 10.1016/0890-8508(92)90007-k. [DOI] [PubMed] [Google Scholar]
- 25.Jones J G, Gardener S, Simon B M, Pickup R W. Antibiotic resistant bacteria in Windermere and two remote upland tarns in the English Lake District. J Appl Bacteriol. 1986;60:443–453. doi: 10.1111/j.1365-2672.1986.tb05090.x. [DOI] [PubMed] [Google Scholar]
- 26.Jones J G, Gardener S, Simon B M, Pickup R W. Factors affecting the measurement of antibiotic resistance in bacteria isolated from lake water. J Appl Bacteriol. 1986;60:455–462. doi: 10.1111/j.1365-2672.1986.tb05091.x. [DOI] [PubMed] [Google Scholar]
- 27.Jovanovic O S, Figurski D H. A potential new gene (tccA) on IncP plasmid RK2 and transposon Tn1721: relationship of its product to the TrwC relaxase/helicase of IncW plasmid R388. Plasmid. 1997;38:220–223. doi: 10.1006/plas.1997.1314. [DOI] [PubMed] [Google Scholar]
- 28.Kayser F H. Evolution of resistance in microorganisms of human origin. Vet Microbiol. 1993;35:257–267. doi: 10.1016/0378-1135(93)90150-6. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi N, Bailey M J. Plasmids isolated from the sugar beet phyllosphere show little or no homology to molecular probes currently available for plasmid typing. Microbiology. 1994;140:289–296. doi: 10.1099/13500872-140-2-289. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Langlois B E, Dawson K A. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl Environ Microbiol. 1993;59:1467–1472. doi: 10.1128/aem.59.5.1467-1472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall B, Tachibana C, Levy S B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983;24:835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980;3:99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- 33.Mizon F M, Gerbaud G R, Leclere H, Chabbert Y A. Présence de plasmids de résistance appartenant un groupe d'incompatabilité C chez Aeromonas hydrophila isolé d'eaux résiduaires. Ann Microbiol. 1978;129B:19–26. [PubMed] [Google Scholar]
- 34.Richards R H, Inglis V, Frerichs G N, Miller S D. Variation in antibiotic resistance patterns of Aeromonas salmonicida isolated from Atlantic salmon Salmo salar L. in Scotland. In: Michel C, Alderman D, editors. Chemotherapy in aquaculture: from theory to reality. Paris, France: Office International des Epizooties; 1992. pp. 276–284. [Google Scholar]
- 35.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sandaa R-A, Enger Ø. Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl Environ Microbiol. 1994;60:4234–4238. doi: 10.1128/aem.60.12.4234-4238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandaa R-A, Enger Ø. High frequency transfer of a broad host range plasmid present in an atypical strain of the fish pathogen Aeromonas salmonicida. Dis Aquat Org. 1996;24:71–75. [Google Scholar]
- 39.Schnabel E L, Jones A L. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl Environ Microbiol. 1999;65:4898–4907. doi: 10.1128/aem.65.11.4898-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith P, Hiney M, Samuelson O. Bacterial resistance to antimicrobial agents used in fish farming: a critical evaluation of method and meaning. Annu Rev Fish Dis. 1994;4:273–313. [Google Scholar]
- 41.Sobecky P A, Mincer T J, Chang M C, Helinski D R. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soriano A C, Castillo J A, Moral C H, Salazar M S, Marcos J Y, Carrasco G N. RFLP-PCR analysis of the aroA gene as a taxonomic tool for the genus Aeromonas. FEMS Microbiol Lett. 1997;156:199–204. doi: 10.1111/j.1574-6968.1997.tb12727.x. [DOI] [PubMed] [Google Scholar]
- 43.Southern E M. Detection of specific sequences among DNA fragments separated by agarose gel electrophoresis. J Mol Biol. 1975;98:503–513. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 44.Tschäpe H, Tietze E, Koch C. Characterization of conjugative R plasmids belonging to the new incompatibility group IncU. J Gen Microbiol. 1981;127:155–160. doi: 10.1099/00221287-127-1-155. [DOI] [PubMed] [Google Scholar]
- 45.Tsuda M, Minegishi K-I, Iino T. Toluene transposons Tn4651 and Tn4653 are class II transposons. J Bacteriol. 1989;171:1386–1393. doi: 10.1128/jb.171.3.1386-1393.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walton J R. Use of antibiotics in veterinary practice. J Med Microbiol. 1992;36:69–70. doi: 10.1099/00222615-36-2-69. [DOI] [PubMed] [Google Scholar]
- 47.Wheatcroft R, Williams P A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981;124:433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Aoki T. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol Immunol. 1992;36:1051–1060. doi: 10.1111/j.1348-0421.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]