Abstract
An Escherichia coli O157:H7 dps::nptI mutant (FRIK 47991) was generated, and its survival was compared to that of the parent in HCl (synthetic gastric fluid, pH 1.8) and hydrogen peroxide (15 mM) challenges. The survival of the mutant in log phase (5-h culture) was significantly impaired (4-log10-CFU/ml reduction) compared to that of the parent strain (ca. 1.0-log10-CFU/ml reduction) after a standard 3-h acid challenge. Early-stationary-phase cells (12-h culture) of the mutant decreased by ca. 4 log10 CFU/ml while the parent strain decreased by approximately 2 log10 CFU/ml. No significant differences in the survival of late-stationary-phase cells (24-h culture) between the parent strain and the mutant were observed, although numbers of the parent strain declined less in the initial 1 h of acid challenge. FRIK 47991 was more sensitive to hydrogen peroxide challenge than was the parent strain, although survival improved in stationary phase. Complementation of the mutant with a functional dps gene restored acid and hydrogen peroxide tolerance to levels equal to or greater than those exhibited by the parent strain. These results demonstrate that decreases in survival were from the absence of Dps or a protein regulated by Dps. The results from this study establish that Dps contributes to acid tolerance in E. coli O157:H7 and confirm the importance of Dps in oxidative stress protection.
Escherichia coli O157:H7 causes hemorrhagic colitis in humans and in some cases may incite hemolytic-uremic syndrome (23, 24). Data from epidemiological investigations indicate that as few as 10 to 100 cells of E. coli O157:H7 per g of raw ground beef are sufficient to cause illness (1, 4, 14). Additionally, person-to-person transmission has occurred in day care facilities, and waterborne transmission has resulted from swimming in contaminated waters (24, 36, 42). Collectively, these and other epidemiological investigations establish that this pathogen has a low infectious dose. Gordon and Small (22) suggested that human pathogens with a low infectious dose that are transmitted by the fecal-oral route are acid tolerant because they must survive passage through the gastric barrier. The acid tolerance of serotype O157:H7 strains of E. coli is further supported by outbreaks involving acidic foods (8, 23) and laboratory studies (7, 15, 37, 46).
Acid tolerance can be classified into three main strategies for bacteria. The first is changes in membrane composition (10, 27), the second is enzymatic or physiological maintenance of internal pH (13, 16, 20, 25, 40), and the third is repair and/or prevention of damage caused to essential cellular components by acidic pH (15, 41, 47). Previous studies with E. coli, Salmonella enterica serovar Typhimurium, and Helicobacter pylori suggest that DNA repair pathways play a role in survival in extreme-acidity conditions such as the gastric barrier (26, 41, 47). It has been shown previously that mutations in DNA repair mechanisms such as recA and uvrB in H. pylori resulted in significant decreases in tolerance for low pH (47), and these results establish the importance of DNA repair systems in survival in acidic conditions.
Damage to DNA can occur at many different sites depending on the reactive substance. Oxidative damage is characterized by the production of hydroxyl radicals that leads to oxidation of sugar and base moieties that can cause strand breaks in DNA (29). Low pH causes DNA damage primarily by removal of purine bases and to a lesser extent by production of double-stranded lesions in the DNA (29). Depurination results in unrepaired DNA and mismatches in repaired sequences, which ultimately can be lethal to the cell (41, 47). DNA repair represents a major strategy for bacteria to remain viable following passage through extreme pH conditions such as the gastric barrier. This emphasizes the need to further characterize DNA protection and repair systems and determine their significance in acid tolerance.
Almirón et al. (2) identified a DNA-binding protein, designated Dps (DNA-binding protein from starved cells), that is produced primarily in stationary-phase cells of E. coli and has been shown to be regulated by ς38, ς70, and OxyR (3, 6, 33). Dps forms spherical dodecamers, homologous to ferritins, that sequester and protect DNA from oxidative stress, nucleases, and UV light (49). Dukan and Touati (18) showed that mutations of recA, recB, and dps in E. coli rendered cells more sensitive to damage from hydroxyl radicals generated by HOCl. This suggests that not only DNA repair, but also DNA protection by dps, is pivotal for survival in extreme conditions. Therefore, the role of dps in protection from acid-mediated DNA damage (depurination) and its contribution to the acid tolerance of E. coli O157:H7 were ascertained. To this end, an E. coli O157:H7 dps null mutant was constructed, and its acid stress and oxidative stress tolerance was compared to that of the parent strain.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were stored in nutrient broth (Difco Laboratories, Detroit, Mich.) supplemented with 10% glycerol at −70°C. Growth of cultures was monitored spectrophotometrically at 600 nm using a Bioscreen Analyzer (Labsystems, Helsinki, Finland). For log-phase cultures, 5 μl of an 18-h culture was used to inoculate 5 ml of Luria-Bertani (LB) broth (43) [with kanamycin for FRIK 47991 and kanamycin and ampicillin for 47991(pSC9915) and 47991(pSC9916)] and incubated with shaking (150 rpm) at 37°C for 5 h (A600 = 0.8). For stationary-phase or late-stationary-phase cultures, LB broth was inoculated as described previously and incubated with shaking (150 rpm) at 37°C for 12 or 24 h (A600 = 1.1), respectively. Transformants and mutants of E. coli were recovered on LB agar or MacConkey sorbitol agar (MSA). Antibiotics (Sigma Chemical Company, St. Louis, Mo.) were added to agar and broth media when appropriate: ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; and kanamycin, 50 μg/ml. Chemicals in buffers and media were obtained from Sigma.
TABLE 1.
Plasmids and E. coli strains used in this study
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab collection |
| ATCC 43895 | stx1 stx2; serotype O157:H7 isolate from hamburger | American Type Culture Collection |
| SY327 λ pir | Δ(lac pro) argE(Am) recA56 nalA Rif(λ pir); carries π protein for R6K γ ori | 38 |
| SM10 λ pir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λ pir oriT of RP4 | 38 |
| FRIK 47991 | FRIK 47; dps::nptI with nptI in opposite orientation from FRIK 47992 | This study |
| FRIK 47992 | FRIK 47; dps::nptI with nptI in opposite orientation from FRIK 47991 | This study |
| Plasmids | ||
| pBR322 | Apr Tcr | Lab collection |
| pUC4K | Apr Kmr; pUC4 with nptI | Pharmacia 39 |
| pCVD402 | R6K γ ori sacB oriT of RP4 | J. Kaper 17 |
| pSC9911 | Aps Tcr; pBR322 with dps | This study |
| pSC9915 | Apr Tcs; pBR322 with dps | This study |
| pSC9916 | Apr Tcs; pBR322 | This study |
| pSC9921 | pSC9911 with dps::nptI | This study |
| pSC9922 | pCVD402 with dps::nptI, but nptI is in opposite orientation from pSC9932 | This study |
| pSC9932 | pCVD402 with dps::nptI, but nptI is in opposite orientation from pSC9922 | This study |
General genetic methods.
Procedures for the isolation of genomic DNA and transformation were carried out as described by Sambrook et al. (43). Plasmid DNA was isolated using the QIAprep Spin Minikit and protocol (Qiagen Inc., Chatsworth, Calif.). Restriction and DNA-modifying enzymes were used as recommended by the manufacturer (New England Biolabs, Beverly, Mass.). DNA fragments were compared to a 1-kb DNA ladder (Promega Corp., Madison, Wis.) and then purified from 1% agarose gels (Gibco BRL, Grand Island, N.Y.) using the Geneclean II kit (Bio 101, Inc., Vista, Calif.). Primary DNA cloning and manipulation were conducted with E. coli DH5α. PCR amplification of DNA was performed using an Amplitron II Thermal Cycler (Barnstead/Thermolyne, Dubuque, Iowa), and conditions were denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min for 30 cycles. All reagents and Taq DNA polymerase were used as recommended by the manufacturer (Perkin-Elmer Co., Foster City, Calif.).
Construction of dps::nptI suicide vector.
The dps gene in pSC9911 was inactivated in vitro by insertion of nptI encoding aminoglycoside 3′-phosphotransferase and confers resistance to kanamycin. The 1.2-kb DNA fragment carrying nptI was isolated from pUC4K (39, 48) and digested with PstI, and the resulting cohesive termini were converted to blunt ends with Klenow fragment. The nptI fragment was inserted into a unique NcoI site present within the open reading frame (ORF) of dps. The resulting construct (pSC9921) containing the 2.0-kb dps::nptI fragment was digested with EcoRI and PstI to liberate the dps::nptI cartridge. The DNA fragment was blunt ended with Klenow fragment and ligated with SmaI-digested pCVD442 (17), forming pSC9922. pCVD442 is a suicide vector containing the R6K origin of replication that requires the λ protein in trans (28) encoded by pir. E. coli SY327 λ pir (38) was transformed with pSC9922. The plasmid pSC9932, which was identical to pSC9922 except that the dps::nptI cartridge was in the opposite orientation, was constructed by following the procedures for pSC9922.
Generation of the dps::nptI mutant.
The suicide vectors, pSC9922 and pSC9932, were used to generate the dps::nptI mutants (FRIK 47991 and FRIK 47992) in E. coli O157:H7 strain ATCC 43895 by homologous recombination (Fig. 1A). Both plasmids contain the RP4 origin of transfer (oriT) (45) and can be conjugally mobilized from donor cells containing the tra gene. Therefore, E. coli SM10 λ pir tra (38, 45) was transformed separately with pSC9922 and pSC9932 and used as a conjugal donor to E. coli O157:H7 strain ATCC 43895. Conjugation was conducted using methods previously described (43) with some modification. The recipient strain ATCC 43895 and donor strain SM10 λ pir tra (containing pSC9922 or pSC9932) were grown overnight on LB agar, removed with a sterile cotton swab, spotted on LB agar, and mixed thoroughly. The donor-recipient mixture was incubated at 37°C for 8 h and then resuspended in 1 ml of saline (0.85% NaCl). Portions of the cell mixture (100 μl) were spread on each of 10 plates of MSA supplemented with kanamycin and sucrose (6%) and incubated overnight at 37°C.
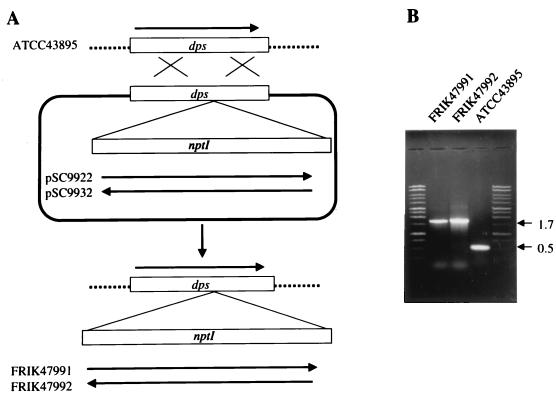
FIG. 1.
Diagram of allelic exchange and confirmation of the dps::nptI mutants. (A) Homologous recombination between chromosomal dps gene from strain ATCC 43895 and pSC9922 or pSC9932. Dashed lines, chromosomal DNA; solid line, plasmid DNA; dps boxes, the target dps gene; nptI boxes, the nptI gene; ×, crossover. (B) PCR analysis of E. coli O157:H7 dps mutant generated by allelic exchange (FRIK 47991 and FRIK 47992). Strain ATCC 43895 is the parent strain for FRIK 47991 and FRIK 47992. Molecular size markers (1-kb ladder [Promega Corp.]) appear in the end lanes of the gel. The oligonucleotides for PCR amplification are presented in Materials and Methods.
The desired transconjugants were selected by their ability to grow on MSA supplemented with kanamycin and sucrose. Strains that were kanamycin resistant and ampicillin sensitive, to confirm the absence of the suicide vector (pSC9922 or pSC9932), were selected for further study. Selected strains were then confirmed for the O157 antigen by slide agglutination (Oxoid, Basingstoke, England) and for the presence of nptI in the dps gene by PCR amplification. The primers, dpskk-1 (5′GCCGGAATTCATGAAATTATGAGTACCGC3′) and dpskk-2 (5′GAAAACTGCAGAATTTATCCAGGTCGCGAGACGACGC3′) were designed to hybridize and amplify the dps ORF. The PCR end products were analyzed by agarose gel electrophoresis to confirm the presence of nptI in dps (1.7-kb fragment) (Fig. 1B).
Complementation of the dps::nptI mutant.
The dps gene (790 bp) and promoter from E. coli O157:H7 strain ATCC 43895 were amplified by PCR and cloned into the EcoRI site of pBR322 to generate pSC9911. The tet gene of pSC9911 was then removed using HindIII and AvaI, treated with Klenow fragment, and blunt end ligated to produce pSC9915 (Table 1). pSC9916 was used as a control and was generated from pBR322 by removing the tet gene. Each plasmid (pSC9915 and pSC9916) was transformed into FRIK 47991 (dps::nptI) using standard methods (43) and used in subsequent acid and hydrogen peroxide challenge studies.
Acid and hydrogen peroxide challenges.
Acid tolerance was assessed in synthetic gastric fluid adjusted to pH 1.8 with HCl (12 N) and filter sterilized as described previously (5). Synthetic gastric fluid was prepared essentially as described by Beumer et al. (9) except that bovine bile was used in place of porcine bile. Cultures in exponential and stationary phases of growth were used to inoculate flasks containing 100 ml of synthetic gastric fluid to achieve a final concentration of ca. 105 CFU/ml. Following inoculation, the flasks were incubated at 37°C with shaking (150 rpm) and samples were removed at appropriate intervals, plated in duplicate on tryptic soy agar using a Model D Spiral Systems plater (Cinncinati, Ohio), and incubated at 37°C. The percent survivors was calculated using the CFU per milliliter as determined immediately after inoculation as 100%. The limit of detection for this method was 10 CFU/ml (12); hence, a maximum decrease of 4 log10 CFU/ml could be detected. Additionally, colonies were randomly tested for each respective phenotype by two methods. The first was by growth in the presence of the respective antibiotic(s), and the second was by PCR amplification of the dps::nptI cartridge and dps complement when applicable.
The assay for survival of log- and stationary-phase cells in the presence of 15 mM hydrogen peroxide was conducted as previously described (34). Hydrogen peroxide was added to cell suspensions (ca. 105 CFU/ml) in 100 ml of phosphate-buffered saline (0.01 M, pH 7.2) and incubated at room temperature (22°C) with shaking (100 rpm). Samples were removed periodically to determine the number of CFU per milliliter as described for acid challenges. All D values were calculated using the formula (Dvalue = −1/slope), where slope represents the linear regression of the data including first and last points.
Cloning and sequencing of dps.
A DNA fragment containing the dps structural gene and upstream regulatory region was amplified from genomic DNA of E. coli O157:H7 strain ATCC 43895 by PCR using a pair of oligonucleotide primers carrying EcoRI or PstI sites on the 5′ ends. The primers (dps-1, 5′CGGAATTCCATAACCATGCAGAATTTCT3′, sense primer, and dps-2, 5′CGGCTGAGCAGCGATGGATTTATTCGAT3′, antisense primer) were designed using the dps sequence of E. coli K-12 (GenBank accession no. X69337) and synthesized (Gibco BRL, Gaithersburg, Md.). The resulting PCR fragment was digested with EcoRI and PstI and ligated into pBR322, previously digested with the same enzymes, to produce pSC9911. The nucleotide sequence of the 790-bp DNA fragment in pSC9911 was determined (University of Wisconsin—Madison Biotechnology Center). Sequence and amino acid comparisons were conducted using BLAST (National Center for Biotechnology Information).
Statistical analyses.
The data reported are the average values from three trials and were analyzed using the t test with SigmaStat (Jandel Scientific, San Rafael, Calif.) software.
Nucleotide sequence accession number.
The nucleotide sequence of dps from E. coli O157:H7 strain ATCC 43895 was deposited in the GenBank database under accession no. AF140030.
RESULTS
Generation and confirmation of the dps::nptI mutant in E. coli O157:H7.
Transconjugants resulting from the conjugation of pSC9922 from SM10 λ pir to O157:H7 strain ATCC 43895 were kanamycin resistant, ampicillin sensitive, and sucrose positive. Strains selected for further study were also positive for the O157 antigen. While allelic exchange between the insert dps::nptI and chromosomal dps can occur by a double crossover, sacB in pSC9922 and pSC9932 encodes levansucrase (21) and selects against the maintenance or integration of these plasmids into the chromosome. Confirmation of a double crossover in which wild-type dps was replaced with the dps::nptI allele was confirmed by PCR.
PCR analysis of genomic DNA from parental strain ATCC 43895 with primers dpskk-1 and dpskk-2 produced a 481-bp fragment (Fig. 1B), whereas genomic DNA from the dps::nptI mutant(s) resulted in an amplified DNA fragment approximately 1.7 kb in length. The 1.7-kb fragment is in agreement with the projected size of the DNA fragment containing wild-type dps (481 bp) and the nptI gene (1.2 kb). FRIK 47991, shown in Fig. 1B, was selected for further study.
Acid tolerance.
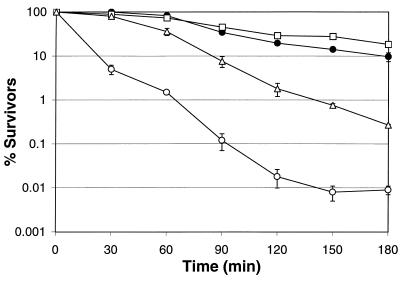
The survival of log-phase cells (5 h, A600 = 0.8) of the parent strain (ATCC 43895) was significantly greater (P < 0.05) than that of the dps::nptI mutant (FRIK 47991) when challenged in synthetic gastric fluid (pH 1.8) (Fig. 2). The DpH 1.8 values for the parent and mutant strains were 157 and 36 min, respectively. The survival of dps::nptI mutants, FRIK 47991 and FRIK 47992, containing nptI in the opposite orientation, did not differ significantly in acid challenges (data not shown). FRIK 47991 was chosen for further study. Complementation of dps::nptI in FRIK 47991 with a functional dps gene (pSC9915) restored acid tolerance to a level equivalent to that of the parent strain. FRIK 47991 containing the control plasmid (pSC9916) displayed survival that was significantly impaired (P < 0.01) compared to that of the parent strain and FRIK 47991 harboring pSC9915.
FIG. 2.
Survival of log-phase (5-h) E. coli O157:H7 parent strain (ATCC 43895) (●), dps::nptI mutant (FRIK 47991) (○), FRIK 47991 with pSC9915 (□), and FRIK 47991 with pSC9916 (▵) in synthetic gastric fluid (pH 1.8). All points represent the means from three independent trials. Error bars represent the standard errors.
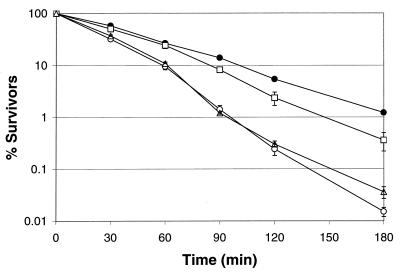
Similar to results with log-phase cells, the survival of early-stationary-phase cells (12 h, A600 = 1.1) of the parent strain and FRIK 47991 containing pSC9915 was significantly greater (P < 0.05) than that of FRIK 47991 and FRIK 47991 containing the control plasmid pSC9916 (Fig. 3). The DpH 1.8 values were 93 min for the parent strain and 71 min for the complemented strain (FRIK 47991 with pSC9915) compared to 49 and 45 min for FRIK 47991 with and without the control plasmid (pSC9916), respectively.
FIG. 3.
Survival of stationary-phase (12-h) E. coli O157:H7 parent strain (ATCC 43895) (●), dps::nptI mutant (FRIK 47991) (○), FRIK 47991 with pSC9915 (□), and FRIK 47991 with pSC9916 (▵) in synthetic gastric fluid (pH 1.8). All points represent the means from three independent trials. Error bars represent the standard errors.
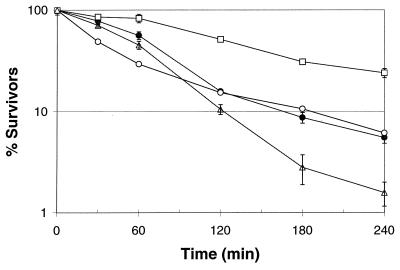
The survival of late-stationary-phase cells (24 h, A600 = 1.1) in synthetic gastric fluid is shown in Fig. 4. The trends in survival of the late-stationary-phase cells were similar to those for log-phase and early-stationary-phase cells with the exception of the dps::nptI mutant (FRIK 47991). The survival of the parent strain, FRIK 47991, and FRIK 47991 with either pSC9915 or pSC9916 was equivalent to or greater than the survival of log- and early-stationary-phase cells (Fig. 2 and 3). FRIK 47991 containing pSC9915 exhibited the lowest rate of decline and had the greatest number of survivors after 4 h of acid challenge.
FIG. 4.
Survival of late-stationary-phase (24-h) E. coli O157:H7 parent strain (ATCC 43895) (●), dps::nptI mutant (FRIK 47991) (○), FRIK 47991 with pSC9915 (□), and FRIK 47991 with pSC9916 (▵) in synthetic gastric fluid (pH 1.8). All points represent the means from three independent trials. Error bars represent the standard errors.
Survival during hydrogen peroxide challenge.
The survival of the parent strain was significantly greater (P < 0.05) than that of FRIK 47991 when challenged in 15 mM H2O2 regardless of the growth phase (Table 2). The maximum survival reported as D values for all strains was with early-stationary-phase cells (12 h). As reported for acid challenges, complementation of dps::nptI with a functional dps gene (pSC9915) restored hydrogen peroxide tolerance to levels equal to or greater than that of the parent strain. The presence of pSC9916 (control plasmid) in FRIK 47991 improved survival slightly, but D values were still significantly less (P < 0.1) than those for the parent strain.
TABLE 2.
Comparison of D values from parent strain (ATCC 43895), dps::nptI mutant (FRIK 47991), and FRIK 47991 with pSC9915 or pSC9916 when challenged in 15 mM H2O2
| Strain | Plasmida |
D value (mean [min] ± SE)
|
||
|---|---|---|---|---|
| Log phase (5 h) | Stationary phase (12 h) | Late stationary phase (24 h) | ||
| ATCC 43895 | None | 23.5 ± 0.8 | 38.2 ± 0.5 | 25.2 ± 0.5 |
| FRIK 47991 | None | 14.9 ± 1.0 | 35.0 ± 0.5 | 19.5 ± 1.4 |
| FRIK 47991 | pSC9915 | 23.6 ± 2.4 | 39.5 ± 1.4 | 28.7 ± 0.7 |
| FRIK 47991 | pSC9916 | 18.4 ± 0.9 | 33.1 ± 2.1 | 23.3 ± 0.1 |
See Table 1.
Cloning and nucleotide sequence of dps.
A 790-bp DNA fragment containing the dps structural gene and upstream regulatory region was amplified by PCR from genomic DNA of E. coli O157:H7 strain ATCC 43895. The nucleotide sequence of the amplified fragment was determined (accession no. AF140030). An ORF starting at nucleotide 276 and ending at nucleotide 779 was identified. The dps gene codes for a 167-amino-acid protein with an estimated mass of 18.7 kDa and a pI of 5.72. The nucleotide sequence of the dps from O157:H7 strain ATCC 43895 was 99% similar (783 of 790 bp) to the dps sequence from E. coli K-12 (GenBank accession no. X69337), and the deduced amino acid sequences from these strains were 100% similar. Of the seven mismatches between the nucleotide sequences of these two strains, two were located in the ORF of dps; however, neither mismatch resulted in a change in the amino acid code. The remaining five nucleotide mismatches were upstream of the ORF, and four of the mismatches occurred in a 12-bp segment that was flanked by inverted repeats (data not shown).
DISCUSSION
Bacteria have evolved with elaborate protection systems to allow survival and/or growth during exposure to acidic environments. The three main defense strategies which protect the cell from acid are changes in membrane composition (10, 27), homeostasis systems for internal pH (13, 16, 20, 25, 40) and pathways for repair-protection of essential cellular components (15, 41, 47). Studies with E. coli, Salmonella, and H. pylori have suggested that DNA repair pathways are important for survival in low-pH conditions such as the gastric barrier (26, 41, 47). Almirón et al. (2) identified a DNA-binding protein (Dps) that is regulated by ς38, ς70, and OxyR and protects DNA from damage (3, 33, 34). Dps forms dodecamers, analogous to ferritins, that interact with DNA to form a stable complex that protects DNA from the hydroxyl radicals formed during oxidative stress (49). Low pH also damages DNA as a result of selective depurination reactions and can cause lesions in double-stranded DNA (29). This study investigated the role of Dps in the acid stress and oxidative stress tolerance of E. coli O157:H7, which epidemiological and laboratory studies demonstrate is particularly tolerant of low-pH conditions (5, 7, 8, 37).
A dps::nptI mutant (FRIK 47991) was generated and used to determine if dps contributes to acid tolerance in E. coli O157:H7. During our evaluation of plasmid constructs to complement the dps::nptI mutant, acid tolerance of the dps mutant FRIK 47991 was further reduced when transformed with pSC9911 (data not shown). In follow-up experiments, the introduction of pBR322 into the parent strain E. coli O157:H7 also decreased acid tolerance (data not shown) and indicated that tetA, which encodes a tetracycline pump (44), decreased acid tolerance (C.-M. Cheng, J. L. Bose, S. H. Choi, and C. W. Kaspar, unpublished observation). Previous studies have documented that the tetracycline resistance gene (tetA) causes pleiotropic effects in addition to directing the efflux of tetracycline from the bacterium (44). Therefore, the tetA gene was removed from pSC9915 (Apr Tcs; used to complement dps::nptI in FRIK 47991) and pSC9916 (Apr Tcs; control plasmid).
Characterization of the survival properties of the parent strain (ATCC 43895) and the dps::nptI mutant (FRIK 47991) demonstrated significant differences during acid and hydrogen peroxide challenges, particularly when log-phase (5-h) cultures were examined. Although dps has been primarily characterized in stationary-phase protection, these results were not unexpected because dps is regulated by ς38, ς70, and OxyR (3, 33). This is also consistent with the growth phase variations in nucleoid composition in log-phase cells observed by Azam et al. (6). Complementation of the dps::nptI mutant with pSC9915 and the restoration of acid stress and oxidative stress tolerance to levels equivalent to or exceeding that of the parent strain demonstrates that Dps or a Dps-regulated protein is responsible for the observed differences in survival.
Early-stationary-phase (12-h) cultures of the parent strain were significantly more tolerant of acid than was the dps::nptI mutant. In both log-phase and 12-h cultures, the parent strain decreased by 1 to 2 log10 CFU/ml after 3 h of acid challenge while FRIK 47991 decreased by ca. 4 log10 CFU/ml. In contrast to the results in acid challenges, 12-h cultures of FRIK 47991 were significantly more tolerant of hydrogen peroxide challenge (ca. 2.5-log10-CFU/ml reduction after 60 min) than were log-phase cultures (4-log10-CFU/ml reduction after 60 min), although the survival of the parent strain was still significantly greater (P < 0.01) than that of FRIK 47991. However, the final numbers (CFU per milliliter) of the parent and dps::nptI mutant after 2 h of hydrogen peroxide challenge were essentially the same (data not shown).
Stationary-phase cells of the parent strain also exhibited increased tolerance for hydrogen peroxide challenge in comparison with log-phase cultures. The increased tolerance of stationary-phase cells for hydrogen peroxide challenge is likely due to the production of catalase. E. coli O157:H7 possess three separate catalase genes (katG, katE, and katP) (11, 32). katG is regulated by OxyR and predominantly produced during log phase, while katE is regulated by rpoS and primarily expressed in stationary phase (32). The regulation of the plasmid-encoded (pO157) catalse (katP) has not been elucidated. Thus, stationary-phase production of catalase (katE) likely provides protection against hydrogen peroxide and compensates for the absence of Dps.
The survival of late-stationary-phase cells (24 h) of the parent strain in acid challenges was similar to that observed with log-phase (5-h) and early-stationary-phase (12-h) cells. There was a significant improvement in the acid tolerance of the dps::nptI mutant in late-stationary-phase cells as viable numbers decreased less than did log- and early-stationary-phase cultures. In fact, the numbers of FRIK 47991 survivors were equivalent to those of the parent strain after 2 h of acid challenge, although there were greater numbers of the parent strain recovered after 30 and 60 min of acid challenge (Fig. 4). The production of other proteins with protective roles in stationary phase, including the DNA-binding proteins encoded by cbpA and rob (6), most likely provides protection from acid in the absence of Dps. The results from hydrogen peroxide challenges with late-stationary-phase cells of the parent strain were similar to those obtained with log-phase cells. The D values from hydrogen peroxide challenges with FRIK 47991 in log phase, early stationary phase, and late stationary phase were 15, 35, and 19 min, respectively. Evidently, the proteins that enhanced acid tolerance in late-stationary-phase cells of FRIK 47991 are less effective in protecting the bacterium from oxidative stress.
As noted above, the complementation of the dps::nptI mutant (FRIK 47991) with pSC9915 increased survival in acid and hydrogen peroxide challenges to a level equivalent to or exceeding that of the parent strain (ATCC 43895). The increased tolerance can be attributed to the multiple copies of dps provided by pSC9915 compared to the single-chromosome-encoded allele found in the parent strain. Transformation of FRIK 47991 with the control plasmid pSC9916 did not restore acid and hydrogen peroxide tolerance; however, in challenges with log-phase cells, FRIK 47991 containing pSC9916 survived better than did FRIK 47991 without the control plasmid. It is possible that the presence of multiple copies of pSC9916 provided some protection against acid and hydrogen peroxide in log-phase cells. This may be explained by the presence of additional nonessential DNA targets that would decrease the rates of depurination from low pH and oxidation of sugar and base moieties from oxygen radicals that target chromosomal DNA.
The sequence homology of dps from E. coli O157:H7 strain ATCC 43895 and the fact that the deduced amino acid sequences were identical to E. coli K-12 demonstrate that the acid tolerance noted for some serotype O157:H7 strains is not due to differences in the ORF of this gene. Studies are in progress to determine if the inverted repeat and nucleotide differences in the upstream region of dps in E. coli O157:H7 influence regulation.
Results from this study demonstrate that dps makes a significant contribution to the acid tolerance of E. coli O157:H7. In addition to acid tolerance, Dps is important in oxidative stress protection as reported previously for non-serotype O157:H7 E. coli (2, 34). It is likely that Dps protects DNA from the deleterious effects of low pH in a manner analogous to oxidative stress protection (49); however, it is possible that Dps influences expression of other genes that protect or repair DNA or provide acid tolerance by another mechanism. Regardless, Dps is a key component of the general stress protection system that is important in the survival of the bacterium.
ACKNOWLEDGMENTS
We thank Jeffrey L. Bose and Barbara Cochrane for technical assistance and Jim Kaper for supplying strains and pCVD442. We are grateful to Chorng-M. Cheng and Jeffery Byrd for helpful discussions and sharing of unpublished data.
The work was supported by grant 96-35201-3430 from the USDA, NRICGP awarded to C.W.K., and the College of Agricultural and Life Sciences, University of Wisconsin—Madison.
REFERENCES
- 1.Abdul-Raouf U M, Beuchat L R, Ammar M S. Survival and growth of Escherichia coli O157:H7 in ground beef as affected by pH, acidulants, and temperature. Appl Environ Microbiol. 1993;59:2364–2368. doi: 10.1128/aem.59.8.2364-2368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almirón M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςs in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. Report on the Escherichia coli O157:H7 outbreak in the western states. Washington, D.C.: Food Safety and Inspection Service, U.S. Department of Agriculture; 1993. [Google Scholar]
- 5.Arnold K W, Kaspar C W. Starvation- and stationary phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azam T A, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamin M M, Datta A T. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besser R E, Lett S M, Weber J T, Doyle M P, Barrett T J, Wells J G, Griffin P M. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA. 1993;269:2217–2220. [PubMed] [Google Scholar]
- 9.Beumer R R, de Vries J, Rombouts F M. Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol. 1992;15:153–163. doi: 10.1016/0168-1605(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 10.Brown J L, Ross T, McMeekin T A, Nichols P D. Acid habituation of Escherichia coli and the potential role of cytoplasmic fatty acids in low pH tolerance. J Food Microbiol. 1997;37:163–173. doi: 10.1016/s0168-1605(97)00068-8. [DOI] [PubMed] [Google Scholar]
- 11.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 12.Byrd J J, Cheville A M, Bose J L, Kaspar C W. Lethality of a heat- and phosphate-catalyzed glucose by-product to Escherichia coli O157:H7 and partial protection conferred by the rpoS regulon. Appl Environ Microbiol. 1999;65:2396–2401. doi: 10.1128/aem.65.6.2396-2401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castanie-Cornet M-P, Penfound T A, Smith D, Elliott J F, Foster J W. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—Western United States, 1992–1993. Morb Mortal Wkly Rep. 1993;42:258–263. [PubMed] [Google Scholar]
- 15.Cheville A M, Arnold K W, Buchrieser C, Cheng C-M, Kaspar C W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBiase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 17.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dukan S, Touati D. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson G P, Creighton R I, Nikolaev Y, Booth I R. Importance of RpoS and Dps in survival of exposure of both exponential- and stationary-phase Escherichia coli cells to the electrophile N-ethylmaleimide. J Bacteriol. 1998;180:1030–1036. doi: 10.1128/jb.180.5.1030-1036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster J W, Hall H K. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J Bacteriol. 1991;173:5129–5135. doi: 10.1128/jb.173.16.5129-5135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gay P, LeCoq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1982;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon J, Small P L. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 739–761. [Google Scholar]
- 24.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 25.Hersh B M, Farooq F T, Barstad D N, Blankenhorn D L, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey E W, Hirshfield I N. Low-pH induced effects on pattern of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan K N, Oxford L, O'Byrne C P. Survival of low-pH stress by Escherichia coli O157:H7: correlation between alterations in the cell envelope and increased acid tolerance. Appl Environ Microbiol. 1999;65:3048–3055. doi: 10.1128/aem.65.7.3048-3055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 29.Lehninger A L, Nelson D L, Cox M M. Principles of biochemistry. 2nd ed. New York, N.Y: Worth Publishers; 1993. [Google Scholar]
- 30.Lin J, Smith M P, Chapin K C, Baik H S, Bennnett G N, Foster J W. Mechanism of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 32.Loewen P C, Switala J, Triggs-Raine B L. Catalase HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 33.Lomovskaya O L, Kidwell J P, Matin A. Characterization of the ς38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol. 1994;176:3928–3935. doi: 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCann M P, Kidwell J P, Martin A. A putative ς factor KatF has a crucial role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead P S, Griffin P M. Escherichia coli O157:H7. Lancet (N Am Ed) 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 37.Miller L G, Kaspar C W. Escherichia coli O157:H7 acid tolerance and survival in apple cider. J Food Prot. 1994;57:460–464. doi: 10.4315/0362-028X-57.6.460. . (Erratum, 57:645.) [DOI] [PubMed] [Google Scholar]
- 38.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 40.Park Y-K, Bearson B, Bang S H, Bang I S, Foster J W. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 41.Raja N, Goodson M, Smith D G, Rowbury R J. Decreased DNA damage by acid and increased repair of acid-damaged DNA in acid-habituated Escherichia coli. J Appl Bacteriol. 1991;70:507–511. doi: 10.1111/j.1365-2672.1991.tb02748.x. [DOI] [PubMed] [Google Scholar]
- 42.Reida P, Wolff M, Pöhls H-W, Kuhlmann W, Lehmacher A, Aleksi S, Karch H, Bockemühl J. An outbreak due to enterohemorrhagic Escherichia coli O157:H7 in a children's day care center characterized by person-to-person transmission and environmental contamination. Zentbl Bakteriol. 1994;281:534–543. doi: 10.1016/s0934-8840(11)80342-7. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Schnappinger D, Hillen W. Tetracycline: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 46.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson S A, Latch R L, Blaser M J. Molecular characterization of the Helicobacter pylori uvrB gene. Gene. 1998;209:113–122. doi: 10.1016/s0378-1119(98)00028-6. [DOI] [PubMed] [Google Scholar]
- 48.Vieira J, Messing J. The pUC plasmids, an M13mp7 derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 49.Wolf S G, Frenkiel D, Arad T, Finkel S E, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]