Abstract
The high-resolution genotyping method of amplified fragment length polymorphism (AFLP) analysis was used to study the genetic relationships between Campylobacter jejuni strains infecting chickens (n = 54) and those causing gastroenteritis in humans (n = 53). In addition, C. jejuni strains associated with the development of Guillain-Barré syndrome (GBS) (n = 14) and Miller Fisher syndrome (MFS) (n = 4), two related acute paralytic syndromes in human, were included. Strains were isolated between 1989 and 1998 in The Netherlands. The AFLP banding patterns were analyzed with correlation-based and band-based similarity coefficients and UPGMA (unweighted pair group method using average linkages) cluster analysis. All C. jejuni strains showed highly heterogeneous fingerprints, and no fingerprints exclusive for chicken strains or for human strains were obtained. All strains were separated in two distinct genetic groups. In group A the percentage of human strains was significantly higher and may be an indication that genotypes of this group are more frequently associated with human diseases. We conclude that C. jejuni from chickens cannot be distinguished from human strains and that GBS or MFS related strains do not belong to a distinct genetic group.
Campylobacter jejuni is an important human pathogen, and it is the most common cause of bacterial gastroenteritis worldwide (24), surpassing Salmonella spp. in most studies. Poultry flocks are highly colonized with C. jejuni, and human infections are commonly associated with contaminated poultry meat (23). These human infections are often self-limiting, although an association of C. jejuni infections with the neurological diseases Guillain-Barré syndrome (GBS) and Miller Fisher syndrome (MFS) has become evident (14, 17). Despite the worldwide prevalence of C. jejuni infections, relatively little is known about the pathogenic characteristics of C. jejuni strains. It is still unknown whether all C. jejuni strains that colonize chickens can be pathogenic for humans and whether all strains can eventually be associated with GBS and MFS. Genetic typing of strains obtained from various sources may elucidate characteristics of C. jejuni strains that are associated with human infections and may result in an improved understanding of the epidemiology of human infections.
For genetic typing of C. jejuni, several methods have been developed. These include pulsed-field gel electrophoresis (PFGE), flagellin PCR-restriction fragment length polymorphism (RFLP) analysis, ribotyping, and randomly amplified polymorphic DNA (RAPD) analysis, which are already in use in a number of laboratories. The advantages and problems of each technique have recently been reviewed by Wassenaar and Newell (28). Recently, we introduced amplified fragment length polymorphism (AFLP) analysis as a high-resolution genotyping method for Campylobacter spp. (5). This method has been applied for typing of several other microorganisms and has the advantage of high levels of discriminatory power, reproducibility, and standardization (22). For AFLP analysis, restriction fragments of chromosomal DNA are selectively amplified by PCR. By using primers containing one or more selective nucleotides extending at the 3′ ends, only a subset of fragments are amplified under stringent PCR conditions (12). AFLP analysis of Campylobacter spp. differentiated C. jejuni from C. coli and was able to subtype strains from both species (5).
Identification of the genetic relationships between C. jejuni strains from poultry and strains infecting humans is necessary for a better understanding of the epidemiology of human infections. In the present study, the AFLP fingerprints of C. jejuni strains from chickens are compared with those of strains obtained from humans with gastroenteritis, and a set of GBS- and MFS-related strains. All strains were isolated between 1989 and 1998 in The Netherlands. AFLP fingerprints were clustered using the Pearson correlation coefficient and the band-based Dice coefficient.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The 53 strains obtained from patients with gastroenteritis were obtained from the two years of a three year case control study among general practicioners in The Netherlands, performed at the National Institute of Public Health and the Environment (RIVM) (4; M. A. S. De Wit, M. P. G. Koopmans, L. M. Kortbeek, W. J. van Leeuwen, A. I. M. Bartelds, and Y. T. H. P. van Duynhoven, submitted for publication). Fifty-four strains were randomly isolated from chickens from geographically dispersed farms in The Netherlands (9, 11; W. Jacobs-Reitsma, C. Becht, J. Bosch, J. van der Plas, and J. A. Wagenaar, 10th Int. Workshop Campylobacter, Helicobacter, Related Organisms, abstr. P38, 1999). From 14 patients with the GBS and 4 patients with the MFS C. jejuni strains were obtained from prospectively cultured stool samples, during a study at the Erasmus University Center in The Netherlands (6). All strains are listed in Table 1.
TABLE 1.
C. jejuni strains used in this study obtained from chickens, patients with gastroenteritis (human), or patients with GBS or MFS
| Strains | Source | Date of isolation
|
Strains | Source | Date of isolation
|
|||
|---|---|---|---|---|---|---|---|---|
| Mo | Yr | Mo | Yr | |||||
| C144a | Chicken | March | 1990 | 96413 | Human | October | 1996 | |
| C350 | Chicken | June | 1990 | 96434 | Human | October | 1996 | |
| C356 | Chicken | June | 1990 | 96442 | Human | October | 1996 | |
| C591 | Chicken | August | 1990 | 96467 | Human | November | 1996 | |
| C690 | Chicken | September | 1990 | 96498 | Human | December | 1996 | |
| C2143 | Chicken | March | 1992 | 96521 | Human | December | 1996 | |
| C2146 | Chicken | March | 1992 | 9746 | Human | January | 1997 | |
| C2150 | Chicken | March | 1992 | 97157 | Human | April | 1997 | |
| C2172 | Human | April | 1992 | 97195 | Human | May | 1997 | |
| C2246 | Chicken | May | 1992 | 97197 | Human | May | 1997 | |
| C2264 | Chicken | May | 1992 | 97198 | Human | May | 1997 | |
| C2345 | Chicken | July | 1992 | 97205 | Human | May | 1997 | |
| C2360 | Chicken | July | 1992 | 97208 | Human | May | 1997 | |
| C2362 | Chicken | July | 1992 | 97222 | Human | May | 1997 | |
| C2412 | Chicken | July | 1992 | 97223 | Human | June | 1997 | |
| C2441 | Chicken | August | 1992 | 97232 | Human | June | 1997 | |
| C2446 | Chicken | August | 1992 | 97248 | Human | June | 1997 | |
| C2450 | Chicken | August | 1992 | 97255 | Human | June | 1997 | |
| C2461 | Chicken | August | 1992 | 97256 | Human | June | 1997 | |
| C2476 | Chicken | August | 1992 | 97258 | Human | June | 1997 | |
| C2481 | Chicken | August | 1992 | 97275 | Human | June | 1997 | |
| C2515 | Chicken | September | 1992 | 97279 | Human | July | 1997 | |
| C2535 | Chicken | September | 1992 | 97294 | Human | July | 1997 | |
| C2555 | Chicken | September | 1992 | 97296 | Human | July | 1997 | |
| C2609e | Chicken | November | 1992 | 97300 | Human | July | 1997 | |
| C2641 | Chicken | November | 1992 | 97384 | Human | September | 1997 | |
| C2651 | Chicken | January | 1993 | 97411 | Human | September | 1997 | |
| da9521 | Chicken | January | 1999 | 97459 | Human | October | 1997 | |
| ha5221 | Chicken | July | 1998 | 97498 | Human | November | 1997 | |
| kc6315 | Chicken | July | 1998 | 97510 | Human | December | 1997 | |
| ba9301 | Chicken | December | 1998 | 97548 | Human | December | 1997 | |
| ia9423 | Chicken | February | 1999 | 9827 | Human | January | 1998 | |
| eb5331 | Chicken | June | 1998 | 9889 | Human | February | 1998 | |
| ga5341 | Chicken | September | 1998 | 98114 | Human | March | 1998 | |
| ca7302 | Chicken | September | 1998 | 98126 | Human | April | 1998 | |
| 106KUb | Chicken | April | 1998 | 98160 | Human | May | 1998 | |
| 141KU | Chicken | June | 1998 | 98161 | Human | May | 1998 | |
| 146KU | Chicken | June | 1998 | 98168 | Human | May | 1998 | |
| 154KU | Chicken | June | 1998 | 98169 | Human | NAf | NA | |
| 156KU | Chicken | June | 1998 | 98171 | Human | June | 1998 | |
| 157KU | Chicken | June | 1998 | 98195 | Human | June | 1998 | |
| 185KU | Chicken | August | 1998 | 98220 | Human | July | 1998 | |
| 159KU | Chicken | June | 1998 | 98251 | Human | August | 1998 | |
| 160KU | Chicken | July | 1998 | GB1d | GBS | November | 1994 | |
| 161KU | Chicken | July | 1998 | GB2 | GBS | NA | 1994 | |
| 170KU | Chicken | July | 1998 | GB3 | GBS | January | 1995 | |
| 174KU | Chicken | July | 1998 | GB4 | GBS | August | 1995 | |
| 177KU | Chicken | July | 1998 | GB5 | GBS | August | 1995 | |
| 179KU | Chicken | July | 1998 | MF6 | MFS | June | 1993 | |
| 190KU | Chicken | August | 1998 | MF7 | MFS | July | 1993 | |
| 192KU | Chicken | August | 1998 | MF8 | MFS | November | 1991 | |
| 201KU | Chicken | September | 1998 | GB11 | GBS | October | 1996 | |
| 231KU1152 | Chicken | October | 1998 | GB13 | GBS | August | 1995 | |
| 228KU1134 | Chicken | October | 1998 | GB14 | GBS | August | 1995 | |
| 96210c | Human | July | 1996 | GB15 | GBS | November | 1996 | |
| 96219 | Human | August | 1996 | GB16e | GBS | September | 1997 | |
| 96243 | Human | August | 1996 | GB17 | GBS | October | 1996 | |
| 96248 | Human | August | 1996 | GB18 | GBS | September | 1998 | |
| 96328 | Human | September | 1996 | GB19 | GBS | October | 1998 | |
| 96357 | Human | September | 1996 | MF20 | MFS | October | 1998 | |
| 96362 | Human | September | 1996 | GB21 | GBS | December | 1998 | |
| 96364 | Human | September | 1996 | |||||
| 96392 | Human | October | 1996 | |||||
| 96402 | Human | October | 1996 | |||||
C. jejuni strains isolated from chickens (12; Jacobs-Reitsma et al., 19th Workshop Campylobacter, Helicobacter, Related Organisms).
C. jejuni strains isolated from chickens (9).
C. jejuni strains isolated from patients with gastroenteritis (3; De Wit et al., submitted).
C. jejuni strains isolated from patients with GBS or MFS (6).
Strains GB16 and C2609 were isolated in Belgium; all other strains were isolated in The Netherlands.
NA, not available.
Bacteria were grown on blood agar plates with 5% sheep blood, at 37°C for 48 h under microaerophilic conditions, with 6% O2, 7% CO2, 80% N2, and 7% H2 using the anoxomat system. Bacteria were stored at −80°C in 10 to 15% glycerol in heart infusion broth.
Isolation of chromosomal DNA.
Cells were scraped from fresh grown plates and washed with 1 ml of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). DNA was isolated using the Puregene kit (Gentra Systems, Minneapolis, Minn.). DNA integrity was checked by agarose gelelectrophoresis, and DNA preparations were stored at −20°C. DNA concentrations were determined with a spectrophotometer and were standardized at 0.1 μg/μl.
AFLP analysis.
Strains were typed with the recently described AFLP method for Campylobacter genotyping according to an adapted protocol of the AFLP microbial fingerprinting method of PE Applied Biosystems (5). In short, chromosomal DNA was digested with HindIII and HhaI and simultaneously ligated with restriction site-specific adapters for 2 h at 37°C. This was followed by a preselective PCR using adapter-specific primers with HindIII (5′-GACTGCGTACCAGCTT) and HhaI (5′-GATGAGTCCTGATCGC). Next, an aliquot is subjected to a selective PCR using a fluorescently labeled HindIII primer that contained an additional A nucleotide at the 3′ end (5′-GACTGCGTACCAGCTTA) and a HhaI primer with an A extension (5′-GATGAGTCCTGATCGCA). The final products were run on a 7.3% denaturing acrylamide sequencing gel for 5 h, performed on an ABI 373A automated DNA sequencer.
Data processing.
After electrophoresis, the fluorescently labeled fingerprints were collected with ABI Genescan software (PE Applied Biosystems). Gels were normalized by using an internal ROX-labeled size standard included in each sample. Densitometric curves were processed with the GelCompar version 4.1 software (Applied Maths, Kortrijk, Belgium). After normalization and background subtraction with mathematical algorithms, the levels of genetic similarity between AFLP patterns were calculated with the Pearson product-moment correlation coefficient (r). Pairwise comparison for band-based Dice analysis were made between all strains with the coefficient of Nei and Li (18) because this method does not infer the direction or weight change of AFLP bands. For cluster analysis of AFLP banding patterns, the unweighted-pair-group method using average linkages (UPGMA) was used (26).
For statistical analysis of the distribution of strains the Fisher exact test (Genestat 5, release 4.1 software) was used.
RESULTS
Interpretation of AFLP fingerprints of C. jejuni.
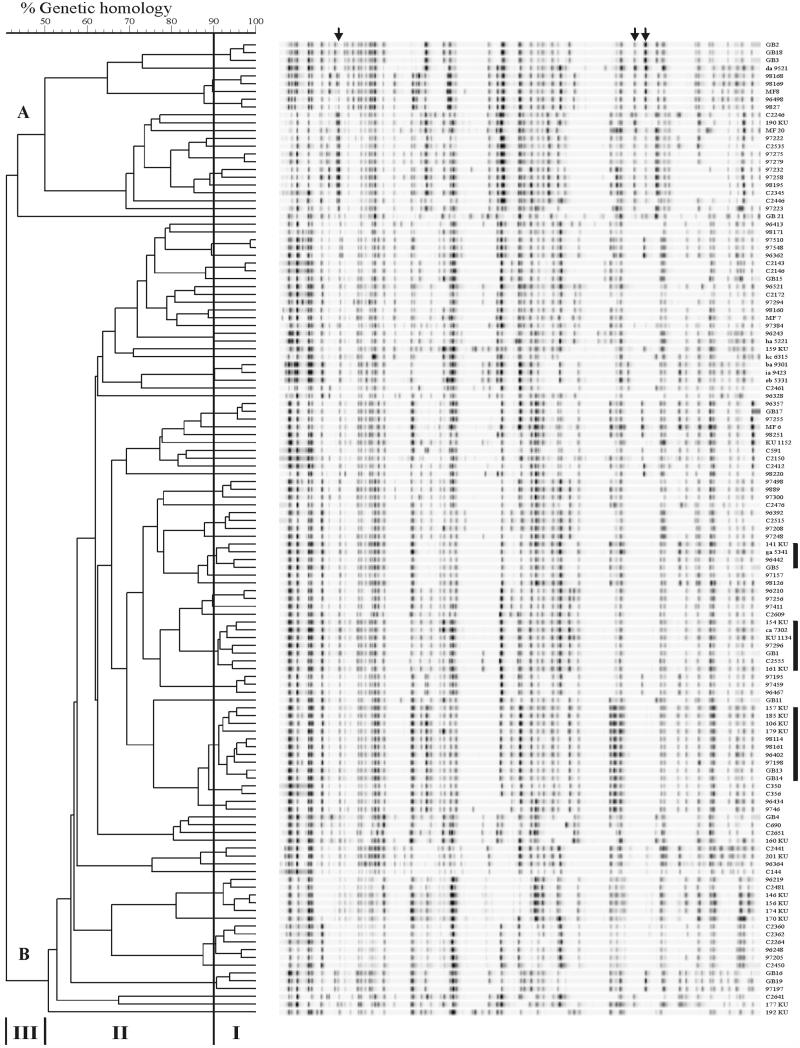
The AFLP method used in this study was recently developed on a small number of strains (5). In this study, highly heterogeneous fingerprints were generated for all 125 C. jejuni (Fig. 1). Fingerprints showed an average of 46 fragments, evenly distributed from 50 to 450 bp. The genetic homologies between banding patterns were analyzed by using the Pearson correlation coefficient to estimate genetic homologies and UPGMA for cluster analysis. Within the dendrogram, three windows of similarity can be seen (5) (Fig. 1). Window I contained patterns with 90 to 100% homology which are therefore considered to be genetically highly related, depending on the branch formation. Three clusters with related fingerprints of strains from chickens and gastroenteritis and GBS-MFS patients were observed, indicated by the black bars in Fig. 1. Window II showed banding patterns with 50 to 90% homology obtained from different strains but also from small clusters of strains that showed some relation in their fingerprints. In some of the clusters strains were present from all three sources that were analyzed.
FIG. 1.
UPGMA dendrogram of AFLP fingerprints from 55 chicken C. jejuni strains, 52 strains from patients with gastroenteritis, and 18 strains related to patients with GBS or MFS. The percentage of genetic homology between banding patterns is indicated. Window I represents small groups of identical fingerprints with 90 to 100% homology. Fingerprints in window II share 50 to 90% homology. Window III displays the low levels of homology (<40%) between the two major groups, A and B, encountered among the studied strains. Arrows indicate the AFLP bands specific to group A. Bars indicate clusters with related fingerprints from all sources.
Window III showed two groups (A and B) of fingerprints that were deeply branched, showing 40% genetic homology (Fig. 1). Strain GB21 clustered separately from these groups. This strain belonged to Penner serotype O:13,65 and showed a reproducible aberrant AFLP fingerprint (6). Group A consisted of 23 strains, while the majority of strains (n = 102) belonged to group B. Two AFLP bands were specific for strains of group A, and one band was specific for a set of strains in group A. These bands are indicated with arrows in Fig. 1. This separation in AFLP groups of C. jejuni was also obvious in the dendrogram of the previously typed small number of strains (5). Protein profiles of a few strains of group A were determined by Peter Vandamme (University of Ghent, Ghent, Belgium), whose results confirmed that these strains belonged to the species C. jejuni subsp. jejuni (25). To assess whether band-based analysis would result in different clustering and discrimination of AFLP patterns, the AFLP fingerprints were analyzed with the band-based Dice correlation coefficient using the algorithms of Nei and Li (18). The band-based analysis was determined with a manual designation of bands and therefore could be biased by individual interpretation differences in band assignment. Nevertheless, with band-based analysis and the use of UPGMA clustering, a dendrogram with identical clustering and discrimination of strains as observed with the correlation-based analysis was obtained (data not shown).
AFLP fingerprints of chicken strains.
The AFLP patterns of C. jejuni strains from chickens were dispersed all over the dendrogram. No specific cluster consisted of only AFLP fingerprints of chicken strains (Fig. 1). Identical AFLP fingerprints were observed between some strains, as is shown in window I of Fig. 1 and in Table 2. Of these strains, C350 and C365 were isolated at the same time at one farm, as well as strains C2143 and C2146. All other strains were isolated at different farms over various years and months (Table 1). AFLP fingerprints of 9 chicken strains were identical to strains from humans with gastroenteritis, and 11 chicken strains shared an AFLP type with strains from gastroenteritis patients or GBS or MFS patients (Table 2).
TABLE 2.
AFLP clusters with genetically related fingerprints (>90% similarity)
| Strain sources | AFLP clusters |
|---|---|
| Chicken + chicken | C2143 + C2146; C350 + C356; 201KU + C2441; ba9301 + ia94223 + eb5331 |
| Gastroenteritis + gastroenteritis | 96498 + 9827; 97275 + 97279; 97232 + 97258; 97510 + 97548; 9889 + 97498 + 97300; 96210 + 97256 + 97411; 97195 + 97459 + 96469; 96434 + 9746 |
| GBS-MFS + GBS-MFS | GB2 + GB18 + GB3 |
| Chicken + gastroenteritis | ha5221 + 96243; C2515 + 96392 + 97208; C2481 + 146KU + 156KU + 174KU + 96219; C2360 + C2362 + C2264 + 96248 + 97205 |
| Gastroenteritis + GBS-MFS | 98168 + 98168 + MF8; 96328 + 96357 + GB17; 97197 + GB16 + GB19 |
| Chicken + gastroenteritis + GBS-MFS | 141KU + ga5341 + 96442 + GB5; 154KU + ca7302 + KU1134 + 161KU + C2555 + GB1 + 96296; 157KU + 185KU + 106KU + 179KU + 98114 + 98161 + 96402 + 97198 + GB13 + GB14 |
Twenty-nine chicken strains showed a unique AFLP fingerprint. The chicken and human strains that grouped together were isolated in all seasons of the year, and only a slight overlap in the isolation data was observed (Table 1).
AFLP fingerprints of human gastroenteritis strains.
AFLP fingerprints of strains from patients with gastroenteritis were also highly diverse. In Table 2 the genetic homology between AFLP fingerprints from human strains and chicken derived strains is shown. The AFLP fingerprints of eight strains were identical to other gastroenteritis strains which were epidemiologically unrelated. Six strains showed AFLP fingerprints that were identical to the fingerprint from the chicken strains (Table 2). Two of these groups consisted of chicken and human strains that were isolated in the summer. Six AFLP types were related to strains from GBS or MFS patients, and AFLP types of six strains grouped in related clusters with strains from all sources (Table 2). Strains in these groups were obtained from all seasons. Nineteen AFLP fingerprints of gastroenteritis strains were unique.
AFLP fingerprints of GBS- and MFS-related C. jejuni strains.
The GBS- and MFS-related strains also showed very heterogeneous AFLP fingerprints that were distributed all over the dendrogram. Strains GB2, GB18, and GB3 displayed identical fingerprints (Fig. 1, window I). Strains GB3 and GB18 belong to the same Penner serotype O:19, strain GB2 is untypeable, and these strains contain identical flaA types (6). AFLP fingerprints of strains GB13 and GB14, obtained from family members of a GBS patient, were indistinguishable. Both strains were obtained in 1995; unfortunately, no strain was obtained from the GBS patient (2).
Strains MF8, GB17, GB16, and GB19 were related to gastroenteritis strains, and four strains (GB5, GB1, GB13, and GB14) clustered with an AFLP type of chicken and gastroenteritis strains (Table 2). Four strains displayed AFLP fingerprints that were not related to other strains.
The number of human strains in group A is 11 gastroenteritis strains and 6 GBS- and MFS-related strains versus 6 chickens strains, and in group B the number is 54 human strains versus 48 chicken strains. Statistical analysis of the distribution of strains within the two groups identified that group A contained significantly more human strains (P = 0.017) (Table 3).
TABLE 3.
Distribution of AFLP fingerprintsa
| Group (n) | No. of strains from (strain source):
|
||
|---|---|---|---|
| Chicken | Gastroenteritis | GBS-MFS | |
| A (23) | 6 | 11 | 6 |
| B (102) | 48 | 42 | 12 |
The proportion of human strains is significantly higher in group A than the number of chicken strains (P = 0.017). The Fisher exact test was applied to assess the significant difference between the proportions of strains in the two groups, by using 2×2 contingency tables.
DISCUSSION
The data presented in this study describe the genetic composition of a set of 125 C. jejuni strains isolated from chickens and humans as determined by AFLP fingerprinting. The main route toward human infection is thought to be the consumption of contaminated poultry products, and the aim of the present study was to establish if the same C. jejuni genotypes occur in both chickens and humans. Furthermore, we searched for specific genotypes that are associated with chicken or human infections only. C. jejuni strains related to the neurological human diseases GBS and MFS were also subtyped in order to establish the genetic relatedness of these strains with strains isolated from chickens. The AFLP fingerprints of all 125 strains appeared to be highly heterogeneous, and no characteristic pattern of strains infecting either chickens or humans was identified. Also, no separate grouping of GBS related strains was obtained.
The high diversity of AFLP fingerprints may be a reflection of the normal genetic diversity that occurs between C. jejuni strains, since several studies have identified diversity among both human and animal strains. With multilocus enzyme electrophoresis analysis, a suitable method for evaluating population genetics of other bacteria, no subgroups of human and animal C. jejuni strains were identified (1). With other typing methods, such as PCR-RFLP analysis of flaA genes, considerable allelic variation of this gene in human and animal strains was observed (20). Also, RAPD analysis could not identify distinct subgroups (13). In addition, Owen et al. (21) showed through a combination of serotyping and PFGE analysis that strains from human cases were highly diverse (21). In contrast, data obtained in Denmark by serotyping (15) and PFGE analysis (19) showed that identical clones of C. jejuni are present among human strains and in strains from chickens and cattle. Instead, our AFLP data on C. jejuni from The Netherlands support the findings that C. jejuni from chickens and humans are genetically diverse and constitute no subpopulations.
A leading risk factor for human campylobacteriosis is the consumption of contaminated poultry meat or cross-contaminated food (24). The AFLP fingerprints of C. jejuni strains from chickens did not belong to a genetically distinct group compared to strains causing human infections. A total of 16 C. jejuni strains (of 67 strains) from human infections shared genotypes with chicken strains, a finding which indicated that a large number of genotypes found in human infections was still not found among the chicken strains. This cannot be explained by a different seasonal distribution of strains, since strains isolated from one season showed predominantly highly diverse AFLP types, and when an overlap of human and chicken strains was observed the strains were obtained from different seasons as well. Therefore, either the overlap in genotypes was missed because of the highly diverse genotypes that are present among the large number of strains found in chickens or strains from sources other than contaminated poultry products contribute to human C. jejuni infections. Other meat-producing animals, including pigs, cattle, and sheep, are also frequently infected with Campylobacter spp. (7). Additional AFLP analysis of more C. jejuni strains from contaminated sources, including those from domestic pets and environmental samples, is needed to establish how AFLP fingerprints from these strains are related to those of human strains.
The AFLP analysis used here permits a high degree of strain differentiation and is of clear value for the determination of subtle interstrain relationships. Perhaps the method is excessively discriminating, but it was established by AFLP analysis that epidemiologically or clonally related strains show identical patterns (5, 27). Strains with minor changes in the flagellin locus were not differentiated by AFLP (5). Comparison of AFLP with PFGE, ribotyping, and flaA PCR-RFLP typing of chickens strains revealed that the differentiation of strains and the identification of related strains is the most effective with AFLP typing (3) and is comparable to PFGE typing when more than one enzyme is used. These data indicate that AFLP analysis correctly reflects a high genetic diversity of chicken and human Campylobacter strains.
Within the dendrogram, two subgroups of C. jejuni with fingerprints with only 40% homology were identified. This separation in fingerprints was also observed when a smaller number of strains were analyzed (5) and was maintained when strains from other geographical sources were compared (data not shown). There were significantly more strains from the gastroenteritis patients and GBS and MFS patients located in group A. The difference in distribution cannot be explained by a difference in host specificity, season of isolation, or virulence of the strains, since both groups consisted of chicken and gastroenteritis and GBS-MFS patient C. jejuni strains that were randomly obtained in The Netherlands (Table 2). However, the significantly higher proportion of human strains in group A is an indication that genotypes of this group are more likely to be associated with human infection. It is remarkable that when the chicken strains were analyzed with PFGE, flaA PCR-RFLP, or ribotyping, this differentiation was not observed (3). Group A contained specific AFLP bands that could indicate potential marker molecules for identification of human pathogenic C. jejuni. However, further analysis of AFLP bands is needed to elucidate what genetic characteristic causes the distinction between C. jejuni AFLP fingerprints and how this is related to human infection.
Based on the serological evidence, the association of certain serotypes of C. jejuni with the development of postinfectious neurological diseases, such as GBS and MFS, now appears established (10, 14). However, much work remains in order to determine how Campylobacter spp. can induce these diseases. Genotyping of serotype O:19 and O:41 strains identified no differences between GBS-related strains and strains isolated from patients without GBS (8, 27). The GBS and MFS strains analyzed in our study represented several distinct serotypes, and no separate genetic lineage of GBS- and MFS-related C. jejuni strains could be defined by AFLP analysis. In addition, the use of other typing methods, including serotyping, flaA typing, RAPD analysis, and PFGE analysis, showed a high diversity of strains, and no specific GBS characteristic genotype could be identified (6). Of course, other yet-unknown factors, such as plasmid-derived factor or the contribution of different gene expressions, may define a type that is related to GBS. Heat-stable antigens of particular C. jejuni serotypes have been implicated as possible pathogenic factors in the development of GBS and MFS (16). However, differences in the manifestations of GBS and MFS may be determined by host factors rather than by specific strain characteristics. A critical issue is which features of C. jejuni are involved in the pathogenesis of GBS and MFS, since no genetic differentiation based on whole genome methods can be made.
With AFLP typing the population of C. jejuni strains from chickens in The Netherlands appeared to be genetically highly diverse. The AFLP genotypes found in strains from chickens showed only limited overlap with genotypes of strains found in human diseases but did not belong to two subpopulations. This may imply that every C. jejuni strain that colonizes chickens may have the potential to cause human infections and even to cause severe postinfectious neurological diseases such as GBS or MFS.
ACKNOWLEDGMENTS
This study was funded by The Dutch Ministry of Agriculture and partly by the Product Boards for Livestock, Meat, and Eggs, Zeist, The Netherlands. C. W. Ang received a grant from the Prinses Beatrix Fonds (95-0518).
We are grateful to Arjen van de Giessen from the National Institute of Public Health and the Environment, Bilthoven, The Netherlands, and to Wilma Jacobs-Reitsma, ID-Lelystad, Lelystad, The Netherlands, for providing the chicken C. jejuni strains. We thank Aline de Koeijer and Michiel van Boven, Department of Immunology, Pathobiology, and Epidemiology, ID-Lelystad, The Netherlands, for the statistical analysis and for critical reading of the manuscript.
REFERENCES
- 1.Aeschbacher M, Piffaretti J-C. Population genetics of human and animal enteric Campylobacter strains. Infect Immun. 1989;57:1432–1437. doi: 10.1128/iai.57.5.1432-1437.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang C W, Van Doorn P A, Endtz H P, Martina I S J, Jacobs B C, Van Koningsveld R, Van der Meche F G A. A single case of Guillain-Barré syndrome in a family with Campylobacter jejuni enteritis. J Neurol. 1998;245:417. doi: 10.1016/s0165-5728(00)00369-6. [DOI] [PubMed] [Google Scholar]
- 3.De Boer P, Duim B, Rigter A, van der Plas J, Jacobs-Reitsma W, Wagenaar J A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Wit M A S, Koopmans M P G, Kortbeek L M, van Leeuwen W J, Vinje J, Bartelds A I M, van Duynhoven Y T P H. Interim report of a study on gastroenteritis in sentinel practices in the Netherlands (NIVEL) 1996–1999. Results of the first two years. RIVM report 216852003. Bilthoven, The Netherlands: RIVM; 1999. [Google Scholar]
- 5.Duim B, Wassenaar T M, Rigter A, Wagenaar J A. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with AFLP fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endtz H P, Ang C W, van den Braak N, Duim B, Price L J, Rigter A, Woodward D L, Rodgers F G, Johnson W M, Wagenaar J A, Jacobs B A, Verbrugh H A, van Belkum A. Molecular characterization of Campylobacter jejuni from patients with Guillain-Barré and Miller Fisher syndrome. J Clin Microbiol. 2000;38:2297–2301. doi: 10.1128/jcm.38.6.2297-2301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fricker C R, Park R W. A two-year study of the distribution of ‘thermophilic’ campylobacters in human, environmental and food samples from the Reading area with particular reference to toxin production and heat-stable serotype. J Appl Bacteriol. 1989;66:477–490. doi: 10.1111/j.1365-2672.1989.tb04568.x. [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto S, Mishu-Allos B, Misawa N, Patton C M, Blaser M J. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997;176:1105–1108. doi: 10.1086/516522. [DOI] [PubMed] [Google Scholar]
- 9.Heuvelink A E, Tilburg J J H C, Voogt N, van Pelt W, van Leeuwen W J, Sturm J M J, van de Giessen A W. Surveillance van bacteriële zoönose-verwekkers bij landbouwhuisdieren. Periode april 1997 tot en met maart 1998. RIVM report 285859 009. Bilthoven, The Netherlands: RIVM; 1999. [Google Scholar]
- 10.Jacobs B C, Hazenberg M P, van Doorn P A, Endtz H P, van der Meche F G. Cross-reactive antibodies against gangliosides and Campylobacter jejuni lipopolysaccharides in patients with Guillain-Barré or Miller Fisher syndrome. J Infect Dis. 1997;175:729–733. doi: 10.1093/infdis/175.3.729. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs-Reitsma W, Bolder N M, Mulder R W A W. Caecal carriage of Campylobacter and Salmonella in Dutch broiler flocks at slaughter: a one-year study. Poultry Sci. 1994;73:1260–1266. doi: 10.3382/ps.0731260. [DOI] [PubMed] [Google Scholar]
- 12.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 13.Madden R H, Moran L, Scates P. Sub-typing of animal and human Campylobacter spp. using RAPD. Lett Appl Microbiol. 1996;23:267–170. doi: 10.1111/j.1472-765x.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 14.Mishu B, Ilyas A A, Koski C L, Vriesendorp F, Cook S D, Mithen F A, Blaser M J. Serologic evidence of previous Campylobacter jejuni infection in patients with the Guillain-Barré syndrome. Ann Intern Med. 1993;118:947–953. doi: 10.7326/0003-4819-118-12-199306150-00006. [DOI] [PubMed] [Google Scholar]
- 15.Moller-Nielsen E, Engberg J, Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immun Med Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 16.Moran A P, Appelmelk B J, Aspinall G O. Molecular mimicry of host structures by lipopolysaccharides of Campylobacter and Helicobacter spp.: implications in pathogenesis. J Endotoxin Res. 1996;3:521–531. [Google Scholar]
- 17.Nachamkin I, Mishu-Allos B, Ho T. Campylobacter species and Guillain-Barré Syndrome. J Clin Microbiol. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nei M, Li W-H. Mathematical model for studying genetic variations in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.On S L W, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen R J, Leeton S. Restriction fragment length polymorphism analysis of the flaA gene of Campylobacter jejuni for subtyping human, animal and poultry isolates. FEMS Microbiol Lett. 1999;176:345–350. doi: 10.1111/j.1574-6968.1999.tb13682.x. [DOI] [PubMed] [Google Scholar]
- 21.Owen R J, Slater E, Telford D, Donovan T, Barnham M. Subtypes of Campylobacter jejuni from sporadic cases of diarrhoeal disease in different locations in England are highly diverse. Eur J Epidemiol. 1997;13:837–840. doi: 10.1023/a:1007497005152. [DOI] [PubMed] [Google Scholar]
- 22.Savelkoul P H, Aarts H J, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L, Schouls L, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skirrow M B, Blaser M J. Clinical and epidemiologic considerations. In: Nachamkin I, Tompkins L S, Blaser M J, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 3–9. [Google Scholar]
- 24.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized countries. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 25.Vandamme P, Bot P, Kersters K. Differentiation of Campylobacters and Campylobacter-like organisms by numerical analysis of one-dimensional electrophoretic protein patterns. Syst Appl Microbiol. 1991;14:57–66. [Google Scholar]
- 26.Vauterin L A, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 27.Wassenaar T M, Fry B N, Lastovica A J, Wagenaar J A, Coloe P J, Duim B. Genetic Characterization of Campylobacter jejuni O:41 isolates in relation with Guillain-Barré Syndrome. J Clin Microbiol. 2000;38:874–876. doi: 10.1128/jcm.38.2.874-876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]