Abstract
A competitive reverse transcription-PCR method was developed for the semiquantitation of the expression of genes encoding bicomponent leucotoxins of Staphylococcus aureus, e.g., Panton-Valentine leucocidin (lukPV), gamma-hemolysin (hlgA and hlgCB), and LukE-LukD (lukED). The optimization procedure included RNA preparation; reverse transcription; the use of various amounts of enzymes, antisense primer, and RNA; and the final amplification chain reaction. Reproducible results were obtained, with sensitivity for detection of cDNA within the range of 1 mRNA/104 CFU to 102 mRNA/CFU, depending on the gene. Both specific mRNAs were more significantly expressed at the late-exponential phase of growth. Expression was about 100-fold higher in yeast extract-Casamino Acids-pyruvate medium than in heart infusion medium. Expression of the widely distributed gamma-hemolysin locus in the NTCC 8178 strain was around 10-fold diminished compared with that in the ATCC 49775 strain. Because of the lower level of hlgA expression, the corresponding protein, which is generally not abundant in culture supernatant, should be investigated for its contribution to the leucotoxin-associated virulence. The agr, sar, and agr sar mutant strains revealed a great dependence with regard to leucotoxin expression on the global regulatory system in S. aureus, except that expression of hlgA was not affected in the agr mutant.
Staphylococcal bicomponent leucotoxins are exotoxins consisting of two nonassociated but synergic class S (31 to 32 kDa) and class F (35 kDa) proteins. Among this family of toxins, the Panton-Valentine leucocidin (PVL) is encoded by two contiguous and cotranscribed genes, lukFPV and lukS-PV (31). Another locus encodes γ-hemolysin and comprises three genes: the first two encode class S proteins (HlgA and HlgC), and the third one encodes a class F protein (HlgB). hlgA constitutes an upstream open reading frame, whereas hlgC and hlgB (hlgCB in this text) are cotranscribed (10). Another locus was recently characterized as two cotranscribed class S and class F protein-encoding genes, lukE and lukD (lukED in the text), respectively (16). Production of leucotoxins among Staphylococcus aureus strains was studied by radial gel immunoprecipitation (16, 30), but quantitation of leucotoxin expression by enzyme-linked immunosorbent assay remains difficult because of cross-reactivity due to sequence identity between class S components (55 to 72%) and class F components (71 to 79%), respectively (29). There may be S. aureus strains that produce γ-hemolysin, PVL, and/or LukE-LukD.
Leucotoxin production was associated with infections resulting in furuncles, community pneumonia, and some antibiotic-associated diarrhea (11, 15, 24). These infection-related leucotoxins act as activators of human neutrophils before creating lytic pores sensitive to monovalent cations (34). The leucotoxins were shown to induce an important inflammatory response in vivo in rabbit skin and in rabbit vitreous humor (16, 31, 32). Therefore, to determine their respective roles in pathogenicity and whether expression might influence these roles, it is important to semiquantify, at least, the expression of loci within the leucotoxin family. RNA methods, and reverse transcription (RT)-PCR in particular, were shown to be more sensitive than antibody-based detection methods (3). In contrast to Northern blotting, which has a lower sensitivity (35), RT-PCR allows multiple and simultaneous detection of mRNAs contained in limited amounts of total RNA prepared from tissues or sample volumes. This method is widely used to quantify viruses (human immunodeficiency virus and hepatitis B and C viruses) and cytokine expression in different systems (12, 20). RNA methods are still underutilized for detection (19) or quantitative analysis (14, 18) of bacterial gene expression.
The aim of this work was to develop a semiquantitative and competitive RT-PCR method to compare the expression of the bicomponent leucotoxins under different culture conditions and to explore the dependence of these leucotoxins on the global accessory gene regulator (agr) and staphylococcal accessory regulator (sar) systems (8).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The S. aureus V8 strain (ATCC 49775) produces both γ-hemolysin (HlgA, -B, and -C) and PVL. S. aureus Newman strain (NTCC 8178) produces γ-hemolysin (HlgA, -B, and -C) and LukE-LukD but does not produce PVL. The agr, sar, and agr sar mutant strains derived from Newman were kindly provided by A. L. Cheung (Rockefeller University, New York, N.Y.) (7). The agr, sar, and agr sar mutant strains derived from ATCC 49775 were obtained by transduction with bacteriophage 85 of the Newman agr and sar mutant strains (28). The agr and sar mutant strains were selected for their resistance to tetracycline and erythromycin, respectively, and the disruptions of the agr and sar loci were verified by Southern blotting (33) with specific probes (data not shown).
Escherichia coli Epicurian Coli XL1-Blue Supercompetent cells {E. coli recA1 endA1 thi-1 hsdR17 supE44 relA1 lacZ[F′ proAB lacIqZΔM15 Tn10(tet)]; Stratagene, Amsterdam, The Netherlands} were recipients for the competitor plasmids listed in Table 1.
TABLE 1.
Plasmid competitors used as internal standards for RT-PCR and related materials
| Plasmida | Size (kb) | Properties and construction methodb | Reference |
|---|---|---|---|
| pCU1 | 4.9 | Shuttle vector for E. coli and S. aureus | 1 |
| pCU hlg | 10.3 | pCU1; 5.4-kb ScaI DNA fragment containing hlgA, -B, and -C genes in HincII-PstI sites of pCU1 plasmid | 31 |
| pCU hlgA | 6.9 | pCU1; 2-kb EcoRI-EcoRI DNA fragment encoding hlgA | This study |
| pCU lukED | 8.1 | pCU1; 3.2-kb HindIII DNA fragment encoding lukE and lukD | 16 |
| pCU lukPV | 8.0 | pCU lukS-PV/lukFPV; HindIII-HincII sites containing 3.1-kb HindIII-NruI fragment containing lukS-PV and lukFPV | 31 |
| pGP7 | 7.0 | pCR II lukPV (NdeI°, 80 nt); 80-bp MseI-MseI fragment from pUC19 inserted into NdeI site of pCR II-lukPV (encoding structural lukS-PV and lukFPV genes obtained with TA cloning kit [In vitrogen Corporation]) | This study |
| pGP8 | 10.2 | pCU hlgACB (deletion of nt 2487 to 2598); deletion of pCU-hlgACB at positions 2487 to 2598 (hlgC) obtained by Pfu DNA polymerase (Stratagene) amplification with dedicated primers surrounding the deletion | This study |
| pGP9 | 7.1 | pCU hlgA (EcoRV°, 200 nt); 200-bp AluI-AluI fragment from pCU1 inserted at the unique EcoRV site at position 275 of hlgA | This study |
| pGP10 | 8.35 | pCU lukED (EcoRI°, 250 nt); 250-bp Tsp509I-Tsp509I from pCU1 hlg inserted at the unique EcoRI site at position 1855 of lukD | This study |
Presence of plasmids was maintained in cultures by using 100 μg of ampicillin/ml of medium.
nt, nucleotide.
S. aureus strains were grown in yeast extract-Casamino Acids-pyruvate (YCP) medium (16) (3.0% [wt/vol] yeast extract [Oxoïd Ltd, Basingstoke, England], 2.0% [wt/vol] Bacto Casamino Acids [Difco Laboratories, Detroit, Mich.], 2.0% [wt/vol] pyruvic acid [Sigma-Aldrich, St. Louis, Mo.], 2.5‰ [wt/vol] Na2HPO4, 0.4‰ [wt/vol] KH2HPO4; pH 7.0) or in 2.5% (wt/vol) heart infusion (HI) medium (pH 7.4; Difco Laboratories).
Primers.
Specific primers were deduced from the corresponding nucleotide sequences (Table 2) and were synthesized by Life Technologies (Gaithersburg, Md.).
TABLE 2.
PCR primers used for each template in this study
| Genea | Location | Sequence | Polarity | Size of PCR product (bp) | Size of PCR competitor product (bp) |
|---|---|---|---|---|---|
| lukPV | 2344–2367 | 5′-ATGACTCAGTAAACGTTGTAGAT-3′ | Sense | 520 | 600 |
| X72700a | 2802–2826 | 5′-TCTATCCATTTCACTTTGATAAGT-3′ | Antisense | ||
| hlgA | 170–200 | 5′-ATGATTAAAAATAAAATATTAACAGCAACT-3′ | Sense | 350 | 550 |
| X81586a | 503–527 | 5′-ATCAACATTAGAGTCTTTCGTTTT-3′ | Antisense | ||
| hlgCB | 1904–1926 | 5′-AGCTCTCGAACAACATATTATA-3′ | Sense | 940 | 850 |
| X81586a | 2824–2845 | 5′-CTGCAGCTTTAAGCACTAAAG-3′ | Antisense | ||
| lukED | 1473–1497 | 5′-TAGGCAAATCATCAGTTGCTTCAT-3′ | Sense | 516 | 750 |
| Y13225a | 1967–1989 | 5′-GTAGTTCTGTAACTTTCTTGTTT-3′ | Antisense |
Accession number from the EMBL and GenBank libraries.
Total RNA extractions.
The usual care was taken in handling RNA. Cultures were started with 100 μl of inoculum, grown at 37°C to stationary phase, and dropped in 2-liter Erlenmeyer flasks filled with 120 ml of YCP or HI medium for incubation at 37°C and 180 rpm. Volumes generally corresponding to a range of 5 × 107 to 5 × 109 CFU/ml (1 to 20 ml, depending on growth phase) were serially harvested, and the bacteria were pelleted at 4°C and washed with 1 ml of diethyl pyrocarbonate (DEPC; Sigma-Aldrich)-treated H2O. The washed cell pellet was resuspended in 200 μl of DEPC-treated H2O plus 40 U of RNase OUT (Life Technologies). RNA extraction was achieved with the FastRNA Blue Kit (Bio101, Inc., Vista, Calif.) (6); cells were ground by centrifugation with silica beads associated with chaotropic agents and phenol, according to the manufacturer's recommendations. After isopropanol precipitation, the dried pellet was solubilized in 100 μl of SAFEE (DEPC-treated H2O, 0.5 mM EDTA) and an additional LiCl precipitation step was performed to minimize carbohydrate contamination. The carbohydrate-free pellet was washed twice with 250 μl of SEWS-BLUE (Bio101, Inc.), and the RNA preparation was resuspended in 0.5 mM EDTA (pH 7.5) plus 40 U of RNase OUT and stored in aliquots at −80°C before use.
RNA preparations were also performed using Chomczynski's method (9) (Tri-Reagent RNA preparation [Molecular Research Center, Inc.]). Washed bacteria were first treated with 450 U of lysostaphin (Ambicin L; Applied Microbiology, Inc., New York, N.Y.) for 30 min at 4°C, and RNA was purified according to recommended methods. The RNeasy Mini-Protocol RNA preparation (Qiagen S. A., Courtaboeuf, France) was also tested according to the manufacturer's recommendation, after bacteria were first digested for 10 min with 3 mg of lysozyme (Quantum Biotechnologies, Inc., Montreal, Canada) per ml.
To ensure the complete removal of DNA, RNA aliquots (20 μl) were treated in a 300-μl volume containing 30 μl of 10× DNase I buffer, 20 U of RNase-free DNase (Quantum Biotechnologies, Inc.), and 40 U of RNase OUT for 30 min at 25°C. The reaction was stopped by heating at 65°C for 5 min. RNA was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with isopropanol before being washed twice with 80% ethanol, air dried, resuspended in 50 μl of DEPC-treated H2O plus 20 U of RNase OUT, and stored at −80°C. The quantity and purity of total RNA were deduced from absorbances, gauged by the optical density at 260 nm (OD260) and the OD280. DNA contamination was only detected by RT-PCR with RNA preparations (data not shown) not having an LiCl precipitation prior to DNase treatment, indicating that mucopolysaccharides, which constitute powerful inhibitors for enzymatic reaction (9), have to be eliminated.
Reverse transcription of RNA templates.
cDNA synthesis was performed after optimization of the RT procedure, and the reverse transcription was carried out using Moloney murine leukemia virus (M-MuLV) from Perkin-Elmer (Foster City, Calif.). RNA sample was added to a solution containing 80 pmol of antisense 3′ primer (AP), 20 U of RNase OUT, 1 mM deoxynucleoside triphosphate (dNTP) (Boehringer-Mannheim, Germany), 2 μl of 10× GeneAmp PCR buffer II, 5 mM MgCl2, and 50 U of M-MuLV reverse transcriptase (Perkin-Elmer), and the volume was adjusted to 20 μl by adding DEPC-treated H2O. After a 15-min incubation at 42°C, the reaction was stopped by heating for 5 min at 95°C. The resulting cDNA was stored at −20°C. Parallel incubations were performed without the addition of reverse transcriptase in order to alternatively verify the DNA removal.
PCR.
The amplification step was performed by using previously aliquoted reagents. One microliter of cDNA or RNA solution incubated without RT was added to a 49-μl volume containing 1× Taq buffer (3 mM MgCl2), 0.2 mM dNTP (Boehringer-Mannheim), 0.2 pM concentrations of specific primers, and 2.5 U of Taq DNA polymerase (Life Technologies). The RNA-DNA heteroduplex was denatured for 3.5 min at 94°C. Amplification was achieved by 35 cycles of denaturation for 1.5 min at 92°C, annealing for 1.5 min (at 52°C for lukPV and hlgCB, at 54°C for hlgA, and at 56°C for lukED), and polymerization for 1.5 min at 72°C in a thermocycler (Perkin-Elmer model 9700), and it was ended with 8 min of incubation at 72°C. Samples were stored at −20°C prior to electrophoresis on a 1.2% (wt/vol) small fragment agarose gel (Quantum Biotechnologies, Inc.) in 0.5× TEB buffer (45 mM Tris, 0.6 mM EDTA, 45 mM boric acid [pH 8.3]) and staining with ethidium bromide. The 35 cycles chosen for PCR turned out to be sufficient. In every PCR assay, 15 initial copies of plasmid competitors always provided visible DNA bands on agarose gels.
Specific mRNA quantitation and RT-PCR optimization.
Constant amounts of cDNA corresponding to a determined quantity of CFU were coamplified by PCR with a dilution series of the corresponding competitor (Table 1), ranging from 100 ng to 0.01 fg. Bands of equal intensity on the agarose gel corresponding to amplified cDNA and to the amplified internal standard were assumed to reflect equal amounts of cDNA and the known concentration of competitor DNA from the plasmid. A visual observation of the equivalence of DNA bands was made on agarose gels. When no equivalence was visualized, secondary PCR using middle-range intermediate concentrations of competitors was achieved. Thus, the true equivalence ± 25% is given. For each sample, we performed three competitive PCRs on cDNAs to assess reproducibility. Punctual controls performed on independent RNA extractions and cDNA synthesis corroborated the results. Determinations of the initial number of specific mRNA copies/CFU could then be obtained with the following equation:
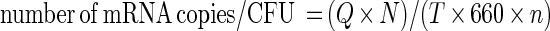 |
with Q being the quantity (in grams) of competitor plasmid, N being Avogadro's number (6.02 × 1023), T being the size of the competitor plasmid (in base pairs), and n being the number of CFU in the initial bacterial culture at such equivalence. The mass of 1 bp was assumed to be 660 Da.
The three mRNA extraction methods cited above were compared for efficiency. Total RNA was extracted from 3 × 109 CFU of S. aureus ATCC 49775 grown in YCP medium. After RT-PCR, 1 specific hlgCB mRNA/80 CFU was detected with the Fast-Prep and Tri-Reagent procedures but no signal was obtained with the Qiagen S. A. procedure (data not shown). The Fast-Prep method was retained because it is rapid, more standardized, and apparently efficient for S. aureus.
During the optimization, three kinds of M-MuLV RT were tested: M-MuLV from New-England Biolabs or Perkin-Elmer, and SuperScript II reverse transcriptase (Life Technologies). The RNA preparation described above was submitted to reverse transcription by each enzyme, according to the manufacturers' recommendations. Simultaneous competitive PCR allowed detection of 1 hlgA mRNA/20 CFU with the SuperScript II reverse transcriptase or the M-MuLV from Perkin-Elmer, but only 1 mRNA/200 CFU was detected in the case of the M-MuLV from New England Biolabs (data not shown). Furthermore, the M-MuLV from Perkin-Elmer was found to be more efficient when working with low yields of mRNAs. It was retained for subsequent analyses.
Different parameters of the reverse transcription procedure (i.e., various amounts of total RNA, AP, and M-MuLV [Perkin-Elmer]) were tested in combination to optimize and to standardize the RT-PCR procedure. Total RNA extraction was performed on S. aureus ATCC 49775 grown in YCP medium to a bacterial density of 109 CFU/ml. The RNA concentration was determined before lukPV reverse transcription was carried out. The combination of 0.5 μg of total RNA, 1.6 pM AP, and 50 U of M-MuLV from Perkin-Elmer was used in further experiments, since it allowed detection of 8 mRNA/CFU ± 20% whereas other conditions were more expensive and appeared to be less sensitive (up to 10 ± 3 mRNA/10 CFU).
RESULTS
Sensitivity of RT-PCR.
Amplifications of cDNA and competitive internal standard were found to proceed with equal efficiency, independently of the PCR phase, up to the plateau (13). To determine the sensitivity of the RT-PCR, a culture in YCP medium of the Newman strain (NTCC 8178) was harvested at 9 × 108 CFU/ml, and total RNA was purified. Competitive RT-PCR allowed quantification of 1 hlgCB mRNA/25 CFU. Then, serial dilutions of DNA-free RNA preparation, which corresponded to concentrations of from 1.3 × 108 to 1.3 × 104 CFU/50 μl, were carried out and were submitted to hlgCB-specific reverse transcriptions. PCR on cDNA equivalent to from 5.2 × 106 to 5.2 × 102 CFU per 2-μl aliquot was performed with 100-μl reaction mixtures. Positive signals for the hlgCB amplified fragments were visualized for 15-μl aliquots, corresponding to a sensitivity of 2.5 × 105 CFU. Taking into account the initial titer of 1 mRNA/25 CFU, the results corresponded to a sensitivity at least as low as 1 hlgCB mRNA/60 CFU for the whole RT-PCR.
Expression in YCP and HI media of lukPV, hlgA, and hlgCB from S. aureus V8 (ATCC 49775).
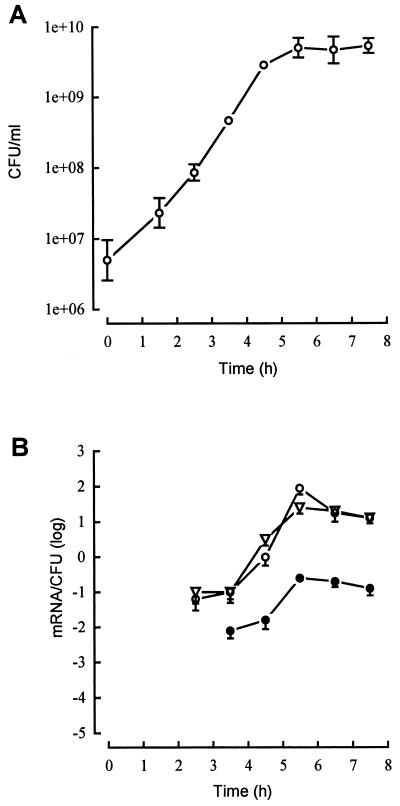
Iterative RNA preparations at chosen times of the growth in YCP medium of ATCC 49775 were submitted to lukPV, hlgA, and hlgCB RT-PCR. The semiquantitative curves show the number of mRNA per CFU relative to the passage of time (Fig. 1 and 2). Expression at 37°C of lukPV mRNAs increased 103-fold (Fig. 1), from 40 ± 12 mRNA/600 CFU at a bacterial density of 6 × 107 CFU/ml (2.5 h of culture) to 90 ± 30 mRNA/CFU at a bacterial density of 4.2 × 109 CFU/ml (5.5 h of culture). Concomitantly, expression of hlgA increased 30-fold to reach 25 ± 5 mRNA/100 CFU and the expression of hlgCB increased 300-fold to reach 23 ± 7 mRNA/CFU. Transcription of the lukP and lukV genes reached the highest level at the late-exponential growth phase, as was observed for the two other transcriptional units. Expression of hlgCB was close to lukPV, except for a significant difference in its optimal expression, which was threefold lower. The abundance of hlgA transcripts was low compared to hlgCB and lukPV transcripts, with an optimal expression 300-fold lower than that of lukPV and 100-fold lower than that of hlgCB. During 2 h of the stationary-growth phase, where the bacterial density remained almost constant, the expression of the transcription units decreased to a limited extent, not more than 10-fold.
FIG. 1.
Growth dependence of the transcription of lukPV (○), hlgA (●), and hlgCB (▿) in wild-type S. aureus ATCC 49775 grown in YCP medium. The expression of the genes encoding staphylococcal leucotoxins was semiquantified by RT-PCR. (A) Growth curve. (B) Semiquantitation curves of the expression of lukPV, hlgA, and hlgCB. The data represent the means ± standard deviations (error bars) from three RT-PCRs.
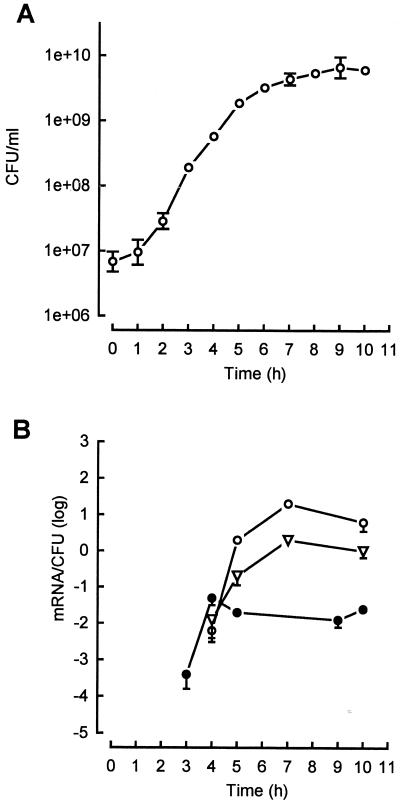
FIG. 2.
Growth dependence of the transcription of lukED (○), hlgA (●), and hlgCB (▿) in wild-type S. aureus Newman (NTCC 8178) grown in YCP medium. The expression of the genes encoding staphylococcal leucotoxins was semiquantified by RT-PCR. (A) Growth curve. (B) Semiquantitation curves of the expression of lukED, hlgA, and hlgCB. The data represent the means ± standard deviations (error bars) from three RT-PCRs.
Leucotoxin mRNA transcription of ATCC 49775 in HI medium remained basal during growth whatever the transcription unit considered. Expressions increased between 1.6 × 108 CFU/ml (3 h of culture) and 7.5 × 108 CFU/ml (4 h of culture), from a minimum of 10 ± 5 mRNA/106 CFU to a maximum of 40 ± 10 mRNA/104 CFU. As mentioned above, expression of the considered mRNAs decreased by about 10-fold during the beginning of the stationary-phase growth.
Expression in YCP and HI media of lukED, hlgA, and hlgCB from S. aureus Newman (NTCC 8178).
In YCP medium, expression of hlgA was detected at a bacterial density of 9 × 107 CFU/ml, with 40 ± 12 mRNA observed per 105 CFU (Fig. 2A), which increased 100-fold until a bacterial density of 2.7 × 108 CFU/ml was reached and remained stable at the beginning of the stationary-growth phase. Expression of hlgCB varied by 100-fold and appeared to be quite delayed, compared to that of hlgA, being optimal at a bacterial density of 4 × 109 CFU/ml. Finally, the Newman strain appeared to be a better producer of lukED mRNA than of those specific for hlgCB and hlgA, with 20 ± 6 mRNA/CFU at a density of 4 × 109 CFU/ml, though expression decreased threefold for the stationary-growth phase.
In HI medium, expression of leucotoxins remained very low, never exceeding 100 ± 25 mRNA/104 CFU, and hlgCB appeared to be strongly repressed compared to its expression in the YCP medium. Leucotoxin expression remained detectable from the mid-log phase on but did not increase more than 10-fold.
Comparison of leucotoxin expression in agr, sar, and agr sar mutant strains derived from ATCC 49775 and Newman and grown in YCP medium.
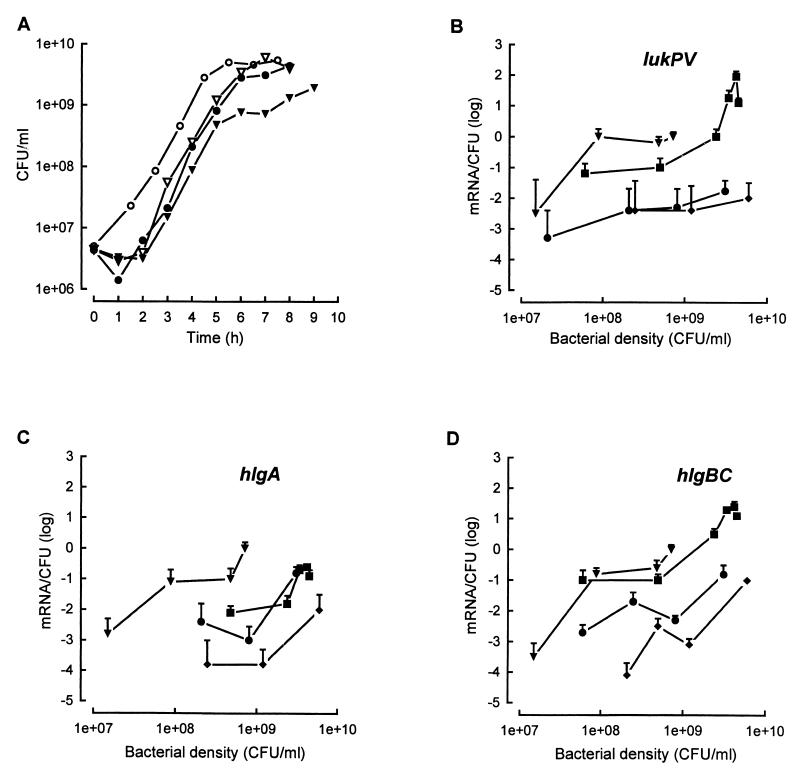
To assess the regulation of leucotoxins by the global agr sar regulatory system, wild-type and agr, sar, and agr sar mutant ATCC 49775 and NTCC 8178 strains were grown in YCP medium at 37°C before leucotoxin mRNAs were semiquantified. The semiquantitative curves show the dependence of the number of mRNA per CFU on bacterial density (Fig. 3 and 4). The bacterial density of the ATCC 49775 agr sar mutant strain was quite inferior to those of the other mutant or wild-type strains during the stationary phase. Semiquantitative analysis for bacterial densities of ≥109 CFU/ml was not obtained.
FIG. 3.
Expression of lukPV, hlgA, and hlgCB of S. aureus ATCC 49775 (wild type) (■) and agr (●), sar (⧫), agr sar (▾) mutants of the same strain grown in YCP medium. (A) Growth curves. (B, C, and D) Semiquantitative curves specific for lukPV, hlgA, and hlgCB, respectively. The data represent means ± standard deviations (error bars) from three RT-PCR.
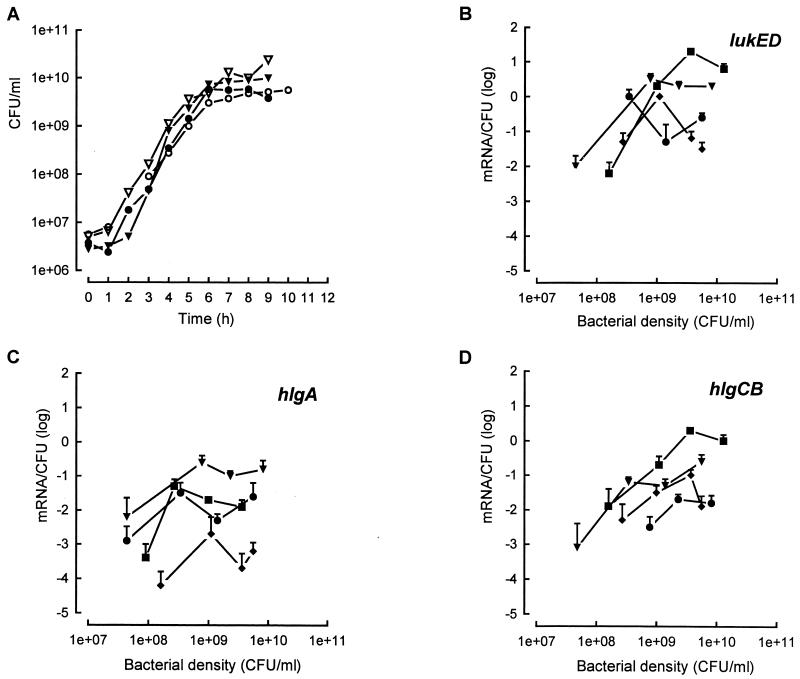
FIG. 4.
Expression of lukED, hlgA, and hlgCB of S. aureus Newman NTCC 8178 wild-type strain (■) and agr (●), sar (⧫), and agr sar (▾) mutant strains grown in YCP medium. (A) Growth curves. (B, C, and D) Semiquantitative curves specific for lukED, hlgA and hlgCB, respectively. The data represent means ± standard deviations (error bars) from three RT-PCR analyses.
Significant variations in the expression of leucotoxins occurred according to the bacterial density and the mutated strain. Expression of lukPV appeared to be the most sensitive, as it remained insignificant in ATCC 49775 agr and sar mutant strains. Nevertheless, it was detected at a bacterial density of 107 CFU/ml and increased to 100 ± 30 mRNA/100 CFU at a bacterial density of 9 × 107 CFU/ml for the agr sar mutant strain (Fig. 3B). Expression of hlgCB (Fig. 3D) in the ATCC 49775 agr and sar mutant strains was comparable to that observed for lukPV in the same mutated strains, though its expression increased at higher bacterial densities. As observed for the two other loci in the agr sar mutant, hlgCB was also derepressed during the exponential-growth phase to amounts of 600 ± 120 mRNA/600 CFU at a bacterial density of 7 × 108 CFU/ml. Expression of hlgA did not appear to be very sensitive to the agr mutation. It was significantly close between the agr mutant strain and the wild-type strain at a bacterial density of 3 × 108 CFU/ml (30 ± 6 mRNA/600 CFU for each), while for the sar mutant strain, the expression level was significantly less than that for either the agr mutant or the wild type. Expression of hlgA in the agr sar mutant strain appeared to be derepressed and significant at 600 ± 180 mRNA/600 CFU at a bacterial density of 8 × 108 CFU/ml, as was observed for lukPV.
Compared to the ATCC 49775 strain, similar results were obtained for the mutant strains derived from Newman (Fig. 4). For the lukED and hlgCB loci, expression was significantly diminished by 10- to 100-fold in both the agr and sar mutants. Expression of lukED remained significant, at 100 ± 15 mRNA/100 CFU for a bacterial density of 109 CFU/ml in the sar mutant (Fig. 4B). Again, as shown in Fig. 4C, the hlgA expression in the Newman agr mutant was not affected compared to that in the ATCC 49775 agr mutant (Fig. 3C) and was comparable to the wild type. Finally, expression of all loci in the Newman sar agr double mutant was detected at a lower bacterial density than was the case for the wild type (Fig. 4B, C, and D). This expression increased to a maximum, reached at around 109 CFU/ml, and then remained stable. Expression of hlgA in the agr sar mutant was around 10-fold higher than it was in the wild type. Finally, the expression of lukPV, hlgCB, and lukED was dependent on both agr and sar expression whatever the strain studied.
DISCUSSION
Compared to the cumulative detection of protein expression and production by radial immunoprecipitation or Northern blotting, RT-PCR methods constitute sensitive and real-time alternatives. A competitive RT-PCR has been optimized for the detection of staphylococcal leucotoxin mRNAs. Because of the short half-life of bacterial RNAs (6) and the need for reproducibility, a standardized procedure for the preparation of total RNAs was used. In this work, the competitive RT-PCR test using plasmid competitors provided a semiquantitative approach, with a sensitivity of 60 CFU expressing one specific mRNA, which is in the range reported (14) in a study that used the gyr invariant reporter gene and provided relative variations of mRNA expression compared to the reporter.
Expression of the staphylococcal leucotoxins from S. aureus grown in YCP medium was detected at very low yields at the beginning of the exponential-growth phase but increased by about 3 log units from the mid-log phase to the late-exponential growth phase. Then expression remain significant, but at a lower level, at the beginning of the stationary growth. The possible increase of ribonucleases, the nonsaturating conditions of enzymes for gene transcription at the stationary phase, or the privileged efficiency of expression for other genes might explain such observations (17). However, the expression of hlgA appeared to be 100-fold lower than that of lukPV, hlgCB, or lukED, which effectively results in a lower yield of protein recovered from bacterial cultures (31). Similar results were obtained whatever the two strains tested.
When the bacteria were grown in HI medium, expression of the toxins remained basal, not exceeding 10 ± 2.5 mRNA/3,000 CFU, showing that expression depends on regulation factors which are themselves dependent on environment conditions. Similar observations were made for the control of the expression of hla (alpha-hemolysin gene) with variations of salt concentrations (NaCl and KCl concentrations) of the medium (26). This observation may also suggest the presence of antagonist molecules in growth media or the presence of regulatory mechanisms in the bacteria.
To illustrate this observation, dependence of the expression of the leucotoxins on the global agr sar accessory system (7, 21) was investigated with agr- and sar-defective mutants of ATCC 49775 and NTCC 8178 S. aureus strains. The results (Fig. 3 and 4) showed that expression of lukPV, lukED, hlgA, and hlgBC depends on the sar locus for the two strains, whereas hlgA seemed to be the only locus not sensitive to the agr mutation. For the sensitive loci, the control exerted by both the agr and sar loci results in a 10- to 100-fold increase of expression that is maximal at the late-exponential growth phase. In the sar- and agr-defective mutants, the lack of control by sarA and sarB expression products (4, 23) and by agrA, -B, -C, -D, and RNA III expression products, via P2 and P3 promoters (2, 23), led to the detection of an early and significant expression of leucotoxins. The sar locus was demonstrated to control partially and positively the agr locus by the DNA binding protein SarA (8), but it may also directly control genes encoding exoproteins (7). The lack of sar and agr expression products may effectively result in the absence of control of toxin expression. A recently characterized sigma factor in S. aureus (36) acts on shock proteins (22) but was also reported to act on the sar locus, by increasing sarA expression and, consequently, the expression of alpha-hemolysin (5). Therefore, absence of both the agr and sar controls would result in a lack of regulation, in the case of independence of leucotoxin genes with another mechanism. Differences in expression of staphylococcal leucotoxins in the agr sar double mutants may reflect differences in the control by other mechanisms, e.g., the sigma factor. Only hlgA in the ATCC 49775 agr sar mutant did not show a strong increase compared to that in the NTCC 8178 agr sar mutant. Differences in the sequences of the corresponding promoters (11, 31) and genes may be responsible for such discrepancies; for the loci encoding the leucotoxins discussed, promoter regions do not show great sequence identity, except for a GNA(T)TAAA sequence located 35 to 31 bases upstream of the ATG codons (29). Unfortunately, the molecular mechanisms of the regulation of staphylococcal exoprotein expression remain poorly studied (25). Goerke et al. (14) recently showed that the expression of alpha-toxin (hla) was independent of the agr locus in vitro and in vivo and that the agr activity seemed to be nonessential in cystic fibrosis lung infections. The authors postulated that in vivo, other regulatory mechanisms apart from agr were involved. A potential element in the regulation of gene expression is the variable stability of mRNAs according to the locus and to the growth rate of the bacteria (17). The half-life of the ompA transcript in E. coli is reduced from 17 min in fast-growing cells to 4 min in slow-growing cells, due to the variable stability of the 5′ end of the ompA mRNA. This could affect the regulation rate of the regulatory system.
In conclusion, as clinical S. aureus strains may encode from two (hlgA and hlgCB) to four (hlgA, hlgCB, lukPV, and lukED) leucotoxin loci and combinations of the corresponding class S and class F proteins result in leucotoxins with specific biological activities (16, 29), semiquantitation of their expression provides insights concerning their respective and possible involvement in bacterial virulence. hlgA appears to be the least expressed of the proteins studied. As toxin concentrations result from their cumulative expression, it may be hypothesized that HlgA is not as abundant as the other leucotoxin components. This is regularly the case upon purification of this protein (31). However, expression remains highly dependent on environmental conditions (e.g., growth media) and on the strain genetic equipment. Sensitivity to antibiotics of the gene encoding alpha-toxin secreted by S. aureus was reported in in vitro studies (27). Inappropriate antimicrobial therapy might, therefore, result in a temporal upregulation of virulence factors, leading to undesirable lesions. Semiquantitation of virulence factors in in vivo experimental models would be a suitable tool for the evaluation of antimicrobial therapeutics.
ACKNOWLEDGMENTS
We thank A. L. Cheung (Rockefeller University) for the kind gift of the S. aureus Newman agr, sar, and agr sar mutant strains. We appreciated the excellent technical assistance of V. Wattelet. We thank N. Boord for English improvement.
This work was supported by grant EA 1318 from the French “Direction de la Recherche et des Etudes Doctorales” (DRED).
REFERENCES
- 1.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K D, Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 2.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caudai C, Padula M G, Bastianoni I, Valensin P E, Shyamala J H, Boggiano C A, Almi P. Antibody testing and RT-PCR results in hepatitis C virus (HCV) infection: HCV-RNA detection in PBMC of plasma viremia-negative HCV-seropositive persons. Infection. 1998;26:151–154. doi: 10.1007/BF02771840. [DOI] [PubMed] [Google Scholar]
- 4.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung A L, Chien Y T, Bayer A S. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated sarA expression in Staphylococcus aureus. Infect Immun. 1999;67:1331–1337. doi: 10.1128/iai.67.3.1331-1337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 7.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien Y T, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Mackey K. Modification of the TRI Reagent™ procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. BioTechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- 10.Cooney J, Kienle Z, Foster T J, O'Toole P W. The gamma-hemolysin locus of Staphylococcus aureus comprises three linked genes, two of which are identical to the genes for the F and S components of leukocidin. Infect Immun. 1993;61:768–771. doi: 10.1128/iai.61.2.768-771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couppié P, Cribier B, Prévost G, Grosshans E, Piémont Y. Leucocidin from Staphylococcus aureus and cutaneous infections: an epidemiological study. Arch Dermatol. 1994;130:1208–1209. doi: 10.1001/archderm.130.9.1208. [DOI] [PubMed] [Google Scholar]
- 12.Delassus S. Quantification of cytokine transcripts using polymerase chain reaction. Eur Cytokine Netw. 1997;8:239–244. [PubMed] [Google Scholar]
- 13.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goerke C, Campana S, Bayer M G, Döring G, Botzenhart K, Wolz C. Direct quantitative analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun. 2000;68:1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravet A, Rondeau M, Harf-Monteil C, Grunenberger F, Monteil H, Scheftel J M, Prévost G. Predominant Staphylococcus aureus isolated from antibiotic-associated diarrhea is clinically revelant and produces enterotoxin A and the bicomponent toxin LukE-LukD. J Clin Microbiol. 1999;37:4012–4019. doi: 10.1128/jcm.37.12.4012-4019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gravet A, Colin D A, Keller D, Girardot R, Monteil H, Prévost G. Characterization of a novel structural member, LukE-LukD, of the bi-component staphylococcal leucotoxins family. FEBS Lett. 1998;436:202–208. doi: 10.1016/s0014-5793(98)01130-2. [DOI] [PubMed] [Google Scholar]
- 17.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 18.Hellyer T J, DesJardin L E, Hehman G L, Cave M D, Eisenach K D. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:290–295. doi: 10.1128/jcm.37.2.290-295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. Transcription analysis of the gene tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem. 1997;244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 20.Klein S A, Ottmann O G, Ballas K, Dobmeyer T S, Pape M, Weidmann E, Hoelzer D, Kalina U. Quantification of human interleukin 18 mRNA expression by competitive reverse transcriptase polymerase chain reaction. Cytokine. 1999;11:451–458. doi: 10.1006/cyto.1998.0424. [DOI] [PubMed] [Google Scholar]
- 21.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 22.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C J. An experimental vaccine that targets staphylococcal virulence. Trends Microbiol. 1998;6:461–463. doi: 10.1016/s0966-842x(98)01404-8. [DOI] [PubMed] [Google Scholar]
- 24.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132. [DOI] [PubMed]
- 25.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 26.Ohlsen K, Koller K-P, Hacker J. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun. 1997;65:3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohlsen K, Ziebuhr W, Koller K-P, Hell W, Wichelhaus T A, Hacker J. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 1998;42:2817–2823. doi: 10.1128/aac.42.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phonimdaeng P, O'Reilly M, Nowlan P, Bramley A J, Foster T J. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol. 1990;4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 29.Prévost G. The bi-component staphylococcal leucocidins and gamma-hemolysins (toxins) In: Alouf J E, Freer J H, editors. The comprehensive sourcebook of bacterials toxins. 2nd ed. London, England: Academic Press, Inc.; 1999. pp. 402–418. [Google Scholar]
- 30.Prévost G, Couppié P, Prévost P, Gayet S, Petiau P, Cribier B, Monteil H, Piémont Y. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J Med Microbiol. 1995;42:237–245. doi: 10.1099/00222615-42-4-237. [DOI] [PubMed] [Google Scholar]
- 31.Prévost G, Cribier B, Couppié P, Petiau P, Supersac G, Fink-Barbançon V, Monteil H, Piémont Y. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siqueira J A, Speeg-Schatz C, Freitas F I S, Sahel J, Monteil H, Prévost G. Channel-forming leucotoxins from Staphylococcus aureus cause severe inflammatory reactions in a rabbit eye model. J Med Microbiol. 1997;46:1–9. doi: 10.1099/00222615-46-6-486. [DOI] [PubMed] [Google Scholar]
- 33.Southern E. Detection of specific sequence among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 34.Staali L, Monteil H, Colin D A. The pore-forming leukotoxins from Staphylococcus aureus open Ca2+ channels in human polymorphonuclear neutrophils. J Membr Biol. 1998;162:209–216. doi: 10.1007/s002329900358. [DOI] [PubMed] [Google Scholar]
- 35.Wang A M, Doyle M V, Mark F. Quantification of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternative sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]