Abstract
Xanthan-modifying enzymes are powerful tools in studying structure-function relationships of this polysaccharide. One of these modifying enzymes is xanthan lyase, which removes the terminal side chain residue of xanthan. In this paper, the cloning and sequencing of the first xanthan lyase-encoding gene is described, i.e., the xalA gene, encoding pyruvated mannose-specific xanthan lyase of Paenibacillus alginolyticus XL-1. The xalA gene encoded a 100,823-Da protein, including a 36-amino-acid signal sequence. The 96,887-Da mature enzyme could be expressed functionally in Escherichia coli. Like the native enzyme, the recombinant enzyme showed no activity on depyruvated xanthan. Compared to production by P. alginolyticus, a 30-fold increase in volumetric productivity of soluble xanthan lyase was achieved by heterologous production in E. coli. The recombinant xanthan lyase was used to produce modified xanthan, which showed a dramatic loss of the capacity to form gels with locust bean gum.
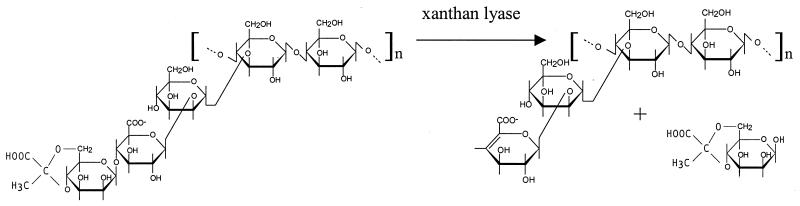
Xanthan is an extracellular polysaccharide produced by Xanthomonas campestris that is widely applied as a thickener of aqueous solutions and as a stabilizer of emulsions, foams, and particulate suspensions (12). Xanthan consists of pentasaccharide repeating units: the β-1,4-glucan backbone is replaced on alternate glucosyl residues with a trisaccharide side chain consisting of α-mannose, β-glucuronic acid, and β-mannose (see Fig. 1). The inner mannosyl residues are usually acetylated, whereas about 30% of the terminal mannosyl residues are pyruvated.
FIG. 1.
Action of pyruvated mannose-specific xanthan lyase on the pentasaccharide repeating unit of xanthan. Acetyl groups are omitted for clarity.
Also, xanthans carrying truncated side chains, produced by mutants of X. campestris, have been described (3, 18). Studies of these variant xanthans showed that truncation of the side chain affects the rheological properties of xanthan. The absence of the terminal side chain mannosyl residue results in weaker viscosifier than xanthan (10), whereas the absence of both the terminal mannosyl and the glucuronyl residue results in a viscosifier superior to that of xanthan (7). These truncated side chain xanthans are interesting polysaccharides, both from a scientific and a practical point of view. They are, however, produced at low yields, especially the polytrimer (19). Furthermore, the chain length of the backbone may differ from native xanthan, hampering correct comparison of xanthans with different side chains. Enzymatic modification of the side chains would be a preferable method to obtain tailor-made xanthans, as the effect of a step-by-step removal of side chain residues can be studied, leaving the backbone unaffected.
Mixed or pure bacterial cultures that grow on xanthan generally produce a mix of xanthan-degrading enzymes (4, 6, 8, 14, 17). One of these degradative enzymes that is potentially useful for xanthan modification is xanthan lyase. Xanthan lyase removes the terminal mannosyl residue via β elimination, yielding a free mannose and a tetrasaccharide repeating unit (Fig. 1). Both nonspecific (16) and pyruvated mannose-specific xanthan lyases (1, 6, 14) have been described. The resulting polytetrasaccharide differs from the polytetrasaccharide produced by X. campestris mutants: xanthan lyase-modified xanthan bears a 4,5-ene-glucuronic acid instead of a glucuronic acid as a terminal side chain residue. So far, no amino acid sequences of xanthan lyases have been published and hence no information exists on homology or relatedness with other polysaccharide-degrading enzymes.
Previously, a xanthan-degrading bacterium, Paenibacillus alginolyticus strain XL-1, was isolated, and the pyruvated mannose-specific xanthan lyase produced by this organism was purified and characterized (14). In this report, we present the first nucleotide sequence of a gene encoding xanthan lyase. The gene encoding pyruvated mannose-specific xanthan lyase from P. alginolyticus XL-1 was cloned, sequenced, and functionally expressed in Escherichia coli.
MATERIALS AND METHODS
Strains, plasmids, and DNA manipulations.
For DNA isolation, P. alginolyticus strain XL-1 was cultured at 30°C on mineral salts medium (14) supplemented with 5 g of mannose/liter and 0.05 g of filter-sterilized yeast extract/liter. E. coli strains XL1-Blue MRF′ (Stratagene, La Jolla, Calif.) and BL21(DE3) (Novagen, Madison, Wis.) were used for cloning and expression studies. E. coli strains and transformants were cultured at 37°C on Luria-Bertani (LB) medium (10 g of tryptone/liter, 5 g of yeast extract/liter, and 10 g of NaCl/liter), supplemented with the proper antibiotic when required.
The ZAP Express vector (Stratagene) was used for construction of a genomic library of P. alginolyticus XL-1. The vector pUC19 (21) was used for construction of sublibraries of genomic P. alginolyticus DNA. The vector pGEM-T Easy (Promega, Madison, Wis.) was used for cloning of PCR products. The vector pET28a (Novagen) was used for heterologous expression of the xanthan lyase-encoding gene.
Plasmid DNA was isolated from E. coli using Qiagen miniprep spin columns (Westburg BV, Leusden, The Netherlands). Agarose-trapped DNA was extracted using the Qiaex II gel extraction kit (Westburg BV). Plasmid DNA was introduced into E. coli by electroporation using a Bio-Rad gene pulser. Genomic DNA of P. alginolyticus XL-1 was isolated using the cetyltrimethylammoniumbromide procedure (2). Other standard molecular biology techniques were carried out according to the method of Sambrook et al. (15).
Construction of a genomic library of P. alginolyticus XL-1 in E. coli.
Genomic DNA of P. alginolyticus XL-1 was partially digested with Bsp143I (an isoschizomer of Sau3A) (MBI Fermentas, Vilnius, Lithuania), and fragments of 4 to 14 kb were isolated after agarose gel electrophoresis. The purified fragments were ligated to a BamHI-digested and calf intestine alkaline phosphatase-treated ZAP Express vector (Stratagene) according to the suppliers' instructions. After transfection of the packaged ligation mix, a primary phage library containing 14,500 PFU was obtained. After amplification and mass excision of the phage library, the resulting pBK-CMV-phagemid library was transfected to E. coli XL-1 Blue MRF′ and plated on LB agar containing ampicillin (100 μg/ml) as well as X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) for blue-white screening. It appeared that 75% of the colonies contained an insert with an average size of 4.5 kb. If a genome size of 4.2 Mb is assumed for P. alginolyticus XL-1 (as for Bacillus subtilis), the primary library represented 12 times the genome.
PCR experiments.
PCRs were carried out in an automated thermal cycler (Perkin-Elmer). Standard reaction conditions used were 10 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, and ending with 10 min at 72°C. Primer R1 (5′-CTG CGC TTC CGC GGT AAG CG-3′) was designed on the reverse strand of the xalA gene, near the start codon. This primer was used, in combination with primer M13 (5′-CCC AGT CAC GAC GTT GTA AAA CG-3′) or M13 reverse (5′-AGC GGA TAA CAA TTT CAC ACA GG-3′), to obtain the sequence upstream of the xalA gene, including the start of the gene (see Results for details). Primer C5 (5′-GAA TTC ATG TCG GAT GAG TAT GAT ACG CTG C-3′) was designed on the 5′ end of the xalA gene, starting at the codon for the N-terminal Ser residue (boldface) of the mature protein. An EcoRI site (underlined) was introduced for cloning purposes; a start codon (in italics) was included for expression in vectors lacking a transcription start. Primer C7 (5′-AAG CTT CCC TCC CCA AAG CTG C-3′) was designed on the reverse complement strand of the xalA gene, upstream of the putative terminator sequence, including a HindIII site (underlined) for cloning purposes. Degenerate oligonucleotide primers are listed in Table 1.
TABLE 1.
Amino acid sequences of the N terminus and internal peptides of purified pyruvated mannose-specific xanthan lyase from P. alginolyticus XL-1 and nucleotide sequences of the derived degenerate primers
| Peptide | Sequencea | Degenerate oligonucleotide primerb |
|---|---|---|
| N terminus | SDEYDTLRLKWRDM (residues 1–14) | D2: 5′-GAY GAR TAY GAY ACV TTR C-3′ |
| 15 | SXISSENSIGT (residues 324–334) | D5: 5′-TCS GAR AAY TCS ATY GGH AC-3′; D7c: 5′-GTD CCR ATS GAR TTY TC-3′ |
| 17 | TSAQVSSYASNPNISVL (residues 626–642) | D12: 5′-CTR ATR TTN GGR TTR CTN GC-3′ |
| 25 | TPGGTTNYLXVDLR (residues 49–62) | |
| 28 | ENTLNVVGVNFX (residues 653–664) |
X can be either cysteine or tryptophan. Cysteine was not determined with the method used. Tryptophan was masked by diphenylthiourea, which is formed from phenylisothiocyanate in the Edman degradation. Underlined amino acids were used to design degenerate primers. Residue numbers refer to the position in the deduced amino acid sequence of the mature enzyme encoded by the xalA gene (1 = the N-terminal residue).
D = (A + T + G); H = (A + T + C); N = (A + C + T + G); R = (A + G); S = (C + G); V = (A + C + G); Y = (C + T).
Primer D7 is complementary to D5.
Determination of internal amino acid sequences of xanthan lyase.
P. alginolyticus XL-1 xanthan lyase was produced and purified as described previously (14), followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The Coomassie-stained xanthan lyase band was cut from the gel and transferred to an Eppendorf tube for in-gel digestion. Briefly, washing was carried out in 0.2 M ammonium bicarbonate containing 50% acetonitrile. The protein was reduced with dithiothreitol and alkylated with iodoacetamide followed by in-gel digestion with 0.5 to 3 μg of trypsin (Promega, modified) in 0.2 M ammonium bicarbonate overnight at 37°C. The tryptic peptides were extracted using acetonitrile in 0.1% trifluoroacetic acid, first at 60% and then at 40%. The peptide extract was separated and fragments were isolated on polyvinylidene difluoride membrane for sequence analysis using a microblotter system (Perkin-Elmer). Edman degradation of polyvinylidene difluoride-bound peptides was carried out with a Procise cLC sequencer (Perkin-Elmer).
Expression of xanthan lyase in E. coli BL21(DE3).
Plasmid pEXL1 (pET28a carrying the xalA gene as a 2.7-kb EcoRI/HindIII fragment) was transformed to a λDE3 lysogen of E. coli BL21 for expression testing. Strain BL21(DE3) harbors the T7 RNA polymerase gene required for transcription of the target gene in pET28a. Both the T7 RNA polymerase and the target gene are under the control of the lac repressor (lacI), thus allowing tight control of the expression of the target gene. For expression of the xalA gene, BL21(DE3) carrying pEXL1 was cultured on 50 ml of LB medium containing 50 μg of kanamycin/ml. At an optical density at 600 nm of 0.5, 1 mM IPTG was added for induction, and the culture was incubated for another 3 h. The cells were harvested and the cell pellet was resuspended in 5 ml of 15-mM potassium phosphate, pH7. After sonication, the cell extract was assayed for xanthan lyase activity.
Xanthan lyase assay.
Xanthan lyase activity was determined as described previously (14), by incubating xanthan lyase with xanthan and measuring the increase of A235, the increase of reducing sugars (11), or the increase of thiobarbituric acid-reactive material (20).
Protein electrophoresis.
SDS-PAGE was carried out according to the method of Laemmli (9) using a Hoeffer Mighty Small system (Pharmacia). For estimation of molecular mass, prestained precision standards (10 kDa to 250 kDa; Bio-Rad) were used. Gels were stained with Coomassie brilliant blue.
Rheological measurements.
Rheological measurements on xanthan-locust bean gum (LBG) gels were performed in a Bohlin CVO rheometer using a C14 cup-and-bob geometry. Gels were formed by mixing solutions of 5 g of xanthan (native or modified)/liter and 5 g of LBG/liter (in 15 mM potassium phosphate buffer, pH7). Modified xanthan was prepared by incubating 10 ml of a solution of 5 g of xanthan/liter in 15 mM potassium phosphate (pH 7) with 500 μl of cell extract of an induced BL21(DE3) culture harboring pEXL1 for 2 h at 30°C. After transfer to the rheometer, the gel was heated from 25 to 80°C and subsequently cooled from 80 to 25°C. After the gel was allowed to set at 25°C for 15 min, the gel was submitted to a stress sweep regimen from 0.08 to 3 Pa at a frequency of 1 Hz to determine the rigidity (elastic modulus, G′) and the viscosic aspect (viscous modulus, G") of the molecular bonding in the gel.
Nucleotide sequence accession number.
The DNA sequence of xalA has been deposited in the EMBL/GenBank/DDBJ database under the accession number AF242413.
RESULTS
Amino acid sequences of xanthan lyase peptides and synthesis of a specific P. alginolyticus xanthan lyase probe by PCR.
Xanthan lyase was purified and both N-terminal and internal amino acid sequences were determined (Table 1). Degenerate primers, designed from these amino acid sequences, were used in PCRs with genomic P. alginolyticus XL-1 DNA as a template to obtain xanthan lyase-specific DNA fragments.
The combination of primers D2 and D7 yielded a 1-kb PCR product (product A), whereas primers D5 and D12 yielded a 0.9-kb PCR product (product B). As D5 and D7 were designed from the same peptide, but complementary to each other, the amplified sequences must be overlapping parts of the xanthan lyase-encoding gene. PCR products A and B were cloned in pGEM-T Easy to obtain pGA and pGB, respectively. The nucleotide sequences of the inserts of pGA and pGB were determined. The deduced amino acid sequences contained the peptide sequences, including the amino acid residues that were omitted in the primer design (see Table 1). Furthermore, the sequence of peptide 25 was found in the deduced amino acid sequence of PCR product A. These results strongly suggested that the cloned fragments contained parts of the gene encoding pyruvated mannose-specific xanthan lyase. The insert of plasmid pGA was excised with EcoRI, and the resulting 1-kb fragment was isolated. The insert fragment was used as a template in a PCR with primers D2 and D7 using digoxigenin-labeled nucleotides to obtain a specific digoxigenin-labeled xanthan lyase probe.
Isolation of the P. alginolyticus xanthan lyase-encoding gene.
Initially, we attempted to isolate the xanthan lyase-encoding gene from the P. alginolyticus XL-1 genomic library, both by expression screening and by colony hybridization with the specific xanthan lyase probe. For expression screening, the library was plated on LB medium solidified with a mixture of xanthan and LBG. As purified xanthan lyase liquefies xanthan-LBG gels (unpublished observations), these plates should be liquefied by colonies expressing xanthan lyase. Although liquefying colonies were obtained, none of these expressed xanthan lyase, based on activity measurements. Also, screening by colony hybridization proved to be unsuccessful: a total of 22,000 colonies were screened but no positive clones could be detected. As the complete xanthan lyase-encoding gene could not be obtained from a representative genomic library, it was decided to construct sublibraries to try to obtain the gene, if not in its entirety, then in parts.
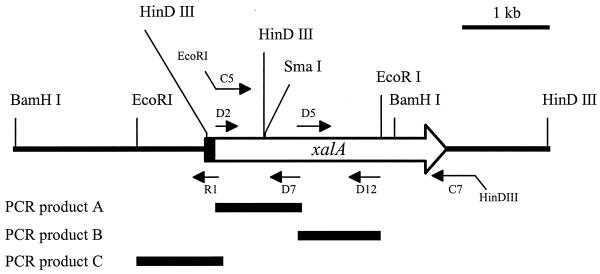
Genomic DNA of P. alginolyticus XL-1 was digested with several restriction enzymes, and the resulting fragments were separated on an agarose gel. After Southern blotting and hybridization with the probe, a 4.3-kb BamHI fragment, a 2.9-kb EcoRI fragment, and two HindIII fragments (0.6 and 3.6 kb) were visible. The latter observation was in accordance with the nucleotide sequence of PCR fragment A, which contained a HindIII site at 556 bp from the 5′ end. Genomic BamHI fragments (fraction of 4.2 to 4.4 kb), EcoRI fragments (fraction of 2.7 to 3.0 kb), and HindIII fragments (fractions of 0.5 to 0.7 kb and 3.5 to 3.7 kb) were isolated and ligated into the corresponding sites of pUC19. After transformation, colony hybridization was carried out with the xanthan lyase probe to determine which clones carried the DNA fragments of interest. No hybridizing transformants containing a BamHI or an EcoRI fragment could be detected. Both HindIII fragments, however, could be obtained from the corresponding sublibraries. Plasmid DNA was isolated, and the complete nucleotide sequences of the HindIII fragments were determined. Based on the nucleotide sequences and the Southern blot analysis, a restriction map was constructed (Fig. 2).
FIG. 2.
Schematic representation of the genomic DNA fragment carrying the P. alginolyticus XL-1 xalA gene. The xalA gene is indicated by an open arrow; the part of the gene encoding the signal peptide is in black. Primer R1 was used to obtain the sequence upstream of the 5′ HindIII site in the xalA gene. Primers D2 and D7 yielded PCR product A, which was used as a template to obtain a specific xanthan lyase probe. Primers D5 and D12 yielded PCR product B. PCR product C was obtained from a reaction on the ligation mixture of the EcoRI sublibrary, using primers R1 and M13 reverse. Primers C5 and C7 were used to synthesize the part of the gene encoding mature xanthan lyase for heterologous expression.
The sequence obtained from the two HindIII fragments encoded mature xanthan lyase as well as a large part of the signal sequence. The start of the gene, however, was missing. From Fig. 2 it can be observed that the hybridizing EcoRI and BamHI fragments should contain the missing part of the gene. These fragments could not, however, be isolated from the sublibraries. Apparently, the introduction of plasmids containing these inserts, which carry a truncated xanthan lyase gene with the complete signal sequence, as well as probably a promoter region, is lethal to E. coli. Therefore, it was attempted to amplify a part of the EcoRI fragment by PCR. A reaction was carried out on the ligation mixture of the EcoRI sublibrary, using specific primers for the xanthan lyase-encoding gene (R1) and the regions of pUC19 flanking the insertion site (M13 or M13 reverse). The primer combination R1 and M13 reverse yielded the expected 1-kb product (product C). The complete nucleotide sequence of this PCR product was determined. The sequence overlapped the sequence of the xanthan lyase-encoding gene by 200 bp and contained the missing part of the gene, as well as about 800 bp upstream of the start codon.
Nucleotide sequence of the xanthan lyase-encoding gene.
From the two genomic HindIII fragments and PCR product C, the complete xanthan lyase-encoding gene and its flanking regions could be reproduced (see “Nucleotide sequence accession number” above). The sequence revealed a putative promoter sequence, a putative ribosome-binding site, and a 2,811-bp open reading frame starting with an ATG start codon and ending with TAA. A putative terminator sequence (inverted repeat) was observed 60 bp downstream of the stop codon. The open reading frame, designated xalA, encoded a 100,823-Da protein, of which the first 36 amino acids represent a Bacillus-like signal sequence (13). The signal peptide displays three positively charged residues (Arg) in the N-terminal region, a 20-amino-acid hydrophobic core terminated by two helix-breaking Pro residues, and a signal peptidase recognition site (Ala-X-Ala). Behind this putative cleavage site, the N terminus of the mature enzyme should start, which is indeed identical to the determined N-terminal amino acid sequence of purified P. alginolyticus XL-1 xanthan lyase. Furthermore, all determined internal peptide sequences (see Table 1 for residue numbers) could be observed in the deduced amino acid sequence, confirming that the proper gene was obtained.
The mature xanthan lyase consists of 900 amino acids with a calculated molecular mass of 96,887 Da. A comparison of the deduced amino acid sequence of xalA with sequences in the SwissProt database revealed homology with other polysaccharide lyases, i.e., hyaluronidases and chondroitinases. The observed homologies were, however, very low. The highest scores were 36% identity in a 690-amino-acid overlap for Streptomyces griseus hyaluronidase (accession number BAA78618) and 33% identity in a 770-amino-acid overlap for a putative lyase secreted by Streptomyces coelicolor (accession number CAA19982). P. alginolyticus XL-1 xanthan lyase did not show any activity on hyaluronic acid or chondroitin.
Functional expression of mature xanthan lyase in E. coli.
The xalA gene, without the signal sequence, was cloned into the expression vector pET28a in order to prove the functionality of the gene. The oligonucleotides C5 and C7 were designed to synthesize the gene by PCR while introducing the appropriate restriction sites (i.e., EcoRI and HindIII, respectively) for cloning the gene into pET28a. A reaction with these primers on genomic P. alginolyticus XL-1 DNA yielded the expected 2.7-kb fragment, which was cloned in pGEM-T Easy to obtain pGXL1. Unfortunately, the restriction sites to be used for cloning were also present in the xalA gene itself (see Fig. 2). Thus, to avoid the necessity of partial restriction with EcoRI and HindIII, pGXL1 was digested with EcoRI/SmaI and with SmaI/HindIII. The EcoRI/SmaI fragment containing the 5′ part of the xalA gene and the SmaI/HindIII fragment containing the 3′ part were isolated and ligated into EcoRI/HindIII-digested pET28a to obtain plasmid pEXL1. After transformation and amplification of pEXL1 in E. coli XL1-Blue MRF′, the plasmid was transformed to E. coli BL21(DE3) for expression analysis. The BL21(DE3) transformants were plated on LB medium, supplemented with 1 mM IPTG and 50 μg of kanamycin/ml and solidified by xanthan and LBG. The gel was liquefied, confirming that xanthan lyase was expressed.
Induced BL21(DE3) cells carrying pEXL1 were harvested, and the cell pellet was sonicated. Upon incubation of the soluble fraction of the sonicated cells with xanthan, xanthan lyase activity could be demonstrated by measuring the increase of A235, the increase of reducing sugars, and the increase of thiobarbituric acid-reactive material. The soluble xanthan lyase activity in the cell extract, as quantified by measuring A235, amounted to 8.3 U/ml. A total soluble xanthan lyase activity of 41.5 U was obtained from one 50-ml culture. Also, the insoluble fraction of the cell extract, resuspended in 15 mM potassium phosphate buffer, contained a trace of xanthan lyase activity (about 2% of total activity). This activity was doubled after overnight incubation at 4°C, suggesting that the recombinant enzyme was partially captured in inclusion bodies that resolubilized and became active upon incubation in buffer.
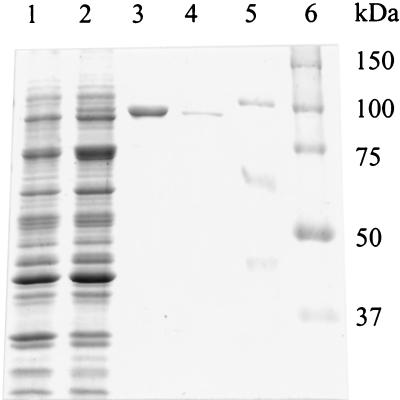
SDS-PAGE analysis (Fig. 3) showed a 100-kDa band, corresponding to recombinant xanthan lyase, appearing after induction in the cell extract of E. coli carrying pEXL1 (lane 2). Also, the insoluble fraction of the cell extract (lane 3) contained this band in a huge amount, confirming that at least 95% of the xanthan lyase was deposited as inclusion bodies. The molecular mass of the recombinant xanthan lyase as observed in lane 2 is slightly higher than the molecular mass of native xanthan lyase (lane 4). This can be explained by the pET28a-borne N-terminal His tag that is attached to the recombinant xanthan lyase. The estimated molecular mass of mature xanthan lyase is in accordance with the molecular mass predicted from the deduced amino acid sequence (97 kDa). Previously, however, a molecular mass of 85 kDa was reported (14). This difference can be attributed to the 94-kDa marker (phosphorylase b, lane 5) that was initially used as a reference in estimating the molecular mass of xanthan lyase. As can be observed from Fig. 3, phosphorylase b migrates more slowly than the 100-kDa marker of the Bio-Rad precision protein standard (lane 6). Thus, the molecular mass of xanthan lyase was previously estimated to be smaller than 94 kDa, whereas its actual size is 97 kDa.
FIG. 3.
SDS-PAGE gel of fractions of uninduced and induced E. coli BL21(DE3) cells carrying pEXL1. Lane 1, uninduced cells, total cell protein; lane 2, induced cells, cell extract (soluble fraction); lane 3, induced cells, cell extract (insoluble fraction; the relative amount loaded is 1/10 of that loaded in lanes 1 and 2); lane 4, native purified xanthan lyase; lane 5, marker proteins (phosphorylase b, 94 kDa; bovine serum albumin, 67 kDa; ovalbumin, 43 kDa); lane 6, Bio-Rad precision protein standards.
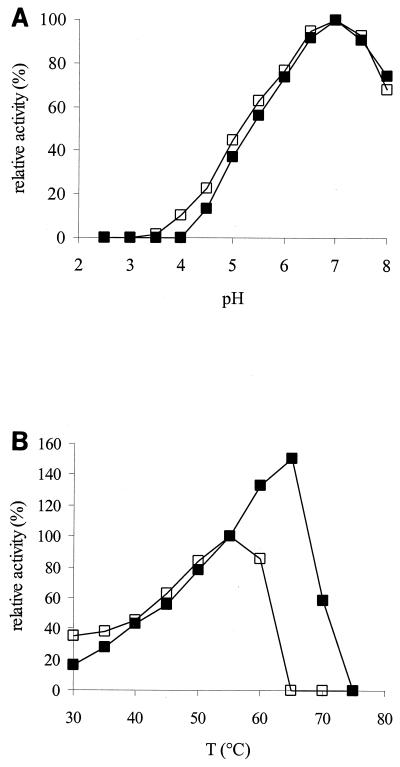
Like the purified P. alginolyticus enzyme, the recombinant xanthan lyase showed no activity on depyruvated xanthan. As shown in Fig. 4A, the pH profiles of native and recombinant xanthan lyase are identical. The temperature profiles, however, are different (Fig. 4B). Recombinant xanthan lyase still shows activity at 65°C whereas native xanthan lyase rapidly loses activity above 55°C. Considering that purified native xanthan lyase is not stable above 45°C (14), the results suggest that the recombinant xanthan lyase is slightly more stable.
FIG. 4.
(A) Relative activity of native xanthan lyase (purified, open symbols) and recombinant xanthan lyase (cell extract, closed symbols) at different pH values, on xanthan in McIlvaine buffer. (B) Relative activity of native xanthan lyase (purified, open symbols) and recombinant xanthan lyase (cell extract, closed symbols) at different temperatures, on xanthan in 15 mM potassium phosphate buffer, pH 7. The activity of both enzymes was set at 100% at the conditions under which the native xanthan lyase was most active. T, temperature.
Modification of xanthan with recombinant xanthan lyase.
To illustrate an effect of recombinant xanthan lyase on the physical properties of xanthan, xanthan-LBG gels were prepared with unmodified xanthan and xanthan lyase-modified xanthan. After preparing the gels, the elastic modulus (G′) and the viscous modulus (G") of the gels were determined. The G′ value, which is a measure for the “rigidity” of the gel, was 35 for the gel prepared with unmodified xanthan. The gel prepared with xanthan lyase-modified xanthan, however, had a G′ value of only 1.4. Based on this value, the mixture of modified xanthan and LBG could hardly be classified as a gel. Consequently, tan δ (= G"/G′), which is a measure for the “fluidity” of the gel, is much higher for the modified xanthan-LBG gel (1.0 as opposed to 0.12 for the unmodified xanthan-LBG gel), confirming the visual observation that this gel has a far more liquid character than the other gel.
DISCUSSION
This report describes the first nucleotide sequence of a xanthan lyase-encoding gene. The gene, encoding a pyruvated mannose-specific xanthan lyase of P. alginolyticus XL-1, was designated xalA, as probably a second xanthan lyase with a different substrate specificity is also produced by this strain (14). Comparison of the deduced amino acid sequence of xanthan lyase to sequences in the SwissProt database showed only slight homologies to other polysaccharide lyases, indicating that this enzyme constitutes a new family of polysaccharide lyases.
The mature xanthan lyase, devoid of its signal sequence, was functionally expressed in E. coli. The N terminus of mature xanthan lyase was identical to the deduced N terminus located behind the putative signal peptidase cleavage site, confirming the start of the mature enzyme. Furthermore, the internal peptide sequences obtained from native purified pyruvated mannose-specific xanthan lyase were all present in the deduced amino acid sequence of xalA. The recombinant enzyme was, like the P. alginolyticus XL-1 enzyme, not active on depyruvated xanthan. Furthermore, native and recombinant xanthan lyases showed identical pH profiles. The temperature profiles, however, indicated that the recombinant xanthan lyase is a bit more stable than the native xanthan lyase. The increased stability can probably be attributed to a stabilizing effect of proteins from the cell extract that were present in the case of recombinant xanthan lyase. Alternatively, the His tag may stabilize the enzyme, or the conformation of the enzyme may be slightly different in the heterologous host. The deduced amino acid sequence predicted a 97-kDa protein, which was in accordance with the observed molecular mass of native as well as recombinant xanthan lyase on SDS-PAGE (Fig. 3).
The undisrupted xanthan lyase-encoding gene could not be isolated from a representative genomic library. In addition, fragments containing the 5′ terminus of the gene could not be obtained from various sublibraries. These results suggest that the 5′ end of the xanthan lyase-encoding gene in combination with the DNA upstream of the start codon (i.e., the promoter sequence and the ribosome-binding site) cannot be rescued in E. coli. Possibly, the signal sequence linked to the (truncated) xanthan lyase protein is expressed, which may be lethal to E. coli, e.g., by blocking the protein export system or accumulation in the cell membrane. Nevertheless, the remainder of the xalA gene could be obtained by PCR, using a ligation mixture for the construction of one of the sublibraries as a template.
Although most of the recombinant xanthan lyase was captured in inclusion bodies, the production of soluble xanthan lyase by recombinant E. coli was greatly enhanced compared to that by P. alginolyticus XL-1. A 50-ml culture of E. coli BL21(DE3) harboring pEXL1 yielded 40 U of soluble xanthan lyase, whereas 2 liters of P. alginolyticus XL-1 culture yielded only 50 U (14). Thus, recombinant production of xanthan lyase provides the possibility of producing relatively large amounts of enzyme. Furthermore, no extensive purification is required, as no interfering xanthan-affecting enzyme activities are produced by E. coli. This makes the recombinant xanthan lyase an excellent tool for studying structure-function relationships of xanthan. This was illustrated by preparing xanthan lyase-modified xanthan and determining its ability to form gels with LBG. Whereas native xanthan formed a firm gel with LBG, modified xanthan showed hardly any interaction. This radical change in functional properties was caused by the action of a single enzyme that removes only the pyruvated terminal side chain residues. The availability of xanthan lyase opens the possibility for detailed further studies of the physical properties of xanthan lyase-modified xanthan. Furthermore, xanthan lyase treatment can be followed by further modifications. These could be enzymatic to obtain a (partial) polytrimer using enzymes like unsaturated glucuronyl hydrolase (4,5-ene-β-glucuronidase) (5) or chemical modification at the double bond in the 4,5-ene-β-glucuronyl residue.
ACKNOWLEDGMENTS
We thank Katja Grolle, Food Physics Group, Wageningen University, for assistance with the rheological measurements and Tony van Kampen, Laboratory of Molecular Biology, Wageningen University, for assistance in determining nucleotide sequences. The N terminus of xanthan lyase was determined by R. Amons of the Sylvius Laboratory, Leiden, The Netherlands.
This research project was financially supported by ABON (the Association of Biotechnological Research Centres in The Netherlands).
REFERENCES
- 1.Ahlgren J A. Purification and characterization of a pyruvated-mannose-specific xanthan lyase from heat-stable, salt-tolerant bacteria. Appl Environ Microbiol. 1991;57:2523–2528. doi: 10.1128/aem.57.9.2523-2528.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1991. [Google Scholar]
- 3.Betlach M R, Capage M A, Doherty D H, Hassler R A, Henderson N M, Vanderslice R W, Marellia J D, Ward M B. Genetically engineered polymers: manipulation of xanthan biosynthesis. In: Yalpani M, editor. Industrial polysaccharides: genetic engineering, structure/property relations and applications. Amsterdam, The Netherlands: Elsevier; 1987. pp. 35–80. [Google Scholar]
- 4.Cadmus M C, Slodki M E, Nicholson J J. High-temperature, salt-tolerant xanthanase. J Ind Microbiol. 1989;4:127–133. [Google Scholar]
- 5.Hashimoto W, Kobayashi E, Nankai H, Sato N, Miya T, Kawai S, Murata K. Unsaturated glucuronyl hydrolase of Bacillus sp. GL1: novel enzyme prerequisite for metabolism of unsaturated oligosaccharides produced by polysaccharide lyases. Arch Biochem Biophys. 1999;368:367–374. doi: 10.1006/abbi.1999.1305. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto W, Miki H, Tsuchiya N, Nankai H, Murata K. Xanthan lyase of Bacillus sp. strain GL1 liberates pyruvylated mannose from xanthan side chains. Appl Environ Microbiol. 1998;64:3765–3768. doi: 10.1128/aem.64.10.3765-3768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassler R A, Doherty D H. Genetic engineering of polysaccharide structure: production of variants of xanthan gum in Xanthomonas campestris. Biotechnol Prog. 1990;6:182–187. doi: 10.1021/bp00003a003. [DOI] [PubMed] [Google Scholar]
- 8.Hou C T, Barnabe N, Greaney K. Biodegradation of xanthan by salt-tolerant aerobic microorganisms. J Ind Microbiol. 1986;1:31–37. doi: 10.1128/aem.52.1.37-44.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Levy S, Schuyler S C, Maglothin R K, Staehelin L A. Dynamic simulations of the molecular conformations of wild type and mutant xanthan polymers suggest that conformational differences may contribute to observed differences in viscosity. Biopolymers. 1996;38:251–277. doi: 10.1002/(sici)1097-0282(199602)38:2<251::aid-bip10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 11.Miller G L. Use of dinitrosalicylic acid for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 12.Morris V J. Bacterial polysaccharides. In: Stephen A M, editor. Food polysaccharides and their applications. New York, N.Y: Dekker; 1995. pp. 341–375. [Google Scholar]
- 13.Nagarajan V. Protein secretion. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. [Google Scholar]
- 14.Ruijssenaars H J, de Bont J A M, Hartmans S. A pyruvated mannose-specific xanthan lyase involved in xanthan degradation by Paenibacillus alginolyticus XL-1. Appl Environ Microbiol. 1999;65:2446–2452. doi: 10.1128/aem.65.6.2446-2452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Sutherland I W. Xanthan lyases—novel enzymes found in various bacterial species. J Gen Microbiol. 1987;133:3129–3134. doi: 10.1099/00221287-133-11-3129. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland I W. Polysaccharide lyases. FEMS Micobiol Rev. 1996;16:323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 18.Tait M I, Sutherland I W. Synthesis and properties of a mutant type of xanthan. J Appl Bacteriol. 1989;66:457–460. [Google Scholar]
- 19.Vanderslice R W, Doherty D H, Capage M A, Betlach M R, Hassler R A, Henderson N M, Ryan-Graniero J, Tecklenburg M. Genetic engineering of polysaccharide structure in Xanthomonas campestris. In: Crescenzi V, Dea I C M, Paoletti S, Stivala S S, Sutherland I W, editors. Biomedical and biotechnological advances in industrial polysaccharides. New York, N.Y: Gordon and Breach; 1989. pp. 145–156. [Google Scholar]
- 20.Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. I. identification. J Biol Chem. 1959;234:705–709. [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]