Abstract
Xylose metabolism, a variable phenotype in strains of Lactococcus lactis, was studied and evidence was obtained for the accumulation of mutations that inactivate the xyl operon. The xylose metabolism operon (xylRAB) was sequenced from three strains of lactococci. Fragments of 4.2, 4.2, and 5.4 kb that included the xyl locus were sequenced from L. lactis subsp. lactis B-4449 (formerly Lactobacillus xylosus), L. lactis subsp. lactis IO-1, and L. lactis subsp. lactis 210, respectively. The two environmental isolates, L. lactis B-4449 and L. lactis IO-1, produce active xylose isomerases and xylulokinases and can metabolize xylose. L. lactis 210, a dairy starter culture strain, has neither xylose isomerase nor xylulokinase activity and is Xyl−. Xylose isomerase and xylulokinase activities are induced by xylose and repressed by glucose in the two Xyl+ strains. Sequence comparisons revealed a number of point mutations in the xylA, xylB, and xylR genes in L. lactis 210, IO-1, and B-4449. None of these mutations, with the exception of a premature stop codon in xylB, are obviously lethal, since they lie outside of regions recognized as critical for activity. Nevertheless, either cumulatively or because of indirect affects on the structures of catalytic sites, these mutations render some strains of L. lactis unable to metabolize xylose.
Xylose metabolism has been described for a wide array of microorganisms. Extensively characterized xylose-metabolizing (Xyl+) bacteria include Escherichia coli (24, 46), Lactobacillus spp. (3, 4, 26–28), Bacillus spp. (16, 33, 38), and Staphylococcus xylosus (44). Free xylose can be transported via low-affinity symporters (XylE or XylT) or high-affinity binding-protein dependent systems (XylFGH) (1, 6, 9, 41). Xylose isomerase (xylA gene) then converts the aldose sugar to xylulose, which is phosphorylated by xylulokinase (xylB gene). Further metabolism of xylulose-5-phosphate occurs via the pentose-phosphate or phosphoketolase pathways. Xylose metabolism is induced by xylose, mediated via XylR. In Salmonella and E. coli strains, XylR is an activator when xylose is present (42, 46). In gram-positive organisms, XylR is a repressor which is inactivated when xylose binds (13, 26, 27, 37, 44, 45). The catabolite repression of xylose metabolism by glucose is mediated primarily through the CcpA protein (40).
Lactococcus lactis subsp. cremoris and almost all L. lactis subsp. lactis strains cannot metabolize xylose. These organisms have undergone intense selection for use in dairy fermentations, but plants are thought to be their original ecological niche because L. lactis subsp. lactis isolates have been recovered from many different plants (22, 34, 35). Also, the plant isolate Lactobacillus xylosus has been reclassified as L. lactis subsp. lactis (39). However, L. lactis subsp. cremoris strains are almost exclusively dairy associated (22).
We discovered xylose metabolic genes from both plant (Xyl+) and dairy (Xyl−) isolates of L. lactis. The sequencing of xylRAB from L. lactis strains IO-1, 210, and B444-9 is described here. The induction of xylose isomerase and xylulokinase activities from these three strains grown on different carbon sources was measured. Activity measurements in conjunction with the discovery of a xylR gene suggest that a positive regulatory mechanism is present in lactococcal strains. Mutations which could account for the lack of xylose isomerase and xylulokinase activity were observed in L. lactis subsp. lactis 210. Further downstream of xylRAB are genes encoding a mutarotase, xylanase, and xyloside transporter (K. A. Erlandson, S. Delamarre, and C. A. Batt, unpublished results), whose presence collectively suggests that ancestors of these strains were fully able to degrade and metabolize xylan.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation.
Strains and vectors are described in the Table 1. L. xylosus was reclassified as L. lactis ssp. lactis based on immunological characteristics, 16S sequences, and fatty acid composition (39) and is referred to as L. lactis B-4449 here. All strains were routinely cultivated at 30°C in M17 medium (Difco, Detroit, Mich.) with 0.5% glucose or xylose. E. coli strains were cultured with aeration at 37°C in Luria broth (LB; Sigma, St. Louis, Mo.), with 100 μg of ampicillin (Sigma) or 50 μg of carbenicillin (Sigma) per ml where appropriate.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genetic markers | Source |
|---|---|---|
| Strains | ||
| L. lactis subsp. lactis 210 | Xyl− | J. Kondo (Marschall Products) |
| L. lactis subsp. lactis IO-1 | Xyl+ | P. Stansbury (University of Hertfordshire, Hertfordshire, United Kingdom) |
| L. lactis subsp. lactis NRRL B-4449 (formerly L. xylosus) | Xyl+ | A. J. Sinskey (Massachusetts Institute of Technology) |
| E. coli TG1 | supE hsdD5 thi Δ(lac− proAB) (F′ traD36 proAB laclq lacZ ΔM15) | |
| E. coli JM109 | el4−recA1 endA1 gyrA96 thi-1 hsDR17 supE44 relA1 Δ(lac− proAB) (F′ traD36 proAB laclqlacZΔM15) | |
| E. coli DH5αF′ | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF) U169 deoR[80dlacΔ(lacZ)M15] | |
| Plasmids | ||
| pUC19 | Apr; lazZ′ | New England Biolabs (Beverly, Mass.) |
| pGEM-T | Apr; lacZ′ | Promega (Madison, Wis.) |
| pET19d | Apr | Novagen (Madison, Wisc.) |
Sequencing strategies.
A combination of strategies was used to sequence the xyl operon genes (Table 2). First, the L. lactis B-4449 xylose isomerase gene was sequenced in two fragments. Degenerate xylA PCR primers (NH-1 and WG-1) were designed based on the published N-terminal sequence of the L. xylosus XylA (48) and an amino acid sequence (WGGREG, amino acids [aa] 188 to 193) conserved among bacteria including E. coli, Salmonella enterica serovar Typhimurium, Lactobacillus pentosus, and Bacillus spp. A cloned portion of the resulting 540-bp fragment was hybridized to a 2.4-kb EcoRI/HindIII insert in a chromosomal minilibrary. A portion of this fragment encoded the first 311 aa of XylA. The sequences of the 3′ end of the xylA gene and the 5′ end of xylB were obtained via inverse PCR with primers 7 and 5.
TABLE 2.
Summary of primers
| Primer | Sequence (5′-3′)a | Location (nucleotides) | Comment |
|---|---|---|---|
| WG-1 | CCYTCRCGRCCGCCCCA | 1913–1897b | L. lactis B-4449 xylA PCR |
| NH-1 | ACNAARACNAARAAYATGTTY | 1378–1399b | L. lactis B-4449 xylA PCR |
| 5 | TGATGCCAATCTTGG | 2220–2234b | L. lactis B-4449 xylA inverse PCR |
| 7 | TCTGGTAGGTCTTCTCCTA | ∼1.6 kg upstream of xylA | L. lactis B-4449 xylA inverse PCR |
| BFX | CGGGATCCATGTTCATTCTCTCCTAATCAC | 1171–1192c | xylA PCR and Northern primer |
| HRX | CCCAAGCTTGGTTCCTAAGT | 2702–2691c | xylA PCR and Nothern primer |
| BFX-rev | GTGATTAGGAGAGAATGAACAT | 1192–1171c | xylR-xylB inverse PCR |
| HRX-rev | TTAGGAATTGACTTAGGAACC | 2681–2702c | xylR-xylB inverse PCR |
| 210ip820-rev | TGAGTAACAAGCACCCGGAACC | 3544–3523c | xylB inverse PCR |
| R210xb-rev | TAGAAGCGGTCACTAAAGC | 4011–4029c | xylB inverse PCR |
| Ip412-rev | AGAAATTGCTGGATAAACTGG | 2964–2945c | xylB inverse PCR |
| M13-40 | GTTTTCCCAGTCACGAC | NAd | Vector sequencing primer |
| M13rev(ht) | GCTTTAGTGACCGCTTCTA | NA | Vector sequencing primer |
R = A or G; Y = C or T; H = A or C or T; N = any nucleotide.
Nubmering corresponds to L. lactis B-4449 xyl-xyn operon (including sequence upstream of xylR); GenBank accession no. AF092042.
Numbering corresponds to L. lactis IO-1 xyl-xyn operon (including upstream sequence); GenBank accession no. AF092041.
NA, not available.
PCR primers (BFX and HRX) designed from the L. lactis B444-9 xylA and 5′ xylB sequences allowed us to amplify the corresponding 1.5-kb product from L. lactis strains IO-1 and 210. The PCR product was sequenced after ligation into pUC19 and transformation into E. coli JM109. Three inverse PCR reactions with primer pairs BFX-rev–HRX-rev, 210ip820-rev–R210xb-rev, and ip412-rev–R210xb-rev (Table 2) were used to walk upstream of xylA to obtain the xylR nucleotide sequence and to walk downstream of xylA to obtain the xylB sequence from L. lactis 210. PCR primers were designed from the 210 sequences to determine the xylR and xylB sequences from L. lactis IO-1 and L. xylosus.
Inverse PCR.
Chromosomal DNA was extracted from lactococci according to the method described by Kim and Batt (21). The chromosomal DNA was digested using a variety of restriction enzymes (New England Biolabs, Beverly, Mass.). The enzyme reactions were inactivated by incubation at 65°C for 15 min or by phenol extraction. Ligation reactions contained restricted DNA at a series of concentrations from 25 to 750 ng, together with 1 U of T4 DNA ligase (Gibco, Gaithersburg, Md.), 10 μl of 5× ligation buffer (Gibco), and sterile distilled H2O, for a final reaction volume of 50 μl. Ligations were incubated for 1 h at 37°C and heat inactivated at 65°C for 20 min. The resulting self-ligated DNA was then precipitated with NaCl and ethanol and then resuspended in sterile distilled H2O to give a final concentration of 5 ng/μl for use in PCR.
PCR amplification.
PCR's were carried out in 1× PCR buffer (20 mM Tris-HCl [pH 8.3], 50 mM KCl) with a final volume of 50 μl. Reactions contained 1 μl of self-ligated chromosomal DNA or crude cell lysate prepared according to the method of Czajka and Batt (7), 50 pmol of each primer, 100 mM concentrations of each deoxynucleotide triphosphate, 1 U of AmpliTaq DNA Polymerase (Perkin-Elmer, Foster City, Calif.), and 1.5 mM MgCl2. A Hybaid TR1 or Perkin-Elmer 2400 thermocycler was programmed with a 4-min hold at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55 to 65°C (depending on the primer Tm), and 1 min at 72°C, followed in turn by a 10-min hold at 72°C. For inverse PCR the extension time at 72°C was increased to 2 min.
Cloning.
The xylA genes from the three strains were cloned prior to sequencing. Standard (pUC19) and T-overhang (pGEM-T) vectors were used with a variety of host strains that allow blue-white screening, as indicated in Table 1. Ligations were performed according to the methods of Promega (Madison, Wis.) for pGEM-T or Sambrook et al. (36). Standard methods were used to prepare and transform competent cells (36).
Sequencing and sequence analysis.
The L. lactis B444-9 xylA gene was sequenced manually according to the double-stranded DNA method of Wang (49). Sequenase (v. 2.0; Amersham, Piscataway, N.J.), [α-32P]dATP, and dideoxynucleotides were used, together with the universal M13 primers (−40 and reverse) or internal primers. The other genes were sequenced by the Cornell University BioResource Center using an ABI Prism 373A Stretch automated sequencer. Several xylA PCR clones from each strain were analyzed to confirm the sequences. All other sequences were obtained directly from PCR products. Sequences were analyzed using several programs from the Lasergene software suite (EditSeq v. 3.12, SeqMan v. 3.53, and MegAlign v. 3.05; DNASTAR, Inc., Madison, Wis.). We also performed BLASTX searches (2) of the GenBank database with our nucleotide sequences to identify homologous genes.
Xylose isomerase assays.
The cysteine carbazole method was used to measure xylose isomerase activity as indicated by the appearance of xylulose (10). Reagents were purchased from Sigma. Stationary phase M17 xylose- or glucose-grown cultures (100 ml) were harvested, resuspended in 4 ml of 50 mM Tris-HCl (pH 7.5) containing 1 mM dithiothreitol, and lysed by sonication. Sample absorbances were measured at 500 nm with an LKB Biochrom Ultraspec II spectrophotometer (Biochrom, Cambridge, United Kingdom) after 20 min at room temperature. One unit of enzyme activity is expressed as 1 nmol of d-xylulose produced per min.
Xylulokinase assays.
Xylulose formation was measured using a method modified from Shamana and Sanderson (43) that couples xylulokinase to pyruvate kinase and l-lactate dehydrogenase. Cell lysates (containing 80 mg of protein) were prepared by sonication as described above. The oxidation of NADH was monitored by measuring the absorbance at 340 nm with a Beckman DU 640 spectrophotometer (Beckman Instruments, Fullerton, Calif.). One unit of enzyme activity was expressed as 1 nmol of d-xylulose consumed per min.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the L. lactis B444-9, L. lactis IO-1, and L. lactis 210 xylRAB genes are AF092042, AF092041, and AF092040, respectively.
RESULTS
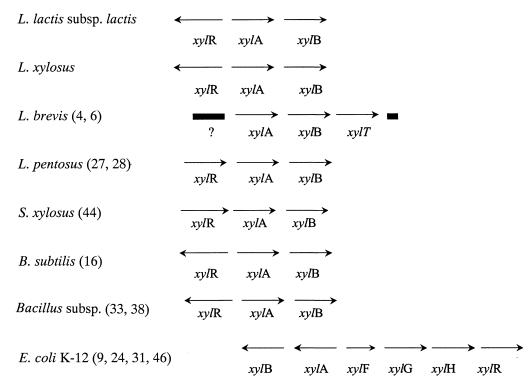
Three xylose metabolism genes (xylRAB) were sequenced from three lactic acid bacteria, including one Xyl− strain. A combination of PCR products generated with degenerate conserved, homologous, and inverse primers was assembled to give a total of 4.2 kb of sequence from L. lactis subsp. lactis IO-1 and L. lactis subsp. lactis B444-9 and 5.4 kb of sequence from L. lactis subsp. lactis 210 (Fig. 1).
FIG. 1.
Organization of the xyl locus in various bacteria. The xylR gene encodes a regulatory protein, xylA encodes a xylose isomerase, xylB encodes a xylulose kinase; xylF encodes a xylose-binding protein, xylT encodes a xylose transporter, xylG encodes an ATP-binding protein, and xylH encodes a membrane transporter (ABC-type xylose transport). ?, Sequenced open reading frame of unknown function. References are given in parentheses.
Xylose isomerase.
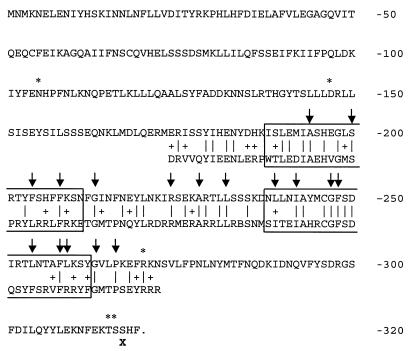
Xylose isomerase is encoded by a 1,320-bp gene in L. lactis strains IO-1, 210, and B444-9. Figure 2 shows the IO-1 XylA sequence with conserved and similar residues indicated based on an alignment of 22 group II xylose isomerase sequences, including our three sequences. These xylose isomerases are grouped together because of their catalytic metal preference and size, which ranges from 438 to 465 aa (25, 48, 50). A total of 96 aa (21%) are identical for all 22 bacteria. An additional 118 residues (26%) are similar or identical for 90% of the strains. All of the group II XylAs conserved 17 active-site residues (Fig. 2) determined for the group I Streptomyces rubiginosus XylA (50–52). Although there are conserved residues throughout the group II protein, the most highly conserved region, aa 179 to 345, is in the middle of the protein. This region includes 12 S. rubiginosus active-site residues. The carboxyl terminus of XylA is the least conserved among the group II proteins. The XylA sequences of L. lactis B444-9, L. lactis IO-1, and L. lactis 210 are very similar (Fig. 2). Only 2 aa differ between the Xyl+ strains L. lactis IO-1 and L. lactis B444-9 (Lys407→Glu and His416→Tyr). These residues are not conserved among the other XylAs. Strain 210 differs from IO-1 at six residues: Met202→Arg, Asp218→Tyr, Ser247→Ala, Ala275→Val, Ser388→Thr, and Lys407→Glu. Three of the six residues (i.e., residues 202, 218, and 247) are similar in the other group II XylAs, and residue 275 is conserved in 20 of the 22.
FIG. 2.
L. lactis IO-1 XylA sequence with residues conserved among 22 group II XylA sequences indicated. The group II xylose isomerase sequences examined were from L. lactis IO-1 (this work), L. lactis 210 (this work), L. lactis B-4449 (this work), L. brevis (GenBank accession no. AF045552), L. pentosus (M57384), S. xylosus (X57599), B. licheniformis (Z80222), B. megaterium Z71474), B. subtilis (U66480), Bacillus sp. strain LW2 (L12967), Thermotoga neapolitana (L38994), Thermoanaerobacter spp. strain (U21678), Thermoanaerobacter thermosulfurogenes (J05650), Thermoanaerobacter saccharolyticum (L09699), Thermoanaerobacter thermosaccharolyticum (M91248), Thermoanaerobacter thermohydrosulfuricus (D00756), Thermoanaerobacter ethanolicus (AF001974), E. coli (X04691), Klebsiella pneumoniae (X61059), Klebsiella aerogenes (Swiss-Prot accession no. P29442), and H. influenzae (GenBank accession no. U32791). Conserved residues are indicated as follows: bold underlined residues, identical for all 22 strains; underlined residues, identical or similar for 20/22 strains; ∗ (above sequence), difference between L. lactis IO-1, 210, and/or B-4449; ↓, conserved S. rubiginosus active site residue, according to Whitaker (50).
Xylose isomerase activity.
Xylose isomerase activities for crude extracts of L. lactis strains IO-1, B444-9, and 210 were measured as described in Materials and Methods. The specific activity for IO-1 cells grown on xylose was 464 U/mg of protein, a value almost fourfold greater than that for B444-9 (127 U/mg of protein). Glucose-grown IO-1 and B444-9 xylose isomerase activities averaged 12.8 U/mg of protein. L. lactis 210 had little xylose isomerase activity whether grown on xylose (<6 U/mg of protein) or glucose (<12 U/mg of protein).
Xylulokinase.
L. lactis strains IO-1 and B444-9 have a 1,506-bp xylB gene located 66 bp downstream of the xylA translation stop codon. The L. lactis 210 xylB gene is 67 bp downstream of xylA. Xylulokinase sequences, which range in size from 475 to 511 aa, are available from 14 of the 22 XylA group II organisms. Figure 3 displays the IO-1 XylB sequence with conserved and similar residues indicated, based on an alignment of the 14 sequences. The XylB sequences are less conserved than the XylA sequences, because only 45 residues (10%) are identical for all 14 strains. An additional 62 aa are conserved and 51 aa are similar for 85% of the strains. Similar and identical residues are found throughout the XylB protein, but homologous regions with clusters of conserved residues are small and concentrated in the N-terminal half of the protein.
FIG. 3.
L. lactis IO-1 XylB sequence with residues conserved among 14 XylB sequences indicated. The xylulokinase sequences examined were from L. lactis IO-1 (this work), L. lactis 210 (this work), L. lactis B-4449 (this work), L. brevis (GenBank accession no. AF045552), L. pentosus (M57384), B. licheniformis (Z80222), B. subtilis (U66480), B. megaterium (Z71474), S. xylosus (X57599), T. ethanolicus (AF001974), E. coli (X04691), K. pneumoniae (X61059), K. aerogenes (Swiss-Prot accession no. P29444), and H. influenzae (GenBank accession no. U32791). Conserved residues are indicated as follows: bold underlined residues, identical for all 14 strains; underlined residues, identical or similar for 12/14 strains; ∗ (above sequence), difference between L. lactis IO-1, 210, and/or B-4449; X (underneath sequence), premature 210 stop codon. The FGGY kinase signature sequences (PROSITE patterns PS00933 and PS00445) are boxed.
The L. lactis strain IO-1 and B-4449 XylB sequences, like their XylA sequences, are very similar. However, the 210 xylB sequence includes a premature translation stop codon corresponding to residue 154 (indicated in Fig. 3). The stop codon is immediately after an apparent −1 frameshift in a run of six (instead of seven) adenine residues. Resequencing this region confirmed the frameshift. Premature termination of translation is the likely explanation for the lack of significant xylulokinase activity of strain 210 (see below). If the stop codon were ignored, only seven residues (aa 200 to 202, 230, 290, 448, and 496) differ between L. lactis strains IO-1, B444-9, and 210 (Fig. 3). None of these differences correspond to conserved XylB residues. Aside from the stop codon, strain 210 differs from the Xyl+ strains at residue 290 (cysteine instead of tyrosine).
Xylulokinase activity.
Xylulokinase activities for L. lactis strains IO-1, 210, and B444-9 were determined using a coupled enzyme assay as described in Materials and Methods. As with the xylose isomerase activities, the xylulokinase activity of IO-1 surpassed that of B444-9. The magnitude of the difference was much less, however, with 53.8 U/mg of protein for IO-1 compared to 41.4 U/mg of protein for B444-9. The enzyme activities for glucose-grown strains IO-1 or B-4449 were two to three times lower than when grown on xylose (24.9 and 12.4 U/mg, respectively). L. lactis 210 had a very low level of xylulokinase activity whether grown on xylose (7.8 U/mg) or on glucose (6.8 U/mg).
Xylose regulator.
In all three strains, the 1,280 bp of sequence upstream of xylA included a 950-bp open reading frame, divergently oriented and 104 bp upstream from xylA, which we have termed xylR. The XylR protein is not homologous to XylRs from other gram-positive bacteria such as Bacillus megaterium and L. pentosus. Instead, it is moderately homologous to proteins such as RhaS (21% identical and 40% similar over 250 aa), the E. coli rhamnose metabolism regulator in the AraC/XylS family. This family includes other carbohydrate operon regulators such as melR (from E. coli), rafR (Pediococcus pentosaceus), and xylR (E. coli) which regulate melibiose, raffinose, and xylose metabolism, respectively. Gallegos et al. (12) have recently redefined this family to include over 100 proteins, with a C-terminal 99-aa stretch of homology constituting the DNA-binding domain of the regulator proteins. This region contains two helix-turn-helix (HTH) motifs. Although our XylR is only moderately homologous to individual AraC family proteins, a PFSCAN search (15, 29) of the PROSITE profile database revealed that it was significantly homologous to the 99-aa AraC family consensus region (Fig. 4). The IO-1 XylR sequence matched 37 of the 99 residues and was similar to 17 residues. Gallegos et al. identified 17 residues in the 99-aa stretch that are highly conserved (shared by more than 60% of the family member sequences). Thirteen of these residues are conserved by the IO-1 XylR.
FIG. 4.
L. lactis IO-1 XylR sequence comparison with HTH-AraC family 2 consensus (PROSITE profile PS01124). Conserved residues are indicated as follows: |, identical to HTH-AraC family 2 consensus; +, similar to HTH-AraC Family 2 consensus; ↓, residues conserved in >60% of AraC/XylS family (see reference 12 for details); X, (underneath sequence), premature 210 stop codon. Boxed residues are HTH motifs 1 and 2 (see the text). Residues which differ between L. lactis strains IO-1, 210, or B-4449 are indicated by an asterisk above the sequence.
Eight residues differ among the three lactococcus or lactobacillus strains. L. lactis 210 has a Asn106→Ser substitution outside of the AraC family consensus region and an Phe269→Ile substitution very near the C terminus of the consensus region. L. lactis B-4449 has an Asp147→His substitution. Also, due to an additional adenosine at the end of a run of six adenosines, the 5 aa at the C terminus of XylR are not conserved in the L. lactis 210. The strain 210 xylR gene encodes 2 aa after the frameshift before a premature stop codon (indicated in Fig. 4) results in a protein that is 3 aa shorter than that of L. lactis strains IO-1 and B-4449.
DISCUSSION
Xylose metabolism has been described and characterized in lactic acid bacteria, but it is rare in L. lactis. We have discovered a metabolic pathway for the metabolism of xylose to xylulose-5-phosphate in L. lactis. Xylose metabolism requires the products of the xylR, xylA, and xylB genes. Although the three strains we studied possess sequences for these xyl genes, xylose metabolism in L. lactis is a function of the source of the strains. L. lactis subsp. lactis 210 is a dairy starter culture strain and cannot metabolize xylose, whereas L. lactis subsp. lactis B-4449 and L. lactis subsp. lactis IO-1 (which were isolated from a vegetation sample and a kitchen drain in Japan, respectively) can metabolize xylose. Recently, other Xyl+ L. lactis strains have been isolated from minimally processed fruit and vegetable products, which further supports the link between functional xylose metabolism and habitat (20).
Other than their differences in habitat, L. lactis B-4449, L. lactis IO-1, and L. lactis 210 are closely related. The ribotype patterns of L. lactis strains IO-1 and 210 have similarity values to the L. lactis B-4449 pattern of 0.69 and 0.72, respectively (data not shown). The l-lactate dehydrogenase sequences of the three strains also differ by less than 1% compared to 12 and 17% dissimilarities to Streptococcus thermophilus and Lactobacillus casei, respectively (11).
Although the xylose isomerases of our two Xyl+ strains differed by only 2 aa, the specific activity of IO-1 was almost fourfold greater than that of B-4449 when both strains were grown on xylose. The two residues are not conserved among the other XylAs nor are they in obvious proximity to any active-site residues. Northern analysis indicates that the xylA gene is transcribed at similar levels in both IO-1 and B-4449 (data not shown). Also, although the xylA gene is transcribed, L. lactis 210 did not have significant xylose isomerase activity. Its sequence differs by only 6 aa from IO-1. Three of these residues, R202, Y218, and V275, are located in highly conserved regions and differ from the group II XylA consensus (L/M202, D/E218, and A/S275; Fig. 2). The nucleotide sequence of the xylR-xylA intergenic region is identical in strains IO-1, 210, and B-4449, so it is more likely that differences in activity among the three strains are due to differences in protein structure rather than transcription levels. However, all of the conserved xylose isomerase active-site residues identified (50) are intact in L. lactis IO-1, L. lactis 210, and L. lactis B-4449. Site-directed mutagenesis studies are in progress in our laboratory to characterize the functional importance of the differences between the L. lactis strain IO-1, 210, and B-4449 XylAs.
Differences in xylulokinase activities were also measured among L. lactis strains IO-1, 210, and B-4449. The xylulokinase activity of L. lactis 210 is very low regardless of the carbon source (7.8 U/mg of protein when xylose grown versus 6.8 U/mg when glucose grown). Although the strain 210 xylB gene is transcribed (Erlandson et al., unpublished), a premature translation stop codon (corresponding to residue 154) would almost certainly prevent formation of active xylulokinase.
As with xylose isomerase, xylose-grown IO-1 had more xylulokinase activity than B-4449. Their XylB sequences differed at six residues, none of which are conserved among the compared XylBs. XylB is part of a large, extensively characterized ATPase superfamily of proteins which includes other sugar kinases, eukaryotic heat shock and stress-70 proteins, and actin (17, 18). There are 127 structurally equivalent residues shared among the family members which fold into two symmetrical domains (each with a βββαβαβα topology) to form an ATP- and substrate-binding cleft (17). Using the algorithm of Rost and Sander (32), the secondary structure of XylB was predicted (data not shown). The sequence differences of IO-1 versus B-4449 did not map to a β-sheet or α-helix. However, residue 230 (lysine in IO-1, glutamate in B-4449) is 10 residues from Asp239 (Asp245 of E. coli glycerol kinase), a key residue conserved throughout the superfamily that is thought to act as a general catalytic base (30). Residue 230 is also 3 aa from a sequence block thought to be involved in hinge motion during protein conformational change (5). In addition, residue 448 (phenylalanine in IO-1, leucine in L. xylosus) is located six residues after a second sequence block putatively involved in hinge motion (5). Therefore, differences in the predicted three-dimensional structure of the IO-1 XylB could account for its greater activity relative to B-4449.
Xylulokinase and xylose isomerase activities decreased substantially when L. lactis strains IO-1 and B-4449 were grown on glucose. Catabolite repression by glucose is a phenomenon common to all xylose metabolizers (13, 14, 31, 33). We could not find cis-acting cre sites upstream of xylA or xylB. However, it is likely that lactococci have a homologue to the cre sequences identified in Bacillus spp., L. pentosus, and S. xylosus that bound the catabolite regulator CcpA (19, 26, 40, 45).
The XylR sequences of L. lactis strains IO-1, 210, and B-4449 are moderately homologous to AraC/XylS family proteins, including the E. coli and Haemophilus influenzae XylR activator proteins. The AraC/XylS family profile (PROSITE entry PS01124) consists of 99 aa, usually toward the C-terminal end of the protein, encompassing two HTH motifs. The region is thought to constitute the DNA-binding domain of the proteins. Our XylR is homologous over the entire AraC family profile, corresponding to aa 174 to 272 of the IO-1 sequence (Fig. 4). The xylR genes from all other gram-positive bacteria (B. subtilis, B. megaterium, B. licheniformis, L. pentosus, and S. xylosus) fall into a different family of transcriptional regulators and sugar kinases, the ROK family (47). The repressor proteins in the ROK family (PROSITE entry PS01125) have an HTH motif in the N-terminal portion of the protein and a 300-residue domain with a central glycine-rich region, although the rest of the consensus pattern is not highly conserved. The ROK XylR protein has been shown to bind a tandem repeat, designated xylO, near the xylA transcription start site (8, 13, 23, 27, 37, 45). This xylO site does not appear to be present upstream of xylA in our three strains. It is unusual that our XylR is homologous to the AraC family proteins because all of these proteins are activators, except the E. coli cellibiose repressor (CelD) and the bifunctional E. coli AraC arabinose and Yersinia pestis YbtA pesticin and yersiniabactin regulators (which activate or repress depending on the effectors). In general, xylose regulation is positive in the case of gram-negative organisms but negative in gram-positive organisms (16, 23, 42, 46). Our sequence homology suggests a positive regulatory mechanism different from that previously described for gram-positive xylose metabolism.
What will be the fate of the xyl genes in dairy L. lactis strains? We speculate that the xylose metabolic genes in dairy L. lactis strains are becoming pseudogenes because there is no selective pressure to maintain xylose metabolism in the dairy environment. Deleterious mutations in the xyl genes have accumulated in L. lactis 210, and dairy starter L. lactis subsp. cremoris strains have even more extensive mutations to the xyl genes than L. lactis subsp. lactis 210 (data not shown). Interestingly, L. lactis subsp. cremoris strains have never been isolated from plant or other environmental samples, despite thorough searching (22, 34, 35). It is also possible that the xyl genes are evolving new catalytic functions in dairy lactococcal strains. To examine this, the activity of the isomerase and kinase enzymes would need to be tested against alternative substrates and/or mutants isolated with increased activity against these substrates.
In conclusion, we have discovered the genetic potential in L. lactis for xylose metabolism encoded by the xylRAB genes. These genes are organized in what could be a single xylAB operon, potentially regulated by xylR. The Xyl− dairy starter culture strain L. lactis 210 has experienced a loss of function in an environment no longer selective for xylose metabolism. Plant environmental isolates such as L. lactis B-4449 and L. lactis IO-1 retain the ability to metabolize xylose.
REFERENCES
- 1.Ahlem C, Huisman W, Neslund G, Dahms A S. Purification and properties of a periplasmic d-xylose-binding protein from Escherichia coli K-12. J Biol Chem. 1982;257:2926–2931. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bor Y-C. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1993. [Google Scholar]
- 4.Bor Y-C, Moraes C, Lee S-P, Crosby W, Sinskey A J, Batt C A. Cloning and sequencing the Lactobacillus brevis gene encoding xylose isomerase. Gene. 1992;114:127–131. doi: 10.1016/0378-1119(92)90718-5. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase and galactokinase families. Prot Sci. 1992;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaillou S, Bor Y-C, Batt C A, Postma P W, Pouwels P H. Molecular cloning and functional expression in Lactobacillus plantarum 80 of xylT, encoding the D-xylose-H+ symporter of Lactobacillus brevis. Appl Environ Microbiol. 1998;64:4720–4728. doi: 10.1128/aem.64.12.4720-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czajka J, Batt C A. Verification of causal relationships between Listeria monocytogenes isolates implicated in food-borne outbreaks of listeriosis by randomly amplified polymorphic DNA patterns. J Clin Microbiol. 1994;32:1280–1287. doi: 10.1128/jcm.32.5.1280-1287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahl M K, Degenkolb J, Hillen W. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J Mol Biol. 1994;243:413–424. doi: 10.1006/jmbi.1994.1669. [DOI] [PubMed] [Google Scholar]
- 9.Davis E O, Henderson P J F. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J Biol Chem. 1987;262:13928–13932. [PubMed] [Google Scholar]
- 10.Dische Z, Borenfreund E. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J Biol Chem. 1951;192:583–587. [PubMed] [Google Scholar]
- 11.El Khal W. M.S. thesis. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 12.Gallegos M, Schlief R, Bairoch A, Hofman K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gartner D, Degenkolb J, Ripperger A E, Allmansberger R, Hillen W. Regulation of the Bacillus subtilis W23 xylose utilization operon: interaction of the Xyl repressor with the xyl operator and the inducer xylose. Mol Gen Genet. 1992;232:415–422. doi: 10.1007/BF00266245. [DOI] [PubMed] [Google Scholar]
- 14.Ghangas G S, Wilson D B. Isolation and characterization of the Salmonella typhimurium xylose regulon. J Bacteriol. 1984;157:158–164. doi: 10.1128/jb.157.1.158-164.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribskov M, McLachlan A D, Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci USA. 1987;84:4355–4359. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastrup S. Analysis of the Bacillus subtilis xylose regulon. In: Ganesan A T, Hoch J A, editors. Genetics and biotechnology of bacilli. Vol. 2. New York, N.Y: Academic Press, Inc.; 1988. pp. 79–83. [Google Scholar]
- 17.Hurley J H. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 18.Hurley J H, Faber H R, Worthylake D, Meadow N D, Roseman S, Pettigrew D W, Remingon S J. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 19.Jacob S, Allmansberger R, Gartner D, Hillen W. Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet. 1991;229:189–196. doi: 10.1007/BF00272155. [DOI] [PubMed] [Google Scholar]
- 20.Kelly W J, Davey G P, Ward L J. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int J Food Microbiol. 1998;45:85–92. doi: 10.1016/s0168-1605(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 21.Kim S G, Batt C A. Heterologous expression and stability of the Escherichia coli β-galactosidase gene in Streptococcus lactis by translation fusion. Food Microbiol. 1988;5:59–73. [Google Scholar]
- 22.Klijn N, Weerkamp A H, De Vos W M. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl Environ Microbiol. 1995;61:788–792. doi: 10.1128/aem.61.2.788-792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreuzer P, Gartner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989;171:3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawlis V B, Dennis M S, Chen E Y, Smith D H, Henner D J. Cloning and sequencing of the xylose isomerase and xylulokinase genes of Escherichia coli. Appl Environ Microbiol. 1984;47:15–21. doi: 10.1128/aem.47.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, Bhatnagar L, Saha B C, Lee Y E, Takagi M, Imanaka T, Bagdasarian M, Zeikus J G. Cloning and expression of the Clostridium thermosulfurogenes glucose isomerase gene in Escherichia coli and Bacillus subtilis. Appl Environ Microbiol. 1990;56:2638–2643. doi: 10.1128/aem.56.9.2638-2643.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lokman B C, Heerikhuisen M, Leer R J, van den Broek A, Borsboom Y, Chaillou S, Postma P, Pouwels P H. Regulation of expression of the Lactobacillus pentosus xylAB operon. J Bacteriol. 1997;179:5391–5397. doi: 10.1128/jb.179.17.5391-5397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lokman B C, Leer R J, van Sorge R. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol Gen Genet. 1994;245:117–125. doi: 10.1007/BF00279757. [DOI] [PubMed] [Google Scholar]
- 28.Lokman B C, Van Santen P, Verdoes J C, Kruse J, Leer R J, Posno M, Pouwels P H. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol Gen Genet. 1991;230:161–169. doi: 10.1007/BF00290664. [DOI] [PubMed] [Google Scholar]
- 29.Lüthy R, Xenarios I, Bucher P. Improving the sensitivity of the sequence profile method. Prot Sci. 1994;3:139–146. doi: 10.1002/pro.5560030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettigrew D W, Smith G B, Thomas K P, Dodds D C. Conserved active site aspartates and domain-domain interactions in regulatory properties of the sugar kinase superfamily. Arch Biochem Biophys. 1998;349:236–245. doi: 10.1006/abbi.1997.0444. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld S A, Stevis P E, Ho W Y. Cloning and characterization of the xyl genes from Escherichia coli. Mol Gen Genet. 1984;194:410–415. doi: 10.1007/BF00425552. [DOI] [PubMed] [Google Scholar]
- 32.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 33.Rygus T, Scheler A, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch Microbiol. 1991;155:535–542. doi: 10.1007/BF00245346. [DOI] [PubMed] [Google Scholar]
- 34.Salama M S, Musafija-Jeking T, Sandine W E, Giovannoni S J. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus including Lactococcus lactis subspecies cremoris. J Dairy Sci. 1995;78:1004–1017. [Google Scholar]
- 35.Salama M S, Sandine W E, Giovannoni S J. Isolation of Lactococcus lactis subsp. cremoris from nature by colony hybridization with rRNA probes. Appl Environ Microbiol. 1993;59:3941–3945. doi: 10.1128/aem.59.11.3941-3945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Scheler A, Hillen W. Regulation of xylose utilization in Bacillus licheniformis: Xyl repressor-xyl-operator interaction studied by DNA modification, protection and interference. Mol Microbiol. 1994;13:505–512. doi: 10.1111/j.1365-2958.1994.tb00445.x. [DOI] [PubMed] [Google Scholar]
- 38.Scheler A, Rygus T, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus licheniformis-encoded regulon for xylose utilization. Arch Microbiol. 1991;155:526–534. doi: 10.1007/BF00245345. [DOI] [PubMed] [Google Scholar]
- 39.Schleifer K H, Kraus J, Dvorak C, Klipper-Balz R, Collins M D, Fischer W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol. 1985;6:183–195. [Google Scholar]
- 40.Schmiedel D, Hillen W. Contributions of XylR, CcpA and cre to diauxic growth of Bacillus megaterium and to xylose isomerase expression in the presence of glucose and xylose. Mol Gen Genet. 1996;250:259–266. doi: 10.1007/BF02174383. [DOI] [PubMed] [Google Scholar]
- 41.Schmiedel D, Kintrup M, Kuster E, Hillen W. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol Microbiol. 1997;23:1053–1062. doi: 10.1046/j.1365-2958.1997.2881654.x. [DOI] [PubMed] [Google Scholar]
- 42.Shamanna D K, Sanderson K E. Genetics and regulation of d-xylose in Salmonella typhimurium LT2. J Bacteriol. 1979;139:71–79. doi: 10.1128/jb.139.1.71-79.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamanna D K, Sanderson K E. Uptake and catabolism of d-xylose in Salmonella typhimurium LT2. J Bacteriol. 1979;139:64–70. doi: 10.1128/jb.139.1.64-70.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sizemore C, Buchnere E, Rygus T, Witke C, Goetz F, Hillen W. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon. Mol Gen Genet. 1991;227:377–384. doi: 10.1007/BF00273926. [DOI] [PubMed] [Google Scholar]
- 45.Sizemore C, Wieland B, Gotz F, Hillen W. Regulation of Staphylococcus xylosus xylose utilization genes at the molecular level. J Bacteriol. 1992;174:3042–3048. doi: 10.1128/jb.174.9.3042-3048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song S, Park C. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol. 1997;179:7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Titgemeyer F, Reizer J, Reizer A, Saier M H. Evolutionary relationships between sugar kinases and transcription repressors in bacteria. Microbiology. 1994;140:2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 48.Vangrysperre W, VanDamme J, Vanderkerckhove J, DeBruyne C K, Cornelis R, Kerster-Hilderson H. Localization of the essential histidine and carboxylate group in d-xylose isomerases. Biochem J. 1990;265:699–706. doi: 10.1042/bj2650699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y. Double-stranded DNA sequencing with T7 polymerase. BioTechniques. 1988;6:843–845. [PubMed] [Google Scholar]
- 50.Whitaker R D. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 51.Whitaker R D, Cho Y, Cha J, Carrell H L, Glusker J P, Karplus P A, Batt C A. Probing the roles of active site residues in d-xylose isomerase. J Biol Chem. 1995;270:22895–22906. doi: 10.1074/jbc.270.39.22895. [DOI] [PubMed] [Google Scholar]
- 52.Whitlow M, Howard A J, Finzel B C, Poulos T L, Winborne E, Gilliland G L. A metal-mediated hydride shift mechanism for xylose isomerase based on the 1.6 Å Streptomyces rubiginosus structures with xylitol and d-xylose. Proteins Struct Funct Genet. 1991;9:153–173. doi: 10.1002/prot.340090302. [DOI] [PubMed] [Google Scholar]