Abstract
We examined the genotypic diversity of 64 Bradyrhizobium strains isolated from nodules from 27 native leguminous plant species in Senegal (West Africa) belonging to the genera Abrus, Alysicarpus, Bryaspis, Chamaecrista, Cassia, Crotalaria, Desmodium, Eriosema, Indigofera, Moghania, Rhynchosia, Sesbania, Tephrosia, and Zornia, which play an ecological role and have agronomic potential in arid regions. The strains were characterized by intergenic spacer (between 16S and 23S rRNA genes) PCR and restriction fragment length polymorphism (IGS PCR-RFLP) and amplified fragment length polymorphism (AFLP) fingerprinting analyses. Fifty-three reference strains of the different Bradyrhizobium species and described groups were included for comparison. The strains were diverse and formed 27 groups by AFLP and 16 groups by IGS PCR-RFLP. The sizes of the IGS PCR products from the Bradyrhizobium strains that were studied varied from 780 to 1,038 bp and were correlated with the IGS PCR-RFLP results. The grouping of strains was consistent by the three methods AFLP, IGS PCR-RFLP, and previously reported 16S amplified ribosomal DNA restriction analysis. For investigating the whole genome, AFLP was the most discriminative technique, thus being of particular interest for future taxonomic studies in Bradyrhizobium, for which DNA is difficult to obtain in quantity and quality to perform extensive DNA:DNA hybridizations.
Due to their nitrogen-fixing symbiosis with soil bacteria collectively named rhizobia, legumes play an important role in the nitrogen cycle, especially in the tropics. They are used to restore or increase soil fertility of degraded soils in intercropping systems, as green manure, and to produce medicinal and commercial by-products. In Senegal (West Africa), many native legumes are important plant resources for many purposes, and they are the only alternatives to the costly and pollutive mineral nitrogen fertilizers used in agriculture. Many of these legumes are well adapted to the local arid climatic conditions. As drought and soil erosion progress, they are still present as colonizers. The family Leguminosae is considered to be of tropical or subtropical origin, and many of the recently proposed new rhizobial species originate from leguminous plants from these zones that had not been previously investigated.
After several prospections around the country, we focused on 27 native nodulated leguminous plants species belonging to the genera Abrus, Alysicarpus, Bryaspis, Chamaecrista, Cassia, Crotalaria, Desmodium, Eriosema, Indigofera, Moghania, Rhynchosia, Sesbania, Tephrosia, Zornia, which play an important ecological role and have agronomic potential in arid regions. As very little or no information concerning their associated rhizobia were available so far, we obtained 71 nodule isolates from these legumes in different regions in Senegal and first characterized them by sodium dodecyl sulfate-polyacrylamide gel electrophoresis whole-cell protein profiles and by 16S amplified ribosomal DNA restriction analysis (ARDRA) fingerprint analysis (9). Our main conclusion was that the strains were diverse and belonged to five phylogenetic subgroups inside Bradyrhizobium, but further genotypic characterization of these strains was needed to precisely place them in the general classification.
Taxonomically, rhizobia comprise six genera, Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium, Azorhizobium (for a review, see reference 44), and Allorhizobium (8). They constitute a phylogenetically heterogeneous group, and their taxonomy is being reexamined. Some rhizobia are more closely related to clinical bacteria, like Afipia, Blastobacter, and other nonsymbiotic bacteria, like Mycoplana and Bartonella, than to other rhizobia (39, 43, 44). In addition, several new rhizobial groups have been described, but have not yet been classified or named (for reviews, see references 24 and 44). In particular, the precise taxonomical status of many Bradyrhizobium sp. strains isolated from various legumes is not clarified (9, 10, 25, 26, 27, 34). For this genus (Bradyrhizobium), several authors have reported the lack of consistency between results obtained by different taxonomic techniques (10, 20, 32, 45).
A need for a confident strategy to investigate diversity among Bradyrhizobium populations was claimed, specifically including molecular methods (9, 34). 16S-23S rRNA intergenic gene spacer (IGS; corresponding to the spacer between 16S and 23S rRNA genes) sequences exhibit a large variability and are useful to identify genomic groups at the intraspecific level (4, 16, 22). Moreover, PCR-restriction fragment length polymorphism (PCR-RFLP) of IGS has been reported to be a useful fingerprinting method to characterize bacterial strains, with a higher discriminating power than the 16S ARDRA method. It has been applied to known rhizobial species, such as Rhizobium leguminosarum, Rhizobium “hedysari,” Sinorhizobium meliloti, and Rhizobium galegae (6, 22, 31, 33), and also tropical rhizobia strains (18) and Bradyrhizobium strains from the Canary Islands (37).
The amplification fragment length polymorphism (AFLP) technique (38, 46) is a highly discriminating fingerprinting method, based on the selective PCR amplification of certain restriction fragments from a digest of total genomic DNA. The technique involves three steps: (i) restriction of the DNA with two enzymes and ligation of oligonucleotide adapters, (ii) selective amplification of sets of restriction fragments, and (iii) polyacrylamide gel electrophoresis of the amplified fragments. Originally developed for plant genome studies, this technique has also been used to characterize various bacterial species (for a review, see reference 5). As this technique investigates the whole genome, and as the results were reported to be in good agreement with those obtained by DNA:DNA hybridization, it could be a good alternative to the latter, which is especially difficult to perform with Bradyrhizobium.
Here, 64 Bradyrhizobium sp. strains from small Senegalese legumes were further characterized genotypically by PCR-RFLP analysis of the IGS region between 16S and 23S rRNA genes and by AFLP analysis, including representative strains for Bradyrhizobium species and groups reported in the past (9, 10, 25, 26, 40). The results are compared with previous 16S ARDRA grouping, and taxonomic resolution levels of the three techniques are discussed.
MATERIALS AND METHODS
Bacterial strains and isolation procedures.
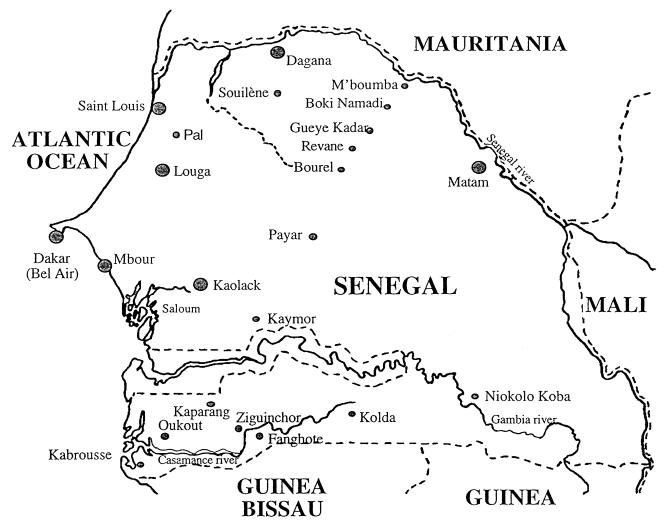
The strains used in this study are listed in Table 1, and their places of isolation are indicated on Fig. 1. Bacterial strains were grown as previously described (9, 40).
TABLE 1.
Strains used
| Straina | Other strain designation | Host plant or origin | Geographical originb | Reference or source | IGS PCR-RFLP grouping | AFLP grouping |
|---|---|---|---|---|---|---|
| ORS 18 | LMG 15159 | Alysicarpus ovalifolius | Dakar Bel-Air (CS) | 9 | VIII | 17 |
| ORS 23 | LMG 15161 | Tephrosia villosa | Dakar Bel-Air (CS) | 9 | VIII | 17 |
| ORS 28 | LMG 15164 | Indigofera tinctoria | Mbour (CS) | 9 | I | 36 |
| ORS 29 | LMG 15165 | Indigofera tinctoria | Mbour (CS) | 9 | I | 43 |
| ORS 30 | LMG 15166 | Indigofera hirsuta | Dakar Bel-Air (CS) | 9 | I | 36 |
| ORS 31 | LMG 15167 | Indigofera tinctoria | Pal (NS) | 9 | VIII | 17 |
| ORS 84 | LMG 15174 | Indigofera astragalina | Payar (CS) | 9 | I | 37 |
| ORS 85 | LMG 15175 | Indigofera astragalina | Payar (CS) | 9 | I | 47 |
| ORS 86 | LMG 15176 | Tephrosia purpurea | Bourel (Ferlo, NS) | 9 | XIII | 40 |
| ORS 87 | LMG 15177 | Tephrosia purpurea | Bourel (Ferlo, NS) | 9 | XIII | 40 |
| ORS 88 | LMG 15178 | Tephrosia purpurea | Bourel (Ferlo, NS) | 9 | XIII | 40 |
| ORS 89 | LMG 15179 | Tephrosia purpurea | Bourel (Ferlo, NS) | 9 | XIII | 34 |
| ORS 90 | R-2196 | Tephrosia sp. | Bourel (Ferlo, NS) | 9 | XIII | 34 |
| ORS 523 | R-2194 | Indigofera senegalensis | Dakar Bel-Air (CS) | 9 | VIII | NDc |
| ORS 935 | LMG 15269 | Rhynchosia minima | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 936 | LMG 15270 | Rhynchosia minima | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 937 | LMG 15271 | Rhynchosia minima | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 938 | LMG 15272 | Rhynchosia minima | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 976 | R-2197 | Indigofera senegalensis | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 977 | LMG 15273 | Indigofera senegalensis | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 978 | LMG 15274 | Indigofera senegalensis | Gueye Kadar (Ferlo, NS) | 9 | V | 20 |
| ORS 979 | LMG 15275 | Indigofera senegalensis | Gueye Kadar (Ferlo, NS) | 9 | VI | 46 |
| ORS 980 | LMG 15276 | Indigofera senegalensis | Gueye Kadar (Ferlo, NS) | 9 | VI | 46 |
| ORS 984 | LMG 15279 | Indigofera senegalensis | Mboumba (Ferlo, NS) | 9 | Sepd | Sep |
| ORS 986 | LMG 15280 | Indigofera senegalensis | Mboumba (Ferlo, NS) | 9 | VI | 46 |
| ORS 987 | LMG 15281 | Indigofera senegalensis | Mboumba (Ferlo, NS) | 9 | VI | 46 |
| ORS 1216 | LMG 15300 | Indigofera senegalensis | Boki Namadi (Ferlo, NS) | 9 | ND | Sep |
| ORS 1217 | LMG 15301 | Indigofera senegalensis | Boki Namadi (Ferlo, NS) | 9 | VI | 46 |
| ORS 1218 | R-2200 | Indigofera senegalensis | Boki Namadi (Ferlo, NS) | 9 | VI | 46 |
| ORS 1219 | LMG 15302 | Indigofera senegalensis | Boki Namadi (Ferlo, NS) | 9 | VI | 46 |
| ORS 1220 | LMG 15303 | Indigofera senegalensis | Boki Namadi (Ferlo, NS) | 9 | VI | 46 |
| ORS 1228 | LMG 15304 | Indigofera astragalina | Payar (CS) | 9 | I | 47 |
| ORS 1229 | LMG 15305 | Indigofera astragalina | Payar (CS) | 9 | I | 37 |
| ORS 1810 | LMG 15242 | Crotalaria lathyroïdes | Kabrousse (Casamance, SS) | 9 | Sep | Sep |
| ORS 1811 | LMG 15243 | Crotalaria goreensis | Kabrousse (Casamance, SS) | 9 | Sep | Sep |
| ORS 1812 | LMG 15365 | Abrus stictosperma | Fanghote (Casamance, SS) | 9 | XII | 42 |
| ORS 1813 | LMG 15244 | Crotalaria hyssopifolia | Kabrousse (Casamance, SS) | 9 | XI | Sep |
| ORS 1814 | LMG 15245 | Crotalaria hyssopifolia | Fanghote (Casamance, SS) | 9 | XI | 19 |
| ORS 1815 | LMG 15246 | Crotalaria hyssopifolia | Fanghote (Casamance, SS) | 9 | Sep | Sep |
| ORS 1816 | LMG 15247 | Crotalaria hyssopifolia | Fanghote (Casamance, SS) | 9 | I | 38 |
| ORS 1817 | LMG 15366 | Eriosema glomeratum | Oukout (Casamance, SS) | 9 | Sep | Sep |
| ORS 1818 | LMG 15248 | Indigofera microcarpa | Oukout (Casamance, SS) | 9 | Sep | Sep |
| ORS 1819 | LMG 15249 | Crotalaria retusa | Kabrousse (Casamance, SS) | 9 | I | 38 |
| ORS 1820 | LMG 15250 | Indigofera hirsuta | Fanghote (Casamance, SS) | 9 | I | 38 |
| ORS 1823 | LMG 15367 | Indigofera hirsuta | Wouring (Niokolokoba, SS) | 9 | XI | 19 |
| ORS 1824 | LMG 15253 | Indigofera hirsuta | Wouring (Niokolokoba, SS) | 9 | XI | 19 |
| ORS 1826 | LMG 15255 | Alysicarpus glumaceus | Fanghote (Casamance, SS) | 9 | XI | 19 |
| ORS 1827 | LMG 15256 | Alysicarpus glumaceus | Fanghote (Casamance, SS) | 9 | III | 44 |
| ORS 1828 | LMG 15368 | Alysicarpus ovalifolius | Niokolokoba (SS) | 9 | V | 20 |
| ORS 1831 | LMG 15369 | Alysicarpus rugosus | Fanghote (Casamance, SS) | 9 | IV | 45 |
| ORS 1832 | LMG 15258 | Bryaspis lupulina | Kagnout (Casamance, SS) | 9 | VIII | 41 |
| ORS 1836 | LMG 15261 | Crotalaria glaucoïdes | Kaparang (Casamance, SS) | 9 | XI | 35 |
| ORS 1838 | LMG 15263 | Sesbania rostrata | Kaolack (CS) | 9 | VIII | 41 |
| ORS 1844 | LMG 15266 | Chamaecrista sp. | Karounate (Casamance, SS) | 9 | I | 38 |
| ORS 1845 | LMG 15267 | Moghania faginea | Kaparang (Casamance, SS) | 9 | III | 44 |
| ORS 1847 | LMG 15268 | Zornia glochidiata | Fanghote (Casamance, SS) | 9 | Sep | Sep |
| ORS 1848 | LMG 15373 | Indigofera hirsuta | Kaparan (Casamance, SS) | 9 | Sep | 35 |
| ORS 1849 | LMG 15374 | Indigofera hirsuta | Fanghote (Casamance, SS) | 9 | XI | 19 |
| ORS 1894 | LMG 15696 | Indigofera hirsuta | Kolda (Casamance, SS) | 9 | I | 47 |
| ORS 1896 | LMG 15697 | Cassia absus | Kolda (Casamance, SS) | 9 | IV | 45 |
| ORS 1898 | LMG 15699 | Tephrosia bracteolata | Kolda (Casamance, SS) | 9 | I | 37 |
| ORS 1899 | LMG 15700 | Indigofera stenophylla | Kolda (Casamance, SS) | 9 | I | 43 |
| ORS 1903 | LMG 15702 | Tephrosia villosa | Kolda (Casamance, SS) | 9 | I | 37 |
| ORS 1905 | LMG 15703 | Tephrosia bracteolata | Kolda (Casamance, SS) | 9 | I | 33 |
| Bradyrhizobium sp. (Faidherbia) | ||||||
| ORS 101 | LMG 10664 | Faidherbia albida | Senegal | 10 | V | 20 |
| ORS 103 | LMG 10665 | Faidherbia albida | Dakar Bel-Air (CS) | 10 | V | 20 |
| ORS 110 | LMG 10666 | Faidherbia albida | Louga (NS) | 10 | XIII | Sep |
| ORS 112 | LMG 10668 | Faidherbia albida | Louga (NS) | 10 | XIII | 33 |
| ORS 117 | LMG 10673 | Faidherbia albida | Louga (NS) | 10 | XIII | 33 |
| ORS 121 | LMG 10677 | Faidherbia albida | Louga (NS) | 10 | XIV | 29 |
| ORS 130 | LMG 10686 | Faidherbia albida | Louga (NS) | 10 | XIV | 29 |
| ORS 133 | LMG 10689 | Faidherbia albida | Louga (NS) | 10 | XIII | Sep |
| ORS 156 | LMG 11951 | Faidherbia albida | Djinaki, Senegal | 10 | ND | 19 |
| ORS 166 | LMG 10709 | Faidherbia albida | Casamance (SS) | 10 | X | 31 |
| ORS 162 | LMG 10705 | Faidherbia albida | Casamance (SS) | 10 | X | 31 |
| ORS 163 | LMG 10706 | Faidherbia albida | Oussouye (Casamance, SS) | 10 | X | 31 |
| ORS 170 | LMG 10713 | Faidherbia albida | Bayotte (Casamance, SS) | 10 | XII | 42 |
| ORS 174 | LMG 10717 | Faidherbia albida | Badiana (Casamance, SS) | 10 | XIII | 33 |
| ORS 180 | LMG 10719 | Faidherbia albida | North Senegal | 10 | ND | 37 |
| ORS 184 | LMG 10723 | Faidherbia albida | Keur Momar Sarr (Guiers Lake, NS) | 10 | Sep | Sep |
| ORS 187 | LMG 10726 | Faidherbia albida | Dagana (NS) | 10 | XI | 20 |
| ORS 188 | LMG 10727 | Faidherbia albida | Dagana (NS) | 10 | V | 20 |
| Bradyrhizobium sp. (Aeschynomene) | ||||||
| ORS 277 | LMG 12186 | Aeschynomene sensitiva | Elinkine (Casamance, SS) | 25 | Sep | Sep |
| ORS 278 | LMG 12187 | Aeschynomene sensitiva | Elinkine (Casamance, SS) | 25 | Sep | Sep |
| ORS 304 | LMG 8069 | Aeschynomene elaphroxylon | Guiers Lake (NS) | 1 | VIII | 17 |
| ORS 306 | LMG 8300 | Aeschynomene indica | Guiers Lake (NS) | 1 | XV | 28 |
| ORS 324 | LMG 8295 | Aeschynomene afraspera | Keur Maktar (NS) | 25 | XVI | 6 |
| ORS 347 | LMG 10298 | Aeschynomene afraspera | Tobor (Casamance, SS) | 2 | Sep | Sep |
| ORS 352 | LMG 15384 | Aeschynomene afraspera | Senegal | 25 | XVI | 6 |
| ORS 353 | LMG 15385 | Aeschynomene afraspera | Senegal | 25 | XVI | 6 |
| ORS 358 | LMG 10303 | Aeschynomene nilotica | 25 | ND | 34 | |
| BTAi1 | LMG 11795 | Aeschynomene indica | United States | 25 | XV | 28 |
| Bradyrhizobium japonicum | ||||||
| Bonnier 3.1 | LMG 4252 | Glycine max | II | 15 | ||
| Erdman 1BOa2 | LMG 4262 | Albizia julibrissin | ND | Sep | ||
| Erdman 3c3a1 | LMG 4265 | Ulex europaeus | ND | Sep | ||
| USDA 59 | LMG 4271 | Glycine max | North Carolina | II | 15 | |
| Erdman 314a8 | LMG 4272 | Pueraria lobata | ND | Sep | ||
| Erdman 316n10 | LMG 4274 | Vigna unguiculata | Sep | Sep | ||
| NZP 5533 | LMG 6136 | Glycine max | United States | VII | 12 | |
| NZP 5549T | LMG 6138T | Glycine max | Japan | 17 | II | 15 |
| USDA 135 | LMG 8321 | Glycine max | Iowa | V | 20 | |
| USDA 110 | Glycine max | Florida | VII | 12 | ||
| Bradyrhizobium elkanii | ||||||
| NZP 5531T | LMG 6134T | Glycine max | Maryland | 19 | XI | 32 |
| NZP 5532 | LMG 6135 | Glycine max | Wisconsin | 19 | XI | 32 |
| USDA 61 | Glycine max | North Carolina | XI | ND | ||
| Bradyrhizobium liaoningense | ||||||
| SFI 2281T | LMG 18230T | Glycine max | China | 42 | IX | 16 |
| SFI 2062 | LMG 18231 | Glycine max | China | 42 | IX | 16 |
| Bradyrhizobium sp. | ||||||
| NZP 2314 | LMG 6129 | Lotus pedunculatus | Australia | 26 | ND | 39 |
| USDA 3002 | LMG 8888 | Acacia decurens | Brazil | 26 | Sep | Sep |
| BR 6011 | LMG 9514 | Lonchocarpus costatus | Brazil | 26 | ND | Sep |
| BR 3606 | LMG 9959 | Acacia mollissima | Brazil | 26 | XI | Sep |
| BR 3621 | LMG 9966 | Acacia mangium | Espirito Santo, Brazil | 26, 27 | XI | 48 |
| BR 4406 | LMG 9980 | Enterolobium ellipticum Benth. | Rio de Janeiro, Brazil | 26, 27 | ND | Sep |
| BR 8406 | LMG 10018 | Dalbergia nigra Allem. | Rio de Janeiro, Brazil | 26, 27 | ND | 48 |
| INPA 9A | LMG 10029 | Derris sp. | Brazil | 26 | ND | Sep |
| TAL 1127 | LMG 10295 | Cajanus cajan | ND | 36 | ||
| ORS 58 | LMG 10663 | Dalbergia melanoxylon | Senegal | 9 | ND | 17 |
| MAR 1505 | LMG 14304 | Vigna unguiculata | 34 | ND | Sep | |
| MAR 1587 | LMG 14311 | Arachis hypogaea | 34 | ND | Sep | |
| MSDJ 718 | Lupinus luteus | France | 21 | Sep | 39 |
Original strain number, or as received. BR, strains from the CNPBS/EMBRAPA, Centro Nacional de Pesquisa em Biologia do Solo, Seropedica 23851, Rio de Janeiro, Brazil/Emprasa Brasiliera de Pesquisa Agropequaria; BCCM/LMG, Bacteria Collection, Laboratorium voor Microbiologie, Ghent, Belgium; MAR, Soil Productivity Research Laboratory, Marondera, Zimbabwe; MSDJ, Institut National de la Recherche Agronomique (INRA), Microbiologie des Sols, Dijon, France; INPA, National Institute of Amazonia Research, Manaus, Brazil; NZP, Culture Collection of the Department for Scientific and Industrial Research, Biochemistry Division, Palmerston North, New Zealand; ORS, ORSTOM Collection, Institut de Recherche pour le Développement, BP 1386, Dakar, Senegal; R-, Research collection of the Laboratorium voor Microbiologie, Ghent, Belgium; SFI, Soils and Fertilizers Institute, Chinese Academy of Agricultural Sciences, Beijing, People's Republic of China; TAL, Nitrogen Fixation in Tropical Agricultural Legumes (NifTAL), University of Hawaii, Paia; USDA, U.S. Department of Agriculture, Beltsville, Md.
CS, central Senegal; NS, north Senegal; SS, south Senegal.
ND, not determined.
Sep, separate.
FIG. 1.
Sites of sampling in Senegal.
PCR amplification of IGS (16S-23S rDNA) region.
Total DNA was prepared as previously described (9). Alternatively, for some strains, the PCR amplification was done from bacterial cell preparation, prepared as follows: cells were grown on yeast extract-mannitol agar slants (36) for 72 h at 28°C and then washed with sterile distilled water. The cell suspension was adjusted to an optical density (620 nm) of 0.5 by dilution in water. An aliquot of 100 μl of this cell suspension was pelleted, and cells were resuspended in 100 μl of sterile Milli-Q water (Milli-Q plus; Millipore, Saint-Quentin-Yvelines, France) and lysed with 100 μl of 10 mM Tris-HCl (pH 8.2) and 13 μl of proteinase K (1 mg/ml) (Merck-Belgolabo, Overijse, Belgium) during 2 h at 55°C. Then, the cells were boiled for 10 min to denature the enzyme. The PCR was carried out with 250 ng of DNA or with 10 μl of bacterial cell suspension as template DNA.
Primers FGPS1490 (28) and FGPS132′ (29) were used to amplify the IGS regions. FGPS1490 corresponds to conserved sequences in the 3′ part of the 16S rDNA gene right next to the IGS (corresponding to the Escherichia coli numbering positions 1525 to 1541), and reverse primer FGPS132′ corresponds to the beginning 5′ part of the 23S rDNA (corresponding to the E. coli numbering positions 115 to 132). PCR was performed as described by Laguerre et al. (22).
PCR-amplified DNA was visualized by Hoefer HE 33 Mini Submarine electrophoresis (Amersham Pharmacia Biotech) at 70 V for 30 min with 5 μl of the amplified mixture on 2% (wt/vol) Biozym DNA Agarose (Biozym, Landgraaf, The Netherlands) in TAE buffer (40 mM Tris-acetate, 2 mM EDTA, 20 mM acetic acid, pH 8.0) containing 0.5 mg of ethidium bromide/ml.
RFLP analysis.
Aliquots (10 μl) of PCR products were digested with 5 U of restriction endonuclease in 20-μl reaction volumes by using the manufacturer's recommended buffer and incubation conditions. The following restriction enzymes were used: AluI and NdeII (Boehringer Mannheim Biochemica, Brussels, Belgium), DdeI (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands), and HaeIII, HhaI, HinfI, MspI, and RsaI (New England Biolabs Inc., Leusden, The Netherlands). Restricted DNA was analyzed by horizontal electrophoresis in 4% Nusieve 3:1 or Metaphor agarose (FMC, Rockland, Maine) gels. DNA molecular-weight-marker VIII (Boehringer Mannheim) was used as standard for gel calibration. Electrophoreses were carried out at 80 V for 4 h with standard gels (11 by 14 cm or 10 by 15 cm) on a Bethesda Research Laboratories Horizon 11-14 apparatus or with a DNA Sub Cell unit (15-by-10-cm tray) (20 wells; Bio-Rad Laboratories NV, Nazareth-Eke, Belgium).
The gels were stained and photographed as described by Heyndrickx et al. (12).
The scanned gel images were digitized and stored in a computer as described by Heyndrickx et al. (12).
Pattern analysis was performed using the Gel Compar software (35) version 4.2 (Applied Maths, Kortrijk, Belgium), and an unweighted pair group method with averages (UPGMA) dendrogram was constructed using the Dice similarity coefficient (SD).
AFLP analysis.
The experimental protocol used was modified from that of Vos et al. (38) as described by Huys et al. (14) and was essentially similar to that used in the study of Willems et al. (40). In short, 1 μg of total genomic DNA was digested by two restriction enzymes. Double-stranded oligonucleotide adapters with a single-stranded overhang homologous to the 5′ and 3′ ends generated during restriction were ligated to the DNA fragments. The ligated DNA fragments were amplified by PCR using primers complementary to the adapter and restriction site sequence with additional selective nucleotides at their 3′ end. The amplified products were separated by polyacrylamide gel electrophoresis, and the resulting banding pattern was revealed through autoradiography. Bordetella holmesii strain LMG 15945 was used as the reference strain because AFLP analysis of this strain generated a banding pattern displaying evenly distributed and well-separated bands over the entire length of the gel.
The cluster analysis of AFLP patterns was done by analyzing the scanned profiles on a personal computer with the Gel Compar program, version 4.2, using the Dice coefficient and the UPGMA clustering algorithm.
RESULTS
IGS PCR-RFLP analysis. (i) Cluster analysis of IGS RFLP patterns.
The 16S-23S rDNA IGS region of 104 Bradyrhizobium strains was amplified by PCR and restricted with eight endonucleases. One to six DNA fragments were generated by each restriction enzyme. For each strain, the patterns obtained with the eight enzymes were combined, resulting in 70 different combinations referred to as IGS rDNA types (Table 2).
TABLE 2.
Different rDNA IGS types and restriction patterns determined by PCR-RFLP analysis of the rDNA IGS regions
| rDNA IGS clusters and their composition | rDNA IGS typea | Sizeb of IGS PCR product (bp) | Different restriction patterns typesc of rDNA IGS digested with:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AluI | DdeI | HaeIII | HhaI | HinfI | MspI | NdeII | RsaI | |||
| Cluster I | 1 | 825 | 2 | 2 | 2 | 31 | 3 | 2 | 2 | 2 |
| 2 | 824 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 2 | |
| 3 | 820 | 2 | 2 | 2 | 4 | 3 | 2 | 3 | 2 | |
| 4 | 823 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 3 | |
| 5 | 816 | 2 | 2 | 2 | 3 | 3b | 2 | 2 | 8 | |
| 6 | 802 | 2 | 2 | 2 | 31 | 2 | 2 | 2 | 5 | |
| 7 | 820 | 2 | 2 | 2 | 2 | 14 | 2 | 4 | 2 | |
| 8 | 825 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| 9 | 825 | 2 | 2 | 2 | 3 | 2 | 2 | 4 | 2 | |
| Cluster II | 10 | 821 | 2b | 2 | 2 | 28 | 2 | 9b | 27b | 17 |
| B. japonicum | 11 | 825 | 22 | 23 | 9 | 28 | 2 | 9b | 27b | 17 |
| Cluster III | 13 | 900 | 4 | 22 | 2 | 3 | 16 | 20 | 5 | 6 |
| Cluster IV | 18 | 820 | 4 | 22 | 30 | 11 | 21 | 35 | 3 | 5 |
| 19 | 850 | 4 | 22 | 30 | 11 | 2 | 36 | 37 | 5 | |
| Cluster V | 24 | 850 | 12 | 6 | 9 | 13 | 6 | 10 | 18 | 7 |
| Bradyrhizobium sp. (F. albida) | 25 | 854 | 12 | 19 | 9 | 2 | 6 | 10 | 18 | 7 |
| B. japonicum | 26 | 820 | 12 | 19 | 9 | 2 | 6 | 10 | 16 | 7 |
| 27 | 860 | 12 | 6 | 9 | 11 | 6 | 10 | 18 | 8 | |
| 28 | 850 | 12 | 6 | 36 | 11 | 2 | 10 | 18 | 8 | |
| 29 | 864 | 12 | 6 | 10 | 2 | 6 | 10 | 18 | 7 | |
| 30 | 853 | 12 | 6 | 10 | 32 | 6 | 10 | 18 | 7 | |
| 31 | 900 | 12 | 6 | 10 | 2 | 2 | 10 | 18 | 7 | |
| 32 | 900 | 12 | 6 | 10 | 39 | 2 | 10 | 18 | 8 | |
| Cluster VI | 33 | 804 | 11 | 6 | 34 | 40 | 2 | 29 | 15 | 5 |
| 34 | 862 | 11 | 6 | 11 | 12 | 5 | 11 | 15 | 6 | |
| 35 | 862 | 29 | 6 | 11 | 5 | 5 | 29 | 31 | 2 | |
| Cluster VII | 36 | 870 | 10 | 6 | 9 | 11 | 2 | 12 | 17 | 5 |
| B. japonicum | 37 | 880 | 10 | 6 | 9 | 11 | 6 | 12 | 17 | 6 |
| Cluster VIII | 38 | 900 | 7 | 3 | 5 | 23 | 5 | 6 | 7 | 5 |
| 39 | 920 | 7 | 3 | 5 | 6b | 6 | 6 | 7 | 6 | |
| 40 | 913 | 7 | 3b | 6 | 6 | 6 | 6 | 7 | 6 | |
| 41 | 842 | 7 | 3 | 5 | 23 | 19 | 5b | 7b | 4 | |
| Bradyrhizobium sp. (A. elaphroxylon) | 42 | 950 | 8 | 3 | 5 | 7 | 19 | 5 | 7 | 4 |
| Cluster IX | 44 | 780 | 5 | 2 | 15 | 45 | 2 | 10 | 4 | 5 |
| B. liaoningense | ||||||||||
| Cluster X | 45 | 845 | 16 | 7 | 12 | 14 | 5 | 13 | 14 | 10 |
| Bradyrhizobium sp. (F. albida) | ||||||||||
| Cluster XI | 46 | 850 | 15 | 20 | 14 | 20 | 5 | 15 | 13 | 12 |
| 47 | 824 | 13 | 21 | 14 | 34 | 5 | 15 | 15 | 12 | |
| Bradyrhizobium sp. (A. mangium) | 48 | 843 | 13 | 21 | 14 | 18 | 6 | 15 | 12 | 12 |
| Bradyrhizobium sp. (A. mollissima) | 49 | 862 | 14 | 10 | 15 | 18 | 6 | 17 | 12 | 11 |
| Bradyrhizobium sp. (F. albida) | 50 | 864 | 14 | 20 | 14 | 18 | 6 | 16 | 12 | 11 |
| B. elkanii | 51 | 860 | 14 | 20 | 15 | 8 | 2 | 16 | 12 | 11 |
| 52 | 870 | 15 | 20 | 14 | 18 | 5 | 15 | 13 | 11 | |
| 53 | 852 | 23 | 10 | 15 | 18 | 6 | 17 | 12 | 11 | |
| Cluster XII | 54 | 822 | 19 | 8 | 13 | 16 | 6 | 14 | 22 | 11 |
| Bradyrhizobium sp. (F. albida) | 55 | 800 | 19 | 8 | 13 | 15 | 6 | 14 | 22 | 11 |
| Cluster XIII | 57 | 866 | 21 | 4 | 7 | 8 | 7 | 7 | 9 | 6 |
| 58 | 850 | 20 | 4 | 8 | 9 | 7 | 7 | 10 | 6 | |
| Bradyrhizobium sp. (F. albida) | 59 | 840 | 20 | 4 | 8 | 9 | 7 | 8 | 10 | 6 |
| 60 | 840 | 20 | 4 | 8 | 9 | 7 | 8 | 10 | 5 | |
| Cluster XIV | 61 | 880 | 21 | 5 | 8 | 9 | 6 | 9 | 11 | 6 |
| Bradyrhizobium sp. (F. albida) | 62 | 862 | 21 | 5 | 8 | 31 | 6 | 9 | 11 | 6 |
| Cluster XV | 63 | 1,000 | 9 | 14 | 22 | 25 | 12 | 22 | 8 | 15 |
| Bradyrhizobium sp. (A. indica) | 64 | 1,000 | 9 | 14 | 23 | 25 | 12 | 23 | 8 | 15 |
| Cluster XVI | 66 | 1,038 | 17 | 11 | 19 | 22 | 10 | 19 | 19 | 13 |
| Bradyrhizobium sp. (A. afraspera) | 67 | 950 | 17 | 11 | 18 | 21 | 9 | 19 | 20 | 18 |
| Separate | 12 | 780 | 2 | 2 | 31 | 2 | 2 | 2 | 34 | 2 |
| 14 | 795 | 3 | 2 | 2 | 37 | 2 | 2 | 4 | 21 | |
| 20 | 870 | 31 | 2 | 30 | 23 | 5 | 20 | 26 | 5 | |
| 21 | 832 | 4 | 28 | 32 | 31 | 21 | 31 | 3 | 20 | |
| 22 | 900 | 34 | 13 | 2 | 7 | 5 | 33 | 37 | 13 | |
| 23 | 920 | 32 | 29 | 2 | 23 | 23 | 26 | 35 | 13 | |
| 56 | 880 | 13 | 20b | 30 | 15 | 6 | 35 | 38 | 23 | |
| 70 | 923 | 25 | 17 | 26 | 20 | 10 | 26 | 28 | 13 | |
| Bradyrhizobium sp. (A. decurens) | 15 | 814 | 3 | 2 | 3 | 5 | 4 | 2 | 3 | 3 |
| B. japonicum | 16 | 840 | 37 | 33 | 3 | 20 | 4 | 9 | 18 | 2 |
| Bradyrhizobium sp. (L. luteus) | 17 | 806 | 5 | 23 | 3 | 28 | 4 | 3 | 3 | 5 |
| Bradyrhizobium sp. (A. afraspera) | 43 | 913 | 27 | 15 | 24 | 7 | 14 | 24 | 26 | 16 |
| Bradyrhizobium sp. (A. sensitiva) | 65 | 1,006 | 28 | 25 | 27 | 30 | 12 | 28 | 30 | 15 |
| Bradyrhizobium sp. (A. sensitiva) | 68 | 934 | 26 | 12 | 21 | 23 | 13 | 21 | 23 | 6 |
| Bradyrhizobium sp. (F. albida) | 69 | 910 | 25 | 9 | 7 | 17 | 8 | 2 | 29 | 13 |
An rDNA IGS type corresponds to one combination of restriction patterns obtained with eight restriction enzymes.
The size of the PCR product corresponds to the mean size of the undigested PCR product estimated by summing the sizes of the restriction fragments. Results of Bradyrhizobium strains from small legumes are in bold.
A restriction pattern type refers to the pattern obtained for one strain with one restriction enzyme.
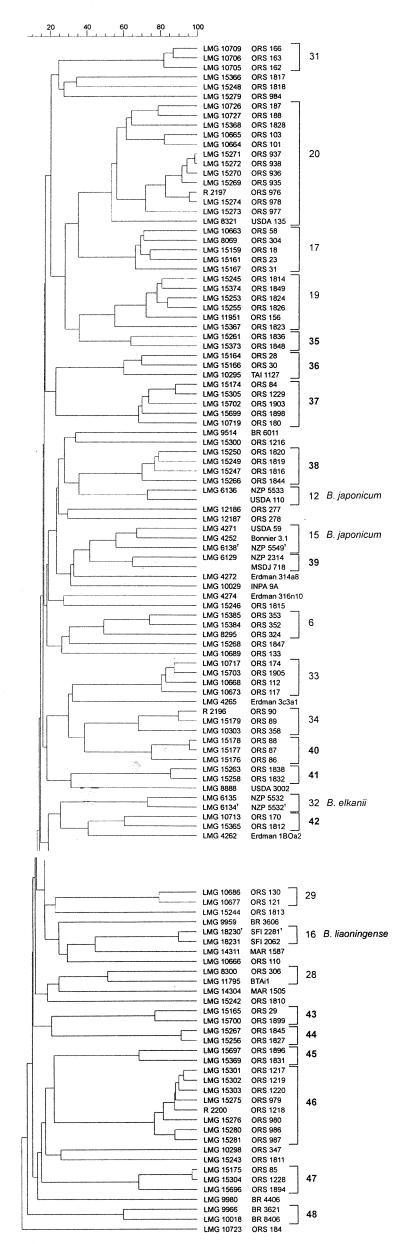
A dendrogram (Fig. 2) was constructed based on the UPGMA algorithm by analyzing the similarity between the restriction fragments with the software Gel Compar, version 4.2 (35).
FIG. 2.
Dendrogram (UPGMA) obtained by numerical analysis comparison of the normalized and combined IGS patterns by PCR-RFLP analysis. Clusters of strains were delineated above 70% similarity.
At a correlation coefficient of about 70%, the different IGS rDNA types formed 16 clusters (I to XVI), 6 of which together contained the majority of the strains studied (clusters I, V, VI, VIII, XI, and XIII). Fifteen strains occupied separate positions, namely eight Senegalese strains from small legumes (ORS 1811, ORS 1847, ORS 1810, ORS 1815, ORS 1817, ORS 1818, ORS 1848, and ORS 984), strain LMG 8888 (from Brazil), Bradyrhizobium japonicum strain LMG 4274, Bradyrhizobium sp. (Lupinus luteus) strain MSDJ 718, two photosynthetic Bradyrhizobium sp. (Aeschynomene sensitiva) strains (ORS 277 and ORS 278), Bradyrhizobium sp. (Aeschynomene afraspera) strain ORS 347, and Bradyrhizobium sp. (Faidherbia albida) strain ORS 184.
The three reference strains of Bradyrhizobium elkanii grouped in cluster XI, together with two strains from Brazil, BR 3606 and BR 3621, the latter having a 16S rDNA sequence close to that of Bradyrhizobium elkanii (3, 10). Cluster XI also included Bradyrhizobium sp. (F. albida) strain ORS 187 and seven strains from small legumes.
The two Bradyrhizobium liaoningense reference strains formed cluster IX.
Except for LMG 4274 (separate position, see above), B. japonicum reference strains were found in three clusters, namely II, V, and VII. Clusters II and VII consisted of only B. japonicum strains, the type strain (LMG 6138T, hybridization group I [13]) being in cluster II, and USDA 110 (hybridization group Ia [13]) being in cluster VII. Cluster V included B. japonicum strain LMG 8321, three Bradyrhizobium sp. (F. albida) strains, and eight strains from small legumes.
Clusters X and XIV consisted of only Bradyrhizobium sp. (F. albida) strains. Clusters XV and XVI consisted of only photosynthetic Bradyrhizobium sp. (Aeschynomene) strains. Of the two nonphotosynthetic strains from Aeschynomene included in the study, one occupied a separate position in the dendrogram (ORS 347, see above) and the other (ORS 304) was grouped in cluster VIII together with strains from small legumes. Clusters I, III, IV, and VI consisted only of strains from small legumes. Half of the strains of cluster I originated from south Senegal and half were from three Indigofera species in central Senegal. Clusters III and IV consisted of strains isolated from south Senegal, while all strains of cluster VI were isolated from north Senegal. Other strains from small legumes were found in clusters V (see above), VIII, XI (with B. elkanii, see above), XII, and XIII [half Bradyrhizobium sp. (F. albida) strains and half Bradyrhizobium sp. (Tephrosia purpurea) strains].
Most of the strains forming each of the clusters V and XI are strains from small legumes of the same geographical origin, north Senegal and south Senegal, respectively.
Consequently, there is some correlation between the clustering of the PCR-RFLP IGS rDNA patterns and the geographic origin of the strains.
In total, no obvious relationship was apparent between the PCR-RFLP IGS rDNA clustering and the host-plant origin or host range (9) of the strains.
(ii) Size of IGS PCR products.
We estimated the sizes of IGS PCR products by summing the sizes of the restriction fragments. All strains produced a single PCR product ranging from 780 to 1,038 bp, depending on the strain (Table 2). The differences observed in the size of the PCR products could be in part explained by the presence of several tRNA genes varying in number and type in the IGS regions (16, 22, 23).
PCR products from the strains of B. japonicum were quite diverse in size. B. japonicum strains USDA 110 and LMG 6136 (cluster VII) each produced an amplification product of around 900 bp, which is in agreement with a report of Laguerre et al. (22). B. japonicum strains LMG 4252, LMG 6138T, LMG 4271 (cluster II), and LMG 8321 (cluster V) produced single amplification products of 821 to 825 bp. B. japonicum LMG 4274 (Fig. 1, separate position in the dendrogram) produced a PCR product with an intermediate size of 840 bp.
The three B. elkanii strains (cluster XI) studied produced PCR products of almost identical size (860 to 864 bp).
The two strains of B. liaoningense (cluster IX) and one strain from a small legume, LMG 15243 (with a separate pattern), showed the smallest PCR product (780 bp).
PCR products of Aeschynomene photosynthetic strains were the largest, and sizes ranged between 934 and 1,038 bp. The two nonphotosynthetic strains of Aeschynomene included in the study yielded PCR products of 913 and 950 bp.
The sizes of the PCR products for the strains of F. albida were also diverse, ranging from 800 to 910 bp, and only four of the strains yielded PCR products of the same size.
The strains from small legumes produced single bands ranging from 780 to 923 bp. In general, the strains from small legumes exhibiting PCR products of approximately the same size grouped in the same cluster; exceptions are strains of cluster VIII (Fig. 2) exhibiting PCR products of heterogeneous sizes (842 to 920 bp).
AFLP analysis. (i) Optimization of the AFLP technique for Bradyrhizobium strains.
Several enzyme combinations were tested to determine the most suitable one, i.e., one producing a large number of fragments (30 to 50) of many different lengths resulting in an evenly distributed banding pattern. For Bradyrhizobium organisms which have a high GC percentage in their genomes, the combination of TaqI (T/CGA) and ApaI (GGGCC/C) proved the most useful.
In the same way, four combinations of primers with different selective bases were tested, and primers 5′-GACTGCGTACAGGCCCG-3′ and 5′-GATGAGTCCTGACCGAA-3′ were retained (characters in bold were the selective bases).
(ii) Numerical analysis of Bradyrhizobium AFLP patterns.
In our AFLP study, we included 63 strains from small legumes, 14 reference strains representative for B. japonicum, B. elkanii, and Bradyrhizobium liaoningense, 18 Bradyrhizobium sp. (F. albida) and 10 Bradyrhizobium sp. (Aeschynomene) strains representative for AFLP groups 6, 16, 17, 19, 20, 28, 29, 31, 33, and 34 described elsewhere (40), and 13 other Bradyrhizobium sp. strains from various host plants (10, 21, 26, 27, 34).
As 13 of the clusters corresponded to previous AFLP groups 6, 12, 15, 16, 17, 19, 20, 28, 29, 31, 32, 33, and 34 (40), we named them accordingly and we numbered our new clusters from 35 to 48. Similarities between AFLP patterns were calculated using the Dice coefficient, and the strains were then grouped by UPGMA cluster analysis (Fig. 3).
FIG. 3.
Dendrogram obtained by UPGMA analysis of AFLP patterns of Senegalese isolates, some representative strains from Brazil, from F. albida, and from Aeschynomene, and the reference strains of Bradyrhizobium. The clusters were delineated at above 50% similarity. The clusters 6, 12, 15, 16, 17, 19, 20, 28, 29, 31, 32, 33, and 34 correspond to previously described AFLP clusters (42). The new groups identified in this study (35 to 48) are in bold.
Considerable profile heterogeneity between strains was revealed, and at a similarity coefficient of 50% (which is commonly used to delineate AFLP clusters in other bacterial groups [14, 15] and in Bradyrhizobium [40]), 27 groups could be distinguished.
B. japonicum reference strains were found in different clusters. Five B. japonicum strains formed groups 12 (including USDA 110) and 15 (including the type strain, LMG 6138). B. japonicum strains LMG 4262, LMG 4265, and LMG 4272 had separate positions, and strain LMG 8321 grouped in cluster 20, which mainly consisted of new isolates from Rynchosia minima and Indigofera senegalensis and of three Bradyrhizobium sp. (F. albida) strains.
The two strains of B. elkanii formed cluster 32. The two B. liaoningense strains formed cluster 16.
Each of the 13 Bradyrhizobium sp. strains from various host plants (10, 21, 26, 27, 34) occupied separate positions in the dendrogram, except for the two of them forming cluster 48.
Of strains from small legumes, 19 grouped in previously described groups 17, 19, 20, 33, and 34 (40), 9 occupied separate positions, and 35 formed new clusters 35, 36, 37, 38, 40, 41, 42, 43, 44, 45, 46, and 47. Clusters 37 and 42 contained Bradyrhizobium sp. (F. albida) strains LMG 10719 and LMG 10713, which were previously described as separate (40). Cluster 36 consisted of two Senegalese strains and one Bradyrhizobium sp. (Cajanus cajan) strain.
In general, AFLP clusters consisted of strains isolated either from north Senegal or from south Senegal.
Except for one strain, all the strains from small legumes grouping in cluster 20 were isolated from the same place in north Senegal. The strains forming cluster 40 were isolated from the same host plant and had the same geographic origin.
So, a relationship may exist between AFLP grouping and the geographic origin of the strains.
On the other hand, there is no evident relationship between the plant origin or nodulation host range (9) of the strains and AFLP grouping except for cluster 46, which consisted of strains exclusively isolated from the same host plant.
DISCUSSION
For Bradyrhizobium, many groups have been identified during recent years, but their taxonomic status has remained unclear due to the inconsistency of the results obtained by different taxonomic methods. There is a need to use several methods, preferably genotypic ones, to draw more reliable taxonomic conclusions.
In a previous report, we isolated and performed initial characterization of 71 nodule isolates from small legumes in Senegal by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of total proteins and by 16S ARDRA and compared them to a number of Bradyrhizobium reference strains. We concluded that these Bradyrhizobium strains were diverse and formed seven phylogenetic groups. Here, we continued the study of these strains by two genomic techniques, PCR-RFLP analysis of the IGS region between 16S and 23S rRNA genes and AFLP analysis. We focused on the applicability and comparison of the taxonomic resolution level of these two techniques to characterize Bradyrhizobium strains and help elucidate taxonomic problems encountered in this genus. Both IGS PCR-RFLP and AFLP analyses confirmed that Bradyrhizobium strains from small legumes are diverse.
By IGS PCR-RFLP using eight restriction enzymes, the 104 strains studied produced 70 types of combined restriction profiles, forming 16 groups. By AFLP analysis, the strains formed 27 groups. Table 3 shows comparative results obtained with essentially the same strains by the two techniques IGS PCR-RFLP and AFLP analyses and also by previous 16S ARDRA (9). There is very good agreement between results from the three techniques, except for the two Bradyrhizobium sp. (F. albida) strains ORS 187 and ORS 110. The 16S ARDRA and AFLP results for ORS 187 are consistent, but not with the IGS results; the 16S ARDRA and IGS results for ORS 110 do not match. In most cases, each AFLP cluster contained strains showing the same or very similar rDNA IGS types and belonging to one IGS PCR-RFLP cluster.
TABLE 3.
Comparison of results from 16S ARDRA and AFLP and PCR-RFLP analyses of 16S-23S rDNA IGS region
| ARDRA groupsa | IGS rDNA PCR-RFLP cluster | IGS rDNA PCR-RFLP type(s) | AFLP cluster(s) |
|---|---|---|---|
| A, B, E, F | I | 2 | 33 |
| 3 | 47 | ||
| 4, 5 | 38 | ||
| 6, 7, 9 | 37 | ||
| 8 | 43 | ||
| II | 10, 11 | 15 | |
| IV | 18, 19 | 45 | |
| V | 25, 26, 27, 28, 29, 30, 31, 32 | 20 | |
| VII | 36, 37 | 12 | |
| VIII | 38 | NDb | |
| 39, 41 | 17 | ||
| 40 | 41 | ||
| IX | 44 | 16 | |
| XI | 53 | 20 | |
| XIII | 58 | Sepc | |
| XV | 63, 64 | 28 | |
| XVI | 66, 67 | 6 | |
| Sep | 12, 14, 21, 22 | Sep | |
| 17 | 39 | ||
| 20, 23 | Sep | ||
| C, G | VI | 33, 34, 35 | 46 |
| X | 45 | 31 | |
| XI | 46 | 48 | |
| 47 | Sep | ||
| 48 | 19, 35 | ||
| 50, 51 | 32 | ||
| 52 | Sep | ||
| XII | 54, 55 | 42 | |
| XIII | 57 | 34, 40, Sep | |
| 60 | 33 | ||
| XIV | 61, 62 | 29 | |
| Sep | 21, 22, 70 | Sep | |
| 56 | 35 | ||
| Sep | ND | ND | Sep |
| I | 1 | 36 | |
| ND | I | 1 | 36 |
| III | 13 | 44 | |
| V | 24 | 20 | |
| VIII | 42 | 17 | |
| XI | 49d | ND | |
| XIII | 58, 59 | 33 | |
| Sep | 15, 16, 43, 65, 68, 69 | Sep |
Group designations are according to those of Doignon-Bourcier et al. (9).
ND, not determined.
Sep, separate.
The IGS PCR-RFLP type 49 corresponds to B. elkanii reference strain USDA 61.
The strains belonging to the 16S ARDRA groups A, B, E, and F (B. japonicum lineage) belonged to 43 IGS PCR-RFLP types (corresponding to 12 main IGS PCR-RFLP clusters) and to 17 AFLP clusters. Interestingly, strains of 16S ARDRA group A, exclusively consisting of photosynthetic strains, were also separate from nonphotosynthetic strains in AFLP and IGS PCR-RFLP groups. The strains belonging to 16S ARDRA groups C and G (B. elkanii lineage) belonged to 20 IGS PCR-RFLP types (corresponding to six IGS PCR-RFLP main clusters) and 10 AFLP clusters.
This confirms, as expected from literature dealing with other bacterial groups, that IGS PCR-RFLP (4, 22) and AFLP (15) analyses are more discriminative than 16S ARDRA for Bradyrhizobium and do almost differentiate at the strain level. Moreover, it has been reported that the AFLP method was more efficient for assessing intrapathovar diversity of the genus Pseudomonas than the rapid amplified polymorphic DNA method (7).
For screening purposes, fine discriminating genotypic techniques such as IGS PCR-RFLP and AFLP are recommended; IGS PCR-RFLP has some advantages because the experimental protocol is sure, simple, and similar to that used for other studies on the 16S rDNA gene and is less laborious than the AFLP technique. Our results agreed with the study of Leblond-Bourget et al. (23) on Bifidobacterium species, showing that IGS rDNA regions are useful for rapid identification or intraspecific phylogenetic studies, while 16S rRNA sequences are a good tool to infer interspecific links (11).
As expected from the literature (3, 9, 13, 19, 30, 40, 41, 45), our results show some heterogeneity among B. japonicum reference strains. Four of the B. japonicum reference strains included, LMG 4262, LMG 4265, LMG 4272, and LMG 4274, were of different plant origins, Albizzia, Ulex, Pueroria, and Vigna, respectively. They all occupied separate positions by both IGS PCR-RFLP and AFLP analyses. On the contrary, a majority of the B. japonicum reference strains originating from Glycine max clustered in two main groups, IGS II/AFLP 15 and IGS VII/AFLP 12. One exception is B. japonicum strain LMG 8321 grouping together with isolates from small legumes and from F. albida in IGS V/AFLP 20. By the two techniques, and also by 16S ARDRA (9), B. japonicum strain USDA 110 grouped separately from the B. japonicum type strain LMG 6138. This result corroborated other studies based on DNA:DNA hybridizations (13) and 16S rDNA analysis (3, 37).
By the three techniques, the B. elkanii strains grouped together; moreover, by AFLP analysis, they formed a separate group.
By ARDRA, both B. liaoningense strains were found in the same group as B. japonicum type strain LMG 6138; here, they were distinct from B. japonicum and formed a separate group by both IGS PCR-RFLP and AFLP analyses.
Of the eight restriction enzymes used for IGS PCR-RFLP in our study, HinfI and RsaI were the least discriminative. Except for three strains, LMG 15268, LMG 8888, and LMG 4274, all the strains could be differentiated by using the combination of the IGS PCR-RFLP patterns obtained with two enzymes, NdeII and MspI.
No clear relationship could be evidenced between plant origin, host range (9), and the grouping of strains by AFLP and IGS PCR-RFLP analyses. On the contrary, we found a certain degree of relationship between AFLP and IGS PCR-RFLP groups and the geographical origins of the strains, especially between north and south Senegal, corresponding to different ecological regions. Senegal is a constrasting country, from its northern part being in the sahelian zone (dry steppe receiving 100 to 300 mm rainfall per year) to the southern part being in the guineo-soudanian zone (deciduous forest receiving 800 to 1,200 mm of rainfall per year). In general, groups mainly consisted of isolates either from north Senegal (IGS groups V, VI, XIII, and XIV corresponding to AFLP groups 20, 29, 33, 34, 40, and 46) or south or central Senegal (IGS groups I, III, IV, X, XI, and XII corresponding to AFLP groups 19, 31, 35, 36, 37, 38, 42, 43, 44, 45, and 47). However, this is not a general rule since some exceptions exist, like IGS group VIII and AFLP group 33 consisting of strains originating from everywhere in Senegal.
Here, we extended the screening of our Bradyrhizobium strain collection by AFLP analysis. The AFLP technique has been described as having a similar discriminating power as DNA:DNA hybridizations, making it a potentially useful taxonomic tool for species delineation for a number of different bacterial groups (7, 14, 15). This result is not unexpected since this method studies the whole bacterial genome, resulting in a profile with many bands (15). DNA:DNA hybridizations remain required to fully evaluate the discriminative power of the technique in the case of Bradyrhizobium.
ACKNOWLEDGMENTS
This work was supported by the BRG (Bureau des Resources Génétîques) and by the Commission of the European Communities (STD3 program, contracts TS2 0169-F, TS3*CT920047, and TS3*CT93-0232 [DG12 HSMU], and BRIDGE program, contracts BIOT-CT91-0263 and BIOT-CT91-0294).
F.D.-B. is indebted to the French Ministry of Education for a Ph.D. research grant. M.G. is indebted to the Fund for Scientific Research-Flanders (Belgium) for research and personnel grants. A.W. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral research fellow.
REFERENCES
- 1.Alazard D. Stem and root nodulation in Aeschynomene spp. Appl Environ Microbiol. 1985;50:732–734. doi: 10.1128/aem.50.3.732-734.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alazard D. La nodulation caulinaire dans le genre Aeschynomene. Ph.D. thesis. Lyon, France: University Claude Bernard-Lyon I; 1991. [Google Scholar]
- 3.Barrera L L, Trujillo M E, Goodfellow M, Garcia F J, Hernandez-Lucas I, Davila G, van Berkum P, Martinez-Romero E. Biodiversity of bradyrhizobia nodulating Lupinus spp. Int J Syst Bacteriol. 1997;47:1086–1091. doi: 10.1099/00207713-47-4-1086. [DOI] [PubMed] [Google Scholar]
- 4.Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16s/23s ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- 5.Blears M J, de Grandis S A, Trevors H L, Trevors J T. Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications. J Ind Microbiol Biotechnol. 1998;21:99–114. [Google Scholar]
- 6.Brunel B, Rome S, Ziani R, Cleyet-Marel J C. Comparison of nucleotide diversity and symbiotic properties of Rhizobium meliloti populations from annual Medicago species. FEMS Microbiol Ecol. 1996;19:71–82. [Google Scholar]
- 7.Clerc A, Manceau C, Nesme X. Comparison of randomly amplified polymorphic DNA with amplified fragment length polymorphism to assess genetic diversity and genetic relatedness within genospecies III of Pseudomonas syringae. Appl Environ Microbiol. 1998;64:1180–1187. doi: 10.1128/aem.64.4.1180-1187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lajudie P, Laurent-Fulele E, Willems A, Tork U, Coopman R, Collins M D, Kersters K, Dreyfus B L, Gillis M. Description of Allorhizobium undicola gen. nov. sp. nov. for nitrogen-fixing bacteria efficiently nodulating Neptunia natans in Senegal. Int J Syst Bacteriol. 1998;48:1277–1290. doi: 10.1099/00207713-48-4-1277. [DOI] [PubMed] [Google Scholar]
- 9.Doignon-Bourcier F, Sy A, Willems A, Torck U, Dreyfus B, Gillis M, de Lajudie P. Diversity of bradyrhizobia from 27 tropical leguminosae species native of Senegal. Syst Appl Microbiol. 1999;22:647–661. doi: 10.1016/S0723-2020(99)80018-6. [DOI] [PubMed] [Google Scholar]
- 10.Dupuy N, Willems A, Pot B, Dewettinck D, Vandenbruaene I, Maestrojuan G, Dreyfus B, Kersters K, Collins M D, Gillis M. Phenotypic and genotypic characterization of bradyrhizobia nodulating the leguminous tree Acacia albida. Int J Syst Bacteriol. 1994;44:461–473. doi: 10.1099/00207713-44-3-461. [DOI] [PubMed] [Google Scholar]
- 11.Fox G E, Wisotzeky J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 12.Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods. 1996;26:247–259. [Google Scholar]
- 13.Hollis A B, Kloos W E, Elkan G H. DNA:DNA hybridization studies of Rhizobium japonicum and related Rhizobiaceae. J Gen Microbiol. 1981;123:215–222. [Google Scholar]
- 14.Huys G, Kersters I, Coopman R, Janssen P, Kersters K. Genotypic diversity among Aeromonas isolates recovered from drinking water production plants as revealed by AFLP™ analysis. Syst Appl Microbiol. 1996;19:428–435. [Google Scholar]
- 15.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 16.Jensen M A, Webster J A, Strauss N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan D C. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow growing root nodule bacteria from leguminous plants. Int J Syst Bacteriol. 1982;32:136–139. [Google Scholar]
- 18.Khbaya B, Neyra M, Normand P, Zerhari K, Filali-Maltouf A. Genetic diversity and phylogeny of rhizobia that nodulate Acacia spp. in Morocco assessed by analysis of rRNA genes. Appl Environ Microbiol. 1998;64:4912–4917. doi: 10.1128/aem.64.12.4912-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuykendall L M, Saxena B, Devine T E, Udell S E. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol. 1992;38:501–503. [Google Scholar]
- 20.Ladha J K, So R B. Numerical taxonomy of photosynthetic rhizobia nodulating Aeschynomene species. Int J Syst Bacteriol. 1994;44:62–73. [Google Scholar]
- 21.Laguerre G, Allard M R, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laguerre G, Mavingui P, Allard M R, Charnay M P, Louvrier P, Mazurier S I, Rigottier-Gois L, Amarger N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol. 1996;46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 25.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol. 1999;65:3084–3094. doi: 10.1128/aem.65.7.3084-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreira F, Gillis M, Pot B, Kersters K, Franco A A. Characterization of rhizobia from different divergence groups of tropical Leguminosae by comparative polyacrylamide gel electrophoresis of their total proteins. Syst Appl Microbiol. 1993;16:135–146. [Google Scholar]
- 27.Moreira F M S, Haukka K, Young J P W. Biodiversity of rhizobia isolated from a wide range of forest legumes in Brazil. Mol Ecol. 1998;7:889–895. doi: 10.1046/j.1365-294x.1998.00411.x. [DOI] [PubMed] [Google Scholar]
- 28.Navarro E, Simonet P, Normand P, Bardin R. Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Arch Microbiol. 1992;157:107–115. doi: 10.1007/BF00245277. [DOI] [PubMed] [Google Scholar]
- 29.Ponsonnet C, Nesme X. Identification of Agrobacterium strains by PCR-RFLP analysis of pTi and chromosomal regions. Arch Microbiol. 1994;161:300–309. doi: 10.1007/BF00303584. [DOI] [PubMed] [Google Scholar]
- 30.Scholla H M, Moorefield J A, Elkan H E. DNA homology between species of the rhizobia. Syst Appl Microbiol. 1990;13:288–294. [Google Scholar]
- 31.Selenska-Pobell S, Evguenieva-Hackenberg E, Radeva G, Squartini A. Characterization of Rhizobium “hedysari” by RFLP analysis of PCR amplified rDNA and by genomic PCR fingerprinting. J Appl Bacteriol. 1996;80:517–528. doi: 10.1111/j.1365-2672.1996.tb03251.x. [DOI] [PubMed] [Google Scholar]
- 32.So R B, Ladha J K, Young J P W. Photosynthetic symbionts of Aeschynomene spp. form a cluster with bradyrhizobia on the basis of fatty acid and rRNA analyses. Int J Syst Bacteriol. 1994;44:392–403. doi: 10.1099/00207713-44-3-392. [DOI] [PubMed] [Google Scholar]
- 33.Terefework Z, Nick G, Suomalainen S, Paulin L, Lindström K. Phylogeny of Rhizobium galegae with respect to other rhizobia and agrobacteria. Int J Syst Bacteriol. 1998;48:349–356. doi: 10.1099/00207713-48-2-349. [DOI] [PubMed] [Google Scholar]
- 34.Van Rossum D, Schuurmans F P, Gillis M, Muyotcha A, Van Verseveld H W, Stouthamer A H, Boogerd F C. Genetic and phenotypic analyses of Bradyrhizobium strains nodulating Peanut (Arachis hypogaea L.) roots. Appl Environ Microbiol. 1995;61:1599–1609. doi: 10.1128/aem.61.4.1599-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 36.Vincent J M. A manual for the practical study of root nodule bacteria. International Biological Programme handbook no. 15. Oxford, United Kingdom: Blackwell Scientific Publ. Ltd.; 1970. [Google Scholar]
- 37.Vinuesa P, Rademaker J L W, de Bruijn F J, Werner D. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol. 1998;64:2096–2104. doi: 10.1128/aem.64.6.2096-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willems A, Collins M D. Phylogenetic analysis of rhizobia and agrobacteria based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1993;45:706–711. doi: 10.1099/00207713-43-2-305. [DOI] [PubMed] [Google Scholar]
- 40.Willems A, Doignon-Bourcier F, Coopman R, Hoste B, de Lajudie P, Gillis M. AFLP fingerprint analysis of Bradyrhizobium strains isolated from Faidherbia albida and Aeschynomene species. Syst Appl Microbiol. 1999;23:137–147. doi: 10.1016/S0723-2020(00)80055-7. [DOI] [PubMed] [Google Scholar]
- 41.Wong F Y K, Stackebrandt E, Ladha J K, Fleischman D E, Date A R, Fuerst J A. Phylogenetic analysis of Bradyrhizobium japonicum and photosynthetic stem-nodulating bacteria from Aeschynomene species grown in separated geographical regions. Appl Environ Microbiol. 1994;60:940–946. doi: 10.1128/aem.60.3.940-946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L M, Ge C, Cui Z, Li J, Fan H. Bradyrhizobium liaoningense sp. nov. isolated from the root nodules of soybean. Int J Syst Bacteriol. 1995;45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 43.Yanagi M, Yamasato K. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett. 1993;107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 44.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 45.Young J P W, Downer H L, Eardly B D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zabeau M, Vos P. Selective restriction fragment amplification: a general method for DNA fingerprinting. Publication no. 0 534 858 A1. Munich, Germany: European Patent Office; 1993. [Google Scholar]