Abstract
The composition of predominant soil bacteria during grassland succession was investigated in the Dutch Drentse A area. Five meadows, taken out of agricultural production at different time points, and one currently fertilized plot represented different stages of grassland succession. Since fertilization and agricultural production were stopped, the six plots showed a constant decline in the levels of nutrients and vegetation changes. The activity of the predominant bacteria was monitored by direct ribosome isolation from soil and temperature gradient gel electrophoresis of reverse transcription (RT)-PCR products generated from bacterial 16S rRNA. The amounts of 16S rRNA of 20 predominant ribosome types per gram of soil were monitored via multiple competitive RT-PCR in six plots at different succession stages. These ribosome types mainly represented Bacillus and members of the Acidobacterium cluster and the α subclass of the class Proteobacteria. The 20 16S rRNA molecules monitored represented approximately half of all bacterial soil rRNA which was estimated by dot blot hybridizations of soil rRNA with the Bacteria probe EUB338. The grasslands showed highly reproducible and specific shifts of bacterial ribosome type composition. The total bacterial ribosome level increased during the first years after agricultural production and fertilization stopped. This correlated with the collapse of the dominant Lolium perenne population and an increased rate of mineralization of organic matter. The results indicate that there is a true correlation between the total activity of the bacterial community in soil and the amount of bacterial ribosomes.
The agricultural overproduction of the last decades in Western Europe resulted in the release of more and more land from agricultural management, and many meadows providing hay for cattle feeding were left as unfertilized grassland. Further management of these areas was aimed at restoring the former species-rich vegetation by nonextensive hay making at moderate frequency. A well-studied model system to monitor this process is the Drentse A grassland research area. In the history of the Drentse A grasslands three different periods can be distinguished based on changing hay-making and fertilization practices (21). Until the 1930s, these grasslands showed a species-rich vegetation and were cut once or twice a year for hay production without application of chemical fertilizers. Then the agricultural use was intensified by increasing the cutting frequency and raising the hay production by applying artificial mineral fertilizers, resulting in domination by high-yield grass species and an overall decrease in species richness. Since the late 1960s part of the land has been released from agricultural production to restore the former species-rich vegetation by taking off hay only once a year without any fertilizer application. Today, different stages of this succession can be observed, since over the years more and more plots were added to the restoration management process. At the time of sampling, the plots selected for this study were still fertilized (1997) or had been taken out of production in 1991, 1990, 1985, 1972, or 1967. From long-term observations of vegetation and soil properties in permanent plots, it is known that these plots indeed represent the temporal successional sequence (3). A constant reduction in the levels of nutrients in the soil was driven by the vegetation, since nutrients like nitrogen, potassium, or phosphate were removed with the biomass during the yearly cutting and taking off of hay. This process resulted in unfavorable conditions for fast-growing species with high nutrient demand, like Lolium perenne. With the decline of this dominant high-yield vegetation, an increasing diversity of plant species could be observed (22). All these effects were documented by studying the successional changes in plant community composition, but the impact of this process on the bacterial community in soil remained widely unexplored. In one study a reduction in culturable ammonium-oxidizing bacteria during grassland succession was observed, and this reduction was correlated to the reduced availability of nitrogen in the soil (31). However, the effect of the grassland succession on the bacterial community in general remained unclear, since a vast majority of soil bacteria must be considered unculturable (2). For instance, in soils all over the world, bacteria of the Acidobacterium cluster are abundantly detected by molecular markers (15) but remain uncultured. Such bacteria can be detected by extracting nucleic acids directly from soil samples and identifying the nucleotide sequences of PCR-amplified 16S rRNA genes (35). The predominant bacteria in Drentse A grasslands were previously identified on the basis of the main bacterial 16S rRNA sequences in the soils (11) and mainly represented Bacillus-related organisms and members of the α subclass of the class Proteobacteria (α-Proteobacteria), the Acidobacterium cluster (15), the order Verrucomicrobiales (36), and the uncultured peat actinobacteria (23). In this study, the shifts of the bacterial community were monitored at the level of major bacterial taxa by quantitative dot blot hybridization (29). Moreover, changes in specific 16S rRNA quantities were determined by using multiple competitive reverse transcription (RT)-PCR (12) to monitor shifts of the predominant ribosome types.
MATERIALS AND METHODS
Field site.
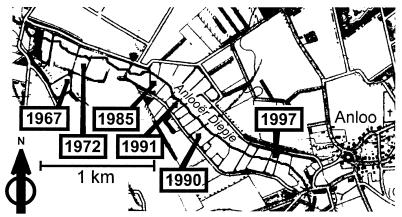
The Drentse A long-term grassland research area in The Netherlands (6°41′E, 53°03′N) is a stretch of grassland meadows on a glacial sand plain along the Anlooër Diepje Brook. The soil is a loamy fine sand of peaty appearance, is normally quite wet, and has a high organic matter content, usually 10 to 15% (31). The climate is Atlantic with a mean annual temperature of 8.5 to 9.0°C and 800 to 850 mm of rain year−1 (6). Soil humidity and pH were estimated as described by Stienstra et al. (31). The water content was quite variable over the fields, ranging from 16 to 37% with a mean of 28% ± 7%. The pH of the soils was between 4.2 and 4.9. Six plots were selected (Fig. 1). For each plot eight mixed samples from sites approximately 10 m apart were taken (approximately 4 by 2 grid). Each mixed sample consisted of five 50-g soil cores that were taken with a drill (depth, 0 to 10 cm) at 1-m distances and were transferred into sterile sample bags. To equalize local variation within a plot, two distant mixed samples were pooled by sieving (2-mm mesh) and mixing single samples (5 g each). Finally, each plot was represented by four pooled samples. Compared to these sieved and pooled samples, undistorted soil crumbs which included the original amount of root material gave the same temperature gradient gel electrophoresis (TGGE) fingerprints reproducibly in space and time for one plot (9). Therefore, preparation of four pooled samples was sufficient for each plot.
FIG. 1.
Map of the Drentse A area. The plots are located near the Anlooër Diepje Brook and are separated by channels and hedges.
Preparation of rRNA.
Several rRNA standards were prepared by rRNA extraction from laboratory cultures by using the following strains: Arthrobacter atrocyaneus DSM 20127, Bacillus benzoevorans DSM 6385, Escherichia coli NM 522, and Sinorhizobium meliloti DSM 1981. All strains were grown in culture as described by the distributors (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; Promega, Madison, Wis.) and used for rRNA extraction as previously described (12). The amount of extracted rRNA was estimated spectrophotometrically (model DU50 spectrophotometer; Beckman, Fullerton, Calif.). A solution of 1 μg of rRNA per ml and subsequent twofold serial dilutions were prepared in glycerol-Tris buffer (50% glycerol, 10 mM Tris-HCl; pH 8.0) and used as standards for quantitative dot blot hybridization or multiple competitive RT-PCR (only E. coli).
Soil rRNA was prepared by ribosome isolation from Drentse A soil samples as previously described (7, 10). Briefly, the bacteria in 1 g of soil (four pooled samples from each of the six plots) were lysed in ribosome buffer by bead beater treatment. Differential centrifugations separated the ribosome suspension from soil particles, humic acid contaminants, and cell debris. After precipitation of the ribosomes by ultracentrifugation, the rRNA was purified by DNase digestion, phenol extractions, and ethanol precipitations. Solutions of rRNA were prepared in glycerol-Tris buffer in a final volume of 100 μl, representing 10 mg (dry weight) of soil μl−1.
Quantitative dot blot hybridization.
Taxon-specific quantification of rRNA was done by dot blot hybridization with soil rRNA and rRNA standards for Bacteria, α-Proteobacteria, and high- and low-G+C-content gram-positive bacteria from pure cultures (see above). The soil rRNA was prepared from 24 pooled soil samples (4 samples per plot). The total amounts of bacterial rRNA per gram of soil have been estimated with the Bacteria-specific EUB338 probe (1) and the E. coli standard rRNA. The mean values were the 100% reference values used to calculate the multiple competitive RT-PCR data, including the group-specific probe signals. Probe ALF1b and the S. meliloti standard rRNA have been applied to quantify rRNA of α-Proteobacteria (16). Probe HGC was specific for rRNA of high-G+C-content gram-positive bacteria (32) like the A. atrocyaneus standard rRNA. The LGC-b probe and the B. benzoevorans standard rRNA have been applied to quantify gram-positive bacteria with low G+C contents (19). Dot blot hybridization experiments were performed on Hybond N+ membranes (Amersham, Slough, England). Using a standard protocol (25), 10 μl of rRNA per dot was applied and immobilized by baking for 30 min at 120°C. The 24 soil rRNA samples represented 100 mg of soil each, and the bacterial rRNA standards were applied in eight different amounts (500, 250, 100, 50, 25, 10, 5, and 2 ng per dot). Oligonucleotide probes were 5′ labeled by using phage T4 polynucleotide kinase (Promega) and 30 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham). For hybridization 4 μl of labeled probes was used. Prehybridization, hybridization, and stringent washing steps were performed as described by Manz et al. (16) or by Meier et al. for the LGC-b probe (19). A detection screen (Molecular Dynamics, Sunnyvale, Calif.) was incubated for 3 h with the hybridized membrane, and the probe signals were detected with a PhosphorImager SF (Molecular Dynamics). Quantification was performed with the image analysis software ImageQuant V.3.3 (Molecular Dynamics). A linear relationship between blot signal strength and rRNA amount was calculated for the standard rRNA by linear regression. With this standard line the soil rRNA signals were transformed into micrograms of rRNA per gram of soil.
Multiple competitive RT-PCR.
The multiple competitive RT-PCR was performed with an rTth DNA polymerase kit (Perkin-Elmer, Norwalk, Conn.). RT reaction mixtures (10 μl) contained 10 mM Tris-HCl (pH 8.3), 90 mM KCl, 1 mM MnCl2, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, 750 nM primer L1401 (20), 2.5 U of rTth DNA polymerase, soil rRNA, and E. coli rRNA standard. A joint master mixture for five RT was prepared, 5 μl of soil rRNA was added, and the mixture was divided and placed in five reaction tubes to ensure that each reaction mixture contained the same amount of soil rRNA (representing 10 mg of soil). The E. coli rRNA standards, representing different amounts (2, 1, 0.5, 0.25, and 0.125 ng of rRNA), were added. After incubation for 15 min at 68°C, 40 μl of a PCR additive containing 10 mM Tris-HCl (pH 8.3), 100 mM KCl, 0.75 mM EGTA, 0.05% Tween 20, 3.75 mM MgCl2, 50 μM dATP, 50 μM dCTP, 50 μM dGTP, 50 μM dTTP, and 190 nM primer U968-GC (20) was added. Amplification was performed in a GeneAmp PCR System 2400 thermocycler (Perkin-Elmer) by using 35 cycles of 94°C for 10 s, 56°C for 20 s, and 68°C for 40 s. The E. coli standards have previously been demonstrated to equally coamplify with the Drentse A sequences by using primers L1401 and U968-GC (12).
The Diagen TGGE system (Diagen, Düsseldorf, Germany) was used for sequence-specific analysis by TGGE (24) after multiple competitive RT-PCR. Electrophoresis took place along a temperature gradient from 37 to 46°C at a fixed current of 9 mA (about 120 V) for 16 h in 1× TA buffer (40 mM Tris-acetate, pH 8.0). The gel (200 by 190 by 0.8 mm) was composed of 6% (wt/vol) acrylamide, 0.1% (wt/vol) bisacrylamide, 8 M urea, 20% (vol/vol) formamide, and 2% (vol/vol) glycerol in 1× TA buffer. Silver-stained gels were scanned with a JX-330 flatbed scanner with a transparency lid (Sharp Electronics, Mahwah, N.J.) and were analyzed with image analysis software (MolecularAnalyst/PC fingerprinting software; Bio-Rad, Hercules, Calif.). Background correction was done with a standard technique of the software by following the rolling-circle principle.
Twenty different bands of the TGGE fingerprints were analyzed. They were known by sequence and were checked for representing only one sequence via V6 hybridization (8). The E. coli bands and the corresponding environmental ribosome types with the most similar signal strength were quantified by estimating the pixel volumes (PV) of the band images. The original rRNA amount (M) of the environmental ribosome types (R) was calculated as follows: MR = PVR × PVE. coli−1 × ME. coli. Since the soil rRNA input per reaction mixture represented 10 mg of an original soil sample, the individual rRNA amounts per gram of soil could be calculated. The absolute rRNA values were transformed to relative quantities to overcome rRNA extraction bias (12), because the ribosome isolation method used was expected not to release all ribosomes from the soil (7).
RESULTS
Quantitative dot blot hybridization.
The Bacteria-specific EUB338 probe was used to determine the amount of bacterial rRNA per gram (dry weight) of soil in six plots representing different stages of succession in the Drentse A grassland soil (Fig. 1). Compared to the 1997 plot, more than twofold higher rRNA yields were obtained from the 1991 plot and also slightly increased yields were obtained from the 1990 and 1985 plots (Table 1). The use of taxon-specific probes also allowed us to identify the most active bacterial groups in Drentse A grassland soils. In comparison to the Bacteria-specific EUB338 probe, Firmicutes with low G+C contents were detected as the dominant major taxon by the LGC-b probe. Approximately half of all bacterial ribosomes in Drentse A grassland soils appeared to be from this taxon. Calculated as a part of the EUB338 signal, the ALF1b probe for α-Proteobacteria and the HGC probe for Firmicutes with high G+C contents each detected approximately 20% of all bacterial ribosomes. The ALF1b, HGC, and LGC-b probe signals showed comparable ratios for all plots. Apparent grassland succession tendencies could not be identified on this taxonomic level.
TABLE 1.
Estimation of the total and taxon-specific relative bacterial rRNA contents of soil
| Plot | Years without fertilization | Concn of Bacteria rRNA (μg g of soil−1)a | Relative rRNA yield (% of EUB338 probe yield)
|
||
|---|---|---|---|---|---|
| α-Proteobacteria | Low-G+C-content Firmicutes | High-G+C-content Firmicutes | |||
| 1997 | 0 | 1.1 (0.3)b | 18.2 (11.4) | 47.1 (14.4) | 13.1 (2.6) |
| 1991 | 6 | 2.5 (0.6) | 19.2 (8.2) | 56.6 (2.9) | 22.4 (3.1) |
| 1990 | 7 | 1.6 (0.5) | 29.4 (6.1) | 40.0 (5.4) | 14.9 (2.8) |
| 1985 | 12 | 1.7 (0.4) | 26.1 (5.2) | 44.3 (11.2) | 20.6 (4.2) |
| 1972 | 25 | 1.1 (0.5) | 20.0 (4.0) | 53.0 (7.5) | 25.4 (9.0) |
| 1967 | 30 | 1.3 (0.2) | 20.8 (5.4) | 50.3 (15.3) | 18.6 (7.7) |
Determined with probe EUB338.
The values in parentheses are standard deviations (n = 4).
Multiple competitive RT-PCR.
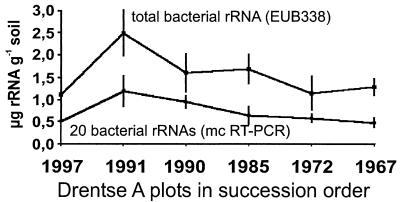
Since no obvious response to grassland succession was observed at the level of major bacterial taxa, the distribution of the main ribosome types was determined sequence specifically by multiple competitive RT-PCR for TGGE fingerprints. The absolute quantities of rRNA of the 20 most prominent sequences per gram of soil were determined (Table 2) and found to represent approximately 50% of all bacterial rRNA, as quantified by the EUB338 probe (Fig. 2). The data indicated that within the first years after fertilization was stopped, the amount of bacterial rRNA increased about twofold and subsequently decreased.
TABLE 2.
Multiple competitive RT-PCR results for the 20 ribosome types in the six plots
| Ribosome type | rRNA concn (ng g of soil−1)
|
|||||
|---|---|---|---|---|---|---|
| 1967 plot | 1972 plot | 1985 plot | 1990 plot | 1991 plot | 1997 plot | |
| DA001 | 99 (12)a | 77 (7) | 86 (20) | 120 (14) | 149 (18) | 66 (8) |
| DA007 | 15 (4) | 11 (4) | 17 (2) | 29 (3) | 28 (3) | 9 (4) |
| DA008 | 1.3 (0.6) | 14 (6) | 14 (3) | 23 (8) | 30 (4) | 10 (2) |
| DA011 | 11 (2) | 7 (2) | 8 (1) | 10 (1) | 19 (4) | 10 (4) |
| DA022 | 24 (6) | 31 (4) | 39 (13) | 37 (10) | 48 (5) | 16 (3) |
| DA032 | 36 (12) | 65 (10) | 78 (8) | 63 (13) | 122 (23) | 56 (17) |
| DA036 | 16 (5) | 61 (12) | 56 (6) | 51 (4) | 96 (18) | 36 (5) |
| DA040 | 5 (1) | 18 (5) | 36 (14) | 42 (15) | 52 (10) | 34 (8) |
| DA054 | 59 (7) | 65 (12) | 64 (16) | 75 (5) | 110 (22) | 47 (11) |
| DA056 | 26 (9) | 34 (8) | 24 (5) | 25 (3) | 33 (3) | 13 (3) |
| DA057 | 27 (9) | 13 (2) | 16 (5) | 24 (7) | 42 (11) | 20 (5) |
| DA066 | 3.8 (0.4) | 3.6 (0.7) | 3.9 (1.2) | 8 (2) | 19 (8) | 24 (7) |
| DA067 | 5 (1) | 16 (2) | 18 (2) | 25 (3) | 54 (16) | 27 (7) |
| DA079 | 66 (13) | 73 (16) | 74 (22) | 122 (10) | 120 (24) | 60 (15) |
| DA101 | 32 (5) | 38 (15) | 38 (8) | 48 (13) | 65 (11) | 26 (4) |
| DA111 | 12 (1) | 14 (4) | 17 (6) | 5 (1) | 22 (7) | 8 (2) |
| DA115 | 19 (3) | 18 (7) | 23 (9) | 48 (5) | 34 (11) | 27 (2) |
| DA116 | 16 (3) | 11 (1) | 16 (5) | 20 (2) | 25 (2) | 9 (2) |
| DA122 | 1.3 (0.2) | 5 (1) | 10 (1) | 15 (3) | 20 (4) | 7 (3) |
| DA136 | 4.4 (1.3) | 7 (2) | 8 (2) | 12 (3) | 27 (10) | 15 (5) |
The values in parentheses are standard deviations (n = 4).
FIG. 2.
Comparison of rRNA quantification by dot blot hybridization and multiple competitive (mc) RT-PCR. The upper graph shows the total bacterial rRNA yield per gram (dry weight) of soil as estimated by dot blot hybridization with the EUB338 probe. The lower graph represents the sums of the single values for the 20 ribosome types as calculated by multiple competitive RT-PCR. The vertical bars indicate the standard deviations (n = 4).
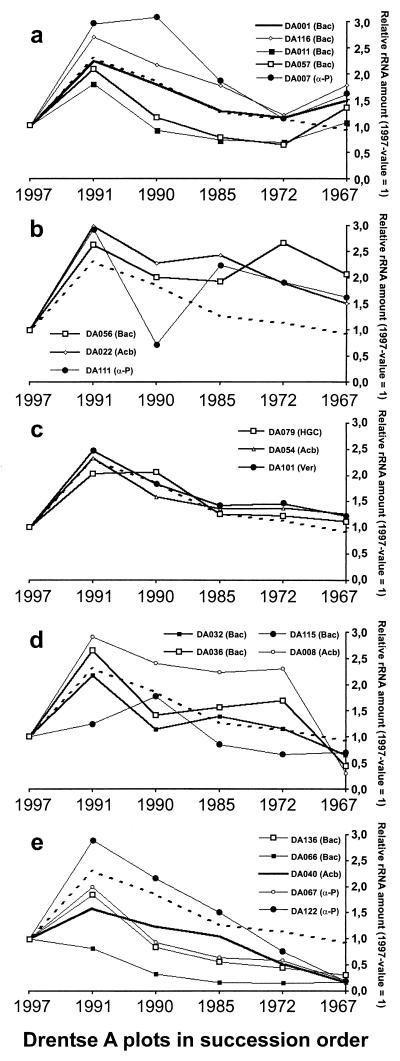
The individual responses of the ribosomes gave a more specific picture (Fig. 3). All individual rRNA amounts were normalized to the 1997 value to highlight the specific tendencies. The particular changes in rRNA level allowed definition of five categories. The first category includes the signals of increased intensity in the TGGE fingerprints of the 1967 plot. These signals represented the ribosomes that increased in the latest stage of succession (Fig. 3a). This positive response was demonstrated for the α-Proteobacterium DA007 and four Bacillus-like 16S rRNA. The second category of ribosomes showed an intermediate positive tendency followed by a steady high level (Fig. 3b). Here we find one representative each of the prominent taxa Bacillus, α-Proteobacteria, and the Acidobacterium cluster. Three of the strongest TGGE bands represented the third group of signals, having similar relative intensities in all fingerprints and not clearly deviating from the general tendency (Fig. 3c). Here we find the representative of the Verrucomicrobiales, peat actinobacteria, and one representative of the Acidobacterium cluster. Ribosomes of the fourth category followed an indistinct tendency, finally ending at a low level in the 1967 plot (Fig. 3d). Here we find again a member of the Acidobacterium cluster and three Bacillus relatives. Finally, the TGGE signals that appeared to be most intense in the 1997 plot (Fig. 4) represented the ribosomes which clearly decreased during grassland succession (Fig. 3e). Here we find two representatives each of the taxa Bacillus and α-Proteobacteria and one representative of the Acidobacterium cluster. All the rRNA levels were drastically decreased in the 1967 plot, while the 1997 plot and the 1967 plot had comparable total rRNA amounts (Fig. 2).
FIG. 3.
Normalized multiple competitive RT-PCR results for the 20 ribosome types in the six plots. All 1997 values were defined as 1, and all other data points were calculated by using these baseline values. The dotted graphs represent the average rRNA levels during grassland succession based on the sum of the values for all 20 ribosome types (Fig. 2). The 16S rRNA clusters are indicated in parentheses: Bac, Bacillus; HGC, high-G+C-content gram-positive bacteria; Acb, Acidobacterium cluster; Ver, Verrucomicrobium cluster; α-P, α-Proteobacteria. (a) Ribosome types with increasing relative rRNA amounts at the later stages of grassland succession; (b) ribosome types with increased relative rRNA amounts at the intermediate stage of grassland succession; (c) rRNA levels with minute deviations from the average; (d) ribosome types with inconstantly decreasing rRNA levels; (e) dramatically declining rRNA levels during grassland succession. The vertical bars indicate the standard deviations (n = 4).
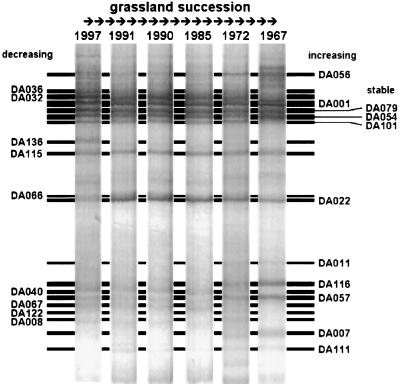
FIG. 4.
Representative TGGE fingerprints of RT-PCR products obtained with primers U968-GC and L1401 for the six plots in successional order from the initial situation in the 1997 plot to the advanced succession stage of the 1967 plot. The ribosome types whose levels decreased during succession are indicated on the left side, and the ribosome types whose levels increased are indicated on the right side. Three ribosome types remained stable without obvious relative changes.
DISCUSSION
Multiple competitive RT-PCR with soil rRNA.
The U968-GC–L1401 primer pair has been demonstrated previously to equally amplify 16S rRNAs from the E. coli standard and the 20 cloned 16S ribosomal DNA (rDNA) amplicons of this study in kinetic PCR for the sequences concerned, in limiting-dilution PCR with soil DNA, and finally, in simulations of (multiple) competitive RT-PCR assays with defined rRNA standards and artificial rRNA mixtures (10). However, it should be kept in mind that some (hitherto unknown) abundant bacterial rRNAs may be neglected by the PCR primers used. Nevertheless, this possible bias could be of only limited extent, since the 20 sequences investigated represented approximately half of all bacterial rRNA extracted from the soil (Fig. 2). The absolute values (Table 2) are used not for comparison of the different sequences but in relation to the total rRNA yield (Fig. 2 and 3). This ratio is more reliable, because with the ribosome isolation protocol used for the recalcitrant soil matrix and the unavoidable purification measures, a considerable loss of yield was inherent (7). As demonstrated previously (10), application of relative quantification procedures clearly improved the reproducibility of the method.
Impact of grassland succession on the soil bacteria.
This study was aimed at providing insight into the effects of grassland succession on the composition of the soil bacterial community in the Drentse A grasslands. The plots in this area represented different stages of temporary grassland succession and served as a suitable model system for long-term vegetation changes in the Drentse A grasslands without fertilization (21). The sampling sites were located along a brook and provided some heterogeneity in soil quality, as demonstrated by the water content. Nevertheless, the quite homogeneous TGGE fingerprints generated from soil 16S rRNA did not indicate heterogeneity of microbial habitats. The bacterial ribosomes directly extracted from soil samples were used for identification of the bacteria present (by 16S rRNA sequences) and as an indicator of bacterial activity (by determination of the number of ribosomes per gram of soil). According to Ward et al. (35), the abundance of ribosomes in the environment may be a species-dependent function of individual cells and their growth rates. When the entire bacterial community is studied, the ribosome abundance reflects the relative contribution of each species to the protein synthesis capacity of the community. As adapted to our approach, we quantified the relative activities of the taxonomic units which represented 20 different 16S rRNAs by their contribution to the protein synthesis capacity of the community but not by their activity per cell. Therefore, the shift of rRNA levels that we detected during grassland succession might have been due to altered proliferation or mortality of cells with unchanged activity or to cell number-independent activity shifts in the populations present. In general, the monitored rRNA levels in Drentse A grasslands approximately doubled a few years after fertilization stopped. This increase in activity probably was correlated to an abrupt change of the vegetation, namely, the collapse of the once dominant L. perenne population. While the L. perenne population faded within a few years after fertilization stopped, the other predominant species, like Yorkshire fog (Holcus lanatus), rough meadow grass (Poa trivialis), and creeping bent (Agrostis stolonifera), could last more than a decade at similar levels before being completely replaced within less than 5 years (22) by increasing populations of keck (Anthriscus sylvestris), common sorrel (Rumex acetosa), and, notably, creeping buttercup (Ranunculus repens). In later succession stages, species like sweet vernal grass (Anthoxanthum odoratum), red fescue (Festuca rubra), and field wood rush (Luzula campestris) appeared. Correlating these vegetation shifts with the changes in the bacterial community is speculative, but a link between the two most dramatic events, the peak of bacterial ribosomes and the disappearance of L. perenne, might be supposed. Decaying plant residues might have increased the nutrient input and supported bacterial activity (27). An increase in the turnover of organic matter was also indicated by earthworm activity. These organisms penetrate soil, transport plant litter under ground, and support bacterial activity in their guts and feces (14). A 1992 study demonstrated that the 1991 plot contained a mean of 308 earthworms per m2, the 1985 plot (which had not been fertilized for 7 years) contained 808 earthworms per m2 and the 1972 plot contained only 233 earthworms per m2 (L. Brussard, G. Tian, R. P. Dick, J. Hassink, A. Stienstra, R. G. M. de Goede, H. Siepel, J. P. Bakker, and H. Olff, poster, Meet. Soil Ecol. Soc., 1993). The same survey also revealed similar peaks for carbon mineralization and microbial biomass. Another study found an increase in nitrogen mineralization in the plot not fertilized for 2 years compared to the plot not fertilized for 7 years (21). The nitrogen mineralization rate increased from 124 to 176 kg ha−1 year−1 and decreased again in older fields. Since all these parameters are linked to bacterial activity, their correlation to the results of the multiple-competitor RT-PCR indicated that there is a dependence between the total activity of bacterial communities in soil and the amount of ribosomes in soil.
Almost all 20 ribosome types exhibited the general tendency of increased ribosome yield in the first years after fertilization was stopped. After the increase in the 1991 plot the rRNA amounts decreased during the subsequent stages of grassland succession. The amounts of some of the 20 ribosome types decreased to approximately 20% of the average for the 1967 plot (Fig. 3e), while the rRNA levels of others increased (Fig. 3a and b), resulting in differences in the TGGE fingerprints for different plots. The five defined categories of response could not be distinguished by the phylogeny of the 16S rRNA and predominant taxa represented. This is in accordance with the results of dot blot hybridization, which indicated that the response to grassland succession is not specific on the level of the major bacterial taxa.
Impact of vegetation on the soil bacteria.
The high spatial reproducibility of TGGE fingerprints for the Drentse A grassland soils was impressive (9) despite the differences in the vegetation of the six plots. Maybe our approach, with resolution on the 16S rRNA level, missed some important community shifts. Identical 16S rRNA sequences might originate from a single strain, from a couple of strains of the same species, or from different, closely related species, but the organisms might exhibit differences in physiology. For instance, the potato brown rot agent Ralstonia solanacearum could be separated from harmless relatives by 23S rRNA but not by 16S rRNA (37), and pathogenic Shigella could not be differentiated from Escherichia by 16S rRNA but could be differentiated by physiology (34). Nevertheless, the homogeneous distribution of identical 16S rRNA sequences at high levels over the large Drentse A area, although not excluding the heterogeneity of physiology, remains to be explained, considering that the microbial community was investigated by using its rRNA, which reflects the activity of the bacteria (33, 35). A direct bacterial response to the vegetation should be expected in the rhizosphere, and grass roots are omnipresent at high densities in the upper layer of grassland soils. It has not been determined which roots were present in the soil cores sampled. Thus, the possibility that grass species prominent on the surface did not contribute equally to the rhizosphere in the upper 10 cm of soil sampled could not be eliminated (but perhaps they contributed equally in deeper layers). Moreover, due to their small size (5 cm in diameter) the soil cores sampled did not necessarily reproducibly represent the plot-specific vegetation. However, the different undisturbed cores yielded almost identical TGGE fingerprints for each plot (9). No known rhizosphere bacteria were detected by 16S rDNA cloning (11). For instance, pseudomonads are well-known rhizosphere bacteria and appear in high numbers in the rhizosphere of L. perenne (17). Nevertheless, rhizosphere γ-Proteobacteria like Pseudomonas appeared to be relatively rare in soils. We are aware of only one case where γ-Proteobacteria were predominant in a rot field containing lots of disposed sugar beets (7). Also, α-Proteobacteria appear at high levels in the rhizosphere. This has been clearly demonstrated for Azospirillum on wheat roots (26) and Rhizobium in the rhizosphere of white clover (Trifolium repens) (17). However, the close relatives of Rhizobium and Pseudomonas, as detected by a culture-independent study of L. perenne and white clover, appeared to be restricted to root-adhering soil particles and especially to the rhizoplane-endorhizosphere fraction (17). Also, white clover is common on Drentse A grasslands, but the prominent Drentse A α-Proteobacteria (DA007, DA067, DA111, DA122) did not show close phylogenetic relationships to known nodule or rhizosphere bacteria. In numerous studies in which no special attention was paid to rhizospheres, abundant soil α-Proteobacteria mostly belonged to uncultured lineages (4, 5, 11, 13, 15, 28; EMBL accession no. AF145805 to AF145880). Therefore, the contribution of the known rhizosphere bacteria to the general bacterial soil community often appears to be surprisingly minute. However, there is one study of grassland soils that detected a couple of relatives of Rhizobium and Bradyrhizobium in a 16S rDNA clone library, perhaps indicating considerable abundance of these groups (18). Nevertheless, it must be remembered that the vast majority of environmental bacteria are not cultivable yet and their physiologies and ecological functions remain completely unknown (30). Although the cultured rhizosphere bacteria appeared to be abundant on roots even in light of culture-independent approaches, there might be more rhizosphere-dependent bacteria belonging to uncultured lineages like the acidobacteria (15).
Conclusions.
Multiple competitive RT-PCR indicated the activity shifts for the predominant soil bacteria during Drentse A grassland succession, while quantitative dot blot hybridization failed to detect differences on a higher taxonomic level. Although the vegetation clearly changed, there was no corresponding drastic reaction of the microbial community. We could quantify reproducible shifts of ribosome levels, but the general composition of the bacterial community remained remarkably stable. Evidence that there is severe competition and major replacement of species, as apparent in the grass vegetation, could not be found.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the European Communities (EC) project “High Resolution Automated Microbial Identification” (EC-HRAMI project BIO2-CT94-3098). The work of A.F. is supported by EC project BIO4-98-0168.
L. Brussaard is especially acknowledged for critically reviewing the manuscript. We also thank the Dutch State Forestry Commission for access to the nature reserve.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of microbial cells without cultivation. Appl Environ Microbiol. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakker J P. Geobotany 14: nature management by grazing and cutting. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1989. [Google Scholar]
- 4.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Goede R G M, Verschoor B C. Nematode fauna of meadows in the Drentse A area, the Netherlands, with different fertilization history. In: de Goede R G M, Bongers T, editors. Nematode communities of northern temperate grassland ecosystems. Landsberg, The Netherlands: Ecomed; 1998. pp. 89–93. [Google Scholar]
- 7.Felske A, Engelen B, Nübel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 9.Felske A, Akkermans A D L. Spatial homogeneity of the most abundant bacterial 16S rRNA molecules in grassland soils. Microb Ecol. 1998;36:31–36. doi: 10.1007/s002489900090. [DOI] [PubMed] [Google Scholar]
- 10.Felske A, Backhaus H, Akkermans A D L. Direct ribosome isolation from soil. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual, supplement 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. p. 1.2.4.1-1.2.4.10. [Google Scholar]
- 11.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felske A, Akkermans A D L, de Vos W M. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl Environ Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soil of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K E. Earthworms: their ecology and relationships with soils and land use. Sydney, Australia: Academic Press; 1985. [Google Scholar]
- 15.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 16.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 17.Marilley L, Hartwig U A, Aragno M. Influence of an elevated atmospheric CO2 content on soil and rhizosphere bacterial communities beneath Lolium perenne and Trifolium repens under field conditions. Microb Ecol. 1999;38:39–49. doi: 10.1007/s002489900155. [DOI] [PubMed] [Google Scholar]
- 18.McCaig A, Glover L A, Prosser J L. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier H, Amann R I, Ludwig W, Schleifer K-H. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 20.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olff H. On the mechanisms of vegetation succession. Ph.D. thesis. Groningen, The Netherlands: State University of Groningen; 1992. [Google Scholar]
- 22.Olff H, Bakker J P. Long-term dynamics of standing crop and species composition after the cessation of fertilizer application to mown grassland. J Appl Ecol. 1991;28:1040–1052. [Google Scholar]
- 23.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the Actinomycete line of descent in different environments and geographic locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 24.Rosenbaum V, Riesner D. Temperature-gradient gel electrophoresis—thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys Chem. 1987;26:235–246. doi: 10.1016/0301-4622(87)80026-1. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Schloter M, Hartmann A. Endophytic and surface colonization of wheat roots (Triticum asetivum) by different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies. Symbiosis. 1998;25:159–179. [Google Scholar]
- 27.Schweizer M, Fear J, Cadisch G. Isotopic (13C) fractionation during plant residue decomposition and its implications for soil organic matter studies. Rapid Commun Mass Spectrom. 1999;13:1284–1290. doi: 10.1002/(SICI)1097-0231(19990715)13:13<1284::AID-RCM578>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 29.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 31.Stienstra A W, Klein Gunnewiek P, Laanbroek H J. Repression of nitrification in soils under climax grassland vegetation. FEMS Microbiol Ecol. 1994;14:45–52. [Google Scholar]
- 32.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wede D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner R. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol. 1994;161:100–106. doi: 10.1007/BF00276469. [DOI] [PubMed] [Google Scholar]
- 34.Wang R F, Cao W W, Cerniglia C E. Phylogenetic analysis and identification of Shigella spp. by molecular probes. Mol Cell Probes. 1997;11:427–432. doi: 10.1006/mcpr.1997.0136. [DOI] [PubMed] [Google Scholar]
- 35.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. In: Marshall K C, editor. Advances in microbial ecology. Vol. 12. New York, N.Y: Plenum Press; 1992. pp. 219–286. [Google Scholar]
- 36.Ward-Rainey N, Rainey F A, Schlesner H, Stackebrandt E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology. 1995;141:3247–3250. [Google Scholar]
- 37.Wullings B A, van Beuningen A R, Janse J D, Akkermans A D L. Detection of Ralstonia solanacearum, which causes brown rot of potato, by fluorescent in situ hybridization with 23S rRNA-targeted probes. Appl Environ Microbiol. 1998;64:4546–4554. doi: 10.1128/aem.64.11.4546-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]