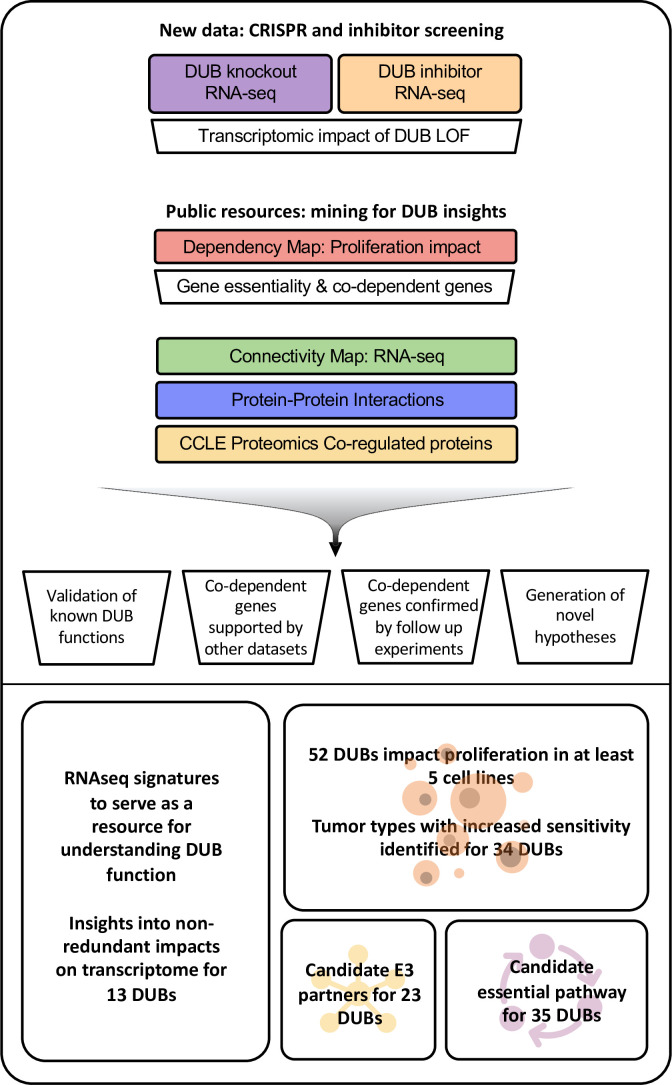
Figure 1. Approach to multi-omics analysis DUBs.
The functional impact of DUB loss was investigated via transcriptomic profiling of cells following CRISPR-Cas9 knockout (purple box); this was compared to signatures for genetic perturbations in the Broad Connectivity Map database of RNA-seq signatures (green). The impact of DUB knockout on cancer cell line proliferation was analyzed using the Broad Dependency Map (red) and compared to other gene knockouts in the dataset; this identified co-dependent genes (sets of genes whose knockout had similar effects on cell proliferation across cell lines). Multiple protein-protein interaction databases and co-expression (correlation in protein abundance across cell lines) in baseline proteomics in Cancer Cell Line Encyclopedia (CCLE; yellow) cell lines were used to provide support for physical or functional interactions among DUBs exhibiting co-dependency. We then used these analyses to explore the impact of DUB knockout on the transcriptome, determine the impact of DUBs on proliferation, and propose E3 ligase interactors and essential functions of DUBs. Acronyms used in figure: knockout (KO), loss of function (LOF), Cancer Cell Line Encyclopedia (CCLE).