Abstract
Wastewater-based epidemiology (WBE) has emerged as a valuable approach for forecasting disease outbreaks in developed countries with a centralized sewage infrastructure. On the other hand, due to the absence of well-defined and systematic sewage networks, WBE is challenging to implement in developing countries like Bangladesh where most people live in rural areas. Identification of appropriate locations for rural Hotspot Based Sampling (HBS) and urban Drain Based Sampling (DBS) are critical to enable WBE based monitoring system. We investigated the best sampling locations from both urban and rural areas in Bangladesh after evaluating the sanitation infrastructure for forecasting COVID-19 prevalence. A total of 168 wastewater samples were collected from 14 districts of Bangladesh during each of the two peak pandemic seasons. RT-qPCR commercial kits were used to target ORF1ab and N genes. The presence of SARS-CoV-2 genetic materials was found in 98% (165/168) and 95% (160/168) wastewater samples in the first and second round sampling, respectively. Although wastewater effluents from both the marketplace and isolation center drains were found with the highest amount of genetic materials according to the mixed model, quantifiable SARS-CoV-2 RNAs were also identified in the other four sampling sites. Hence, wastewater samples of the marketplace in rural areas and isolation centers in urban areas can be considered the appropriate sampling sites to detect contagion hotspots. This is the first complete study to detect SARS-CoV-2 genetic components in wastewater samples collected from rural and urban areas for monitoring the COVID-19 pandemic. The results based on the study revealed a correlation between viral copy numbers in wastewater samples and SARS-CoV-2 positive cases reported by the Directorate General of Health Services (DGHS) as part of the national surveillance program for COVID-19 prevention. The findings of this study will help in setting strategies and guidelines for the selection of appropriate sampling sites, which will facilitate in development of comprehensive wastewater-based epidemiological systems for surveillance of rural and urban areas of low-income countries with inadequate sewage infrastructure.
Keywords: Wastewater-based epidemiology (WBE), Onsite sanitation, SARS-CoV-2 monitoring, National surveillance program, Rural Hotspot based sampling, Urban Drain based sampling, Developing countries
Graphical abstract
1. Introduction
The dynamic nature of SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) infections, with a large number of confirmed cases each day, has rendered individual testing difficult in some situations (Fozouni et al., 2021, Rakib et al., 2021). In this context, wastewater-based epidemiology (WBE) is considered a promising method for population-level disease monitoring (Bivins et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Nemudryi et al., 2020). WBE approach relies on the detection of SARS-CoV-2 genetic materials in wastewater and is widely applied based on the collection of samples from wastewater treatment plants (WWTPs) in countries with centralized sewage networks (Ahmed et al., 2020a; Ahmed et al., 2020b, Ahmed et al., 2020c; Amoah et al., 2022; Arora et al., 2020; Gonzalez et al., 2020; Kumar et al., 2020, 2021a, 2021b; 2022; La Rosa et al., 2020; Sherchan et al., 2020). This approach has not yet been widely applied in countries without centralized sanitation facilities (D'Aoust et al., 2021; Bhattacharya et al., 2021; Haque et al., 2022). Around 90% of sanitation facilities in developing countries are on-site systems and do not fully benefit from current WBE approaches (Sagoe et al., 2019). As the COVID-19 pandemic impacts both urban and rural areas, which rely on onsite sanitation facilities, it has become essential to design a WBE surveillance system for monitoring SARS-CoV-2 in decentralized facilities, particularly in developing countries (Ekumah et al., 2020; Bhattacharya et al., 2021; Vadiati et al., 2022; van der Voorn et al., 2021). A number of previous studies have reported the detection of SARS-CoV-2 genetic materials from non-centralized sewage systems, including river water, airport wastewater, hospital wastewater, market places, and municipal drain wastewater (Haramoto et al., 2020). In Frankfurt, Germany, whole-genome sequencing (WGS) of airport wastewater samples confirmed the arrival of the Omicron variant in November 2021 (Agrawal et al., 2022). The rate of SARS-CoV-2 RNA in hospital wastewater increased similarly to the rising number of hospitalized patients with COVID-19 infection (Acosta et al., 2021). The SARS-CoV-2 RNA genetic materials ORF1ab, N, and S genes were detected in wastewater of fresh markets in Bangkok, Thailand (Thongpradit et al., 2022). COVID-19 genetic materials have been identified in the wastewater drains of the isolation center in Noakhali, Bangladesh (Ahmed et al., 2021; Islam et al., 2021a, 2021b; Jakariya et al., 2021). In addition, the studies by Fongaro et al. (2021) revealed that SARS-CoV-2 genetic materials were absent in the upstream river water, but present in considerable concentrations in the downstream river water with an average of 1.1 × 102 SARS-CoV-2 GC/mL. To overcome the limitations of utilizing the promising WBE system in low-income countries where centralized sanitation structure is absent, hot spots and sewer drain-based WBE surveillance can be the best alternative solution prioritizing prompt proxy measures on infectious and chronic diseases.
COVID-19 continues to be a global challenge as new variants of SARS-CoV-2 emerge and challenge acquired immunity (Haque et al., 2022; Islam et al., 2022). Bangladesh is one of the most populated countries in the world with a population of 165 million (as of 15 April 2022), and population density of 1265 per square kilometer (World meter, 2022). Bangladesh also has been fighting against COVID-19 since the first reported case on March 08, 2020 (IEDCR, 2022; Islam et al., 2021). As of April 13, 2022, the Government of Bangladesh (GoB) has reported 1,952,109 confirmed cases, and 29,124 cases of fatality (IEDCR, 2022). The COVID-19 positivity rate stands at 4.0% with an average case fatality ratio of 1.0% (Reliefweb, 2022). The GoB continues to recommend following COVID-19 regulations such as social distancing, wearing masks in public areas, and maintaining hygiene. It is also laudable that nearly 72% of the total population in Bangladesh has received at least one dose of the COVID-19 vaccine and 49% are vaccinated with the second dose (WHO, 2022). Despite the utmost effort of the regulatory authorities to combat the COVID-19 pandemic, instances of the severity of COVID-19 have been experienced in the rural areas of Bangladesh, due to low coverage of the clinical diagnostic facilities. As an alternative approach, WBE would therefore provide a cost-effective flexible signal as a proxy measure for the prevalence of SARS-CoV-2 at the community level (Bhattacharya et al., 2021; Haque et al., 2022).
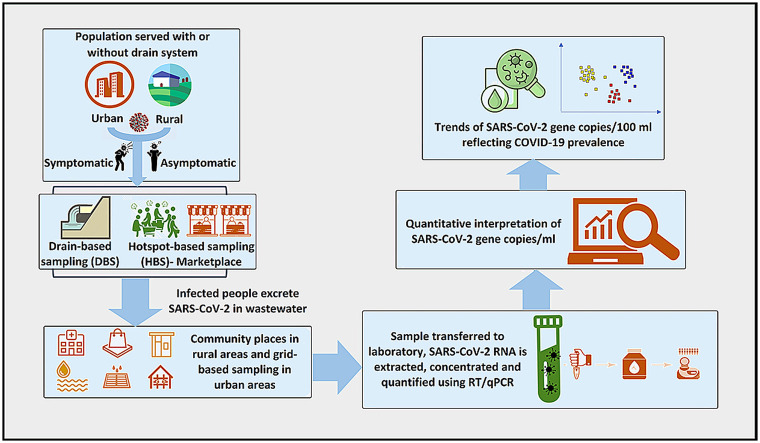
Bangladesh being a developing country primarily relies upon on-site sanitation systems that often lack statutory guidelines for installation and monitoring regulations, causing the wastewater being directly discharged to drain systems, particularly in the urban areas (Hossain et al., 2021). In this regard, the study endeavored to assess sampling sites to detect genetic materials of SARS-CoV-2 in decentralized wastewater systems (Bhattacharya et al., 2021; Haque et al., 2022). The research focused on identifying the sampling sites, which are the best representatives of the community for developing WBE as a tool in areas lacking proper sewage system (Bhattacharya et al., 2021, Vadiati et al., 2022). First, the study developed a drain-based sampling method for urban areas of Bangladesh where only a drain system is available instead of a centralized sewage system. Secondly, the study embarked on a hotspot-based sampling to collect wastewater samples for the rural areas where the drain system is unavailable. The study also proposed suitable wastewater sampling sites for establishing WBE systems in developing countries where proper sanitation facilities are absent.
2. Material and methods
2.1. Selection of sampling sites
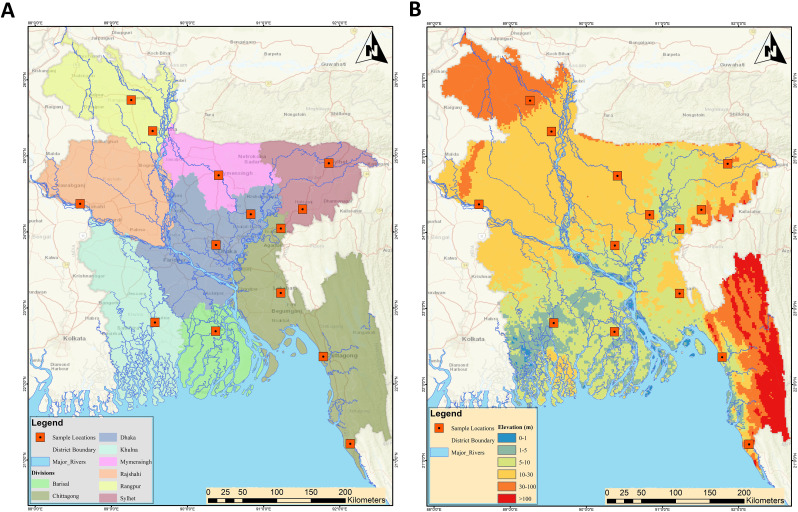
This nationwide WBE surveillance study was carried out in 14 districts covering all eight administrative divisions of Bangladesh. A total of 168 wastewater samples were collected (Fig. 1 ) from urban drains (n = 84) as well as outlets of onsite facilities in rural areas (n = 84) (see Supplementary data; Table ST1 and ST2 for details). We collected repeated samples from the same locations within an interval of two months. The timeline was scheduled as the first round between December 1 to December 5, 2020 whiles the second round was between February 1 to February 5, 2021.
Fig. 1.
Map showing the sampling locations from fourteen districts covering whole Bangladesh rural and urban sites. A) Locations of sampling sites, major rivers, and divisions; B) District boundary and elevation (m).
2.1.1. Drain-based sampling method (DBS)
A drain-based sample (DBS) method was employed for the urban drainage catchment areas including city drain for household wastewater discharges, isolation center of COVID-19 patients, and medical college/hospital drain, (Ahmed et al., 2021).
2.1.2. Hotspot-based sampling (HBS)
Since drainage facilities were not available in rural areas, a hotspot-based sampling (HBS) was carried out at the places with increased transmission risk, more specifically, bus stand/railway stations, community ponds/rivers, and marketplaces were targeted to collect wastewater. We implemented this strategy in all the four study regions, in the northern, southern, central, and northeastern parts of Bangladesh. During sampling, community places such as religious institutions, transportation terminals, community ponds, and canals commonly accessed by the rural population were identified, since the social and cultural traditions of different locations can influence the presence of the genetic markers of the SARS-CoV-2 virus in wastewater.
2.2. Watewater sampling
The wastewater samples were collected from 168 locations in eight divisions capturing the 14 major districts in Bangladesh. The selected districts for sampling were Dhaka, Sylhet, Chittagong, Mymensingh, Rajshahi, Khulna, Barisal, Rangpur, Habigang, Kishorgonj, Brahmanbaria, Gaibandha, and Cox's Bazar, Cumilla. The specific sites were identified to represent the majority of the COVID-19 affected population located in the selected areas for sample collection (see Supplementary data; Figure SF1, SF2 & Table ST1,ST2). Relevant physicochemical and meteorological data were documented using a sample collection form (Supplementary data; Table ST3 & Figure SF3). All wastewater samples were collected at a fixed time in 500 mL autoclaved sampling bottles and carried in the ice-box aseptically. Blank bottles without samples were used to ascertain any contamination during transportation. All the laboratory work and analyses were carried out at the COVID-19 Diagnostic Lab, Department of Microbiology, Noakhali Science and Technology University, (a government laboratory regularly monitored by IEDCR-Institute of Epidemiology, Disease Control and Research and WHO-World Health Organization). Since environmental conditions play an important role in the stability of the SARS-CoV-2 genetic materials, the pH and temperature of samples were measured using a portable pH meter (Milwaukee) and a thermometer (TP-300) and documented in the sample collection forms (Supplementary data; Figure SF3).
2.3. Sample preparation
2.3.1. Concentration process
To remove coarse particles from the samples, we used Whatman filter paper following the concentration technique as described in Ahmed et al. (2021) and Kumar et al. (2020). The wastewater sample (50 mL) was centrifuged at 4500 g for 30 min (Thermo Scientific), followed by filtration of the supernatant using Himedia® 0.22 μm filters. The polyethylene glycol (PEG) technique was used to concentrate each sewage filtrate, where PEG 6000 (80 g/L) and NaCl (17.5 g/L) were mixed in a 25 mL falcon tube and incubated overnight at a shaking speed of 17 °C at 100 rpm. The mixture was centrifuged at 13000 g for 90 min on the following day. After centrifugation, the supernatant was discarded, and the pellet was suspended in 300 μL of RNase-free water.
2.3.2. RNA extraction procedure
A commercially available QIAamp® Viral RNA Mini Kit was used to extract viral RNA in a different laboratory equipped with HEPA environmental air controller within 4–5 h of sample collection. Freshly prepared 560 μL of AVL buffer containing carrier RNA was added to 140 μL concentrated wastewater samples and incubated at room temperature for 10 min followed by addition of 560 μL ethanol (96–100%). 630 μL of the solution was then transferred to the QIAamp Mini column and centrifuged at 8000 rpm for 1 min. Subsequently, the QIAamp Mini column was placed into a clean 2 mL collection tube. Afterwards, 500 μL buffer AW1 was added and centrifuged at 8000 rpm for 1 min. After adding 500 μL buffer AW2, the column was centrifuged at full speed at 14,000 rpm for 3 min. Discarding the old collection tube with the filtrate, the QIAamp Mini column was placed in a new 2 mL collection tube, and centrifuged at full speed for 1 min. Finally, 60 μL buffer AVE was added and incubated at room temperature for 1 min and then centrifuged at 8000 rpm for 1 min to collect the extracted RNA sample. Confirmed COVID-19 positive patient samples (donated by NSTU Diagnostic Lab, GISAID accession ID- EPI_ISL_1626483 to EPI_ISL_16264527) were used as an extraction control. NanoDrop (Thermo Scientific TM Nano Drop 2000 and 2000c, BioRad) was used to determine RNA concentrations, and then stored at −70 °C until further use.
2.4. RT-PCR analysis
Extracted viral RNAs were analyzed without any storage to detect SARS-CoV-2 by RT-qPCR (CFX 96, BioRad) using the RT-PCR kit (Sansure Biotech Inc., China). Technical procedures were carried out as described in the product manual, and the results were interpreted. In brief, we had set the sample layout with RT-PCR protocol covering 45 cycles (Supplementary Table ST4) containing FAM fluorescence select for ORF1ab, ROX for N gene as well as CY5 for human RNase-p gene as an internal control. The standard curve was used to quantify viral copy numbers using the synthetic known copy number of selected genes (2.00E+05 copies/μL) (Supplementary data; Figure SF4).
Before interpretation of RT-PCR results, all test controls (both positive and negative supplied with kit and RNA extracted control) were confirmed, and data from any experiment with failed controls were excluded. We independently validated the Ct value for each sample with another kit (Supplementary data; Figure SF5). These two kits covered two genes of SARS-COV-2, namely ORF1ab and N genes. The Sansure kit uses the ORF1ab, N gene, and RNase-p, while the kit manufactured by BGI uses genes N and ORF1ab. Both the kits used the ORF1ab and N genes, and the results were consistent with the variation of 5% with respect to the presence of the genes (Supplementary data; Figure SF5).
2.5. Statistical analysis
In terms of the abundances of generic components expressed by Ct values, boxplots were generated to compare the data from urban and rural settings. The expression of genetic materials data was fit with a random intercept linear mixed model (LMM) after adjusting drain availability, temperature and regions. This is used to examine overall causes of variation in genetic material properties, i.e., to estimate and interpret variance components (Trabzuni & Thomson, 2014). All clinical data were collected from the dataset of Government COVID-19 positive cases of the Directorate General of Health Services (DGHS) (IEDCR, 2022).
3. Results
3.1. Spatio-temporal distribution of SARS-CoV-2 genetic materials
The spatio-temporal distribution of SARS-CoV-2 genetic materials investigated in the wastewater samples collected from the investigated urban and rural areas are presented in Fig. 2 . During the two different rounds of sampling, a total of 161 samples corresponding to 96% of the sampled wastewaters were found positive for the presence of SARS-CoV-2 genetic materials with the varying number of individual gene markers.
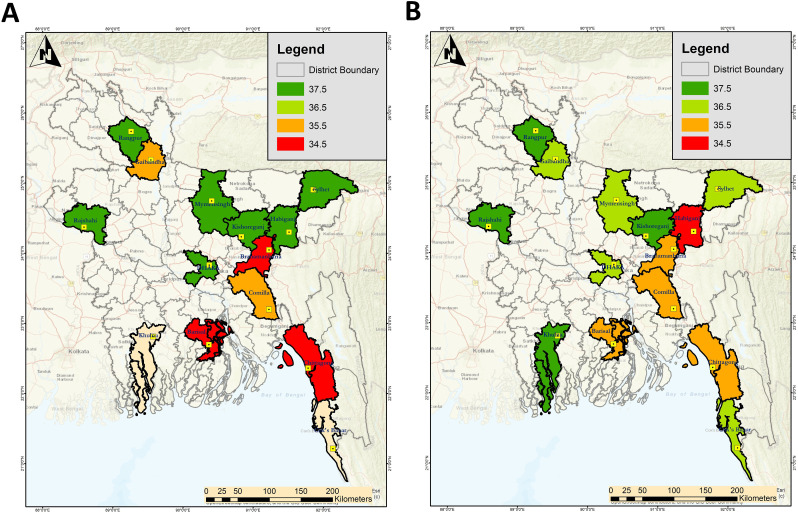
Fig. 2.
Variations in Ct value observed for the wastewater samples collected from different districts of Bangladesh during two sampling phases; A) First-round sampling (December 1–5, 2020); B) Second-round sampling (February 1–5, 2021).
Among the 84 analyzed wastewater samples, 95% of samples were found positive for at least one target of SARS-CoV-2 genes (ORF1ab or N), from 14 districts during the first round of sampling (December 1 to December 5, 2020). The presence of two genes ORF1ab and N genes were found in 21% of samples, the human RNase-p was identified in 12% of samples, and both ORF1ab and human RNase-p were found in 18% of samples (Fig. 3 A). The Ct value noted for the wastewater samples varied within a range between 34.5 and 37.5 (Fig. 2A).
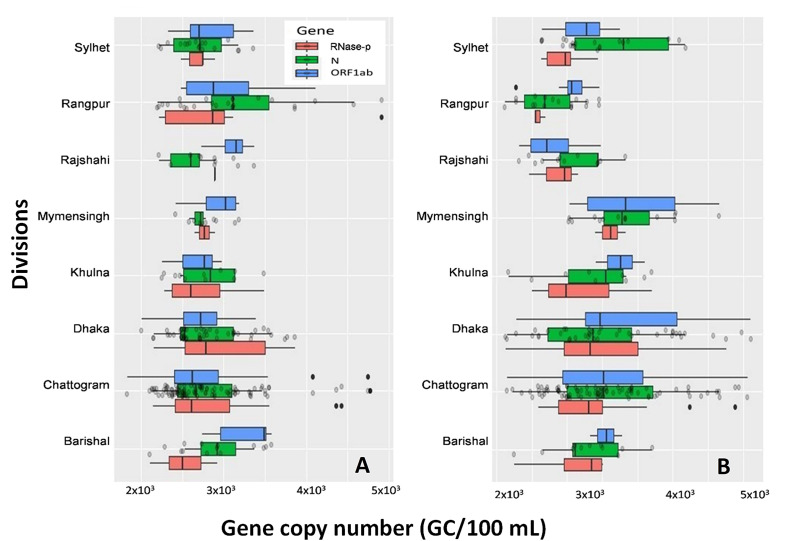
Fig. 3.
Distribution of copy numbers of three SARS-CoV-2 genetic markers in eight divisions. A) First-round sampling from December 1 to December 5, 2020. Chittagong and Rangpur divisions had the lowest range of Ct values for the samples collected during first round sampling. B) Second round sampling from February 1 to 5, 2021. Dhaka, Chittagong, and Mymensingh revealed the lowest Ct values during the second-round sampling.
In the second round of sampling (February 1 to February 5, 2020), 98% of the samples were positive for ORF1ab or N gene. Both ORF1ab and N genes were detected in 10% of samples, N and human RNase-p genes in 19%, and ORF1ab, and human RNase-p genes in 19% of samples. The average Ct value observed in the samples analyzed during the second round was also similar to the first-round samples (Fig. 2B). The presence of SARS-CoV-2 genetic material was thus observed in both urban and rural areas during both rounds (Fig. 3B).
In the second-round wastewater samples, the number of SARS-CoV-2 genes was higher than first-round samples. whereas the number of ORF1ab number genes were highest in both first (62/168) and second (63/168) round samples while the control gene, RNase-p was found to be lowest (Supplementary data; Figure SF6).
3.2. Comparison of the abundance of SARS-CoV-2 genetic materials in wastewater samples between urban and rural areas
For rural areas, the marketplace samples showed a consistent presence of SARS-CoV-2 RNA (Fig. 4 A). In urban areas, COVID-19 isolation centers showed a constant presence of viral RNA genetic materials (Fig. 4B). We detected the lowest Ct values, corresponding to the highest number of gene copies (GC) in the Gaibandha marketplace (Ct: 29.57; 5570 GC/100 mL), while the highest Ct was found in the Mymensingh marketplace (Ct: 37.44; 458 GC/100 mL). The genetic materials of SARS-CoV-2 was found in wastewater samples collected from all the isolation centers, the lowest Ct value in Chittagong Isolation Center (Ct: 26.44; 7440 GC/100 mL) and the highest identified in the Rangpur Isolation Center (Ct: 39.33; 750 GC/100 mL). These findings reveal that the correlation between Ct values and the viral genomic copies follows the prevalence of COVID-19 among the population in selected areas (p = 0.03) as reported by DGHS (IEDCR, 2022).
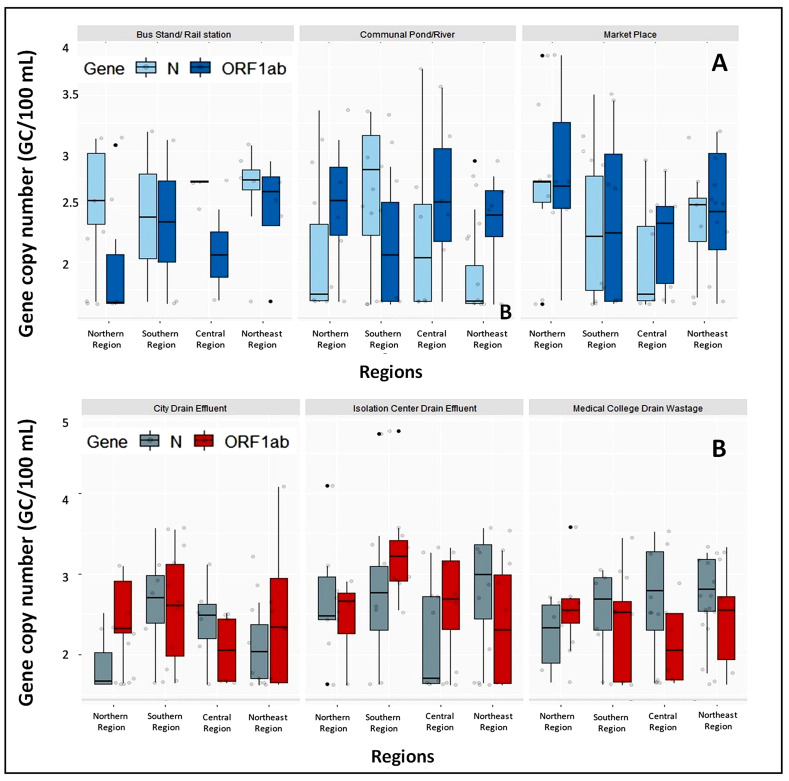
Fig. 4.
A) Comparison of N- and ORF1ab gene copy numbers in the drain outlets of bus stand/rail station, community pond/river, pond/river, and marketplace in rural areas. The marketplace showed consistency of the genetic markers in the rural areas, whereas the bus stands, railway stations, community ponds, and rivers showed a slight variation in the number of SARS-CoV-2 genetic markers different areas. B) Comparison of N and ORF1ab genes in city drain, isolation center, and medical college drain effluents in urban areas. Here the isolation center showed consistency of genetic materials, whereas the city drains and medical college showed a slight variation in four regions.
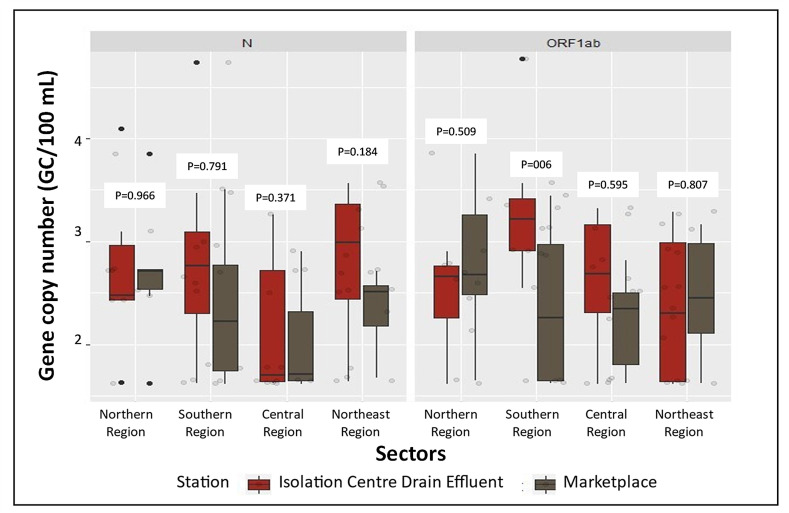
We used a similar procedure for direct testing of genetic materials in the marketplace of rural areas and the isolation centers in urban areas where we did not find any significant changes (Fig. 5 ).
Fig. 5.
Optimized locations of sampling for wastewater-based epidemiological (WBE) surveillance from urban and rural areas based on the observed SARS-CoV-2 viral RNA gene copies. No significant difference was observed in the four regions for isolation center and marketplace except for the ORF1ab genomic copies in the southern region.
3.3. Linear mixed modeling (LMM) for data analysis
We employed six random intercept linear mixed models to fit the expression of genetic materials after adjusting drain availability, region location, and temperature (Table 1 ). The data for the regions without drain was collected from rural marketplaces. In the model, the two rounds of data collection were treated as random. The results show that drain availability has a nearly significant impact on the ORF1ab and N genes (ORF1ab: beta = 0.86, p-value = 0.087 and N: beta = 0.937, p-value = 0.062). A distinct variation was observed in the copy numbers of SARS-CoV-2 ORF1ab and N genes due to geographical characteristics. Despite the fact that there was no statistically significant difference in Ct values between the southern and northern regions, the coefficient of Ct value shows that the southern region (N: beta = −0.988, p-value = 0.180) had lower values. Similarly, the CN in the wastewaters from the central region (ORF1ab: beta = −0.200, p-value = 0.180) and the north-eastern region (ORF1ab: beta = −0.217, p-value = 0.154) do not differ significantly from the northern region. It is also observed that the Ct value based on the ORF1ab expression in the wastewater samples from drained areas was higher by 0.86 units as compared to the samples from areas without drain. When comparing the sampling areas with or without drainage, we found significant difference in ORF1ab and N gene expression values. Furthermore, no significant differences are observed for ORF1ab and N-genes in the studied regions with temperature. The detected SARS-CoV-2 genome copy numbers in marketplace samples correlate well with the prevalent epidemic conditions as revealed by the national database of clinical diagnosis.
Table 1.
Results from the six random-intercept linear mixed models taking expression values of ORF and N genes.
| Comparison Model | Comparison of the prominent SARS-CoV-2 genes |
|||||||
|---|---|---|---|---|---|---|---|---|
| ORF1ab |
N |
|||||||
| Ct value |
Copy Number |
Ct value |
Copy Number |
|||||
| Beta estimate | P-value | Beta estimate | P-value | Beta estimate | P-value | Beta estimate | P-value | |
| Without drain (Reference: with drain) | 0.860 | 0.087 | −0.084 | 0.426 | 0.937 | 0.062 | −0.167 | 0.132 |
| Southern region (Ref: Northern region) | −0.006 | 0.993 | 0.016 | 0.916 | −0.988 | 0.180 | 0.089 | 0.583 |
| Central region (Ref: Northern region) | 0.997 | 0.159 | −0.200 | 0.180 | −0.147 | 0.836 | −0.007 | 0.960 |
| North-Eastern region (Ref: Northern region) | 0.556 | 0.441 | −0.217 | 0.154 | −0.175 | 0.808 | −0.037 | 0.815 |
| Temperature | 0.010 | 0.404 | −0.002 | 0.474 | 0.001 | 0.966 | −0.001 | 0.955 |
3.4. Concordance of SARS-CoV-2 genetic markers in wastewater with confirmed cases of COVID-19 in different districts
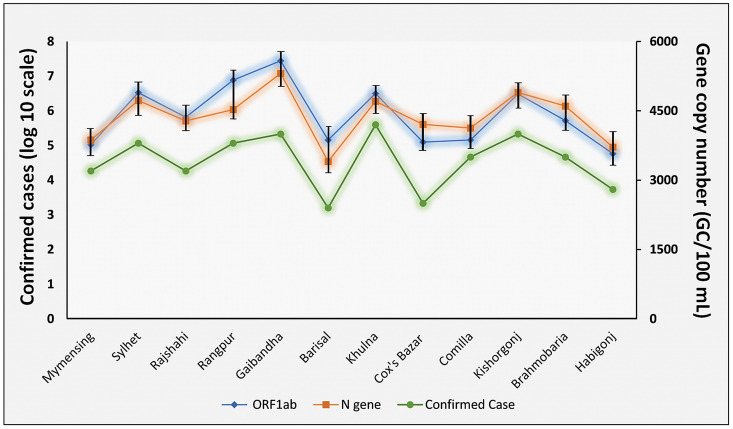
The percentage of confirmed cases was in concordance with the CNs of the wastewater genetic materials of SARS-CoV-2 in different districts during sampling periods. This finding indicated the positive relationship of CN of individual genetic materials with confirmed cases in various study areas (Fig. 6 ). Highest CN (5570/100 mL) were found in marketplace of Gaibandha and the number of confirmed cases was 7343 while the lowest copy number in communal pond (3350/100 mL) correlates with the registered 4960COVID-19 cases in Habigonj.
Fig. 6.
Concordance of the SARS-CoV-2 gene copy numbers in wastewater (expressed as genomic copies of ORF1ab and N genes) with confirmed cases in twelve different districts in Bangladesh. The concentration of SARS-CoV-2 viral genomic copies was higher in the wastewater samples of Rangpur and Gaibandha districts as compared to the samples from other twelve districts. Higher genomic copy counts in wastewater samples also correlated with the clinically confirmed COVID-19 cases. (Confidence Interval (CI) is less than 5%).
4. Discussion
Wastewater-based epidemiology (WBE) is an effective technique for assessing disease prevalence at the community level, as samples can be obtained simply from a free-flowing effluent in a centralized sewage system (Amoah et al., 2022; Prado et al., 2021; Bhattacharya et al., 2021; Haque et al., 2022). However, considering the current global spread of SARS-CoV-2, it is essential to expand the scope of WBE to establish an effective system for regions without a centralized sewage treatment infrastructure (Amoah et al., 2021; Kweinor Tetteh et al., 2020). This study developed a novel WBE system based on sampling onsite sanitation facilities that could be a cost-effective alternative for SARS-CoV-2 management in areas without centralized sewage collection infrastructure.
To simplify the collection of wastewater samples from the complex onsite sanitation systems in rural and urban areas of Bangladesh, we suggest two sampling methods, hotspot-based (rural sites) and drain-based (urban sites) sampling to represent the specific patterns of the prevailing community infection in the specific catchment areas. The drain system in the urban areas is a non-point source where it is difficult to trace back the source of contamination. Hence, the samples were collected from the selected drain catchment areas and places with the maximum concentration of houses, markets, and the possible mass-gathering locations. In the rural areas, the sanitation system is onsite, and there is no such drain system. As a result, the study developed a hotspot-based sampling method where some popular community places were identified. This identification was based on cultural and societal practices of the rural population as they tend to gather and meet in places like religious establishments, markets, restaurants, transport stations, public toilets, etc., which indicated a high possibility of detecting the genetic material. After testing a few communal places, there were overall 96% positive SARS-CoV-2 RNA detections, demonstrating the community wastewater outlets as suitable sampling locations.
In the Central and Southern regions, there was a significant change in SARS-CoV-2 genetic materials in the sampling locations for drain and non-drain portions. It is important to note that the consistent detection of SARS-CoV-2 genetic materials in wastewater samples of the marketplace is due to the continuous gathering of people in clusters around the shops, as well as a general community meet-up place for the local population. In contrast, there was a consistency of genetic materials in the drain effluent of the isolation center in all urban areas due to the visits of numerous patients and their admission due to the clinical diagnosis in these isolation centers. We also found a positive association with the viral load and confirmed cases in all the fourteen observed regions.
The study also reveals that the WBE surveillance system can even be used for community level disease monitoring in areas without a centralized system of wastewater sewage collection. However, the variations in the observed trends of SARS-CoV-2 genetic RNA materials necessitate the analysis of wastewater samples at regular intervals over an extended period. The monitoring of the prevalence of SARS-CoV-2 in communities is possible due to the development of a spatio-temporal monitoring system, which could be utilized to assess the public health scenario (Bertacchini et al., 2020). It has the potential to anticipate how the disease will progress and can serve as a vital tool for community-based early warning systems. The findings are expected to persuade governments worldwide, particularly in developing and low-sanitation countries with predominantly onsite sanitation facilities, to implement wastewater epidemiological surveillance for the SARS-CoV-2 pandemic.
Based on the analysis of wastewater viral load, it can also be concluded that WBE has the ability to provide a more accurate scenario of the prevailing SARS-CoV-2 infections. Lastra et al. (2022) found that a weekly sampling strategy offered adequate quantification. However, due to the limitation of resources it was not possible during this study. Despite the simplicity of WBE sampling and transportation of samples in a timely manner, the procedure of viral RNA concentration and extraction from wastewater from public places was very difficult for low RNA quantities (Pecson et al., 2021; Westhaus et al., 2021). To monitor mutational changes of the SARS-CoV-2 genome and to identify its current and new variants considering national and global perspectives, high-throughput sequencing has been carried out from symptomatic patients but yet to be done from wastewater samples (du Plessis et al., 2021; Hossain et al., 2021, Meredith et al., 2020, Sakib et al., 2021).
5. Conclusion
This is the first study where hot-spot-based and drain-based approaches are suggested for WBE systems in areas with onsite sanitation facilities, through selection and optimization of the best sampling sites for predicting COVID-19 prevalence. Marketplaces play an essential role in every rural community, and everyone visits them at least once a week for the purpose of shopping, socializing, visiting, or doing business. It is therefore apparent that the marketplace is the possible best location to detect the prevalence of COVID-19 in the various regions of the country based on WBE surveillance. The study also stressed the importance of developing an onsite WBE system for detecting SARS-CoV-2 genetic materials in wastewater in areas with on-site sanitation facilities. While comparing the viral genomic components in different wastewater locations in Bangladesh, it was discovered that isolation center wastewater in urban areas had a constant load of viral genetic features. In SARS-CoV-2 isolation facilities, hospital community drains, city drains, communal ponds, and rivers, we discovered sensitive results for detecting SARS-CoV-2 RNA. The study will help the researchers to better understand and monitor the severity of transmission of SARS-CoV-2 and other contagious diseases in areas where onsite sanitation systems include both drain and non-drain components.
Ethics statement
The work did not involve any human subjects or animal experiments.
Author contributions
Md. Jakariya: Conceptualization, Formal analysis, Fund acquisition, Investigation, Methodology, Project administration, Supervision, Roles/Writing - original draft, Writing – review & editing. Firoz Ahmed: Conceptualization, Formal analysis, Funding, Investigation, Methodology, Project administration, Supervision, Roles/Writing - original draft, Writing – review & editing. Md. Aminul Islam: Conceptualization; Investigation; Methodology; Data curation, Formal analysis; Project administration; Visualization, Validation, Writing – original draft, review, and editing. Abdullah Al Marzan: Methodology, Visualization, Roles/Writing - original draft, Writing – review & editing. Mohammad Nayeem Hasan: Formal analysis. Maqsud Hossain: Validation; Visualization, Writing – review & editing. Tanvir Ahmed: Funding acquisition, Project administration, Writing – review & editing. Ahmed Hossain: Writing – review & editing. Hasan Mahmud Reza: Project administration, Validation; Visualization. Foysal Hossen: Validation; Visualization, Writing – review & editing. Turasa Nahla: Roles/Writing - original draft, Writing – review & editing. Mohammad Moshiur Rahman: Writing - review & editing. Newaz Mohammed Bahadur: Writing - review & editing. Md. Tahmidul Islam: Funding acquisition. Md. Didar-ul-Alam: Conceptualization; Project administration. Nowrin Mow: Project administration. Hasin Jahan: Funding acquisition, Project administration. Damia Barcélo: Writing – review & editing. Kyle Bibby: Roles/Writing – review & editing. Prosun Bhattacharya: Conceptualization, Funding, Investigation, Methodology, Project administration, Supervision, Roles/Writing - original draft, Writing – review & editing. All authors critically reviewed and approved the final version of the manuscript. The corresponding authors is responsible for ensuring the accuracy of data and the descriptions which have been agreed by all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by Water Aid Bangladesh, North South University, Dhaka, COVID-19 Diagnostic Lab, Department of Microbiology, Noakhali Science and Technology University (NSTU), Noakhali, Bangladesh, the International Training Network of Bangladesh University of Engineering and Technology (ITN-BUET) - Centre for Water Supply and Waste Management, and KTH Royal Institute of Technology, Sweden. We acknowledge the sincere help and support of the staff and volunteers of NSTU-COVID-19 Diagnostic Lab, Noakhali Science and Technology University, Bangladesh during the different phases of the study. PB and MTI acknowledge the Life Science Technology Platform, Science for Life Laboratory for the seed funding to initiate the wastewater-based epidemiological studies for SARS-CoV-2 in Bangladesh. We would also like to acknowledge the two anonymous reviewers for their critical comments as well as their thoughtful insights, which has significantly improved the manuscript.
Footnotes
This paper has been recommended for acceptance by Da Chen.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envpol.2022.119679.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Acosta N., Bautista M.A., Hollman J., McCalder J., Beaudet A.B., Man L., Waddell B.J., Chen J., Li C., Kuzma D., Bhatnagar S., Leal J., Meddings J., Hu J., Cabaj J.L., Ruecker N.J., Naugler C., Pillai D.R., Achari G., et al. A multicenter study investigating SARS-CoV-2 in tertiary-care hospital wastewater. viral burden correlates with increasing hospitalized cases as well as hospital-associated transmissions and outbreaks. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 Omicron variant at Frankfurt airport but limited spread in the city of Frankfurt, Germany, in november 2021. Microbiol. Resour. Announc. 2022;11(2) doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., Islam M.N., Bahadur N.M., Alam M.D., Reza H.M., Jakariya M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre through wastewater surveillance in Bangladesh. medRxiv. 2020 doi: 10.1101/2020.09.14.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., Islam M.N., Bahadur N.M., Didar-ul-Alam M., Md., Reza H.M., Jakariya M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Abunama T., Awolusi O.O., Pillay L., Pillay K., Kumari S., Bux F. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Mthethwa N.P., Pillay L., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. RT-LAMP: a cheaper, simpler and faster alternative for the detection of SARS-CoV-2 in wastewater. Food Environ. Virol. 2021;13(4):447–456. doi: 10.1007/s12560-021-09489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., Gupta A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020;82(12):2823–2836. doi: 10.2166/wst.2020.540. [DOI] [PubMed] [Google Scholar]
- Bertacchini F., Bilotta E., Pantano P.S. On the temporal spreading of the SARS-CoV-2. PLoS One. 2020;15(10 October) doi: 10.1371/JOURNAL.PONE.0240777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya P., Kumar M., Islam M.T., Haque R., Chakraborty S., Ahmad A., Niazi N.K., Cetecioglu Z., Nilsson D., Ijumulana J., van der Voorn T., Jakariya M., Hossain M., Ahmed F., Rahman M., Akter N., Johnston D., Ahmed K.M. Prevalence of SARS-CoV-2 in communities through wastewater surveillance – a potential approach for estimation of disease burden. Curr. Pollut. Rep. 2021;7:160–166. doi: 10.1007/s40726-021-00178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Alexandria B., Boehm A.B., Joe Brown J., Gianluigi Buttiglieri G., Calabro V., Carducci A., Castiglioni S., CeteciogluGurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., III, Delgado Vela J.D., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-Based Epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- D'Aoust P.M., Towhid S.T., Mercier É., Hegazy N., Tian X., Bhatnagar K., Zhang Z., Naughton C.C., MacKenzie A.E., Graber T.E., Delatolla R. Science of The Total Environment; 2021. COVID-19 Wastewater Surveillance in Rural Communities: Comparison of Lagoon and Pumping Station Samples; p. 149618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Plessis L., McCrone J.T., Zarebski A.E., Hill V., Ruis C., Gutierrez B., Raghwani J., Ashworth J., Colquhoun R., Connor T.R., Faria N.R., Jackson B., Loman N.J., O'Toole Á., Nicholls S.M., Parag K.V., Scher E., Vasylyeva T.I., Volz E.M., et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371(6530):708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekumah B., Armah F.A., Yawson D.O., Quansah R., Nyieku F.E., Owusu S.A., Odoi J.O., Afitiri A.R. Disparate onsite access to water, sanitation, and food storage heighten the risk of COVID-19 spread in Sub-Saharan Africa. Environ. Res. 2020;189 doi: 10.1016/j.envres.2020.109936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Rogovski P., Savi B.P., Cadamuro R.D., Pereira J.V.F., Anna I.H.S., Rodrigues I.H., Souza D.S.M., Saravia E.G.T., Rodríguez-Lázaro D., da Silva Lanna M.C. SARS-CoV-2 in human sewage and river water from a remote and vulnerable area as a epidemiology tool in Brazil. Food Environ. Virol. 2021 doi: 10.1007/s12560-021-09487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D'Ambrosio M., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.-L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.-Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M.A., Wang F., Chen Y., Hossen F., Islam M.A., Hossain M.A., Siddique N., He C., Ahmed F. Bacillus spp. contamination: a novel risk originated from animal feed to human food chains in South-eastern Bangladesh. Front. Microbiol. 2022;12 doi: 10.3389/fmicb.2021.783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain F.E., Islam S., Islam M.A., Islam S., Ahmed F. Detection of virulence genes of APEC (avian pathogenic Escherichia coli) isolated from poultry in Noakhali, Bangladesh. Biores. Commun. 2021 doi: 10.3329/brc.v7i1.54253. [DOI] [Google Scholar]
- Hossain M., Huq T.S., Rahman A., Islam M.A., Tabassum S.N., Hasan K.N., Khaleque A., Hossain M.S., Bahadur N.M., Ahmed F., Reza H.M. Novel mutations identified from whole-genome sequencing of SARS-CoV-2 isolated from Noakhali, Bangladesh. Res. Square. 2021 doi: 10.21203/rs.3.rs-437228/v1. [DOI] [Google Scholar]
- IEDCR . 2022. COVID-19 Dynamic Dashboard for Bangladesh.https://dghs-dashboard.com/pages/covid19.php [Google Scholar]
- Islam A., Sayeed M.A., Rahman M.K., Ferdous J., Shano S., Choudhury S.D., Hassan M.M. Spatiotemporal patterns and trends of community transmission of the pandemic COVID-19 in South Asia: Bangladesh as a case study. Biosaf. Health. 2021;3(1):39–49. doi: 10.1016/J.BSHEAL.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam et al. 2021. Sex-specific Epidemiological and Clinical Characteristics of COVID-19 Patients in the Southeast Region of Bangladesh. MedRxiv. [DOI] [Google Scholar]
- Islam M.A., Haque M.A., Rahman M.A., Hossen F., Reza M., Barua A., Marzan A. Al, Das T., Kumar Baral S., He C., Ahmed F., Bhattacharya P., Jakariya M. A Review on measures to rejuvenate immune system: natural mode of protection against Coronavirus infection. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.837290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakariya M., Ahmed F., Islam M.A., Ahmed T., Marzan A. Al, Hossain M., Reza H.M., Bhattacharya P., Hossain A., Nahla T., Bahadur N.M., Hasan M.N., Islam M.T., Hossen M.F., Alam M. D. ul, Mou N., Jahan H. Wastewater based epidemiology system to detect SARS-CoV-2 genetic material for countries with on-site sanitation facilities: an experience from Bangladesh. medRxiv. 2021 http://medrxiv.org/content/early/2021/08/12/2021.07.30.21261347.abstract 8852000, 2021.07.30.21261347. [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., Joshi C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi M., Shah A.V., Srivastava V., Dave S. Wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: a perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Jiang G., Thakur A.K., Chatterjee S., Bhattacharya T., Mohapatra S., Chaminda T., Tyagi V.K., Vithanage M., Bhattacharya P., Nghiem L.D., Sarkar D., Sonne C., Mahlknecht J. Lead time of early warning by wastewater surveillance for COVID-19: geographical variations and impacting factors. Chem. Eng. J. 2022;441 doi: 10.1016/j.cej.2022.135936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweinor Tetteh E., Opoku Amankwa M., Armah E.K., Rathilal S. Fate of COVID-19 occurrences in wastewater systems: emerging detection and treatment technologies—a review. Water. 2020;12(10):2680. doi: 10.3390/w12102680. [DOI] [Google Scholar]
- Lastra A., Botello J., Pinilla A., Urrutia J.I., Canora J., Sánchez J., Fernández P., Candel F.J., Zapatero A., Ortega M., Flores J. SARS-CoV-2 detection in wastewater as an early warning indicator for COVID-19 pandemic. Madrid region case study. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Meredith G.R., Rakow D.A., Eldermire E.R.B., Madsen C.G., Shelley S.P., Sachs N.A. Minimum time dose in nature to positively impact the mental health of college-aged students, and how to measure it: a scoping review. Front. Psychol. 2020;10 doi: 10.3389/fpsyg.2019.02942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., SARS-CoV-2 Interlaboratory Consortium Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;7(3):504–520. doi: 10.1039/D0EW00946F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Chagas do Vale V.H., Braz R.M.S., de Andrade J., da S.R., Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakib S.H., Masum S., Patwari M.R.I., Fahima R.A., Farhana A., Islam M.A. Design and Development of a low cost Ultraviolet Disinfection system to reduce the cross infection of SARS-CoV-2 in ambulances. IEEE. 2021 doi: 10.1109/ICECIT54077.2021.9641131. [DOI] [Google Scholar]
- Reliefweb . 2022. Bangladesh.https://reliefweb.int/report/bangladesh/bangladesh-epidemiological-highlights-week-1-3-9-jan-2022 Epidemiological Highlights Week 1 (3-9 Jan 2022) [Google Scholar]
- Sagoe G., Danquah F.S., Amofa-Sarkodie E.S., Appiah-Effah E., Ekumah E., Mensah E.K., Karikari K.S. GIS-aided optimisation of faecal sludge management in developing countries: the case of the Greater Accra Metropolitan Area, Ghana. Heliyon. 2019;5(9) doi: 10.1016/j.heliyon.2019.e02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakib M.M.H., Nishat A.A., Islam M.T., Raihan Uddin, M.A., Iqbal M.S., Hossen F.F.B., Ahmed M.I., Bashir M.S., Hossain T., Tohura U.S., Saif S.I., Jui N.R., Alam M., Islam M.A., Hasan M.M., Sufian M.A., Ali M.A., Islam R., Hossain M.A., Halim M.A. Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2. Comput. Biol. Med. 2021 doi: 10.1016/j.compbiomed.2021.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongpradit S., Prasongtanakij S., Srisala S., Kumsang Y., Chanprasertyothin S., Boonkongchuen P., Pitidhammabhorn D., Manomaipiboon P., Somchaiyanon P., Chandanachulaka S., Hirunrueng T., Ongphiphadhanakul B. A simple method to detect SARS-CoV-2 in wastewater at low virus concentration. J. Environ. Publ. Health. 2022;2022:1–7. doi: 10.1155/2022/4867626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D., Thomson P.C. Analysis of gene expression data using a linear mixed model/finite mixture model approach: application to regional differences in the human brain. Bioinformatics. 2014;30(11):1555–1561. doi: 10.1093/bioinformatics/btu088. [DOI] [PubMed] [Google Scholar]
- Vadiati M., Beynagh A., Bhattacharya P., Bandala E.R., Mozafari M. Indirect effects of COVID-19 on the environment: how deep and how long? Sci. Total Environ. 2022;810 doi: 10.1016/j.scitotenv.2021.152255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn T., van den Berg C., Bhattacharya P., Quist J. Never waste a crisis: drawing first lessons from the COVID-19 Pandemic to tackle the water crisis. ACS ES&T Water. 2021;1:8–10. doi: 10.1021/acsestwater.0c00041. [DOI] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 epidemiology and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2022. World Health Organization Bangladesh Morbidity and Mortality Weekly Update (MMWU) No. 10, February 28, 2022.https://cdn.who.int/media/docs/default-source/searo/bangladesh/covid-19-who-bangladesh-situation-reports/who_ban_sitrep_105_20220228.pdf?sfvrsn=4dd0a78d_5 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.