ABSTRACT
Candida albicans is a clinically important polymorphic fungal pathogen that causes life-threatening invasive infections in immunocompromised patients. Antifungal therapy failure is a substantial clinical problem, due to the emergence of an increasing number of drug-resistant isolates. Caspofungin is a common antifungal drug, often used as first-line therapy that inhibits cell wall β-(1,3)-glucan synthesis. In this work, the cell surface of different echinocandin-resistant C. albicans clinical isolates was compared with sensitive isolates and their responses to echinocandin treatment analyzed. Proteomic analysis detected changes in the repertoire of proteins involved in cell wall organization and maintenance, in drug-resistant strains compared to susceptible isolates and after incubation with caspofungin. Moreover, an interaction network was created from the differential expression results. Our findings suggest drug resistance may involve not only a different cell wall architecture, but also a different response to drugs.
KEYWORDS: Candida albicans, drug resistance, caspofungin, proteomics, interaction network
Introduction
Candida albicans is an opportunistic human pathogen, able to switch from commensalism to pathogenicity in response to different cues from the host niches it colonizes [1–3]. A primary determinant of this switch is the cell wall, which plays key roles in pathogenicity and interactions with host defenses [4,5]. The dynamic structure of the cell wall mainly consists of three polysaccharides: chitin, glucan, and mannan [6]. Fungal cell walls are layered structures, with an inner conserved core of chitin and glucan and an outer layer of polysaccharides that varies according to the fungus. Specifically, the wall of C. albicans consists of three different polysaccharides, chitin, β-(1,3)- and β-(1,6)-glucan. Cell wall proteins, often highly mannosylated, form the outermost layer [7,8]. The major class of cell wall proteins are covalently attached to β-(1,6)-glucan by modified Glycosyl Phosphatidyl Inositol (GPI) anchors [7].
Echinocandins are a class of antifungal agents, available for more than a decade, recommended as first-line treatment for many types of Candida infections [9,10]. The fungicidal activity of this class of drugs has been shown in vitro to inhibit the activity of β-(1,3)-glucan synthase, the enzyme required for the synthesis of the glucan layer found in most medically important fungi [11]. Exposure to sub-inhibitory concentrations of these drugs alters the cell wall, increasing chitin synthesis and the exposure of β-(1,3)-glucan on the surface, and phagocytosis by macrophages [12]. Reduced susceptibility to echinocandins has been attributed primarily to point mutations in the GSC1 (FKS1) gene that encodes the catalytic subunit of the glucan synthase complex, which can decrease sensitivity to the drug by several log orders [13,14]. Compromising the integrity of the wall also changes the properties of the plasma membrane, as shown by Kelly and Kavanagh (2010) [15]. They showed caspofungin treatment increased the permeability of the wall, with increased amino acid leakage from the treated cells compared to DMSO-treated cells [16]. Protein release was also increased in caspofungin-treated cells. The released proteins, for example the metabolic enzymes Pgk1, Gpm1, Fba1, and Eno1, are highly immunogenic [17,18], with a possible role in immune response and inflammation in vivo.
The diagnostic “gold standard” for detecting Candida infections is blood culture, with other additional tests based on the detection of circulating polysaccharides from the fungal cell wall or antigens in blood samples [19,20]. However, the diagnosis of antifungal resistance is an issue in the clinic, as it is not routinely performed and requires 48–72 h, which is often too late to influence treatment of the patient [21].
Most of the previous works aimed at biomarker identification focused on identifying azole and echinocandin resistance on Candida species via PCR-based methods [22–25]. In this study, we aimed to characterize the cell walls of caspofungin-resistant isolates and compare them with sensitive isolates, in order to detect differences between the two groups which may be informative for diagnosing drug resistance in C. albicans isolates. The strains were also evaluated for the capacity to form biofilms, an important virulence attribute that contributes to drug-recalcitrant infections. We evaluated the expression of proteins during drug treatment, by mass-spectrometry, with a focus on enzymes involved in cell wall synthesis and maintenance. We performed differential expression (DE) analysis with this proteomics data set. Then, an interaction network was created to examine the relationships between proteins that were differentially expressed in the strains analyzed. Interaction networks have been created using transcriptomic data and applied to study host–pathogen interactions [26,27] and to identify genes involved in the filamentation response [28].
The set of proteins identified by the DE analysis can help to elucidate the responses of C. albicans to echinocandin drugs and give mechanistic insights to the proteins that contribute to cell wall adaptions induced by drug exposure. The detection of proteins that are selectively expressed or more abundant in drug-resistant isolates could potentially be investigated further for a better identification of drug-resistant infections.
Materials and methods
Strains and growth conditions
The isolates of C. albicans used in this study are listed in Table 1. Fungal cells were cultured and maintained according to previous methods [12]. Briefly, they were stored in 25% glycerol at −70ºC and re-cultured in YPD agar (1% [w/v] yeast extract [Oxoid], 2% [w/v] mycological peptone [Oxoid], 2% [w/v] glucose [Fisher Scientific], 2% [w/v] agar [Oxoid]). For overnight cultures, unless indicated otherwise, a single colony of each strain was inoculated into YPD broth (1% [w/v] yeast extract, 2% [w/v] mycological peptone, 2% [w/v] glucose) and incubated overnight at 30 ºC with shaking at 200 rpm. For hyphal induction, cells were grown in RPMI-1640 modified medium (50% [w/v] RPMI-1640 [Sigma-Aldrich Co.], pH 0.8–1.5; 1.65 M MOPS buffer [Melford], pH 7.2; 3.6% [w/v] glucose [Fisher Scientific]; 4.2 mM L-glutamine [Sigma-Aldrich Co.]) with added 20% Fetal Calf Serum (FCS, Gibco) at 37ºC for 6 h with 100 rpm shaking.
Table 1.
Caspofungin MIC against C. albicans isolates.
| Strain ID | Name | IC50CAS (μg/ml) | S/I/Ra | Genotype | Reference |
|---|---|---|---|---|---|
| SC5314 | Ca1 | 0.03–0.06 | S | Wild type | [29] |
| CBS8758 | Ca2 | 0.06–0.125 | S | Wild type | [30] |
| ATCC2091 | Ca3 | 0.06–0.125 | S | Unknown | [31] |
| ATCC76615 | Ca4 | 0.06–0.125 | S | Unknown | [32] |
| B17_009053 | Ca5 | 0.125–0.25 | S | Unknown | Munich, unpublished |
| B17_008835 | Ca6 | 0.06–0.125 | S | Unknown | Munich, unpublished |
| K063-3 | Car1 | 2 | R | GSC1 (FKS1, S645Y)/GSC1 (FKS1, S645Y) | [33] |
| B15_004476 | Car2 | 2-4 | R | Unknown | Munich, unpublished |
| B12_007355_1 | Car3 | 1-2 | R |
GSC1 (FKS1, R1361G)/ GSC1 (FKS1, R1361G) |
Munich, unpublished |
The caspofungin IC50 was calculated after 24h incubation in RPMI-1640 medium in a broth microdilution method according to the CLSI breakpoints. aInterpretive category according to the breakpoints: S=susceptible, I=intermediate, R=resistant.
Antifungal susceptibility testing
The Minimum Inhibitory Concentration (MIC) of caspofungin against C. albicans isolates was determined according to CLSI guidelines [34] and following the protocol described previously by Walker et al. (2008) [35]. Cells were pre-grown overnight in YPD medium at 30 ºC and then diluted to 2 × 106 cells/ml in 2X RPMI-1640 broth (Sigma Aldrich Co.) supplemented with 4.2 mM L-glutamine and grown at 37 ºC. Cells were incubated for 24 h in flat bottomed 96 well plates (Nunc) containing serial dilutions of the drug in sterile water. Caspofungin (Cancidas, Merck and Co. Inc., USA) concentration ranged from 0.016 µg/ml to 16 µg/ml. Following incubation, optical densities were read in a VersaMax microplate reader (Molecular Devices, USA) at 405 nm.
Biofilm formation assay
The capacity of the strains to form biofilms was evaluated by a modified method from Ramage et al. (2001) [36]. Cells were grown overnight in YPD medium at 30 ºC and transferred to a microtiter plate (Nunc) with RPMI-1640 plus 20% Fetal Calf Serum (FCS, Gibco), incubated at 37 ºC for 6, 24, 48 h with 0, 2, or 4 µg/ml caspofungin. Planktonic cells were washed away with PBS (Dulbecco’s Phosphate Buffered Saline [Sigma-Aldrich Co.]), and the cells remaining adhered to the plastic surface (biofilm) were quantified by incubation with 0.05% crystal violet for 20 min. The crystal violet was dissolved in 100% ethanol and the absorbance of the resulting solution was measured, after transfer to a fresh microtiter plate, at 570 nm in a VersaMax microplate reader (Molecular Devices, USA).
Cell wall proteomic analysis
The cell walls were isolated following a published protocol [37] with some modifications. Cells were grown overnight in YPD medium at 30 ºC and transferred to RPMI-1640 at 37 ºC supplemented with 4.2 mM L-glutamine, until exponential phase was reached (OD600 = 0.4 - 0.6). Caspofungin was added to some cultures for 90 min. The caspofungin concentration used was based on the MIC tests, in order to achieve the same percentage of growth (80%) between the different isolates. Cells were then harvested by centrifugation at 3000 x g for 5 min and washed once in 10 mM Tris-HCl (pH 7.5). Mechanical breakage of the cells was accomplished using zirconia/silica 0.5 mm beads (Thistle Scientific) in a FastPrep machine (MP Biomedicals). The cell debris containing cell walls was washed 5 times in 1 M NaCl to remove cytoplasmic contamination, resuspended in buffer (500 mM Tris-HCl buffer [pH 7.5], 2% [w/v] SDS, 0.3 M β-mercaptoethanol, and 1 mM EDTA), boiled 3 times at 100 ºC for 10 min and freeze-dried. The pellets were digested with trypsin according to the PRIME-XS protocol [38]. Mass spectrometry was performed using a Q Exactive Plus (Thermo Fisher Scientific) and tryptic peptides were identified using the MASCOT search engine (Matrix Science) [39]. Analysis of the LC-MS/MS data was carried out with Proteome Discoverer 2.2 software (Thermo Fisher Scientific), with the proteins matched from Candida Genome Database (CGD) (http://www.candidagenome.org/) and a cutoff of at least 2 peptides detected per protein, and their abundance measured by peptide peak areas determined from the extracted ion chromatograms.
Statistical analysis
GraphPad Prism version 5.01 for Windows (GraphPad Software, USA) was used for all the statistical analyses, unless specified. Differences in protein expression between different isolates were calculated using IBM SPSS Statistics (IBM, USA). A general linear model analysis was performed on the ranked values in order to avoid the missing hits in the data. Bonferroni was applied as a post-hoc test at the end of the analysis.
Cell wall network analysis
To build a co-expression network for the cell wall proteome of C. albicans, the LC-MS/MS data were analyzed with two different strategies: a statistical analysis and a network inferential analysis. In order to overcome the problem of the missing values, the data were filtered and randomly imputed using the k-nearest neighbors algorithm. For statistics, a differential expression (DE) analysis was carried out using limma package, part of Bioconductor, an R-based software [40] (Figure3.7). For network inference, a multivariate Poisson log-normal (PLN) model, R-based package was used [41]. Once the network was built and the groups defined, Gene Ontology enrichment was performed in order to name the groups, based on the most abundant terms.
Results
Strain characterization
The susceptibility of nine isolates of C. albicans to caspofungin was evaluated by the broth microdilution method. The minimum inhibitory concentrations of caspofungin are shown in Table 1: six isolates were detected as susceptible (Ca1, Ca2, Ca3, Ca4, Ca5, and Ca6) and three as resistant (Car1, Car2, and Car3) according to CLSI breakpoints [42]. The Ca2, Ca3, Ca4, and Ca6 isolates had the same range of IC50 despite coming from different sources.
Candida spp. infections are often associated with biofilm and biomaterial-related infections [43]. One of the features of biofilms is their reduced antimicrobial susceptibility compared to planktonic cells [44]. In order to assess possible implications of drug resistance in in vivo infections, the biofilm formation capacity of the isolates was evaluated. Cells were grown for 6, 24, 48 and 72 h in the absence of drug, or with 2 or 4 µg/ml caspofungin, at 37 ºC in RPMI-1640 medium, with 20% FCS added, which has been shown to increase biofilm formation [45]. Biomass was then measured through crystal violet absorbance at 570 nm (Figure 1–2). The reference strain of C. albicans Ca1 steadily increased biofilm formation throughout the 72 h in the absence of drug, whereas the addition of 2 or 4 µg/ml caspofungin totally abolished biofilm development (Figure 1). In general, the biofilm biomass of the caspofungin-susceptible isolates was comparable to Ca1 strain (Figure 1), with no substantial biomass increase at 72 h (Figure 1d). The addition of either 2 or 4 µg/ml caspofungin totally abolished the formation of biofilms for all the caspofungin-susceptible isolates (Figure 1a–c, yellow and cyan columns), except for some biomass increase observed at 72 h time point (Figure 1d). The three resistant isolates, except for the 6 h time point (Figure 2a), showed different trends in the presence or absence of the antifungal (Figure 2b–d). Without caspofungin (Figure 2, magenta columns), Car1 isolate produced biofilm similar to its isogenic parental isolate Ca1, whereas the other two resistant isolates did not significantly increase their biomass after the first 6 h time point (Figure 2b–d). The same was observed in the presence of drug (Figure 2, yellow and cyan columns), with Car1 able to reach the same levels of biofilm biomass in the absence of drug (Figure 2, magenta columns). Car2 and Car3 also had similar biofilm growth under the no drug and drug conditions.
Figure 1.
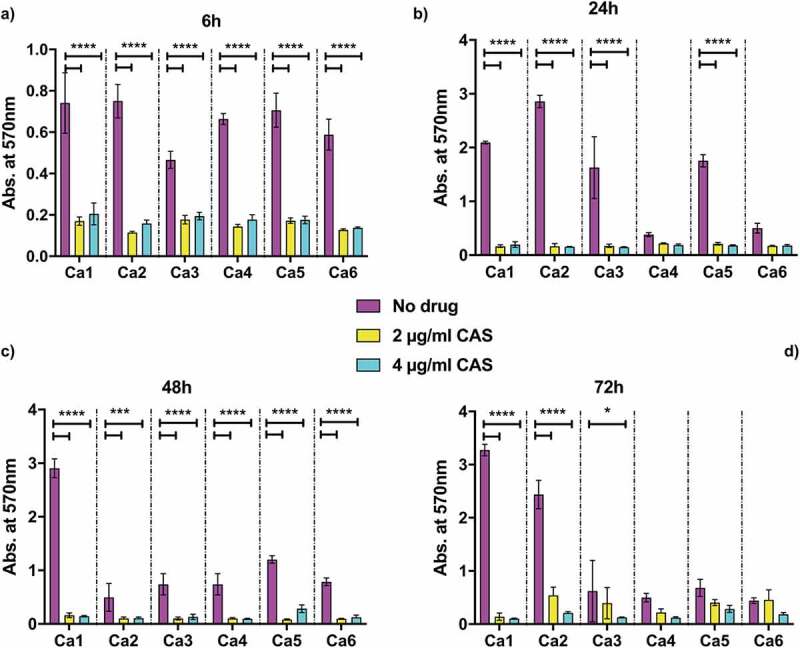
Biofilm formation by caspofungin-susceptible isolates of C. albicans. Absorbance from crystal violet staining of C. albicans Ca1, Ca2, Ca3, Ca4, Ca5, Ca6 isolates grown at 37 C in RPMI-1640 + 20% FCS grown either without drug (magenta) or the addition of 2 μg/ml CAS (yellow) or 4 μg/ml CAS (cyan) and measured at: (a) 6 h; (b) 24 h; (c) 48 h; (d) 72 h. The values are expressed as absorbance at 570 nm. The statistical analysis performed was one-way ANOVA (n = 1 (3 replicates), *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005).
Figure 2.
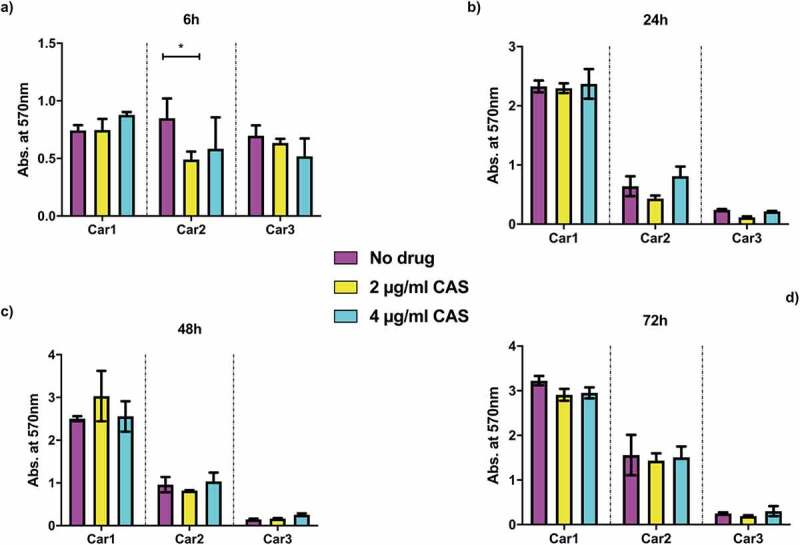
Biofilm formation assay for caspofungin-resistant isolates of C. albicans. Absorbance from crystal violet staining of C. albicans Car1, Car2, and Car3 isolates grown at 37°C in RPMI-1640 + 20% FCS grown either without drug (magenta) or the addition of 2 μg/ml CAS (yellow) or 4 μg/ml CAS (cyan) and measured at: (a) 6 h; (b) 24 h; (c) 48 h; (d) 72 h. The values are expressed as absorbance at 570 nm. The statistical analysis performed was one-way ANOVA (n = 1 (3 replicates), *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005).
Proteomic response to caspofungin
The cell wall proteomes of the isogenic C. albicans strains Ca1 (caspofungin-susceptible) and Car1 (caspofungin-resistant) were then compared. Cells were grown in RPMI-1640 medium at 37 ºC and incubated for 90 min with different caspofungin concentrations based on the ICs (inhibitory concentrations) (shown in Table 1), in order to achieve the same percentage of growth (80%). Incubation of exponentially-growing cells with the drug caused negligible differences in growth between the strains, ensuring the proteome was not altered by factors other than drug effects. Cell walls were isolated according to a modified protocol by Kapteyn et al. (2000) [37]. Tryptic digestion of the extracted walls was carried out, the peptides were analyzed by LC-MS/MS and their sequences identified according to the CGD database. Data analysis was carried out with Proteome Discoverer 2.2 software, using a cutoff of at least 2 peptides detected per protein, with abundances determined using Area Under the Curve measurements. Given the same genetic background, a limited set of proteins was found to be differentially expressed between the two isolates (Figure 3). Pga52 and Pga31, two GPI-anchored proteins of unknown function, were detected in increased abundances in Car1 compared to Ca1 isolate, in the presence (4.9- and 3.6-fold difference, respectively) and absence of caspofungin (13.7- and 5.9-fold difference, respectively). Another GPI-modified cell wall protein, Rbt5, involved in biofilm formation and iron homeostasis, was slightly more abundant in the echinocandin-resistant isolate Car1 without drug (1.7-fold difference) and with drug (5.9-fold difference). Peptides from two proteins of unknown function, C2_04780W_A and C3_07470W_A, were also found in higher amount in the drug-resistant isolate Car1 in the presence (7.2- and 4.4-fold difference, respectively) and absence of echinocandin (3- and 1.4-fold difference respectively). Other proteins were also differentially expressed in the two strains: Slr1, involved in hyphal growth, and Msb2, cell wall damage sensor involved in activation of Cek1 phosphorylation pathway, were more abundant in Car1 with drug (2- and 2.4-fold difference respectively) and slightly less abundant without caspofungin (0.7- and 0.8-fold difference respectively). On the contrary, the cell surface 1,3-beta-glucanosyltransferase Phr2, involved in cell wall remodeling, was detected in higher amounts in Car1 in the absence of caspofungin (3.4-fold difference) than with the drug (0.9-fold difference), the latter due to upregulation of this protein in Ca1 in response to drug treatment. Four proteins were detected in lower amounts for both conditions (presence and absence of drug) in the caspofungin-resistant isolate: the phospholipase Plb5 (0.2- and 0.03-fold difference respectively); the glucose transporters Hgt7 (0.4- and 0.4-fold difference respectively) and Hgt8 (0.3- and 0.4-fold difference respectively); the glucan synthase Gsc1 (Fks1, 0.2-, and 0.2-fold difference respectively).
Figure 3.
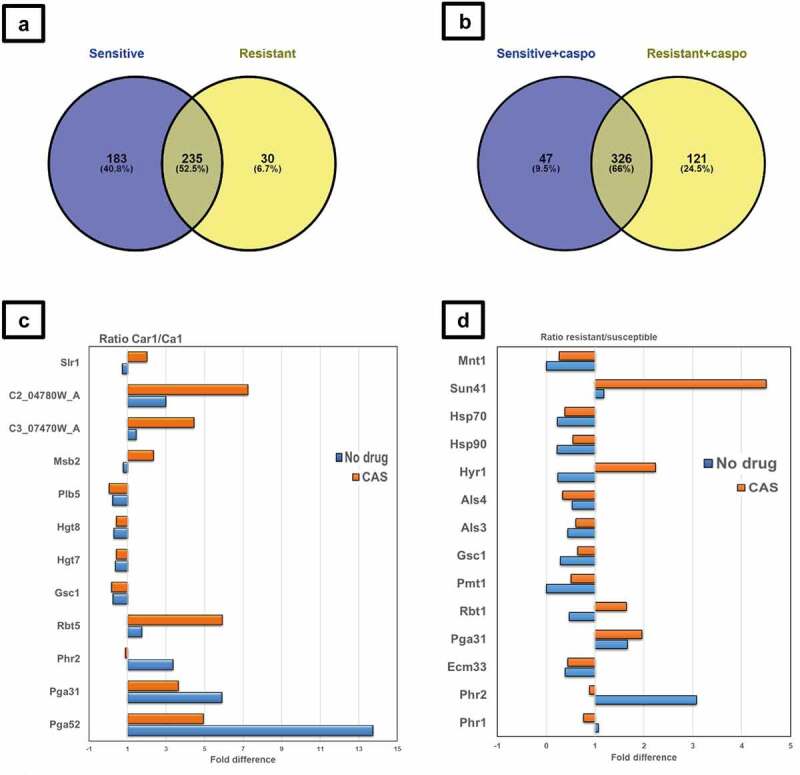
Proteomic analysis of cell wall fractions from C. albicans resistant and susceptible isolates exposed to caspofungin. (a, b) Total number of proteins identified by LC MS/MS in susceptible and resistant isolates in absence (a) and presence (b) of caspofungin in RPMI 1640 medium. (c) Differential expression of relevant proteins identified by LC MS/MS in susceptible Ca1 and resistant Car1 isolates in absence (blue) and presence (orange) of caspofungin in RPMI 1640 medium. The values displayed are the ratios of the averages (n=3) of the peak areas from the LC MS/MS analysis of the two isolates (Car1 and Ca1). The values displayed are the ratios of the log 10 of the averages of the peak areas from the LC MS/MS analysis of the two groups (resistant and susceptible). (d) Differential protein expression for the resistant compared to the susceptible isolates with (orange) and without (blue) caspofungin. The full list is available in File S1 in Supplementary Material. Venn diagrams were created using Venny software (n=1).
Common response to caspofungin
Next, to ascertain whether the differences in the expression of cell surface proteins were strain specific or common among C. albicans isolates, cell walls from seven additional isolates (listed in Table 1) were extracted and analyzed by LC-MS/MS with the same strategy described in the previous section. A total of 842 proteins were detected and 566 of those were represented by at least 2 peptides (File S1 in Supplementary Material). Thirty proteins were found exclusively in the resistant isolates in the absence of drug (Figure 4a), including cell wall proteins (Iff8, Eng1, Exg2, Pra1, Sep7, Plb4.5, Ihd1) but also cytoplasmic and plasma membrane proteins (Erg1, Hgt7, Hgt8, Mid1, Nce102), which are likely to be contaminants of the wall fraction. The 183 proteins detected only in the susceptible strains were mainly cytoplasmic contaminants. Among the 235 proteins in common were proteins involved in cell wall architecture, in particular glucanosyl-transferases such as Bgl2, Phr1, and Phr2, which are responsible for modifications of the glucan chain. Caspofungin caused a decrease, in both groups, in the level of proteins involved in the host defense response and pathogenesis (e.g. Als1, Mp65, Als3) (Figure 4b). Moreover, differences in the abundances of several proteins responsible for cell wall organization and maintenance were measured in the two different conditions by performing a regression analysis and averaging the expression values between the resistant and the susceptible isolates (Figure 3). In particular, large differences were observed for Sun41 (4.5-fold-difference), Mnt1 (0.27), Hyr1 (2.2), Als4 (0.34) in the group of resistant isolates in comparison with the susceptible isolates.
Figure 4.
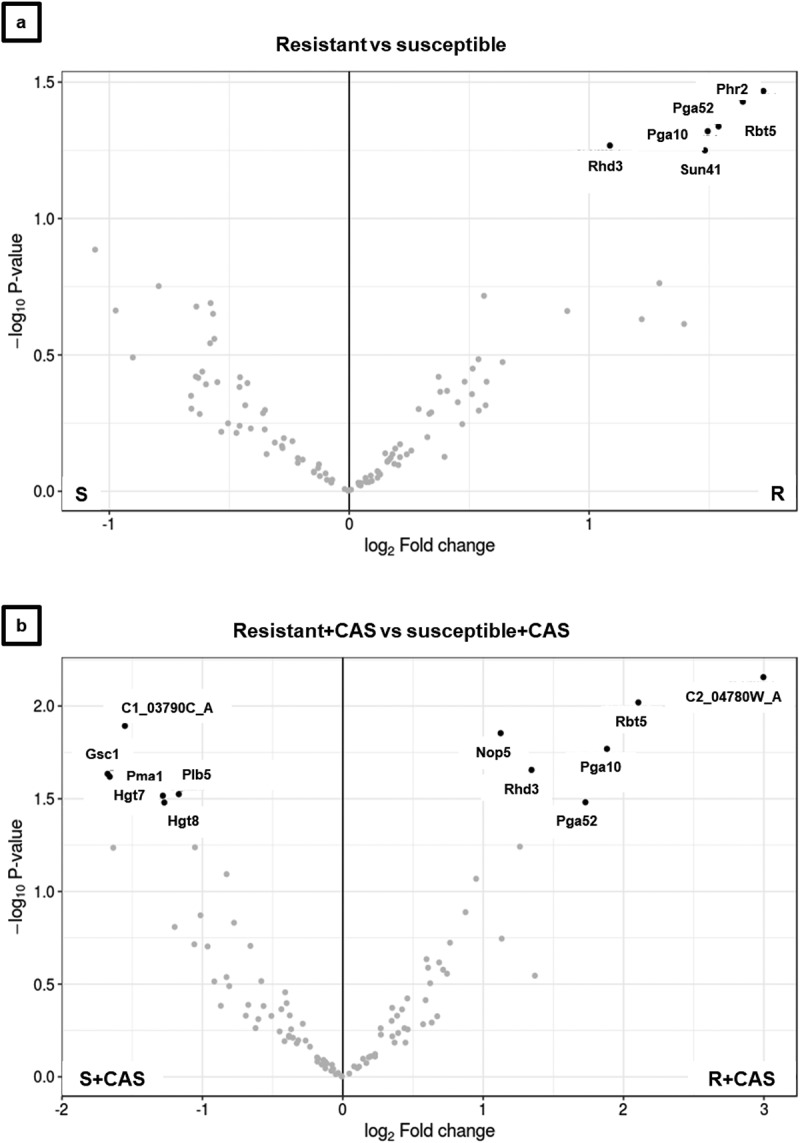
Volcano plots of cell wall proteome comparison of caspofungin -resistant and -susceptible isolate of C. albicans performed by differential expression (DE) analysis. The plots compare fold change and statistical significance of DE for caspofungin-resistant and -sensitive isolates of C. albicans (a) without drug and (b) with drug. The DE analysis was carried out using limma package, part of Bioconductor software. The plots compare fold change and statistical significance of DE for caspofungin-resistant and -sensitive isolates of C. albicans (a) without drug and (b) with drug. The DE analysis was carried out using limma package, part of Bioconductor software.
Interaction network of the cell wall proteome
In order to build an interaction network for the cell wall proteome of C. albicans, the LC-MS/MS data were analyzed with two different strategies: a statistical analysis and a network inferential analysis. Data were filtered, excluding proteins with less than two peptides, and missing values were randomly imputed using the k-NN algorithm. Differential analysis was performed to identify proteins altered in amounts between the two groups of isolates (caspofungin-resistant and caspofungin-susceptible). The results are shown in Figure 5. In the absence of drug, the volcano plot shows a group of proteins differentially expressed (adjusted p-value <0.1; >1log2 fold change difference in the ratios) with higher expression in the caspofungin-resistant compared to the caspofungin-susceptible isolates (Figure 4a). This group included: the cell wall 1,3-beta-glucanosyltransferase Phr2 (1.73 log2 ratio) and glycosidase Sun41 (1.48 log2 ratio); the GPI-anchored proteins Pga52 (1.64 log2 ratio) and Rhd3/Pga29 (1.09 log2 ratio); the proteins related to iron assimilation Rbt5 (1.54log2 ratio) and Pga10 (1.49 log2 ratio).
Incubation with caspofungin increased the number of proteins significantly and differentially expressed between drug-resistant and -susceptible isolates (Figure 4). The group of proteins detected in reduced amounts in the drug-resistant isolates included: the cytoplasmic component C1_03790C_A (−1.55 log2 ratio); the plasma membrane proteins Pma1 (−1.66 log2 ratio), Hgt7 (- 1.28 log2 ratio) and Hgt8 (−1.27 log2 ratio); Gsc1 (Fks1, −1.68 log2 ratio) and the wall enzyme Plb5 (−1.17 log2 ratio). The group of proteins detected in higher amounts in the drug-resistant isolates included: the cytoplasmic component Nop5 (1.12 log2 ratio); the protein of unknown function C2_04780W_A, (3 log2 ratio); and the proteins already detected in no-drug condition (but with increased differences between the group of isolates) Rbt5 (2.11 log2 ratio), Rhd3 (1.34 log2 ratio), Pga10 (1.88 log2 ratio) and Pga52 (1.73 log2 ratio).
A protein co-expression network (Figure 5) was then created considering the connectivity between proteins based on their expression changes in two conditions (± caspofungin) between the two groups of isolates (drug-susceptible and drug-resistant). The green edges of the circles indicate the significant proteins for the DE analysis in all the conditions (adjusted p-value ≤0.1). The lines connecting the circles indicate the positive correlation (red, both increase or both decrease ± caspofungin) or negative correlation (blue, one decreases and the other increases or vice versa). The thickness of the line indicates the strength of the correlation. The histograms inside the circles indicate the expression of each protein related to the normalized average between the two groups of isolates in the two conditions: from left to right, the columns represent susceptible, susceptible + caspofungin, resistant, resistant + caspofungin. The proteins were clustered in seven groups and GO analysis performed. Each group was given a name based on the most abundant GO terms. A general view of the network is presented in Figure 5. A dense net of positive correlations was detected between the ribosomal proteins and the proteins involved in the modulation of the host response. In this group (Figure 5) were highly immunogenic enzymes, such as Pgk1 and Eno1, as well as cell wall-related proteins. Cell wall proteins Rbt5 and Pga10 (which also share a strong positive correlation) were found significantly overexpressed in the resistant isolates in the DE analysis, as well as Rhd3/Pga29, grouped into the ribosomal cluster (Figure 5). The cell wall organization cluster (Figure 5) included proteins involved in modulation of cell wall β–(1,3)-glucan (such as Bgl2, Phr2 and Sun41) and Pga52 that were highly expressed in caspofungin-resistant isolates in absence and presence of drug. A strong positive correlation between Pga52 and Phr2 was noted. Another positive correlation between the cell wall damage sensor, Msb2, and Rbt1, part of the group of proteins involved in adhesion to the host, was detected. Hyphal-associated proteins and adhesins, such as Als1, Hyr1, and Als3 (which shared a strong positive correlation), also belong to this group (Figure 5), as well as the predicted GPI-anchored proteins Sap10 (which has a negative correlation with Hyr1) and Sod4. One of the most heterogenous groups was the germ-tube formation cluster (Figure 5), which included proteins with different functions, ranging from cell wall biosynthetic processes (e.g. Gsc1) to transmembrane transport (e.g. Pma1 and Cdr1), from cell wall adhesins (Als2 and Als4) to cytoplasmic proteins (e.g. Hmo1 and C1_03790C_A). A network of positive correlations of Cdr1 with other membrane proteins, such as Cdr2, Pma1, Ena21, and Sur7, as well as with Gsc1, was noted. Gsc1 (Fks1) and Cdr1 were also involved in a negative correlation with Als4. Another negative correlation was detected between the adhesin Als2 and the unknown protein C2_04780W_A. Another group (Figure 5) is made exclusively of plasma membrane glucose transporters (Hgt6, 7, 8), which did not have particular correlations with members of the other clusters. The last group, G0 (Figure 5), comprised proteins that were not found to have any correlation with any other group nor any protein within this group. Cell wall enzymes belonged to this group (e.g. Cht2, Crh11, Plb5, and Phr1), but also cytoplasmic proteins (e.g. Rpa34 and Rpl10).
Figure 5a.
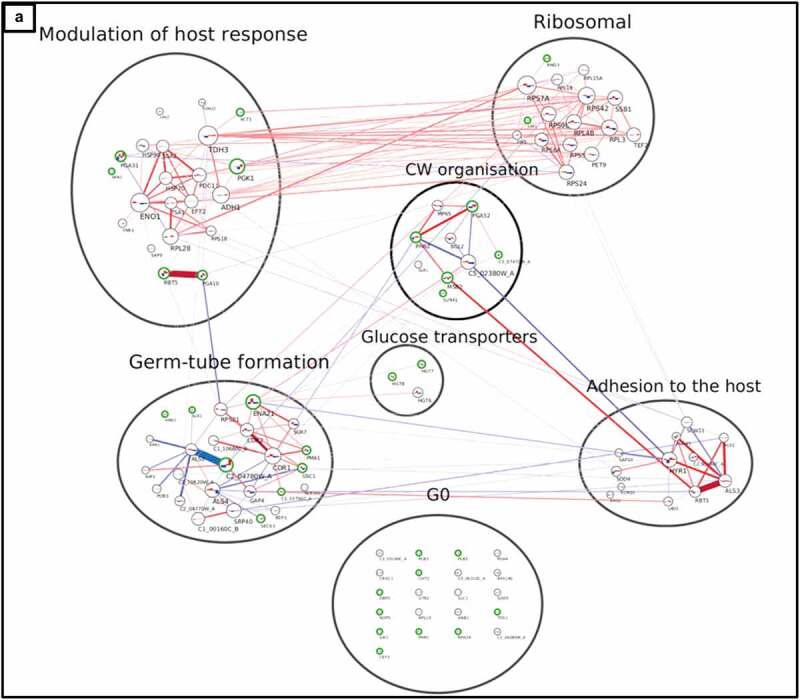
Interaction network of cell wall proteome from C. albicans isolates. The network was created using LC-MS/MS data from cell wall fractions of C. albicans isolates grown in absence and in presence of caspofungin. The green edges of the circles indicate the significant proteins for the DE analysis (adjusted p-value ≤0.1). The lines connecting the circles indicate the positive (red, both increase or decrease+/- caspofungin) or negative interaction (blue, one decreases and the other increases or vice versa). The thickness of the line indicates the strength of the interaction. The histograms inside the circles indicate the expression of the proteins related to the normalized average between the two groups of isolates in the two conditions: from left to right, the columns represent susceptible, susceptible+caspofungin, resistant, resistant+caspofungin. Proteins were clustered in seven groups and the GO analysis performed: (a) general view of the network; (b) ribosomal proteins; (c) cell wall organization; (d) modulation of the host response; (e) G0; (f) adhesion to the host; (g) germ-tube formation; (h) glucose transporters.
Figure 5b.
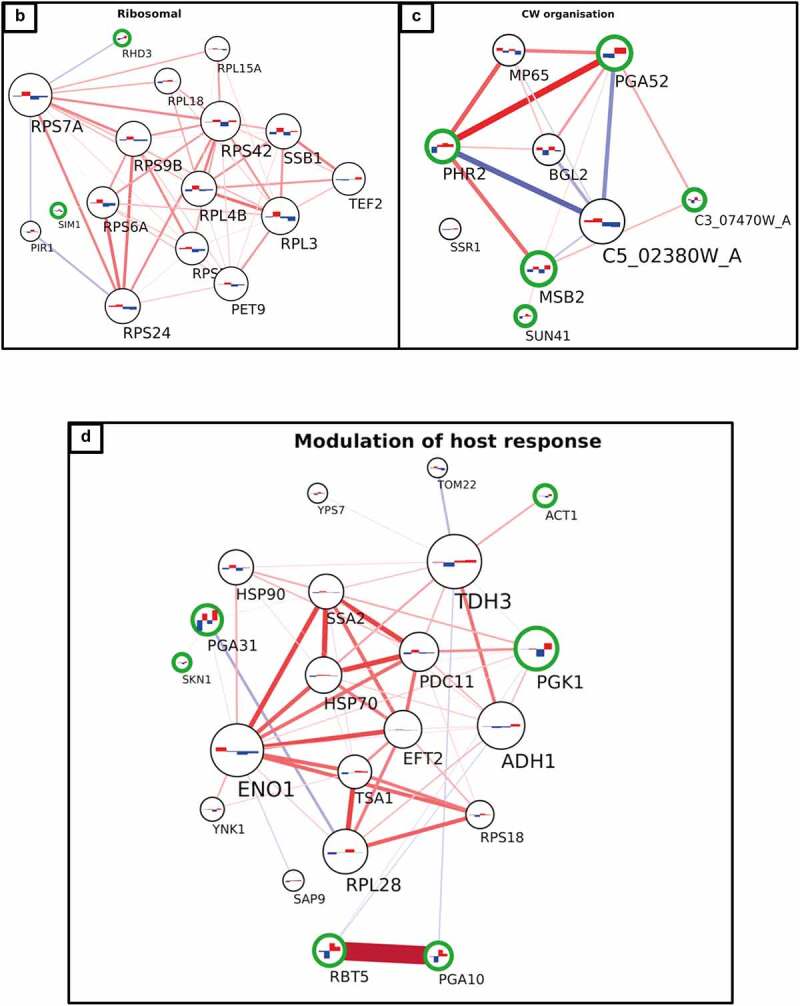
(continued).
Figure 5c.
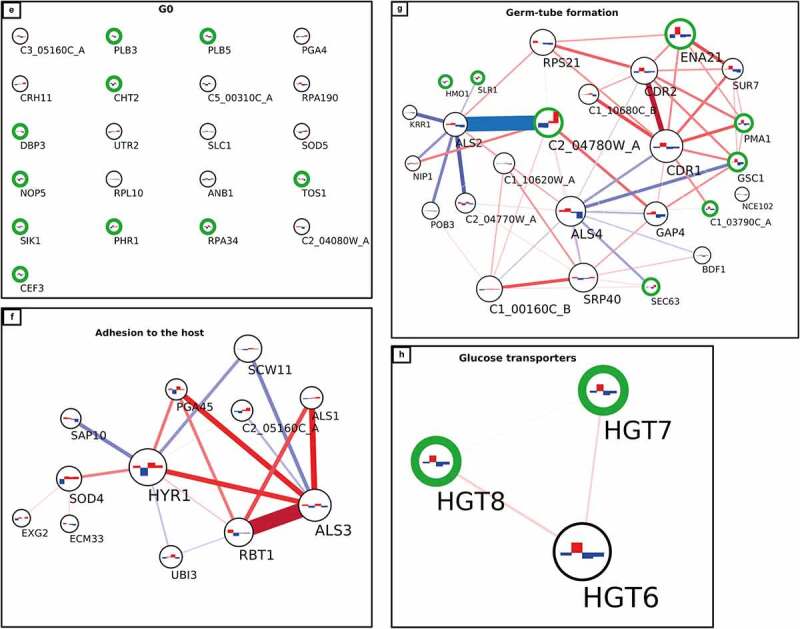
(continued).
Proteins from each group of the network analysis, with a stable difference in the expression between drug-resistant and -susceptible isolates of C. albicans, are listed in Table 2, and are potential diagnostic markers for echinocandin resistance. Moreover, the proteins differentially expressed in response to caspofungin treatment in all isolates, regardless of the isolates’ drug susceptibility are listed in Table 2.
Table 2.
Summary of the groups identified by the integration of a DE analysis and an interaction network built with the proteomics data.
| GO group | Marker | CAS changes |
|---|---|---|
| G1 (CW organization) |
Pga52 | Msb2, C3_07470W_A |
| G2 (Germ-tube formation) |
Gsc1 (Fks1), C2_04780W_A | Slr1 |
| G3 (Modulation of host response) |
Pga10, Rbt5 | Pga31, Skn1 |
| G4 (Adhesion to the host) |
Sap10*, Als1* | Als3* |
| G5 (Ribosomal proteins) |
Rhd3 | Sim1 |
| G6 (Glucose transporters) |
Hgt7,8 | |
| G0 (Ungrouped proteins) |
Plb5 | Cht2, Sik1, Phr1, Rpa34, Tos1 |
The marker column indicates the proteins for each group that were significant for the DE analysis and the CAS changes column the proteins changing in presence of caspofungin for both drug-resistant and –susceptible isolates. *not significant.
Discussion
Treatment options for invasive fungal infections are limited as there are relatively few classes of antifungal drugs available on the market. The main target for current antifungals is plasma membrane ergosterol. Polyene drugs can be fungicidal but toxic to the host, while azoles are fungistatic, hence the fungus is more prone to develop resistance [46]. Echinocandins belong to a different class that target the biosynthesis of cell wall β-(1,3)-glucan [11]. Caspofungin was one of the first echinocandins discovered and patented [47] and is recommended as first-line treatment for Candida infections [10,48]. Despite its fungicidal effect, the occurrence of drug resistance is a problem in the clinic due to acquisition of GSC1 (FKS1) mutations, which decrease the affinity of the drug to the enzyme [49].
In this work, three caspofungin-resistant isolates of C. albicans were characterized and compared to six caspofungin-susceptible isolates, in order to better understand if the resistant isolates had modified call walls and if the two sets of isolates responded differently to the drug. The over-arching aim was to identify possible diagnostic markers for drug resistance.
Car1 is a caspofungin-resistant strain recovered from the kidneys of mice treated with caspofungin, which were infected with the Ca1 strain [33]. Car1 carries the most common hot spot one (HS1) mutation in GSC1 (FKS1) at codon 645 in which serine is replaced by tyrosine. This mutation has been shown to be responsible for a significant increase in the MIC, from 8- to more than 100-fold compared to the wild-type allele, whereas other GSC1 (FKS1) mutations accounted for smaller increases (4- to 30-fold) [13]. The Car3 isolate had a relatively high caspofungin MIC (Table 1) and carried a mutation in HS2 of GSC1 (FKS1, R1361 G), with a nonpolar residue (glycine) substituted for a positively charged one (arginine); this is a mutation more common in C. krusei isolates [50,51].
An important feature of Candida infections in vivo is biofilm formation, which is associated with higher resistance to antifungal drugs. In some cases, biofilms were 1000-fold more resistant to antifungal treatments than planktonic cells [36,58]. The capacity of the nine isolates to form biofilms was assessed in the absence or presence of caspofungin. In general, all the isolates seemed to reach the maximum level of biofilm biomass at the 24 h time point (Figures 1b, 2b). In the absence of drug, the drug-resistant isolates Car1, Car2 had biomass levels comparable to the susceptible isolates after the first time point, with no evident defects in the capacity to form biofilms. The one exception was the Car3 isolate, which did not increase biomass after 6 h (Figure 2b–d). The presence of caspofungin completely inhibited the formation of biofilms in all the drug-susceptible isolates, probably due to the supra-MICs used, causing a decrease in cell viability. In the same conditions, the resistant isolates did not seem to be affected by the presence of the echinocandin. In this case, the amount of drug used was above the IC50 of two out of three isolates, but their biomass levels were comparable to the no-drug condition. This study showed no effect of caspofungin on the capacity to form biofilms by caspofungin-resistant isolates. In contrast, biofilm formation of caspofungin-susceptible isolates was inhibited.
Proteomics has previously shown an increase in the expression, due to caspofungin treatment, of cell wall remodeling enzymes (such as the glucanosyl-transferases Phr1, Phr2) and the Crh family of chitin-glucanosyl-transferases [52,53]. In this work, we also detected altered amounts of several cell wall synthesis and remodeling enzymes in caspofungin-resistant isolates, such as Sun41, Phr1, Phr2, Gsc1, Pmt1, and Mnt1 (Figure 3). Other proteins too were found differentially expressed in the walls of resistant isolates, such as Als3, Als4, Ecm33, and Pga31. Cytoplasmic proteins were detected in increased amount in the susceptible isolates compared to resistant isolates in the absence of drug (Figure 4), which could indicate altered permeability of the wall. Differences in the levels of Gsc1 (Fks1) protein, the echinocandin target, were also detected, with Gsc1 higher in drug-susceptible compared to drug-resistant isolates in the presence and absence of caspofungin (Figure 3 and Figure 3). Previous work showed GSC1 (FKS1) mutations influenced the activity of the enzyme and the echinocandin IC50 for the mutated protein [13], but there was no evidence of different expression patterns. The different Gsc1 (Fks1) levels detected by proteomics could be due to the different genetic backgrounds of the isolates or due to the point mutations affecting protein stability, this has still to be investigated.
These proteomic profiles suggest that cell wall of resistant isolates is likely to be diverse, with altered remodeling and maintenance mechanisms not only upon incubation with drug, but also in normal culture conditions.
The “gold standard” for diagnosing Candida infections is still blood culture, with other additional tests based on the detection of circulating polysaccharides from the fungal cell wall or antigens in blood samples [54]. However, the diagnosis of antifungal resistance is an issue, as it is not routinely performed in the clinic and requires 48–72 h, which is often too late to influence the treatment of the patient [21]. Previous studies focused on molecular and metabolic methods to identify azole resistance [22,55], but no other works focused on biomarkers for diagnosing echinocandin resistance in C. albicans. The network analysis performed in this work identified several proteins that were differentially expressed between resistant and susceptible isolates (Table 2). In particular, future diagnostic approaches using LC-MS/MS should focus on the marker proteins listed in this work. Similar methods were applied for predicting antibiotic resistance in bacteria by the use of MALDI-ToF [56,57], which is not able to identify the specific proteins found differentially expressed in the drug-resistant samples. The protein expression network presented here should be validated on a broader panel of clinical isolates to ensure the identified changes in the cell wall glycoproteome are consistent and common traits in drug-resistant isolates. An interesting possibility is the use of these differentially expressed proteins as markers for antifungal resistance in diagnostic tests that are much quicker to perform. In addition, the highlighted changes in the cell wall proteome may themselves have potential as novel therapeutic targets to block cell wall regeneration mechanisms that reduce drug susceptibility.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Donna MacCallum and Dr. Markus Kostrzewa for providing strains for this work.
Funding Statement
The work was supported by the Horizon 2020 [847507]; H2020 Marie Skłodowska-Curie Actions [H2020-MSCA-ITN-2014-642095]; H2020 Marie Skłodowska-Curie Actions [812969].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
All proteomic data are available at the PRIDE repository accession number: P×D021283.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2081291.
Author contributions
G.B.D.C. and C.A.M. designed the study. G.B.D.C. conducted the experimental work including characterization of the isolates. G.B.D.C. and D.S. performed the proteomic analysis. A.H. and C.L. performed the differential expression analysis and constructed the interaction network. G.B.D.C. and C.A.M. wrote, reviewed, and edited the manuscript.
References
- [1].Dutton LC, Paszkiewicz KH, Silverman RJ, et al. Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. Mol Oral Microbiol. 2016;31(2):136–161. DOI: 10.1111/omi.12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14(4):386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev. 2010;23(2):253–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gow NAR, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15(4):406–412. [DOI] [PubMed] [Google Scholar]
- [5].Hernández-Chávez M, Pérez-García L, Niño-Vega G, et al. Fungal strategies to evade the host immune recognition. J Fungi. 2017;3(4):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gow NAR, Latge J-P, Munro CA. The fungal cell wall: structure, biosynthesis, and function. In: The fungal kingdom. Vol. 5. American Society for Microbiology; 2017. p. 267–292. DOI: 10.1128/microbiolspec.funk-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Plaine A, Walker L, Da Costa G, et al. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 2008;45(10):1404–1414. DOI: 10.1016/j.fgb.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nather K, Munro CA. Generating cell surface diversity in Candida albicans and other fungal pathogens. FEMS Microbiol Lett. 2008;285(2):137–145. [DOI] [PubMed] [Google Scholar]
- [9].Cornely OA, Bassetti M, Calandra T, et al. ESCMID guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect. 2012;18(SUPPL.7):19–37. DOI: 10.1111/1469-0691.12039 [DOI] [PubMed] [Google Scholar]
- [10].Hope WW, Castagnola E, Groll AH, et al. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: prevention and management of invasive infections in neonates and children caused by Candida spp. for Integrated Oncology CIO KölnBonn, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases. 2012. doi: 10.1111/1469-0691.12040 [DOI]
- [11].Douglas CM. Fungal beta(1,3)-D-glucan synthesis.Med Mycol. 2001. [cited 2018 July 5];39(Suppl 1):55–66. Availablse from http://www.ncbi.nlm.nih.gov/pubmed/11800269 [DOI] [PubMed] [Google Scholar]
- [12].Walker LA, Munro CA. Caspofungin induced cell wall changes of Candida species influences macrophage interactions. Front Cell Infect Microbiol. 2020;10:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garcia-Effron G, Park S, Perlin DS. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob Agents Chemother. 2009;53(1):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Garcia-Effron G, Lee S, Park S, et al. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother. 2009;53(9):3690–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kelly J, Kavanagh K. Proteomic analysis of proteins released from growth-arrested Candida albicans following exposure to caspofungin. Med Mycol. 2010;48(4):598–605. [DOI] [PubMed] [Google Scholar]
- [16].Yu ZW, Quinn PJ. Solvation effects of dimethyl sulphoxide on the structure of phospholipid bilayers. Biophys Chem. 1998;70(1):35–39. [DOI] [PubMed] [Google Scholar]
- [17].Bdalla AS, de Hoog GS, Stevens DA, et al. Fructose-Bisphosphate aldolase and pyruvate kinase, two novel immunogens in Madurella mycetomatis. Med Mycol. 2015. DOI: 10.3109/13693786.2011.593005 [DOI] [PubMed] [Google Scholar]
- [18].Mundodi V, Kucknoor AS, Alderete JF. Immunogenic and plasminogen-binding surface-associated alpha-enolase of Trichomonas vaginalis. Infect Immun. 2008;76(2):523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chumpitazi BFF, Lebeau B, Faure-Cognet O, et al. Characteristic and clinical relevance of Candida mannan test in the diagnosis of probable invasive candidiasis. Med Mycol. 2014;52(5):460–469. DOI: 10.1093/mmy/myu018 [DOI] [PubMed] [Google Scholar]
- [20].Prella M, Bille J, Pugnale M, et al. Early diagnosis of invasive candidiasis with mannan antigenemia and antimannan antibodies. Diagn Microbiol Infect Dis. 2005;51(2):95–101. DOI: 10.1016/j.diagmicrobio.2004.08.015 [DOI] [PubMed] [Google Scholar]
- [21].Zhao Y, Perlin DS. Use of novel tools to probe drug resistance in Fungi. In: Handbook of antimicrobial resistance; 2017. p. 385–401. DOI: 10.1007/978-1-4939-0694-9_21. [DOI] [Google Scholar]
- [22].Park S, Perlin DS. Establishing surrogate markers for fluconazole resistance in Candida albicans. Microb Drug Resist. 2005;11(3):232–238. [DOI] [PubMed] [Google Scholar]
- [23].Kordalewska M, Lee A, Zhao Y, et al. Detection of Candida auris antifungal drug resistance markers directly from clinical skin swabs. Antimicrob Agents Chemother. 2019;63(12). DOI: 10.1128/AAC.01754-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao Y, Nagasaki Y, Kordalewska M, et al. Rapid detection of FKS-associated echinocandin resistance in Candida glabrata. Antimicrob Agents Chemother. 2016;60(11):6573–6577. DOI: 10.1128/AAC.01574-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang D, An N, Yang Y, et al. Candida tropicalis distribution and drug resistance is correlated with ERG11 and UPC2 expression. Antimicrob Resist Infect Control. 2021;10(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Amorim-Vaz S, Tran VDT, Pradervand S, et al. RNA enrichment method for quantitative transcriptional analysis of pathogens in vivo applied to the fungus Candida albicans. Mbio. 2015;6(5):e00942-15. DOI: 10.1128/mBio.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Remmele CW, Luther CH, Balkenhol J, et al. Integrated inference and evaluation of host-fungi interaction networks. Front Microbiol. 2015;6:764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martin R, Albrecht-Eckardt D, Brunke S, et al. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One. 2013;8(3). DOI: 10.1371/journal.pone.0058613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans.Genetics. 1993. [cited 2017 Apr 6];134(3):717–728. DOI: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations.Mol Gen Genet. 1984. [cited 2017 Apr 5];198(2):179–182. DOI: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- [31].Morrison CJ, Hurst SF, Bragg SL, et al. Heterogeneity of the purified extracellular aspartyl proteinase from Candida albicans: characterization with monoclonal antibodies and N-terminal amino acid sequence analysis. Infect Immun. 1993. [cited 2019 Aug 1];61(5):2030–2036. DOI: 10.1128/iai.61.5.2030-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Thanos M, Schonian G, Meyer W, et al. Rapid identification of Candida species by DNA fingerprinting with PCR. J Clin Microbiol. 1996. [cited 2019 Aug 1];34(3):615–621. DOI: 10.1128/jcm.34.3.615-621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee KK, MacCallum DM, Jacobsen MD, et al. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother. 2012;56(1):208–217. DOI: 10.1128/AAC.00683-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].M27: reference method for broth dilution antifungal susceptibility testing of yeasts, 4th edition; [cited 2022. Jan 30]. Avalilable from: https://clsi.org/standards/products/microbiology/documents/m27/
- [35].Walker LA, Munro CA, De Bruijn I, et al. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. Cormack BP, ed. PLoS Pathog. 2008;4(4):e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ramage G, VandeWalle K, Wickes BL, et al. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45(9):2475–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kapteyn JC, Hoyer LL, Hecht JE, et al. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol. 2000;35(3):601–611. DOI: 10.1046/j.1365-2958.2000.01729.x [DOI] [PubMed] [Google Scholar]
- [38].Johansson HKL, Svingen T, Boberg J, et al. Calretinin is a novel candidate marker for adverse ovarian effects of early life exposure to mixtures of endocrine disruptors in the rat. Arch Toxicol. 2020;94(4):1241–1250. DOI: 10.1007/s00204-020-02697-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Science M. Mascot search engine | Protein identification software for mass spec data; [cited 2018. Sep 18]. Available from: http://www.matrixscience.com/
- [40].Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. DOI: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chiquet J, Mariadassou M, Robin S.. Variational inference for sparse network reconstruction from count data. 2018.
- [42].Rex JH, Alexander BD, Andes D, et al. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard - third edition. Clin Lab Stand Inst. 2008;(April):1–25. DOI: 10.4319/lo.1981.26.3.0590 [DOI] [Google Scholar]
- [43].Crump JA, Collignon PJ. Intravascular catheter-associated infections.Eur J Clin Microbiol Infect Dis. 2000. [cited 2018 Sep 6];19(1):1–8. DOI: 10.1007/s100960050001. [DOI] [PubMed] [Google Scholar]
- [44].Sardi JCO, Scorzoni L, Bernardi T, et al. Candida species: current epidemiology, pathogenicity, biofilms formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(PART1):10–24. [DOI] [PubMed] [Google Scholar]
- [45].Frade JP, Arthington-Skaggs BA. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses. 2011;54(4):e154–e162. [DOI] [PubMed] [Google Scholar]
- [46].Berkow EL, Lockhart SR. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist. 2017;10:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Black RM, Bouffard FA, Plains S. Of all N. USOO5378804A 11 Patent Number: 5. Vol. 378; 1995. [Google Scholar]
- [48].Cornely OA, Bassetti M, Calandra T, et al. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients for integrated oncology CIO KölnBonn, cologne excel- lence cluster on cellular stress responses in aging-associated diseases. New Microbes New Infect. 2012;18:19–37. [DOI] [PubMed] [Google Scholar]
- [49].Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10(3):121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Park S, Kelly R, Kahn JN, et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother. 2005;49(8):3264–3273. DOI: 10.1128/AAC.49.8.3264-3273.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Desnos-Ollivier M, Bretagne S, Raoux D, et al. Mutations in the fks1 gene in Candida albicans, C. tropicalis, and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European committee on antibiotic susceptibility testing. Antimicrob Agents Chemother. 2008;52(9):3092–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fonzi WA. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans.J Bacteriol. 1999. [cited 2019 Aug 20];181(22):7070–7079. DOI: 10.1128/JB.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pardini G, De Groot PWJ, Coste AT, et al. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J Biol Chem. 2006;281(52):40399–40411. DOI: 10.1074/jbc.M606361200 [DOI] [PubMed] [Google Scholar]
- [54].Consortium O, Gabaldón T, Butler G, et al. Recent trends in molecular diagnostics of yeast infections: from PCR to NGS. FEMS Microbiol Rev. 2019;43(5):517–547. DOI: 10.1093/femsre/fuz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li L, Liao Z, Yang Y, et al. Metabolomic profiling for the identification of potential biomarkers involved in a laboratory azole resistance in Candida albicans. PLoS One. 2018;13(2). DOI: 10.1371/JOURNAL.PONE.0192328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Flores-Treviño S, Garza-González E, Mendoza-Olazarán S, et al. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci Reports 2019 91. 2019;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Maus A, Bisha B, Fagerquist C, et al. Detection and identification of a protein biomarker in antibiotic resistant E. coli using intact protein LC offline MALDI-MS and MS/MS. J Appl Microbiol. 2020;128(3):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rajendran R, Sherry L, Nile CJ, et al. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect. 2016;22(1):87–93. DOI: 10.1016/j.cmi.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomic data are available at the PRIDE repository accession number: P×D021283.


