Abstract
The virulence of a pathogen is dependent on a discrete set of genetic determinants and their well-regulated expression. The ctxAB and tcpA genes are known to play a cardinal role in maintaining virulence in Vibrio cholerae, and these genes are believed to be exclusively associated with clinical strains of O1 and O139 serogroups. In this study, we examined the presence of five virulence genes, including ctxAB and tcpA, as well as toxR and toxT, which are involved in the regulation of virulence, in environmental strains of V. cholerae cultured from three different freshwater lakes and ponds in the eastern part of Calcutta, India. PCR analysis revealed the presence of these virulence genes or their homologues among diverse serotypes and ribotypes of environmental V. cholerae strains. Sequencing of a part of the tcpA gene carried by an environmental strain showed 97.7% homology to the tcpA gene of the classical biotype of V. cholerae O1. Strains carrying the tcpA gene expressed the toxin-coregulated pilus (TCP), demonstrated by both autoagglutination analysis and electron microscopy of the TCP pili. Strains carrying ctxAB genes also produced cholera toxin, determined by monosialoganglioside enzyme-linked immunosorbent assay and by passage in the ileal loops of rabbits. Thus, this study demonstrates the presence and expression of critical virulence genes or their homologues in diverse environmental strains of V. cholerae, which appear to constitute an environmental reservoir of virulence genes, thereby providing new insights into the ecology of V. cholerae.
Vibrio cholerae is known to be an autochthonous inhabitant of brackish waters and estuarine systems (4, 13). Among the 193 currently recognized O serogroups of V. cholerae (43), only serogroups O1 and O139 have caused epidemics of cholera. The other serogroups of V. cholerae, collectively referred to as non-O1 non-O139 serogroups, have not been associated with epidemics but can cause sporadic diarrhea (30) and are ubiquitously distributed in the aquatic environment (22, 26). This sharp distinction between serogroups which can cause cholera and those which are not associated with cholera is related to the observation that more than 95% of the strains belonging to serogroups O1 and O139 produce cholera toxin (CT), which is central to the disease process. In contrast, more than 95% of the strains belonging to non-O1 non-O139 serogroups do not produce CT (15). Another important virulence factor of V. cholerae is the toxin-coregulated pilus (TCP), which is an adhesin that is coordinately regulated with CT production (39). TCP is the only V. cholerae pilus that has been demonstrated to date to have a role in colonization of the gut mucosa of humans (9) and of infant mice (39), the latter being an experimental cholera model.
It has been presumed that CT and TCP are exclusively associated with clinical strains of V. cholerae, notably those belonging to serogroups O1 and O139, whereas reports on the incidence of CT among environmental strains of V. cholerae are rare (24). Similarly, TCP has rarely been reported among environmental strains of V. cholerae, suggesting that TCP is associated only with virulent V. cholerae O1 or O139. Recently, the presence of tcpA in some non-O1 toxigenic strains (8, 32) and in two nontoxigenic, non-O1 non-O139 strains has been published (27).
The genes encoding CT form part of the genome of a lysogenic filamentous bacteriophage, designated CTXφ. The pilus colonization factor TCP is also known to act as a receptor for CTXφ, which can infect nontoxigenic V. cholerae, leading to the emergence of new toxigenic strains (42). The tcpA gene is part of a pathogenicity island of about 39.5 kb known as the V. cholerae pathogenicity island (VPI) (16). The structural features of VPI are suggestive of a bacteriophage origin, and there is at least one report describing the production of a bacteriophage designated VPIφ (17). This supports the current hypothesis that some pathogenic bacteria have evolved from nonpathogenic strains of the same species via horizontal transfer of virulence genes (5).
To understand the ecology of the V. cholerae serogroups associated with cholera, it is important to determine the origin and distribution of virulence genes among environmental strains. In the study reported here, isolation and analysis of unique strains of V. cholerae of environmental origin which possess virulence gene(s) homologues are described. These homologues appear to be variants, or intermediates, in the evolution of virulence genes.
(Part of this paper was presented at the 34th Joint Conference on Cholera and Other Bacterial Enteric Infections Panel, U.S.-Japan Cooperative Medical Sciences Program, held at Shonan Village, Japan, between 30 November and 3 December 1998.)
MATERIALS AND METHODS
Collection and processing of environmental samples.
Water, sediment, and plankton samples collected from three different freshwater lakes and ponds located in the eastern part of Calcutta (longitude 88°20′E, latitude 22°32′N), India, were examined from July 1997 to June 1998, as described previously (3, 25). Water samples were filtered using Whatman no. 1 filter paper and subsequently filtered through a 0.45-μm-pore-size membrane (Millipore Corp., Bedford, Mass.) using vacuum pressure of 15 to 20 lb/in2. The membrane was cut into eight pieces and vortexed in 2 ml of 10 mM phosphate-buffered saline (PBS, pH 7.4) for 3 min. One milliliter of the suspension was added to 10 ml of alkaline peptone water (APW) containing peptone (1%, wt/vol) and NaCl (1%, wt/vol) (pH 8.5) contained in 20-ml screw-cap glass tubes, for enrichment at 37°C with shaking (100 rpm) for 16 to 18 h.
Sediment samples were added to 100 ml of distilled water until the final volume reached 200 ml, mixed well, and allowed to settle. A 10-ml amount of the slurry was centrifuged at 2,000 rpm for 8 min at room temperature to remove particulate matter, and 1 ml of slurry was added to 10 ml of APW (pH 8.5) for enrichment, as described above. Plankton samples were collected using a 20-μm plankton net. The samples were further concentrated using Whatman no. 1 filter paper, with the paper containing the plankton then being washed with 3 ml of PBS. The suspension was homogenized using a glass homogenizer. One milliliter of the homogenized sample was added to 10 ml of APW (pH 8.5) for enrichment. The enriched samples from each of the components, i.e., sediment, water, and plankton, were screened for virulence genes by PCR, as described below.
Isolation of single-cell clones containing virulence genes.
A search for strains possessing the virulence gene(s) was performed when APW (pH 8.5)-enriched samples yielded a positive PCR amplicon for any of the virulence genes of V. cholerae sought in this study. Each sample (20 μl) was streaked on thiosulfate-citrate-bile-sucrose agar (TCBS) (Eiken, Tokyo, Japan) and tellurite taurocholate gelatin agar (TTGA) (trypticase agar base, 10 g; NaCl, 10 g; sodium taurocholate, 5 g; sodium carbonate, 1 g; gelatin, 30 g; agar, 15 g per liter; potassium tellurite, 1% [wt/vol]; pH 8.5) plates. In addition, the enriched samples were also serially diluted and plated on Luria agar (LA; Difco, Detroit, Mich.) supplemented with 1% (wt/vol) NaCl. The rationale for using TCBS, TTGA, and LA concurrently was to search for strains of V. cholerae as well as other heterotrophic bacterial flora which might carry the virulence genes that were being sought. The plates were incubated overnight at 37°C. LA plates which contained 30 to 300 colonies were selected, and each colony was assigned a number. One-third of the colonies were randomly selected for further analysis. A part of each selected colony was inoculated into 2 ml of Luria broth (LB) and grown at 37°C to prepare DNA for confirmation of the presence of the virulence gene by PCR. To prepare template DNA, 1 ml of the culture was centrifuged, resuspended in sterile distilled water, and boiled for 10 min. Similarly, each colony selected from the TCBS and TTGA plates was inoculated into LB, incubated at 37°C with shaking, and processed to obtain template DNA for PCR as described above.
Serology.
The identity of V. cholerae was confirmed as described previously (30). The 24 V. cholerae strains which were found to possess one or another of the virulence genes sought were examined for agglutination by the somatic O antigen serogrouping scheme for V. cholerae developed at the National Institute of Infectious Diseases, Tokyo, Japan (43).
PCR and sequencing.
Three pairs of primers (ctxA, tcpA [classical variant; henceforth designated tcpA-C], and tcpA [El Tor variant; henceforth designated tcpA-E]) were used in the first set of multiplex PCR (18), and two pairs of primers (ctxB and sto [encodes heat-stable enterotoxin]) were used in the second set of multiplex PCR, as described elsewhere (28, 29). The cycling conditions for the PCR assay included an initial denaturation at 94°C for 5 min, followed by 30 cycles of 1.5 min of denaturation at 94°C, 1.5 min of primer annealing at 60°C (for the first set) and 1 min at 55°C (for the second set), and 1.5 min of primer extension at 72°C. PCR assays were also performed to detect the V. cholerae regulatory genes toxR and toxT. The primers used for amplification of toxR and toxT were those described elsewhere (2, 21). Cycling conditions for PCR included an initial denaturation at 94°C for 5 min, 30 cycles of 0.5 min at 94°C (denaturation), 0.5 min at 64°C (primer annealing), and 0.5 min at 72°C (primer extension) for toxR; and 25 cycles of 1 min at 94°C (denaturation), 1 min at 50°C (primer annealing), and 1 min at 72°C (primer extension) for toxT. All PCR assays were performed using an automated thermal cycler (Biometra, Gottingen, Germany).
Sequencing of double-stranded DNA from purified PCR products was carried out using the Taq dye terminator sequencing kit (Perkin-Elmer) and an automated DNA sequencer (ABI Prism 377), following the manufacturer's instructions. Both strands were sequenced using the same forward and reverse primers, which were used for amplifying the classical biotype-specific tcpA. The sequences were aligned using the DNAsis software program (Hitachi), and searches for nearly identical sequences were performed using the Basic Local Alignment Search Tool (BLAST) program available on the National Center for Biotechnology Information network server.
DNA extraction.
A modification of the method of Murray and Thompson (23) was used for DNA extraction. In brief, cells from an 18-h LB culture were collected and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), treated with 10% (wt/vol) sodium dodecyl sulfate and freshly prepared proteinase K (Sigma Chemical Co., St. Louis, Mo.), and incubated at 37°C for 1 h. After incubation, 10% cetyl trimethyl ammonium bromide in 0.7 M NaCl was added and incubated at 65°C for 10 min. The aqueous phase was treated with phenol-chloroform, and the DNA pellet was washed with 70% ethanol. The extracted nucleic acid was suspended in TE and treated with RNase at 37°C for 30 min.
Probes and hybridization.
The presence of virulence-associated genes was confirmed by using specific DNA probes. The ctxA probe consisted of a 540-bp XbaI-ClaI fragment of ctxA cloned in pKTN901 using EcoRI linkers (14). The DNA fragment used as the probe for tcpA in the Southern blot hybridization was generated by PCR using primers described elsewhere (18). The rRNA gene probe consisted of a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pKK3535 (7).
Genomic DNA from representative environmental strains of V. cholerae was digested with the appropriate restriction endonuclease, and the fragments were electrophoretically separated in a 0.8% agarose gel using TAE buffer (40 mM Tris-acetate, 1 mM EDTA). DNA was transferred to a Hybond N+ membrane (Amersham International, PLC, Buckinghamshire, England) using 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) by vacuum transfer (Amersham). The membrane was washed with 10× SSC and dried at room temperature. DNA was covalently immobilized to the membrane using alkali fixation. Southern blotting with probes conjugated to horseradish peroxidase to allow hybridization to be detected with a chemiluminescent substrate (Amersham) was performed as described elsewhere (41). The membrane was washed and exposed to X-ray film (Fuji Film, Fuji, Japan) and developed following the manufacturer's instructions.
The rRNA probe was labeled by random priming with a random primer DNA labeling kit (BRL) and [α-32P]dCTP (3,000 Ci/mmol) (Amersham). Southern blots were hybridized with the labeled probe, and autoradiographs were developed as described elsewhere (7).
Autoagglutination.
The hydrophobicity of V. cholerae is greatly increased in broth culture due to the expression of pili, which cause visible clumping of bacteria, leaving a pellet at the bottom of the tube and a clear supernatant. This phenomenon is known as autoagglutination and has previously been shown to be correlated with the expression of TCP (39). To determine whether environmental V. cholerae strains possessing tcpA-C expressed pili, the strains were grown in LB (pH 6.8) supplemented with 1% (wt/vol) NaCl and incubated at 37°C for 18 h.
Electron microscopy.
Agar media were used to examine the expression of the pilus by the environmental strains of V. cholerae possessing tcpA. The media used included colonization factor antigen (CFA) agar (34) and LB supplemented with 20 g of Bacto agar (Difco) per liter. The strains were incubated at 25°C for 24 or 36 h. Samples were taken from the different agar media at the designated times and processed. Bacterial suspension (5 μl in 10 mM phosphate-buffered saline [pH 7.4]) was deposited on a 300-mesh copper grid coated with a film of pyelovar and stabilized with a thin layer of carbon. After about 1 to 2 min, the excess fluid was blotted and stained with 2% (wt/vol) uranyl acetate for 1 min. Grids were examined using a Philips 420T transmission electron microscope.
Detection of CT by GM1 ELISA.
To detect expression of CT by the environmental strains, the cells were grown either in AKI (containing [per liter] Bactopeptone, 15 g; NaCl, 5 g; yeast extract, 5 g; sodium bicarbonate, 3 g; pH 7.5 [11]) or in YEP (containing [per liter] yeast extract, 4 g; Bactopeptone, 15 g; NaCl, 5 g; pH 7.5 [10]) medium at 37°C, with shaking, for 16 h. After centrifugation, the supernatant was examined for the presence of CT by a monosialoganglioside (GM1) enzyme-linked immunosorbent assay (ELISA) as described by Svennerholm and Holmgren (37). Pure CT (lot no. 19H4022), obtained from Sigma Chemical Co., St. Louis, Mo., was used as the positive control.
Animal passage.
The rabbit ileal loop model was used for animal passage of V. cholerae strain SCE188 (ctxAB+ tcpA), as described previously (19). The cells grown in YEP were introduced into the rabbit ileum and incubated for 18 h. Fluid from the rabbit ileal loop was plated on TCBS, typical cholera organism-like colonies were picked, and their identity was reconfirmed both by biochemical tests and by the multiplex PCR described above. The rabbit-passaged strains were grown in AKI and YEP, and CT from the culture supernatant was measured by GM1 ELISA (37).
Nucleotide sequence accession number.
The nucleotide sequence of the tcpA gene from environmental strain SCE5 of V. cholerae has been deposited with the DNA Data Bank of Japan (DJBB) with accession number AB012946.
RESULTS AND DISCUSSION
Presence of ctxA, tcpA, toxR, and toxT in environmental strains of V. cholerae.
A total of 122 samples (44 water, 34 sediment, and 44 plankton samples) collected from three sites in Calcutta between July 1997 and June 1998 were analyzed by conventional bacteriology and by multiplex PCR assays after enrichment of the samples in APW (pH 8.5). Two multiplex PCR assays were designed to detect five known virulence genes of V. cholerae, including tcpA-E, tcpA-C, ctxA, ctxB, and sto (gene encoding the heat-stable enterotoxin of V. cholerae). Of the 122 enriched APW (pH 8.5) samples analyzed using the two sets of multiplex PCR, five water and four plankton samples examined at different time intervals were positive for either tcpA or ctxAB; none were positive for sto. None of the sediment samples were positive for any of the virulence genes sought. A total of 19 strains of V. cholerae positive for tcpA and another 5 strains positive for ctxA and ctxB were isolated (Table 1). These 24 virulence gene-positive strains of V. cholerae were isolated after examining approximately 4,800 colonies (an average of 200 colonies per search) from either TCBS, TTGA, or LA. Of the 19 environmental V. cholerae strains examined, the size of the tcpA amplicon in 17 strains matched the size of the tcpA amplicon (617 bp) of the reference classical strain (V. cholerae O395), while in 2 strains the size of the tcpA amplicon (471 bp) matched that of the reference El Tor strain (V. cholerae VC20) (Fig. 1). All 19 strains were, however, negative for the 301-bp ctxA and 460-bp ctxB amplicons, indicating that these strains did not have the genetic potential to produce CT. Furthermore, five environmental strains of V. cholerae which were positive for ctxA and ctxB were negative for tcpA with the set of primers used in this study (18). This is contrary to the current assumption that most CT-positive strains are also positive for TCP, since TCP is known to be the receptor for CTXφ infection of V. cholerae. All 24 strains were positive for toxR, a transcriptional activator of many virulence genes in V. cholerae (20, 21). In contrast, toxT was found in only three strains (SCE4, SCE5, and SCE6) positive for tcpA and all five strains positive for ctxAB (Table 1).
TABLE 1.
Details of environmental strains of V. cholerae possessing virulence genes isolated in this study
| Strain | Date of isolation (mo/day/yr) | Source | Serogroup | Ribotypea | Presence of virulence gene:
|
|||
|---|---|---|---|---|---|---|---|---|
| ctxAB | tcpAb | toxR | toxT | |||||
| SCE4 | 7/7/97 | Plankton, freshwater lake | O8 | A | − | + | + | + |
| SCE5 | 7/7/97 | Plankton, freshwater lake | O11 | A | − | + | + | + |
| SCE6 | 7/7/97 | Plankton, freshwater lake | O8 | A | − | + | + | + |
| SCE188 | 12/2/97 | Water, fish farm | O44 | F | + | − | + | + |
| SCE200 | 12/2/97 | Water, fish farm | O44 | ND | + | − | + | + |
| SCE201 | 12/2/97 | Water, fish farm | O44 | ND | + | − | + | + |
| SCE223 | 1/8/98 | Plankton, freshwater pond | O27 | G | + | − | + | + |
| SCE225 | 1/8/98 | Plankton, freshwater pond | O35 | B | − | + | + | − |
| SCE226 | 1/8/98 | Plankton, freshwater pond | O35 | G | − | + | + | − |
| SCE227 | 1/8/98 | Water, fish farm | O35 | B | − | + | + | − |
| SCE228 | 1/8/98 | Plankton, fish farm | O35 | B | − | + | + | − |
| SCE256 | 2/10/98 | Water, fish farm | O42 | C | − | + | + | − |
| SCE257 | 2/10/98 | Water, fish farm | O42 | D | − | + | + | − |
| SCE258 | 2/10/98 | Water, fish farm | O42 | D | − | + | + | − |
| SCE259 | 2/10/98 | Water, fish farm | O42 | D | − | + | + | − |
| SCE260 | 2/10/98 | Water, fish farm | O42 | ND | − | + | + | − |
| SCE261 | 2/10/98 | Water, fish farm | O42 | ND | − | + | + | − |
| SCE263 | 2/10/98 | Water, fish farm | O10 | E | − | + | + | − |
| SCE264 | 2/10/98 | Water, fish farm | O42 | C | − | + | + | − |
| SCE265 | 2/10/98 | Water, fish farm | O42 | C | − | + | + | − |
| SCE340 | 5/5/98 | Plankton, freshwater pond | O69 | H | − | + | + | − |
| SCE341 | 5/5/98 | Plankton, freshwater pond | O69 | H | − | + | + | − |
| SCE354 | 5/19/98 | Water, freshwater pond | O27 | I | + | − | + | + |
| SCE359 | 5/19/98 | Water, fish farm | O8 | J | − | + | + | − |
The various ribotype patterns designated A to J are shown in Fig. 3. ND, not determined.
The tcpA from strains SCE340 and SCE341 belonged to the El Tor variant, while tcpA from the other strains belonged to the classical variant.
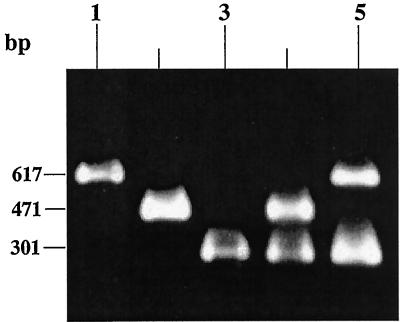
FIG. 1.
PCR analysis of tcpA and ctxA of genomic DNA from representative environmental strains of V. cholerae isolated from enriched APW (pH 8.5) samples. Lane 1, environmental strain of V. cholerae (SCE5) possessing tcpA-C; lane 2, environmental strain of V. cholerae (SCE340) possessing tcpA-E; lane 3, environmental strain of V. cholerae (SCE188) possessing ctxA; lane 4, V. cholerae O1 Ogawa, El Tor (positive control for ctxA and tcpA-E); lane 5, V. cholerae O1, Ogawa, classical (positive control for ctxA and tcpA-C).
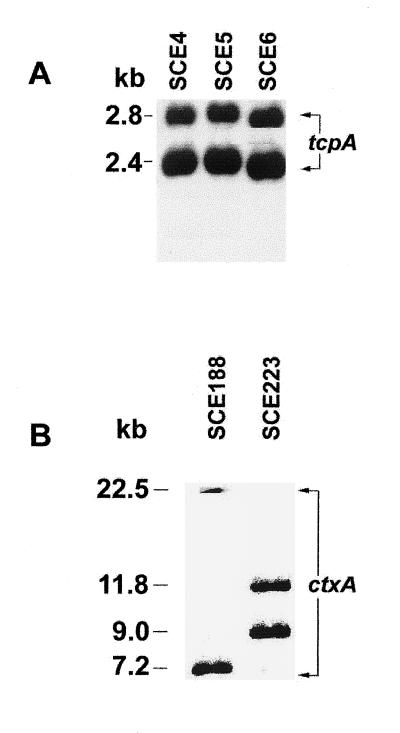
The results of PCR assays were confirmed by Southern hybridization when representative strains positive for tcpA or ctxA were hybridized with the respective probes. Strains positive for tcpA-C (SCE4, SCE5, and SCE6) revealed two fragments after digestion with PstI and probing with tcpA (Fig. 2). Southern blot hybridization using the ctxA probe after digestion of DNA of the representative strains SCE188 and SCE223 with PstI revealed two different restriction patterns, as shown in Fig. 2.
FIG. 2.
Southern blot hybridization of PstI-digested genomic DNA from environmental strains of V. cholerae using tcpA (A) and ctxA (B) probes.
Analysis of ribotypes.
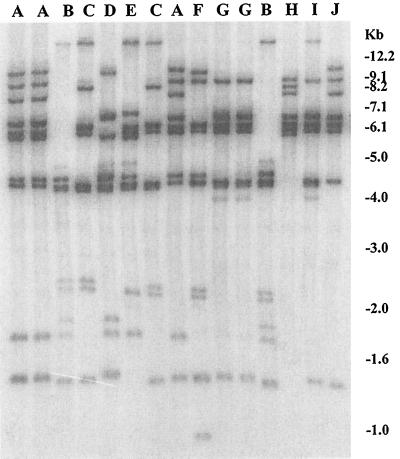
Analysis of BglI restriction patterns of conserved rRNA genes (ribotype) in the environmental strains revealed clonal diversity, and 10 different ribotypes (A through J) were detected (Fig. 3). The distribution of ribotypes among the strains belonging to different serogroups is shown in Table 1. Two strains, SCE4 and SCE5, which belonged to different serogroups (O8 and O11, respectively) belonged to a single ribotype (A). Another O8 strain belonged to a different ribotype (J). Four strains belonging to the O35 serogroup shared two different ribotypes (B and G). Strains belonging to serogroup O42 shared ribotypes C and D. A toxigenic strain, SCE223, shared the same ribotype with a nontoxigenic strain, SCE228. Ribotyping was performed to determine whether strains of V. cholerae isolated from a given APW (pH 8.5) enrichment broth of a particular sample were siblings. For example, strains of V. cholerae with different serogroups (O27 and O35) as well as different ribotypes (B and G) were isolated from an APW (pH 8.5) enrichment of plankton samples collected from a freshwater pond in Calcutta on 8 January 1998. Similarly, APW enrichment of plankton samples collected from a freshwater lake on 7 July 1997 yielded three strains with two different serogroups (O8 and O11) but a single ribotype (A). We also isolated two strains of V. cholerae from an enrichment culture of a sample of water from a fish farm taken on 10 February 1998 which had the same serogroup (O42) and ribotype (C).
FIG. 3.
BglI restriction patterns of rRNA genes in environmental strains of V. cholerae isolated in Calcutta. Ribotype patterns A through J produced by different strains are shown (see Table 1 for details). Numbers indicating the molecular sizes of bands correspond to a 1-kb DNA ladder (Bethesda Research Laboratories).
Nucleotide sequence of tcpA of environmental strains of V. cholerae resembling classical tcpA.
Among several putative colonization factors of V. cholerae, TCP has been shown to be essential for colonization in the infant mouse model as well as in human volunteers (1, 9, 38–40). Thus, the function of TCP in colonization of the human intestinal epithelium is well established, as is its partial homology to type 4 or N-methylphenylalanine pili, the long surface filaments found in a variety of pathogenic bacteria, notably Neisseria gonorrhoeae, Moraxella bovis, Pseudomonas aeruginosa, and Dichelobacter nodosus (6, 10, 35). To determine whether the 617-bp tcpA amplicon was similar to the tcpA of classical V. cholerae epidemic strains, we sequenced this amplified fragment of DNA and determined the extent of similarity between the nucleotide sequences of the amplified DNA and the reported sequence of classical tcpA (6). The nucleotide sequence data of the tcpA-like amplicon of strain SCE5 (V. cholerae serogroup O11), obtained from two sequencing reactions of two independent amplicons, yielded readable sequences of 597 bases, with 97.7% identity to the tcpA gene sequence of the classical V. cholerae O1 (6). Notably, only 14 bases differed. Furthermore, it was found that the derived amino acid sequence of TcpA in environmental strains had an identity of 98.5% when compared with the reported amino acid sequence of TcpA that is found in classical strains of V. cholerae O1, with differences discernible only at positions 104, 144, and 154 (Fig. 4). Homology between the environmental V. cholerae TcpA amino acid sequence and V. cholerae El Tor TcpA sequence (31) was 80.4%, with 39 of the deduced TcpA residues of the environmental strain (SCE5) differing from those of TcpA of the V. cholerae El Tor biotype (Fig. 4).
FIG. 4.
Multiple alignment of pilin amino acid sequence of TcpA-C, TcpA of SCE5, and TcpA-E. The alignment was created by the DNAsis (Hitachi) program. The shaded areas indicate identical residues, while unshaded areas indicate dissimilar residues. The GenBank accession numbers for tcpA-C and tcpA-E are M33514 and U89807, respectively.
The close similarity of most of the tcpA genes found in environmental strains of V. cholerae to the classical V. cholerae tcpA is interesting, despite the fact that the current cholera pandemic is caused by the El Tor biotype. Recent epidemiological data from Bangladesh, where classical V. cholerae existed until 1991 (36), show the absence of this biotype (A. K. Siddique, personal communication). However, the data obtained in this study indicate that a tcpA gene similar to the classical type is present in environmental non-O1, non-O139 V. cholerae strains. An environmental reservoir of tcpA genes of the classical type strongly suggests the possibility of a reemergence of the classical biotype via gene transfer events in the environment. The classical biotype transiently reemerged in Bangladesh in 1983 as the predominant epidemic strain, about 10 years after its apparent replacement by the El Tor biotype (33).
Expression of tcpA.
To determine whether the tcpA genes were expressed, the 19 strains of V. cholerae that were positive for the tcpA amplicon were examined further. It was found that three strains (SCE4, SCE5, and SCE6) exhibited the autoagglutination phenotype when incubated in LB containing 1% NaCl at pH 6.5 with aeration at 37°C. The conditions were different from those reported for expression of classical tcpA, which included growth at 30°C at pH 6.5 in LB (39). Autoagglutination was not observed in any other broth medium or cultural conditions in the other 16 tcpA-positive strains examined. The 16 tcpA-positive but autoagglutination-negative strains were negative for the virulence regulator toxT.
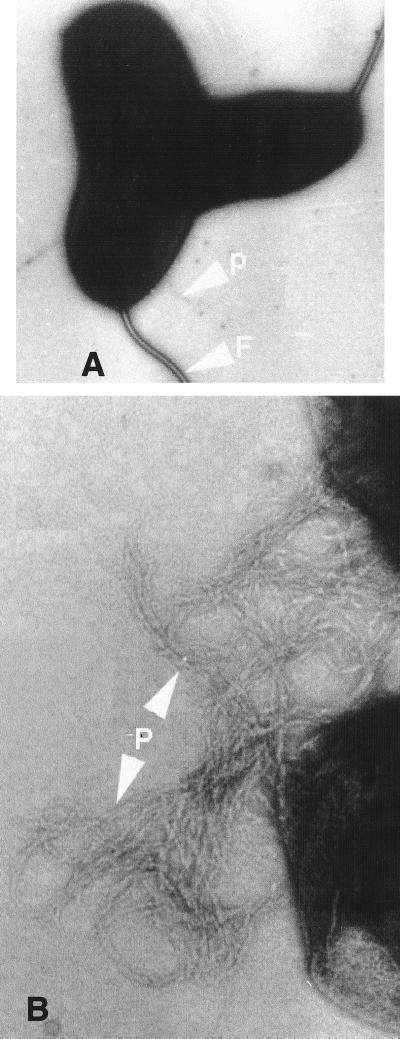
Transmission electron microscopy of negatively stained specimens of SCE5 performed to visualize the pilus revealed that growth on CFA agar for 24 h at 25°C resulted in production of pili attached to the surface of the bacteria (Fig. 5A). Pili in bundles were observed after incubation for 36 h (Fig. 5B). Thus, we were able to demonstrate expression of Tcp by three strains by testing the autoagglutination phenotype and also to visualize pili of SCE5 by electron microscopy. Interestingly, strains positive for both tcpA and toxT showed the autoagglutination phenotype, whereas strains positive for tcpA but negative for toxT did not autoagglutinate. In epidemic strains of V. cholerae, the tcpA gene is located in a 39.5-kb DNA segment along with other physically linked genes involved in Tcp biogenesis (16). It was concluded that the complete VPI is not present in the remaining 16 tcpA-positive environmental strains of V. cholerae.
FIG. 5.
Electron micrographs of pili of environmental V. cholerae strain SCE5. Bacteria were cultured at 25°C for 24 h on CFA agar and negatively stained. (A) Single pilus. Magnification, ×40,000. (B) After 36 h, TCP bundles. Magnification, 147,000. P, pili; F, flagellum.
Expression of CT.
Strains SCE188, -200, and -201 expressed CT in both AKI and YEP media, used for optimal production of CT from El Tor and classical V. cholerae, respectively (11, 12). However, the amount of CT antigen produced by SCE188 and SCE201 was higher in YEP than in AKI, while the yield of CT from SCE200 was the same whether grown in YEP or AKI. Despite possessing DNA fragments with sequences very similar to that of CT genes, two of the environmental isolates, SCE223 and SCE354, did not produce detectable amounts of CT when grown in either YEP or AKI. Passage of strain SCE188 in a rabbit ileum resulted in positive fluid accumulation and isolation of strains that produced twofold more CT than the wild type. This result (Table 2) suggests that selection for strains producing larger amounts of CT in the rabbit ileum occurs in both environmental and epidemic strains (19).
TABLE 2.
Detection of CT produced by environmental strains of V. cholerae in YEP and AKI media by GM1 ELISAa
| Strain or component tested | OD492
|
|
|---|---|---|
| YEP | AKI | |
| SCE188 | 0.76 ± 0.27 | 0.35 ± 0.03 |
| SCE188a | 1.78 ± 0.56 | 0.77 ± 0.20 |
| SCE200 | 0.69 ± 0.24 | 0.69 ± 0.13 |
| SCE201 | 0.89 ± 0.09 | 0.64 ± 0.11 |
| SCE223 | 0.17 ± 0.07 | 0.18 ± 0.10 |
| SCE354 | 0.06 ± 0.01 | 0.06 ± 0.01 |
| VC20 | 2.43 ± 0.12 | 1.81 ± 0.42 |
| SCE18 | 0.11 ± 0.04 | 0.03 ± 0.02 |
| Bufferb | 0.13 ± 0.04 | 0.01 ± 0.12 |
| Medium | 0.17 ± 0.02 | 0.22 ± 0.03 |
| CT (1 ng) | 0.64 ± 0.03 | 0.59 ± 0.08 |
| CT (0.1 ng) | 0.33 ± 0.01 | 0.25 ± 0.02 |
VC20 was a positive control, SCE18 was a negative control, and SCE188a was animal passaged.
Components (buffer, medium, CT) were tested without strains.
Conclusion.
In this study, the occurrence and distribution of selected virulence-associated genes in environmental strains of V. cholerae that had been isolated in Calcutta, India, were demonstrated. These environmental V. cholerae strains were neither O1 nor O139, nor did they carry together the genes for the major virulence factors CT and TCP. Nevertheless, these strains constitute a potential reservoir of virulence genes in the environment. Diverse serogroups of V. cholerae are shown, for the first time, to harbor these genes. What is most exciting is that molecular characterization of microbial ecosystems provides useful information about the ecology of V. cholerae, a bacterium autochthonous to riverine, coastal, and estuarine ecosystems but, at the same time, pathogenic for humans. Environmental studies of V. cholerae have been done with the expectation that V. cholerae strains possessing the entire complement of virulence genes would be isolated. Now it is concluded that virulence genes are dispersed among environmental strains of V. cholerae and may be ferried about, given the fact that most of the virulence genes that were studied are located on mobile elements. Indeed, the potential for “mixing and matching” of genes in the environment or in the human intestine, leading to new pathogenic variants, must now be addressed. Ribotypes of the strains isolated in this study were shared by strains belonging to more than one serogroup, and conversely, a particular serogroup comprised more than one ribotype. Toxigenic strains and nontoxigenic strains belonging to an identical ribotype were also detected, further supporting the hypothesis of gene transfer among vibrios in the environment. Further studies on the ecology and evolution of V. cholerae will surely provide new insights into the epidemiology of cholera.
ACKNOWLEDGMENTS
This work was supported, in part, by the Japan International Cooperation Agency (JICA/NICED Project 054-1061-E-O) and by the National Institutes of Health (grant 1RO1A1392901).
REFERENCES
- 1.Attridge S R, Voss E, Manning P A. The role of toxin-coregulated pili in the pathogenesis of Vibrio cholerae O1 El Tor. Microb Pathog. 1993;15:421–431. doi: 10.1006/mpat.1993.1091. [DOI] [PubMed] [Google Scholar]
- 2.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the toxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty S, Mukhopadhyay A K, Bhadra R K, Ghosh A N, Mitra R, Colwell R R, Shimada T, Yamasaki S, Takeda Y, Berg D E, Nair G B. Proceedings of the 34th Joint Conference on Cholera and Other Bacterial Enteric Infections. The U.S.-Japan Cooperative Medical Sciences Program.; 1998. Molecular ecology of Vibrio cholerae: occurrence and implications of virulence genes in environmental strains of V. cholerae; pp. 15–19. Taiyo-bijutsu, Koto, Tokyo, Japan. [Google Scholar]
- 4.Colwell R R. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 5.Covacci A, Falkow S, Berg D, Rappuoli R. Did the inheritance of a pathogenicity island modify the virulence of Helicobacter pylori? Trends Microbiol. 1997;5:205–208. doi: 10.1016/S0966-842X(97)01035-4. [DOI] [PubMed] [Google Scholar]
- 6.Faast R, Ogierman M A, Stroeher U H, Manning P A. Nucleotide sequence of the structural gene, tcpA, for a major pilin subunit of Vibrio cholerae. Gene. 1989;85:227–231. doi: 10.1016/0378-1119(89)90486-1. [DOI] [PubMed] [Google Scholar]
- 7.Faruque S M, Siddique A K, Saha M N, Asadulghani, Rahman M M, Zaman K, Albert M J, Sack D, Sack R B. Molecular characterization of a new ribotype of Vibrio cholerae O139 Bengal associated with an outbreak of cholera in Bangladesh. J Clin Microbiol. 1999;37:1313–1318. doi: 10.1128/jcm.37.5.1313-1318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh C, Nandy R K, Dasgupta S K, Nair G B, Hall R H, Ghose A C. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR, and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb Pathog. 1997;22:199–208. doi: 10.1006/mpat.1996.0105. [DOI] [PubMed] [Google Scholar]
- 9.Herrington D A, Hall R H, Losonsky G A, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 11.Iwanaga M, Kuyyakanond T. Large production of cholera toxin by Vibrio cholerae O1 in yeast extract peptone water. J Clin Microbiol. 1987;25:2314–2316. doi: 10.1128/jcm.25.12.2314-2316.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwanaga M, Yamamoto K. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J Clin Microbiol. 1985;22:405–408. doi: 10.1128/jcm.22.3.405-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaper J B, Lockman H, Colwell R R, Joseph S W. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 1979;37:91–103. doi: 10.1128/aem.37.1.91-103.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaper J B, Morris J G, Jr, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious diseases. Boca Raton, Fla: CRC Press; 1988. pp. 65–77. [Google Scholar]
- 15.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaolis D K R, Johnson J A, Baily C C, Boedker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 18.Keasler S P, Hall R H. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 19.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 20.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 22.Morris J G., Jr Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- 23.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair G B, Oku Y, Takeda Y, Ghosh A, Ghosh R K, Chattopadhyay S, Pal S C, Kaper J B, Takeda T. Toxin profiles of Vibrio cholerae non-O1 from environmental sources in Calcutta, India. Appl Environ Microbiol. 1988;54:3180–3182. doi: 10.1128/aem.54.12.3180-3182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair G B, Sarkar B L, De S P, Chakraborty M K, Bhadra R K, Pal S C. Ecology of Vibrio cholerae in the fresh water environs of Calcutta. Microb Ecol. 1988;15:203–215. doi: 10.1007/BF02011713. [DOI] [PubMed] [Google Scholar]
- 26.Nalin D R, Daya V, Reid A, Levine M M, Cisneros L. Adsorption and growth of Vibrio cholerae on chitin. Infect Immun. 1979;25:768–770. doi: 10.1128/iai.25.2.768-770.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novais R C, Coelho A, Salles C A, Vincente A C P. Toxin- co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol Lett. 1999;171:49–55. doi: 10.1111/j.1574-6968.1999.tb13411.x. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa A, Takeda T. The gene encoding the heat-stable enterotoxin of Vibrio cholerae is flanked by 123-base-pair direct repeats. Microbiol Immunol. 1993;37:607–616. doi: 10.1111/j.1348-0421.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 29.Olsvik Ø, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth I K, Fields P I. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol. 1993;31:22–25. doi: 10.1128/jcm.31.1.22-25.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramamurthy T, Bag P K, Pal A, Bhattacharya S K, Bhattacharya M K, Sen D, Shimada T, Takeda T, Takeda Y, Nair G B. Virulence patterns of Vibrio cholerae non-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J Med Microbiol. 1993;39:310–317. doi: 10.1099/00222615-39-4-310. [DOI] [PubMed] [Google Scholar]
- 31.Rhine J A, Taylor R K. tcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 32.Said B, Smith H R, Scotland S M, Rowe B. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol Lett. 1995;125:205–210. doi: 10.1111/j.1574-6968.1995.tb07359.x. [DOI] [PubMed] [Google Scholar]
- 33.Samadi A R, Shahid N, Eusuf A, Yunus M, Huq M I, Khan M U, Rahman A S M M, Faruque A S G. Classical Vibrio cholerae biotype displaces El Tor in Bangladesh. Lancet. 1983;i:805–807. doi: 10.1016/s0140-6736(83)91860-3. [DOI] [PubMed] [Google Scholar]
- 34.Sharma D P, Thomas C, Hall R H, Levine M M, Attridge S R. Significance of toxin-coregulated pili as protective antigens of Vibrio cholerae in the infant mouse model. Vaccine. 1989;7:451–456. doi: 10.1016/0264-410x(89)90161-8. [DOI] [PubMed] [Google Scholar]
- 35.Shaw C E, Taylor R K. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect Immun. 1990;58:3042–3049. doi: 10.1128/iai.58.9.3042-3049.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddique A K, Baqui A H, Eusof A, Haider K, Hossain M A, Bashier I, Zaman K. Survival of classic cholera in Bangladesh. Lancet. 1991;337:1125–1127. doi: 10.1016/0140-6736(91)92789-5. [DOI] [PubMed] [Google Scholar]
- 37.Svennerholm A M, Holmgren J. Identification of Escherichia coli heat-labile enterotoxin by means of ganglioside immunosorbent assay (GM1 ELISA) procedure. Curr Microbiol. 1978;1:19–23. [Google Scholar]
- 38.Tacket C O, Taylor R K, Losonsky G, Lim Y, Nataro J P, Kaper J B, Levine M M. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. The use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thelin K H, Taylor R K. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization regulated with cholera toxin. Proc Natl Acad Sci USA. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldor M K, Mekalanos J J. toxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 43.Yamai S, Okitsu T, Shimada T, Katsube Y. Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin, and addition of novel serogroups. Jpn J Assoc Infect Dis. 1997;71:1037–1045. doi: 10.11150/kansenshogakuzasshi1970.71.1037. . (In Japanese.) [DOI] [PubMed] [Google Scholar]