Abstract
Campylobacter jejuni is recognized as a leading human food-borne pathogen. Traditional diagnostic testing for C. jejuni is not reliable due to special growth requirements and the possibility that this bacterium can enter a viable but nonculturable state. Nucleic acid-based tests have emerged as a useful alternative to traditional enrichment testing. In this article, we present a 5′-nuclease PCR assay for quantitative detection of C. jejuni and describe its evaluation. A probe including positions 381121 to 381206 of the published C. jejuni strain NCTC 11168 genome sequence was identified. When this probe was applied, the assay was positive for all of the isolates of C. jejuni tested (32 isolates, including the type strain) and negative for all other Campylobacter spp. (11 species tested) and several other bacteria (41 species tested). The total assay could be completed in 3 h with a detection limit of approximately 1 CFU. Quantification was linear over at least 6 log units. Quantitative detection methods are important for both research purposes and further development of C. jejuni detection methods. In this study, we used the assay to investigate to what extent the PCR signals generated by heat-killed bacteria interfere with the detection of viable C. jejuni after exposure at elevated temperatures for up to 5 days. An approach to the reduction of the PCR signal generated by dead bacteria was also investigated by employing externally added DNases to selectively inactivate free DNA and exposed DNA in heat-killed bacteria. The results indicated relatively good discrimination between exposed DNA from dead C. jejuni and protected DNA in living bacteria.
Campylobacter jejuni has come to be recognized worldwide as a leading cause of diarrheal disease and food-borne gastroenteritis (37). C. jejuni is zoonotic, with many animals serving as reservoirs for human disease (21). Campylobacter cells may enter the environment, including drinking water, through the feces of animals, birds, or infected humans. C. jejuni is susceptible to a variety of environmental conditions that make it unlikely to be metabolically active for long periods of time outside the host (21, 27). However, the organism may remain dormant in water in a state that has been termed “viable but nonculturable” (VNC) (32). These organisms are not able to grow but may survive in the environment for several weeks (3, 4). Contaminated water, raw milk, and poultry appear to be the most common vehicles of transmission of C. jejuni in humans (37). It has been estimated that as little as approximately 500 cells of C. jejuni can cause human illness (2, 31). However, little is known about the virulence factors and the mechanisms of pathogenicity (21).
There are several problems concerning Campylobacter detection, including the low infective dosage numbers and the slow growth rate of the organisms. The traditional methods currently used are time-consuming and laborious, requiring prolonged incubation and selective enrichment to reduce the growth of the background flora to enable biochemical identification. Campylobacter cells may also enter the VNC state due to starvation and physical stress, which may explain the failure of the culture techniques to isolate the organisms from contaminated water samples implicated in outbreaks of infection (14, 32). DNA-based methods such as the PCR have been increasingly used for rapid, sensitive, and specific nonquantitative detection of C. jejuni (25, 40, 46). A quantitative assay, however, is still not available for this organism. Quantitative assays are desirable for diagnostic, legislative, and research purposes. Quantitation is especially important for C. jejuni in relation to the VNC state in elucidating otherwise elusive routes of infection. Among the various quantitative PCR strategies available, those based on real-time monitoring of the amplification reaction are the most accurate (10, 12, 26).
There are, however, still some limitations to nucleic acid-based diagnostics. A major obstacle encountered with the current DNA-based tests is the separation of living and dead microorganisms (11, 18). In order to exploit the full potential of PCR for microbiological diagnosis, there is a great demand for sample preparation methods related to whether the organisms are living, VNC, or dead. Most food decontamination and preservation techniques are aimed on either inactivating or removing potential pathogens (5). The ability of the nucleic acids from the dead cells to generate PCR signals is affected by both the decontamination treatment and the organisms (11, 33). The intrinsic bacterial DNases (11, 36) may also affect the half-life of DNA. It is important to know the stability of nucleic acids in order to interpret whether a potential positive PCR signal is due to living pathogens.
An aspect that has not yet been widely exploited in PCR diagnostics is the physical difference between living and dead cells. The nucleic acids in living cells are protected because the cell walls and membranes are intact. In dead cells, these barriers are compromised and the nucleic acids are thus exposed to compounds added to the sample (24, 30). The differential exposure of DNA in dead cells may be utilized to destabilize or inactivate the exposed nucleic acids, while the nucleic acids within living cells are protected from the treatment by the cell membrane and wall.
In this paper, we describe the development and evaluation of a primer and probe system directed toward a DNA fragment from C. jejuni which can be used in the quantification of C. jejuni using 5′-nuclease chemistry and the ABI Prism 7700 Sequence Detection System from PE Biosystems (Foster City, Calif.). Using this assay and DNA purification by magnetic beads, we investigated the influence of nucleic acids from heat-killed bacteria on the detection of viable C. jejuni. Finally, an approach to the reduction of the background signal generated by DNA in heat-killed bacteria by using external DNases was evaluated. The results presented here provide important background material for the development and interpretation of diagnostic methods for C. jejuni in naturally contaminated water and foods.
MATERIALS AND METHODS
Bacterial strains, media, and cultures.
Thirty-two isolates of C. jejuni, including the C. jejuni type strain, DSMZ 4688T (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), were used to test the specificity of the primers and the probe. The isolates were collected mostly from patients in Norway from 1996 to 1999, but the patients were assumed to have been infected in different parts of the world.
The following bacterial strains were used as negative controls (strains not assigned to any known culture collection are from private collections): C. coli (three strains, including the type strain [DSMZ 4689T]), C. concisus CCUG 13144T (type strain; Culture Collection, University of Göteborg, Göteborg, Sweden), C. curvus CCUG 13146T (type strain), C. fetus subsp. fetus, C. helveticus CCUG 30682T (type strain), C. hyointestinalis subsp. hyointestinalis CCUG 14169T (type strain), C. lari, C. rectus CCUG 20446BT (type strain), C. showae CCUG 30254T (type strain), C. sputorum subsp. bubulis, C. sputorum subsp. sputorum, Actinobacter calcoaceticus, Arcobacter butzleri, A. cryaerophilus CCUG 17801T (type strain), A. skirrowii CCUG 10374T (type strain), Bacillus cereus, B. subtilis ATCC 6633 (American Type Culture Collection, Manassas, Va.), Bacteroides ureolyticus CCUG 7319T (type strain), Brochothrix thermosphacta ATCC 11509T (type strain), Carnobacterium divergens DSMZ 20623T (type strain), C. gallinarum NCFB 2766T (type strain; National Collection of Food Bacteria, Reading, England), Citrobacter freundii NCTC 9750 (type strain; National Collection of Type Cultures, Colindale, London, England), Clostridium perfringens ATCC 27324, Enterobacter aerogenes, E. agglomerans NCTC 9381 (type strain), E. cloacae NCTC 10005 (type strain), Enterococcus faecalis NCDO 2602 (type strain; National Collection of Dairy Organisms, Reading, England), Escherichia coli ATCC 8739, E. coli O157; H7, Flavobacterium odoratum ATCC 4651 (type strain), Flexispira rappini CCUG 23435, Helicobacter fennelliae CCUG 18820T (type strain), H. pylori CCUG 15815 BT (type strain), Lactobacillus acidophilus (type strain), L. casei ATCC 393, L. curvatus DSMZ 20019T (type strain), L. plantarum DSMZ 20174T (type strain), L. sake subsp. sake DSMZ 20017T (type strain), Listeria grayi, L. innocua DSMZ 20649T (type strain), L. ivanovii, L. monocytogenes DSMZ 20600T (type strain), L. seeligeri, L. welshimeri, Micrococcus luteus ATCC 272, Pseudomonas aeruginosa ATCC 15442, Salmonella enterica, S. enterica serovar Kentucky, Shigella sonnei ATCC 11060, Staphylococcus aureus ATCC 25923, Streptococcus pyogenes type 1 ATCC 12344T (type strain), and Yersinia enterocolitica.
All strains were plated on blood agar. The Arcobacter spp. were grown aerobically on Preston broth medium without Preston Campylobacter selective supplement at 30°C. The Bacteroides, Campylobacter, Citrobacter, Flexispira, and Helicobacter spp. were grown microaerobically on Preston broth medium without Preston Campylobacter selective supplement at 37°C. The Clostridium, Enterobacter, Shigella, and Streptococcus spp. were grown anaerobically on brain heart infusion (BHI) broth at 37°C. B. thermosphacta was grown aerobically on BHI broth at 25°C. Enterobacter aerogenes and the Actinobacter, Bacillus, Enterococcus, Escherichia, Listeria, Micrococcus, Pseudomonas, Salmonella, Staphylococcus, and Yersinia spp. were grown aerobically on BHI broth at 30°C. F. odoratum was grown aerobically on BHI broth at 37°C. Lactobacilli were grown aerobically on MRS broth at 25°C, and C. gallinarum was grown aerobically on Trypticase soy yeast medium at 30°C. All agar and media were from Oxoid Ltd., Basingstoke, Hampshire, England. Cultures were serially diluted in Preston broth medium without Preston Campylobacter selective supplement. CFU were enumerated by plating of 0.1 ml of each dilution onto blood agar no. 2, code CM271, with 5% laked horse blood, code SR48, Oxoid Ltd., and microaerobic incubation at 37°C for 2 days.
DNA isolation.
Samples (0.5 ml) of overnight cultures were centrifuged at 6,000 × g for 7 min at 4°C, and the supernatants were discarded. The pellets were stored at −80°C. For DNA isolation, pellets from the C. jejuni type strain were resuspended in 1× TE buffer (10 mM Tris, 1 mM EDTA), pH 8.0. Dynabeads DNA Direct I (Dynal AS, Oslo, Norway), 200 μl, were then added to the suspension of bacteria, and the bacterium-bead suspension was incubated at 65°C for 20 min, followed by incubation at room temperature for another 2 min. DNA bound to magnetic beads was then drawn to the wall of the microcentrifuge tube by a magnet (MPC-E; Dynal AS) for 2 min. The supernatant containing salts, detergent, and cell debris was carefully removed without disrupting the Dynabead-DNA complex. The beads were washed twice with a washing buffer (buffer 2 from the kit). Finally, the DNA was removed from the beads by resuspension in 40 μl of 10 mM Tris HCl, pH 8.0 (buffer 3 from the kit), and incubation at 65°C for 5 min. The beads, now released from the DNA, were collected with the magnet, and the DNA-containing supernatant was transferred to a fresh tube and used directly in the PCR.
TaqMan probe and primer design.
The probe regions used were localized in the completed C. jejuni strain NCTC 11168 genome sequence (http://www.sanger.ac.uk/Projects/C_jejuni/). The Primer Express (version 1.0) ABI Prism (PE Biosystems) was used for the primer-probe design, together with guidelines from PE Biosystems (17). The GCG version of FastA (28) was used to search for similarities to other known sequences.
5′-nuclease-based PCR assay.
Amplification reaction mixtures (50 μl) contained a DNA sample (1 μl); 1× TaqMan buffer A; 5 mM MgCl2; 200 μM each dATP, dCTP, and dGTP; 400 μM dUTP; 0.02 μM C. jejuni-specific probe; 0.3 μM each C. jejuni-specific primer; 1 U of AmpErase uracil N-glycosylase; and 2.5 U of AmpliTaq Gold DNA polymerase. PCR samples and controls were prepared in triplicate. Reaction tubes were MicroAmp Optical tubes, and tube caps were MicroAmp Optical caps. All consumables were supplied by PE Biosystems.
Before amplification, the PCR mixture was heated to 50°C in 5 min to let the uracil N-glycosylase destroy possibly contaminating PCR products and at 95°C for 10 min to denature the template DNA. The amplification profile was 40 cycles of 95°C for 20 s and 60°C for 1 min. Reactions were performed in the ABI Prism 7700 Sequence Detection System (PE Biosystems). Reaction conditions were programmed and data were analyzed on a power Macintosh 4400/20 (Apple Computer, Santa Clara, Calif.) linked directly to the ABI Prism 7700 Sequence Detection System using the SDS 1.6.3 application software (PE Biosystems) as described by the manufacturer. PCR products were detected directly by monitoring the increase in fluorescence from the dye-labeled C. jejuni-specific DNA probe. The TaqMan probe consisted of an oligonucleotide with a 5′ reporter dye and a 3′ quencher dye. The reporter dye carboxyfluorescein was covalently linked to the 5′ end of the oligonucleotide. The fluorescence of the reporter was quenched by 6-carboxy-N,N,N′,N′-tetramethylrhodamine, located at the 3′ end. When the probe was intact, the proximity of the reporter dye to the quencher dye resulted in suppression of the reporter fluorescence. If the probe was cleaved, the reporter and quencher dyes were separated, causing the reporter dye fluorescence to increase. The amplification was plotted as ΔRn, which was the normalized reporter signal (reporter signal minus background), against the number of cycles. A threshold signal was chosen, which intersected the amplification curves in the linear region of the semilog plot. This gave the threshold cycle (CT), which is defined as the PCR cycle where an increase in fluorescence first occurred, for each amplification plot. Different amplifications could then be compared by their respective CTs. The CTs were plotted against log input DNA or cells, which gave standard curves for quantification of unknown samples and the ability to estimate the amplification efficiency of the reaction (10, 29). The PCR product was verified with ethidium bromide-stained 3% agarose gels (SeaPlaque GTG Agarose; FMC BioProducts, Rockland, Maine). Agarose gel electrophoresis was performed essentially as described by Sambrook et al. (35).
Heat treatments.
All experiments were performed in triplicate. Approximately 2.1 × 107 ± 0.4 × 107 cells of the C. jejuni type strain were used in each experiment. The C. jejuni cultures were pelleted at 6,000 × g for 7 min at 4°C, washed, resuspended in water, and transferred to microcentrifuge tubes. The tubes were then incubated at 25, 55, 72, and 100°C, and samples for PCR analysis and plating were investigated at intervals of 5 min, 1 h, 6 h, 24 h, and 5 days. The effect of heat treatment of the cultures and addition of DNase on DNA stability at room temperature was investigated. We used 1.7 × 107 ± 0.8 × 107 cells of the C. jejuni type strain with DNase and 5.0 × 107 ± 0.6 × 107 cells without DNase in these experiments. The cultures were incubated at 20, 55, 72, and 100°C for 5 min and 121°C for 15 min. One set of tubes was then incubated further at room temperature, and samples for PCR analysis were removed at intervals of 5 min, 15 min, and 30 min, 1 h, 6 h, 24 h, and 5 days. Ten units of RQ1 DNase (Promega, Madison, Wis.) and 1× DNase buffer were added to another set of tubes before the incubation at room temperature. Aliquots were analyzed after 5, 15, and 30 min and after 1, 6, and 24 h. For the PCR analysis, DNAs were purified from 10-μl aliquots taken at the respective time points and the subsequent 5′-nuclease PCR assay was performed as described above.
Stability of free DNA versus DNA in intact cells.
Purified DNA from approximately 9.6 × 107 cells was added to a suspension containing 1.1 × 107 ± 0.8 × 107 living cells. The ability of DNase to selectively degrade the free DNA was investigated by addition of 10 U of RQ1 DNase (Promega) and 1× DNase buffer. After 1 h of incubation at room temperature, the cells were pelleted by centrifugation (for separate analysis of the free DNA only) and a 100-μl aliquot of the supernatant was immediately heated to 95°C for 5 min to inactivate the DNase. One microliter of the supernatant was then used in the 5′-nuclease PCR assay.
RESULTS
PCR fragment specificity.
Specific PCR primers and a probe were designed for C. jejuni. The probe region was chosen to optimize specificity and amplification efficiency. The putative primers and probe were constructed using the primer express program, and then these DNA sequences were subjected to a FastA search (28) in the EMBL database (release 60). An 86-bp fragment including positions 381121 to 381206 of the published C. jejuni strain NCTC 11168 genome sequence (http://www.sanger.ac.uk/Projects/C_jejuni/) was identified in these screenings. There were no known sequences in the EMBL database with significant homology to this probe region. The most closely related sequence had 57.7% identity and was located in the putative gene yonO in the complete sequence of B. subtilis strain 168 (16). PCR primers were constructed from the regions including positions 381121 to 381145 (forward) and 381206 to 381185 (reverse), while the probe includes positions 381147 to 381181 (Table 1).
TABLE 1.
Primers and fluorogenic probe for specific detection of C. jejuni
| Probe or primer | Sequence (5′ - 3′) | Denaturation temp (°C)a |
|---|---|---|
| Primers | ||
| Forward | CTG AAT TTG ATA CCT TAA GTG CAG C | 60.4 |
| Reverse | AGG CAC GCC TAA ACC TAT AGC T | 60.3 |
| Probe | TCT CCT TGC TCA TCT TTA GGA TAA ATT CTT TCA CA | 66.6 |
Calculated by the nearest-neighbor algorithm with the Primer Express program (primer concentration, 300 nM; probe concentration, 20 nM; salt concentration, 55 nM).
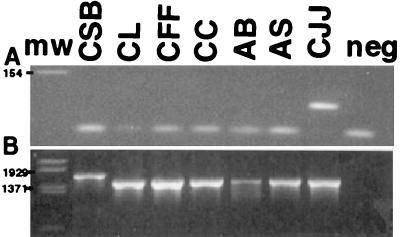
After the probe region was identified on a theoretical basis, the specificity of the selected primers and probes was subjected to an empirical screening. A total of 32 C. jejuni isolates, including the type strain, were tested and found specific to the chosen primers and probe. The specificity of the primers and probe was tested against 13 strains of 11 other Campylobacter species and a set of 41 species belonging to other genera of phylogenetically related or common food-borne organisms and pathogens (see Materials and Methods), all of which were found negative. In these experiments, the quality of the purified DNA was verified through amplification with universal 16S rRNA gene PCR primers (Fig. 1B). In addition, a qualitative PCR with the amplification primers alone was done for selected strains (Fig. 1A). These experiments confirmed that the amplification primers are specific for C. jejuni. Unspecific PCR products other than common artifacts like primer dimers were not detected.
FIG. 1.
Amplification products from the C. jejuni-specific primers (A) and a universal 16S rRNA gene PCR primer pair (39) (B) for a set of Campylobacter strains. The samples were subjected to electrophoresis in 3% (A) and 2% (B) agarose gel at 100 V for 45 min. Ten microliters of the amplification product was loaded in each lane. Lanes: CSB, C. sputorum subsp. bubulis; CL, C. lari; CFF, C. fetus subsp. fetus; CC, C. concisus; AB, A. butzleri; AS, A. skirrowii; CJJ, C. jejuni subsp. jejuni; neg, negative control; mw, molecular weight marker.
Quantitative aspects and detection limits.
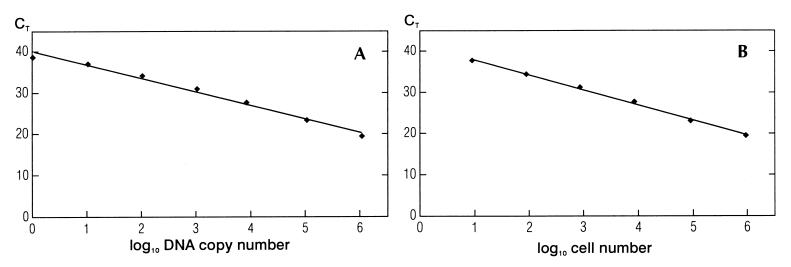
Serial dilutions of purified DNA or cells were made, and two types of standard curves were constructed: DNA standard curves and cell standard curves. For DNA standard curves, DNA isolated from approximately 4.2 × 107 C. jejuni cells was serially diluted 10-fold in 1× TE buffer and subjected to PCR. The standard curve based on the dilutions of DNA showed a linear relationship between log input DNA and threshold cycles (Fig. 2A). The slope of the curve was −3.27, and the square regression coefficient after the linear regression was 0.988. When a new serial 10-fold dilution from the same DNA purification was used in a separate PCR experiment, the slopes in the two runs were almost identical (−3.20 and −3.27). When DNA from a separate isolation was used, the variation in the slopes of the standard curves was no larger than that between different serial dilutions from the same DNA isolation (−3.28 and −3.27) and the square regression coefficient (R2) remained constant. For cell standard curves, approximately 4.2 × 107 cells were serially diluted 10-fold. DNA was isolated from each dilution and subjected to PCR. The standard curve based on six 10-fold dilutions of cells showed a linear relationship between log input cells and the CTs (Fig. 2B). The slope of the curve was −3.66, and the square regression coefficient after the linear regression was 0.997. When the same serial 10-fold dilution of cells (slope, −3.68 versus −3.66) and a new serial 10-fold dilution of cells (slope, −3.46 versus −3.66) were used in separate PCR experiments, the slopes in the different runs were almost identical. When DNA from a separate isolation was used, the variation in the slopes of the standard curves was still small (slope, −3.48 versus −3.66). The square regression coefficient (R2) remained constant during all experiments. Both the DNA standard curves and the cell standard curves showed a higher degree of variability among the triplicates when the amount of template decreased. However, the standard deviations were too small to be indicated in Fig. 2A and B. When the detection limit in the cell standard curves was 1 CFU, the slope of the cell standard curves were similar to that of the DNA standard curves (e.g., −3.23 versus −3.27) and the square regression coefficients were identical, i.e., R2 = 0.988. DNA standard curves showed that the detection limit of the PCR assay was approximately 1 CFU per PCR (Fig. 2A). Cell standard curves showed a detection limit of 10 CFU per PCR (Fig. 2B).
FIG. 2.
(A) 5′-nuclease PCR analysis of serial 10-fold dilutions of C. jejuni DNA. CTs are plotted against the calculated copies of bacterial DNA, i.e., a 10-fold dilution of the bacterial DNA (1.04 × 106 copies/μl). The straight line, which was calculated by linear regression [y = −3.27x (number of cells) + 40.10], shows a square regression coefficient (R2) of 0.988. (B) 5′-nuclease PCR analysis of serial 10-fold dilutions of C. jejuni cells. CTs are plotted against the number of cells of C. jejuni. Template DNA was extracted from samples of cells containing serial 10-fold dilutions from approximately 4.2 × 107 CFU of C. jejuni. The straight line, which was calculated by linear regression [y = −3.66x (number of cells) + 41.54], shows a square regression coefficient (R2) of 0.997.
Effect of heat treatment on C. jejuni DNA stability.
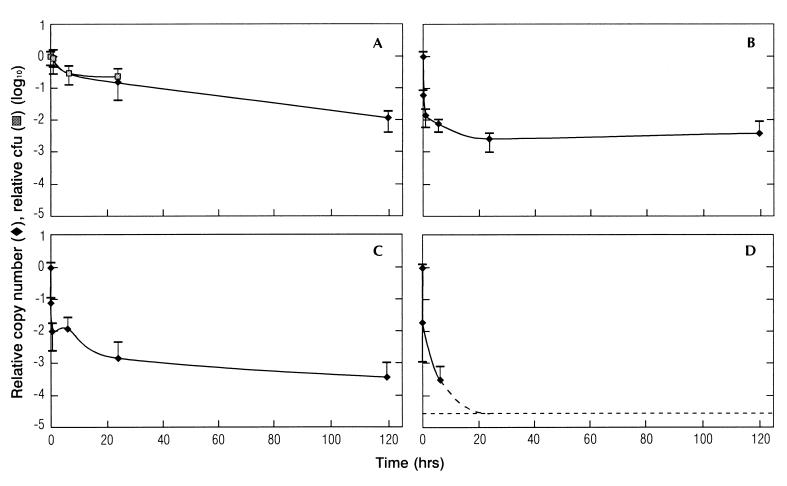
The effect on DNA stability of prolonged exposure to room temperature (approximately 25°C at the time of the experiments) or 55, 72, or 100°C for 5 min or 1, 6, 24, or 120 h was investigated (Fig. 3). Both the colony-forming ability of C. jejuni and the PCR signal generated were determined.
FIG. 3.
Effect of heat treatment on DNA stability. Cells were incubated at 25°C (A), 55°C (B), 72°C (C), and 100°C (D) for up to 5 days, and DNA was quantified by 5′-nuclease PCR assay after 5 min, 1 h, 6 h, 24 h, and 5 days. The amounts of DNA (copy numbers) and numbers of culturable cells (CFU counts) are given relative to the values before heat treatment. The error bars show standard deviations.
The numbers of CFU of cultivable cells at 25°C were 84, 30, and 22% of those at time zero after 1, 6, and 24 h, respectively. There was a relatively good correlation up to 24 h between the CFU counts and the signal generated by the 5′-nuclease PCR for the cells exposed to 25°C (Fig. 3A). The kinetics of both the cell counts and the PCR signal had a relatively short half-life during the first 6 h. From 6 to 24 h, the half-life had stabilized at a lower level. After 5 days, no culturable C. jejuni could be recovered, while the PCR signal was approximately 1% of the signal for the input material.
No viable cells of bacteria exposed to 55°C or greater heat could be recovered. The PCR signal at 55°C was approximately 7% relative to the signal from cells incubated at room temperature at time zero. There was a rapid decline to approximately 1% of the input signal after 1 h. From 1 to 24 h, there was a slower decline to approximately 0.3% of the input signal and after 24 h there was no further reduction in the PCR signal (Fig. 3B). The PCR inactivation kinetics at 72°C was approximately similar to that at 55°C until 24 h. From 24 h on, however, the kinetics resembled that at 25°C (Fig. 3C). Boiling of the sample (100°C) resulted in a 4.5-log reduction of the PCR signal after 6 h relative to the input signal. After 24 h, the detection limit of the assay was reached (Fig. 3D).
Effect of externally added DNases on the stability of DNA in heat-treated cells.
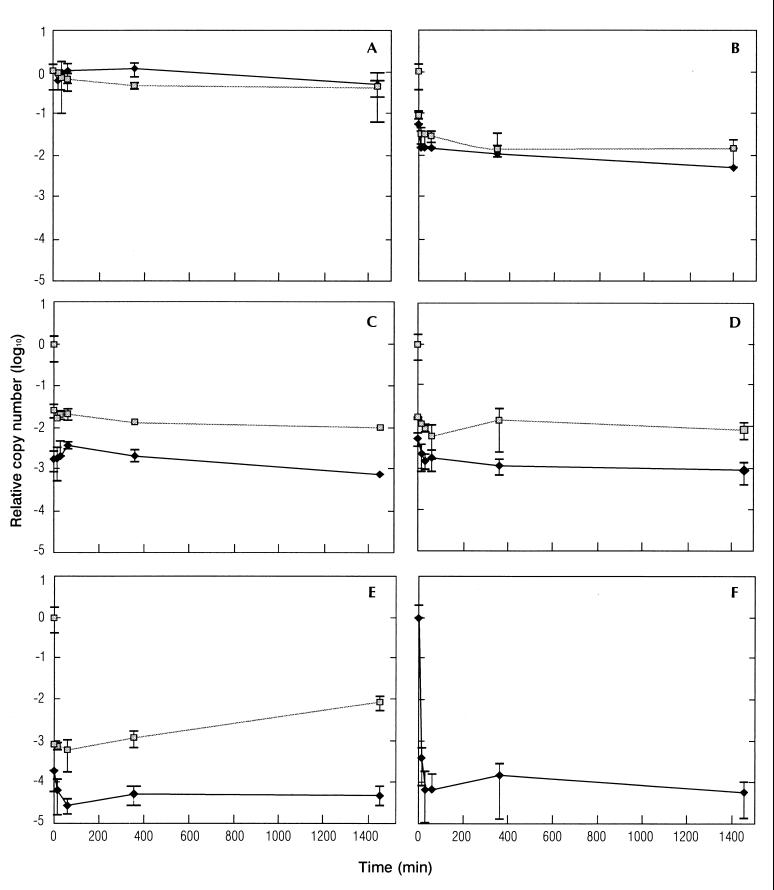
The effect of externally added DNases was compared to control samples to which no DNase was added in cultures incubated at 20, 50, 72, and 100°C for 5 min and at 121°C for 15 min. Results of incubation at room temperature (20°C) after the heat treatments are shown in Fig. 4.
FIG. 4.
Effect of externally added DNase on the stability of DNA in heat-treated cells. Cells were incubated for 5 min at 20°C (A), 55°C (B), 72°C (C), or 100°C (D), or for 15 min at 121°C (E) before the temperature was adjusted to 20°C and DNase was added. DNA was quantified by 5′-nuclease PCR after 5, 15, and 30 min and 1, 6, and 24 h at 20°C in both DNase-treated samples and negative controls. The stability of purified DNA treated with DNase was also investigated (F). The amounts of DNA present with (⧫) and without ( ) DNase treatment, are reported as copy numbers relative to those present before heat treatment. The error bars show standard deviations.
) DNase treatment, are reported as copy numbers relative to those present before heat treatment. The error bars show standard deviations.
There was little difference between the DNase-treated samples and the control at 20 and 55°C (Fig. 4A and B). For the samples incubated at 55°C, the signals with and without DNase stabilized rapidly at 1 to 3% of the input signal (Fig. 4B). For 72, 100, and 121°C, the addition of DNase resulted in a nearly instant 1-log reduction in the signal compared to the control without DNase. No further reductions in the signals were observed for the DNase-treated samples or the controls during the 24 h of incubation after the heat treatments (Fig. 4C, D, and E). For the non-DNase-treated sample incubated at 121°C, the signal increased after 24 h (Fig. 4E); however, after 5 days, the signal was decreasing almost to the same level as after 6 h (result not shown).
The effect of DNase treatment on purified DNA from C. jejuni was also investigated. This DNA was very rapidly degraded, and after 30 min only about 0.01% of the input material was left. This fraction, however, seemed stable for at least 24 h after the treatment (Fig. 4F).
Finally, the influence of Campylobacter cells on the degradation kinetics of purified DNA was investigated. The PCR signals generated from the cells alone, the DNA alone, and the DNA mixed with the cells with and without DNases were determined. The Campylobacter cells did not influence the degradation of the free DNA by the added DNases, and the stability of the free DNA was not affected by the Campylobacter cells when DNase was not added (results not shown).
DISCUSSION
Specific detection and quantification of C. jejuni.
There is a requirement for rapid, quantitative, and accurate measurements of target organisms responsible for food poisoning. In the present study, a 5′-nuclease PCR system was constructed and applied to specifically detect and quantify C. jejuni. The preferred targets for pathogen detection are pathogen determinants. However, the mechanisms by which C. jejuni causes human disease is not completely understood (37). Colonization and/or infection appear to be dependent on intact motility and full-length flagella, and the flagellum gene flaA appears to be essential (22). However, the variation between the different strains was too extensive, so the flaA gene was unsuitable for design of the primers and probe necessary for a 5′-nuclease PCR assay. Empirical data suggest that a gene region in C. jejuni whose function is unknown is specific for this organism (41, 42, 45, 46). This region was compared with the most recently published sequences in the EMBL database (release 60) and the recently completed genome sequence of C. jejuni. The specificity of the constructed primers and probe was tested both by homology searches of nucleotide databases and by screening of a number of C. jejuni strains isolated from patients infected in several parts of the world. No false negatives were recorded among the 32 isolates tested, and no false positives were recorded among the other Campylobacter species or strains belonging to other genera. This demonstrates the high specificity of the designed primer-probe set. Furthermore, the amplification primers alone were also specific for C. jejuni, avoiding potential artifacts in a mixed population due to competition for the amplification primers through amplification of targets from other bacteria.
The square regression coefficients after the linear regressions indicated a good correlation between the amount of template (log input DNA or cells) and the amount of product (represented by the CTs) in the standard curves (R2 = 0.99). The linearity of the standard curves and the fact that the PCR operates with constant efficiency confirm that the assay is well suited for quantitative measurements. The detection limit of the PCR assay was estimated to be approximately 1 CFU/PCR. Our reported limit of detection is similar to those in other reports using a fluorogenic 5′-nuclease PCR assay for endpoint detection. Bassler et al. (1), obtained a detection level of approximately 50 CFU of L. monocytogenes/PCR, while Chen et al. (6) showed a detection limit as low as 2 CFU/PCR from a pure culture of S. enterica serovar Typhimurium.
Quantitative DNA purification was carried out using the DNA Direct system because of both the reproducibility and the simplicity of the protocol (34). The detection limit of the DNA purification method was DNA from 10 cells per PCR. This is good recovery compared to, e.g., standard methods such as extractions with organic solvents (35). Furthermore, the 5′-nuclease assay is especially dependent on pure DNA because the amplification efficiency, and thus the quantification, can be affected by contaminants. No influence on PCR amplification efficiency or inhibition of the PCR was experienced when 12.5% of the purified material was used in the quantitative PCR.
DNA as an indicator of viable C. jejuni.
No real-time quantitative PCR studies have, until now, been performed on the degradation kinetics of DNA from dead bacteria. The few studies that have been done have been qualitative or semiquantitative, and endpoint analyses of PCR amplifications have been employed (11, 18). Generally, the assumption has been that the DNA molecule also persists after the bacteria are dead and thus is not a good marker for the separation of viable and dead bacteria. On the other hand, attempts have been made to use RNA as a living or dead cell marker in several studies (18, 19, 23, 36). However, the conclusions drawn from these experiments are that several assumptions have to be made in order to use RNA as a living or dead cell marker. The targeted gene has to be continuously expressed, the transcript has to be relatively unstable, and finally a specific region has to be identified in the targeted gene. Thus, if possible, it is preferable to use DNA as the target nucleic acid in relation to living versus dead cell studies.
Our data indicate a good correlation between CFU counts and the DNA-targeted 5′-nuclease assay for C. jejuni incubated in pure water at 25°C for up to 24 h (Fig. 3A). This may be due to degradation of DNA by internal DNases in the bacteria which die under these conditions. Furthermore, after 5 days, the 5′-nuclease assay indicated the presence of approximately 1% of the input DNA, while no culturable cells could be recovered. These results are in agreement with an earlier experiment in which C. jejuni strain CB258 was incubated in sterile water at 25°C and both direct viable counts and plate counts were determined. After 6 days, there was a 1- to 2-log reduction in the direct viable counts and a more-than-8-log reduction in the plate counts (20). There was an initial rapid loss of signal in all of the heat-treated samples (55°C, or above). The reason for this is still unknown, but it might be that some of the bacterial DNase activity was left. One can assume that this activity is lost over time, leading to more stable DNA. The generally rapid loss of the DNA signal has important implications for the use of DNA as a viability marker. If the heat treatment history of the sample is known, then it is possible to estimate the likelihood that DNA from bacteria present in the sample prior to the treatment will generate a positive signal.
The ability of DNase to selectively degrade free DNA and DNA in heat-killed Campylobacter to further reduce the signal generated from dead cells was investigated. There were no significant differences between the DNase-treated and untreated samples at 20 or 55°C. The 20°C experiments show that DNA within intact cells is not degraded by externally added DNases and did not result in signal reduction. Although no viable cells could be recovered after the 55°C treatment, this temperature did not seem sufficient to expose the DNA to externally added DNases. For the samples heated to 72, 100, and 121°C, the addition of DNases nearly instantly reduced the amount of template by 1 log compared to the untreated samples. For these temperatures, the major fraction of DNA in the killed cells was not amplified in the assay. After 24 h, the signal from the autoclaved non-DNase-treated sample had increased. This increase may be due to reassociation of the single-stranded DNA. A reassociated double-stranded form may cover the whole amplification region, although each individual single-stranded fragment does not. Then, in the initial phase of the PCR amplification, the individual single-stranded DNA may be extended to cover the whole region.
In conclusion, applying DNase treatment to reduce the noise signal generated by dead bacteria seems promising for samples that have been treated at temperatures above 72°C for 5 min or more. Work is also in progress to further increase the signal-to-noise ratio between living and dead Campylobacter cells through chemical inactivation of DNA in the dead cells (unpublished data).
Comparison of culturing and 5′-nuclease PCR for detection of C. jejuni.
It is evident that traditional culturing results in significant underreporting of potentially infectious C. jejuni. Pearson et al. (27) found the presence of VNC campylobacters in water as the only possibility of transmission to broiler chickens colonized by C. jejuni. Campylobacter in the VNC state may also be more resistant to food processing treatments than cells that can be cultured, especially at low temperatures (20, 32).
In contrast to traditional culturing, VNC C. jejuni may be detected through PCR amplification. However, samples may also test positive although C. jejuni has been inactivated. The direct detection of low cell numbers can also be a problem in food samples. Sensitivity, however, is apparently not a problem with water because bacteria in water samples can be concentrated in several ways (9).
Quantitative detection systems are a requirement when estimating the risk of having infectious Campylobacter in food or water samples. Such risk assessments are also important for future legislative work. As demonstrated in this work, detailed studies of the degradation kinetics of DNA under different processing conditions reveal important information about what effect this DNA has on the detection of viable C. jejuni.
Future developments.
Adaptation of 5′-nuclease technology for quantification of C. jejuni in foods should presumably be feasible. When it comes to naturally contaminated foods, having access to proper protocols for the isolation of bacterial DNA or cells probably will be an important factor. A possible approach may be the use of magnetic beads for specific isolation of the bacteria, followed by isolation of DNA while the bacteria are still attached to the beads. Since paramagnetic beads are easy to manipulate in automated systems (A. Holmberg, A. Deggerdal, and F. Larsen, AMS '95, Third Int. Conf. Automation Mapping DNA Sequencing, abstr. A10, 1995), integrated cell concentration and DNA purification methods should be suited for high-throughput assays. Recently, there also have been efforts to miniaturize 5′-nuclease systems (13, 38) and integrate different processing steps (7, 44). Because of the microscopic size of the beads and the possibilities of performing 5′-nuclease PCR on a nanoliter scale (15), paramagnetic beads and real-time PCR may also be valuable tools in future miniaturized systems.
ACKNOWLEDGMENTS
We are very grateful to Traute Vardund, BABG, National Institute of Public Health, Norway, who provided many of the Campylobacter isolates. We appreciate the advice of Lars Melin, PE Biosystems Sweden, on the design of the primers and probe for C. jejuni.
This work was financed by the Research Levy on certain agricultural products.
REFERENCES
- 1.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black R E, Levine M M, Clements M L, Hughs T P, Blaser M J. Experimental Campylobacter jejuni infections in humans. J Infect Dis. 1988;157:472–480. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J, Hardesty H L, Powers B, Wang W L. Survival of Campylobacter fetus subsp. jejuni in biological milieus. J Clin Microbiol. 1980;11:309–313. doi: 10.1128/jcm.11.4.309-313.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser M J, Taylor D N, Feldman R A. Survival of Campylobacter infections. In: Butzler J-P, editor. Campylobacter infection in man and animals. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 143–161. [Google Scholar]
- 5.Bolder N. Decontamination of meat and poultry carcasses. Trends Food Sci Technol. 1997;5:384–389. [Google Scholar]
- 6.Chen S, Yee A, Griffiths M, Larkin C, Yamashiro C T, Behari R, Paszko-Kolva C, Rahn K, De Grandis S A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol. 1997;35:239–250. doi: 10.1016/s0168-1605(97)01241-5. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Sheldon E L, Wu L, Uribe A, Gerrue L O, Carrino J, Heller M J, O'Connell J P. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 8.Corry J E L, Post P E, Colin P, Laisney M J. Culture media for the isolation of campylobacters. In: Corry J E L, Curtis G D W, Baird R M, editors. Culture media for food microbiology. Amsterdam, The Netherlands: Elsevier Science B.V.; 1995. pp. 129–162. [DOI] [PubMed] [Google Scholar]
- 9.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 11.Herman L. Detection of viable and dead Listeria monocytogenes by PCR. Food Microbiol. 1997;14:103–110. [Google Scholar]
- 12.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibrahim M S, Lofts R S, Jahrling P B, Henchal E A, Weedn V W, Northrup M A, Belgrader P. Real-time microchip PCR for detecting single-base differences in viral and human DNA. Anal Chem. 1998;70:2013–2017. doi: 10.1021/ac971091u. [DOI] [PubMed] [Google Scholar]
- 14.Jones D M, Sutcliffe E M, Curry A. Recovery of viable but non-culturable Campylobacter jejuni. J Gen Microbiol. 1991;137:2477–2482. doi: 10.1099/00221287-137-10-2477. [DOI] [PubMed] [Google Scholar]
- 15.Kalinina O, Lebedeva I, Brown J, Silver J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997;25:1999–2004. doi: 10.1093/nar/25.10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Livak K J, Marmaro J, Flood S J A. Guidelines for designing TaqMan™ fluorogenic probes for 5′ nuclease assays. Perkin-Elmer Research News, ABI PRISM™ Sequence Detection System. Norwalk, Conn: The Perkin-Elmer Corp.; 1995. [Google Scholar]
- 18.McKillip J L, Jaykus L A, Drake M. Nucleic acid persistence in heat-killed Escherichia coli O157:H7 from contaminated skim milk. J Food Prot. 1999;62:839–844. doi: 10.4315/0362-028x-62.8.839. [DOI] [PubMed] [Google Scholar]
- 19.McKillip J L, Jaykus L A, Drake M. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl Environ Microbiol. 1998;64:4264–4268. doi: 10.1128/aem.64.11.4264-4268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medema G J, Schets F M, Van de Giessen A W, Havelaar A H. Lack of colonization of 1 day old chicks by viable, non-culturable Campylobacter jejuni. J Appl Bacteriol. 1992;72:512–516. doi: 10.1111/j.1365-2672.1992.tb01868.x. [DOI] [PubMed] [Google Scholar]
- 21.Nachamkin I. Campylobacter jejuni. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 159–170. [Google Scholar]
- 22.Nachamkin I, Yang X H, Stern N J. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. 1993;59:1269–1273. doi: 10.1128/aem.59.5.1269-1273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norton D M, Batt C A. Detection of viable Listeria monocytogenes with a 5′ nuclease PCR assay. Appl Environ Microbiol. 1999;65:2122–2127. doi: 10.1128/aem.65.5.2122-2127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien M C, Bolton W E. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry. 1995;19:243–255. doi: 10.1002/cyto.990190308. [DOI] [PubMed] [Google Scholar]
- 25.Olsen J E, Aabo S, Hill W, Notermans S, Wernars K, Granum P E, Popovic T, Rasmussen H N, Olsvik O. Probes and polymerase chain reaction of food-borne bacterial pathogens. Int J Food Microbiol. 1995;28:1–78. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 26.Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 27.Pearson A D, Greenwood M, Healing T D, Rollins D, Shahamat M, Donaldson J, Colwell R R. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl Environ Microbiol. 1993;59:987–996. doi: 10.1128/aem.59.4.987-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.PE Biosystems. User bulletin 2: ABI PRISM 7700 Sequence Detection System. Foster City, Calif: PE Biosystems; 1997. [Google Scholar]
- 30.Riedy M C, Muirhead K A, Jensen C P, Stewart C C. Use of a photolabeling technique to identify nonviable cells in fixed homologous or heterologous cell populations. Cytometry. 1991;12:133–139. doi: 10.1002/cyto.990120206. [DOI] [PubMed] [Google Scholar]
- 31.Robinson D A. Infective dose of Campylobacter jejuni in milk. Br Med J. 1981;282:1584. doi: 10.1136/bmj.282.6276.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollins D M, Colwell R R. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossen L, Nørskov P, Holmstrøm K, Rasmussen O F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 34.Rudi K, Kroken M, Dahlberg O J, Deggerdal A, Jakobsen K S, Larsen F. Rapid, universal method to isolate PCR-ready DNA using magnetic beads. BioTechniques. 1997;22:506–511. doi: 10.2144/97223rr01. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sheridan G E, Masters C I, Shallcross J A, MacKey B M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon E B, Hoover D G. Campylobacter jejuni: a bacterial paradox. J Food Safety. 1999;19:121–136. [Google Scholar]
- 38.Taylor T B, Winn-Deen E S, Picozza E, Woudenberg T M, Albin M. Optimization of the performance in silicon-based microstructures. Nucleic Acids Res. 1997;25:3164–3168. doi: 10.1093/nar/25.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanechoutte M, Boerlin P, Tichy H V, Bannerman E, Jager B, Bille J. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int J Syst Bacteriol. 1998;48:127–139. doi: 10.1099/00207713-48-1-127. [DOI] [PubMed] [Google Scholar]
- 40.Waage A S, Vardund T, Lund V, Kapperud G. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl Environ Microbiol. 1999;65:1636–1643. doi: 10.1128/aem.65.4.1636-1643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R-F, Slavik M F, Cao W-W. A rapid PCR method for direct detection of low numbers of Campylobacter jejuni. J Rapid Methods Autom Microbiol. 1992;1:101–108. [Google Scholar]
- 42.Wang R-F, Slavik M F, Cao W-W, Blore P J. Development of DNA probes specific for Campylobacter jejuni. J Rapid Methods Autom Microbiol. 1992;1:83–92. [Google Scholar]
- 43.Wang W-L, Powers B W, Luechtefeld N W, Blaser M J. Effects of disinfectants on Campylobacter jejuni. Appl Environ Microbiol. 1983;45:1202–1205. doi: 10.1128/aem.45.4.1202-1205.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters L C, Jacobson S C, Kroutchinina N, Khandurina J, Foote R S, Ramsey J M. Microchip device for cell lysis, multiplex PCR amplification, and electrophoretic sizing. Anal Chem. 1998;70:158–162. doi: 10.1021/ac970642d. [DOI] [PubMed] [Google Scholar]
- 45.Wesley I V, Sanderson T P, Larson D J, Harmon K M, Andrews J J, Miskimins D W, Zeman D H. Application of multiplex polymerase chain reaction for rapid identification of Campylobacter jejuni and C. coli with reproductive failure. Am J Vet Res. 1997;58:1070–1075. [PubMed] [Google Scholar]
- 46.Winters D K, O'Leary A E, Slavik M F. Rapid PCR with nested primers for direct detection of Campylobacter jejuni in chicken washes. Mol Cell Probes. 1997;11:267–271. doi: 10.1006/mcpr.1997.0116. [DOI] [PubMed] [Google Scholar]