ABSTRACT
Macroautophagy/autophagy is a conserved intracellular degradation pathway that has recently emerged as an integral part of plant responses to virus infection. The known mechanisms of autophagy range from the selective degradation of viral components to a more general attenuation of disease symptoms. In addition, several viruses are able to manipulate the autophagy machinery and counteract autophagy-dependent resistance. Despite these findings, the complex interplay of autophagy activities, viral pathogenicity factors, and host defense pathways in disease development remains poorly understood. In the current study, we analyzed the interaction between autophagy and cucumber mosaic virus (CMV) in Arabidopsis thaliana. We show that autophagy is induced during CMV infection and promotes the turnover of the major virulence protein and RNA silencing suppressor 2b. Intriguingly, autophagy induction is mediated by salicylic acid (SA) and dampened by the CMV virulence factor 2b. In accordance with 2b degradation, we found that autophagy provides resistance against CMV by reducing viral RNA accumulation in an RNA silencing-dependent manner. Moreover, autophagy and RNA silencing attenuate while SA promotes CMV disease symptoms, and epistasis analysis suggests that autophagy-dependent disease and resistance are uncoupled. We propose that autophagy counteracts CMV virulence via both 2b degradation and reduced SA-responses, thereby increasing plant fitness with the viral trade-off arising from increased RNA silencing-mediated resistance.
Abbreviations: AGO1: argonaute1; ANOVA: analysis of variance; ATG: autophagy related; AZD: AZD8055; CMV: cucumber mosaic virus; CaMV: cauliflower mosaic virus; Co-IP: Co-immunoprecipitation; ConA: concanamycin A; CP: coat protein; DAI: days after inoculation; DCL2/DCL4: dicer like 2/ dicer like 4; DMSO: dimethyl sulfoxide; FLUC: firefly luciferase; GFP: green fluorescent protein; GUS: β-glucuronidase; h: hours; NahG: salicylate hydroxylase; NBR1: neighbor of BRCA1; NPR1: non-expressor of pathogensis related 1; PR1: pathogenesis related 1; RDR6: RNA dependent RNA polymerase 6; RFP: red fluorescent protein; RLUC: renilla luciferase; SA: salicylic acid; SGS3: suppressor of gene silencing 3; TuMV: turnip mosaic virus; WT: wild type
KEYWORDS: CMV, host-pathogen trade-off, plant immunity, plant virus, RNA silencing, viral disease, viral effector protein, virus self-attenuation, virus transmission
INTRODUCTION
Macroautophagy/autophagy is a conserved eukaryotic mechanism that is important for cellular homeostasis through its functions as a major catabolic system. In one way, the autophagy process is considered to sequester cytoplasmic portions non-selectively within a vesicular structure called the autophagosome, which subsequently enters the lytic vacuole of plants for degradation and recycling of nutrients [1]. However, the extent to which autophagy operates in a non-selective manner still remains an open question. In parallel, the examples of selective targeting of autophagic substrates by specialized cargo receptors continue to increase [2,3]. The plant autophagic molecular machinery was initially characterized in Arabidopsis thaliana and subsequently recognised in algae, gymnosperms and other angiosperms [4]. Core components of this pathway include numerous ATG (autophagy related) genes, such as ATG5, ATG7 and ATG8. In plants, selective autophagy is known to take part in the turnover of substrates including chloroplasts, peroxisomes, aggregates, ribosomes and proteasomes [5,6]. Selectivity opens the possibility for several highly distinct autophagic processes to operate in parallel and outlines the important challenge to dissect the overall autophagy effect into its specific mechanisms.
Autophagy is induced by numerous environmental conditions, including abiotic stress and nutrient limitation. Its importance for plant adaptation is established in part by the increased sensitivity of autophagy-deficient plants to these conditions [4,7]. Autophagy also plays important roles in plant-pathogen interactions including diseases caused by viruses, bacteria, fungi and oomycetes [8]. These studies support the notion that autophagy is a complex process with several functions operating in parallel to influence the infection outcome. Because pathogens commonly cause severe stress to their host plants, it is not surprising that infected plants show an upregulation of autophagy upon infection [9–13]. This finding outlines the important concept that the interaction between pathogens and their hosts likely co-evolves in the presence of an activated autophagy response. As a consequence, both the pathogen and host could try to utilize the plant autophagy process for their own benefit.
In virus-plant interactions, we can distinguish between two mechanisms that limit disease. Host resistance refers to the ability to suppress virus multiplication whereas tolerance is defined as the ability of the plant to minimize the infection-associated damage as a result of the pathogen infection. Co-adaptation of hosts and viruses has resulted in a range of interactions between antiviral resistance mechanisms and viral counter-strategies, together balancing plant and virus fitness [14,15]. Autophagy has been established as a prominent response pathway to several viral infections in animal systems [16]. An emerging theme from these studies is that viruses have acquired properties to modify autophagy in many ways, including induction, suppression and subversion of its functions. The evolution of such viral properties can be considered to directly reflect the importance of autophagy for virus infection. Recently, fundamental roles for autophagy have been identified in plant viral diseases, including autophagic degradation of viral components, virus-based counteraction, modulation of autophagy as well as autophagy-mediated promotion of plant tolerance [10–12,17,18]. Interestingly, a connection between autophagy and the foremost antiviral defense pathway in plants, RNA silencing, is emerging. Initially, autophagy was suggested to degrade the RNA silencing suppressors of potyviruses (HCpro) and cucumoviruses (2b) [19], and we could show that the autophagy cargo receptor NBR1/Neighbor of BRCA1 degrades HCpro/Helper component proteinase to reduce virus susceptibility and accumulation [10]. Additionally, autophagy degrades the satellite ßC1 RNA silencing suppressor of geminiviruses, AGO1/Argonaute 1, when targeted by the poleroviral RNA silencing suppressor P0, and SGS3/RDR6 (Suppressor of Gene Silencing 3/ RNA Dependent RNA Polymerase 6) in the presence of potyviral VPg/viral protein genome-linked [12,20,21]. Similarly, autophagy degrades SGS3/RDR6 bodies and thereby, reduces resistance against mutant CMV compromised in RNA silencing suppression [22]. Notably, autophagy-based resistance was uncoupled from autophagy-based tolerance against a potyvirus (turnip mosaic virus; TuMV) and a caulimovirus (cauliflower mosaic virus; CaMV) [10,11]. Thus, autophagy shows complex interactions and general potential for regulating plant virus disease.
The defense hormone salicylic acid (SA) can induce autophagy and promotes the early senescence phenotype of autophagy-deficient mutants [23]. The early senescence is accordingly reduced in the SA synthesis mutant AT1G74710/sid2, SA signalling mutant AT1G64280/npr1 and plants expressing the SA-degrading enzyme AT5G33340/NahG [23]. Notably, SA commonly promotes plant virus defense [24,25], but cucumber mosaic virus (CMV; genus cucumovirus) has been found to largely evade SA resistance responses [26] unlike some other viruses [27]. Evasion of SA-induced resistance by CMV is linked to a domain in the virulence protein 2b that is distinct from its well-established function in suppression of antiviral RNA silencing [28,29]. Together, it is plausible that SA could have global roles in plant virus and autophagy interactions, but so far these connections have remained elusive.
In this study, we show that autophagy is induced in an SA-dependent manner during infection with the positive-stranded RNA virus CMV. We found that the viral RNA silencing protein 2b can dampen SA-induced autophagy and is itself subject to autophagic degradation. Because autophagy suppressed CMV RNA accumulation in an RNA silencing- and AGO1-dependent manner, we propose that 2b degradation represents a resistance mechanism that sensitizes CMV to RNA silencing. In a broader context, autophagy and RNA silencing appeared beneficial for CMV infection through the synergistic promotion of plant longevity and viral seed transmission. Our results thereby reveal that autophagy provides resistance by reducing virus accumulation as well as tolerance by decreasing disease severity during CMV infection, and both processes seem to be linked to the autophagic degradation of the major virulence factor 2b.
Results
Autophagy is induced during CMV infection
Owing to the importance of autophagy in various plant virus infections, we set out to determine autophagy regulation of an economically important generalist virus, CMV. Our first interest was to assess whether autophagy levels were altered during infection with the CMV strain PV0187 and used an Arabidopsis line that stably expresses GFP-ATG8a, which is a commonly applied marker for the detection of autophagosomes. The distribution of this marker was altered in CMV infected tissue, including an increased number of smaller GFP-ATG8a-labelled puncta that likely represented autophagosomes (Figure 1A and B). Infected tissue also contained some larger irregular GFP-ATG8a labelled structures that were not observed in healthy tissue (Figure 1A; inset). Treatment with concanamycin A (ConA), an inhibitor of the vacuolar ATPase, was used to stabilize autophagic bodies delivered into the vacuole [11]. The number of GFP-ATGA8a puncta significantly increased upon ConA application in infected tissue (Figure 1A and B), suggesting that autophagy is both activated and completed during CMV infection.
Figure 1.
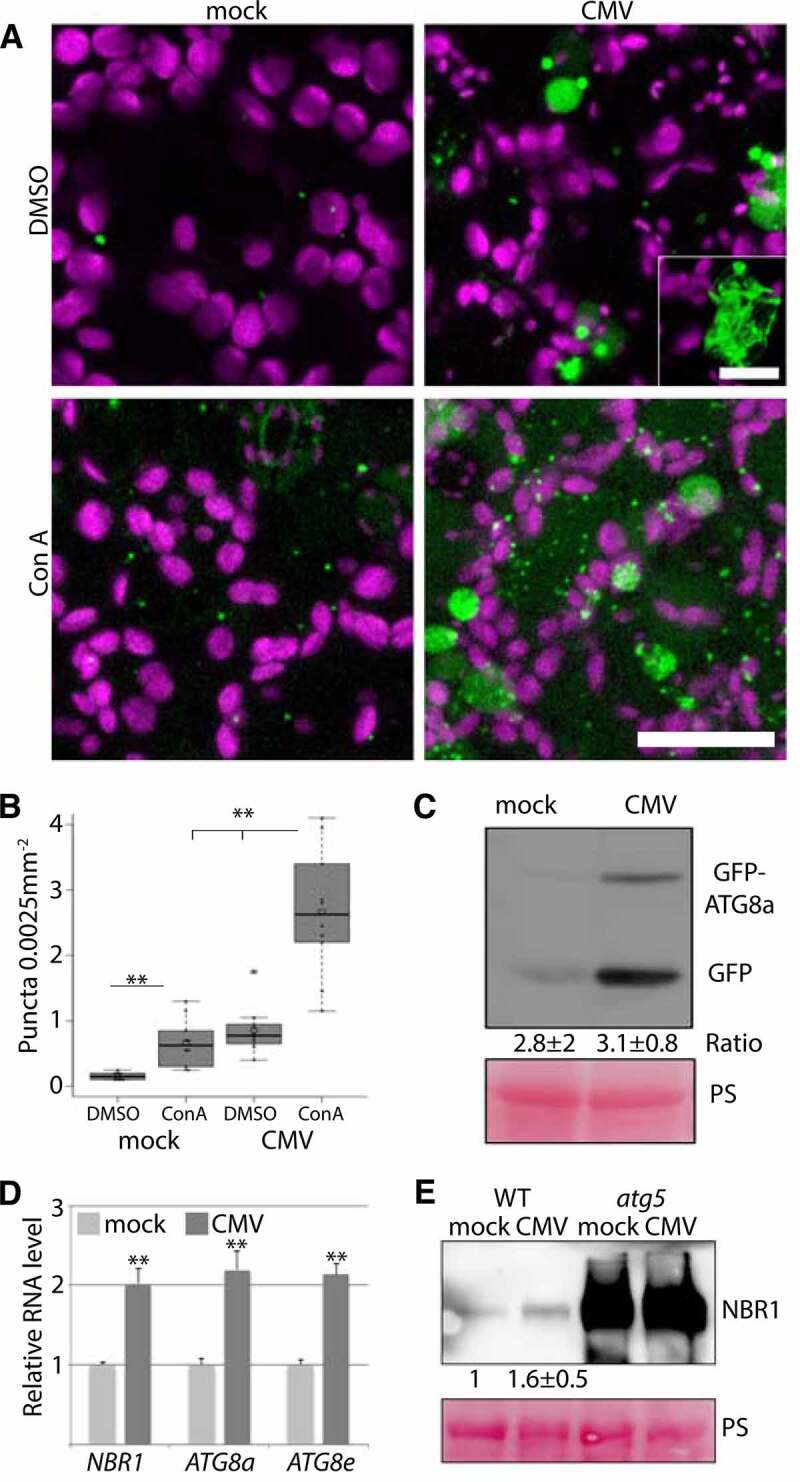
Autophagy is induced during CMV infection. (A) Representative images of the GFP-ATG8a marker in healthy and CMV infected plants with and without concanamycin A (ConA) treatment. Chloroplasts are shown in magenta. Images are confocal Z-stacks. Scale bar: 20 μm. The inset shows a representative of large irregular GFP-ATG8a structures found with CMV. Scale bar: 5 μm. (B) GFP-ATG8a foci were counted from similar images as in (A) using ImageJ. (n= 10). (C) Western blot analysis of free GFP levels derived from GFP-ATG8a in mock and CMV infected plants. Ponceau S (PS) staining verified comparable protein loading. The average and standard deviation from a quantification of the ratio between fusion to free GFP using ImageJ is shown below the western blot, and is based on four independent experiments. (D) Transcript levels for NBR1, ATG8a and ATG8e in mock and CMV infected plants were determined by RT-qPCR. (n= 4). (E) Representative western blot of NBR1 levels in mock and CMV inoculated WT and atg5 plants. Ponceau S (PS) staining verified comparable protein loading. A quantification of NBR1 protein levels using ImageJ from three independent western blots normalized using PS stained Rubisco is shown below the western blot. Statistical significance (*P < 0.05; **P < 0.01) was revealed by Student´s t-test (compared to WT).
The processing of free GFP from the GFP-ATG8 fusion protein is regarded as another reliable proxy for autophagy flux in plants [30]. While free GFP levels were higher in CMV infected tissue compared to the control (Figure 1C), which indeed supported elevated autophagy levels, the GFP-ATG8a fusion form also accumulated to higher levels. Importantly, the ratio of free GFP to fusion form was similar in mock and CMV infected tissue (Figure 1C), consistent with a higher overall level of functional autophagy during infection. Furthermore, we could also observe higher transcript levels of autophagy-related genes AT4G21980/ATG8a, AT2G45170/ATG8e and AT4G24690/NBR1 in CMV infected tissue (Figure 1D), suggesting activation of autophagy already at the transcriptional level. NBR1 delivers cargo to autophagosomes and is degraded in the process, making the NBR1 protein to mRNA ratio another indicator of autophagy activity and flux. Western blot analysis and subsequent quantification suggested that NBR1 protein accumulated in a comparable range with transcript levels in response to CMV (Figure 1E), further supporting that CMV-induced autophagy is functional and completes degradation.
CMV disease is attenuated by autophagy
One important factor influencing virus and plant fitness is the severity of disease. Previously, we showed that biomass loss, as a measure for disease, was severely increased in loss-of-function mutants of the core autophagy gene AT5G17290/atg5 when infected with TuMV or CaMV [10,11]. Since autophagy and defense responses against viruses may involve the SA pathway, we analyzed the interrelationship of autophagy and SA in regulating CMV disease development. When infected with CMV, we observed increased biomass loss in the autophagy mutant atg5 compared to WT already early during infection (10 DAI) (Figure 2A and B). By the later 28 DAI time-point, an apparent senescence phenotype had additionally developed in atg5 (Figure 2C and D). Intriguingly, both the senescence and biomass-loss phenotypes of atg5 were rescued by the SA-biosynthesis mutant sid2, the SA-degrading bacterial enzyme NahG and also by the SA-signalling mutant npr1 (Figure 2A-D). We noted that atg5 sid2 and atg5 npr1, but not atg5 NahG plants, occasionally also developed stronger symptoms (senescence, paleness, lesions).
Figure 2.
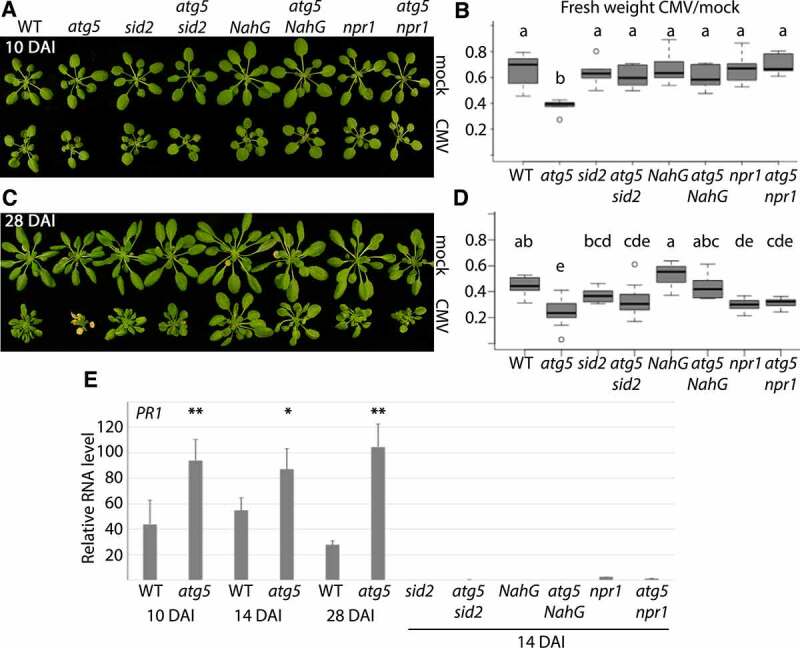
CMV disease is attenuated by autophagy. (A) Representative WT (Col-0), atg5, sid2, atg5 sid2, NahG, atg5 NahG, npr1 and atg5 npr1 plants at 10 DAI (days after inoculation) of CMV compared to mock. (B) The fresh weight ratio of CMV infected to mock plants at 10 DAI. (n<6). Letters indicate genotypes with statistically different levels (ANOVA P < 0.05, Tukey-HSD test). The absolute fresh weight data is presented in (Fig. S1). (C and D) As (A and B) but at 28 DAI. (E) PR1 transcript levels relative to PP2A determined at different time-points of CMV infection in WT and atg5 plants, as well as 14 DAI in the different SA compromised lines (n= 4). The statistical analysis compares WT to atg5 at the different time-points.
We used AT2G14610/PR1 as a marker gene for SA- and NPR1-dependent defense signalling [31]. PR1 expression levels were significantly higher in infected atg5 compared to WT plants at 10, 14 and 28 DAI (Figure 2E). As expected, PR1 expression was severely reduced in NahG, sid2 and npr1 backgrounds when compared to WT and atg5, respectively. This result supported the occurrence of elevated SA responses in atg5, thus positively correlating with increased SA-dependent disease when autophagy is impaired. Together, we suggest that that SA drives CMV disease in autophagy-deficient plants.
Autophagy impacts plant susceptibility and CMV resistance
Having established an evident importance for autophagy in plant tolerance of CMV disease, our next interest was to evaluate potential roles of autophagy in CMV resistance. One relevant aspect of resistance are viral infection rates, which we addressed both in virus inoculation experiments as well as vertical transmission tests. Due to experimental restrictions, we focused our analyses on a single autophagy-deficient mutant AT5G45900/atg7 (comparable with atg5). Our first notion was that the infection rates following mechanical sap inoculation were moderately enhanced in atg7 (Figure 3A). CMV belongs to the plant viruses that can also transmit trans-generationally through seeds in a process termed vertical transmission. Indeed, we found a theoretical transmission frequency of around 2 % (Figure 3B) in WT plants, and by using Gibbs and Gower formulae [32] we further estimated CMV seed transmission to be 1.31 ± 0.8 %, a similar range that has been reported for other CMV strains in Arabidopsis [33]. Notably, we did not detect any seed transmission in atg7 plants, establishing autophagy as essential for vertical transmission of CMV strain PV0187 in Arabidopsis accession Col-0.
Figure 3.
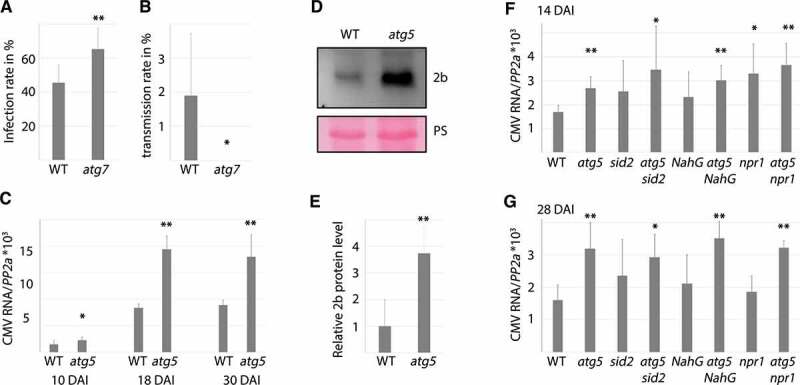
Autophagy impacts various aspects of CMV resistance. (A) CMV infection rate in WT and atg7 plants determined by presence/absence of viral symptoms three weeks after mechanical sap inoculation (8 independent replicates of 9 plants). (B) Vertical transmission of CMV in seeds from infected WT and atg7 plants was estimated by RT-PCR from 10 pools of 20 seedlings per plant (n= 5 independent plants). (C) CMV RNA levels relative to PP2A in WT and atg5 plants determined by RT-qPCR at 10, 18 and 30 DAI (n= 4). (D) A representative western blot of CMV 2b protein levels from infected WT and atg5 plants. Ponceau S (PS) staining verified comparable protein loading. (E) Four independent western blots as in (D) were used for quantification of 2b levels in infected WT and atg5 plants using ImageJ. PS-stained Rubisco was used to normalize loading. (F and G) Relative CMV RNA levels were determined by RT-qPCR at 14 (F) and 28 (G) DAI in WT, atg5, sid2, atg5 sid2, NahG, atg5 NahG, npr1 and atg5 npr1 plants. (n= 4). Statistical significance (**P < 0.01, *P < 0.05) was revealed by Student´s t-test.
Next, we addressed autophagy-dependent virus accumulation at time-points 10, 18 and 30 DAI (Figure 3C). CMV accumulation increased between 10 and 18 DAI, and accumulated to significantly higher levels at all time-points in atg5 plants compared to WT. The availability of an antibody enabled us to also compare CMV 2b protein levels, revealing much stronger 2b accumulation in atg5 than WT (Figure 3D). A quantification suggested that the level of 2b was on average 4-fold enhanced in atg5 (Figure 3E). We also analyzed viral RNA accumulation in atg5, sid2, atg5 sid2, NahG, atg5 NahG, npr1 and atg5 npr1 at an early and later time-point of infection (Figure 3F and G). The single SA mutants showed some variation between experiments, with viral RNA accumulation ranging between WT and atg5 levels. Clearly, disease attenuation by NahG, npr1 and sid2 in atg5 (Figure 2) was not owing to reduced virus accumulation and likewise, increased viral accumulation in atg5 was not owing to SA-dependent senescence as it was essentially similar in atg5 sid2, atg5 NahG and atg5 npr1. In all the experiments, we did not find any evident accumulative effects in the double mutants at the earlier time-points when single SA-mutants frequently accumulated more viral RNA than WT. Thus, autophagy and SA seem to act to some extent in the same resistance pathway at earlier stages of infection. Together, these results uncouple disease severity from resistance under autophagy deficiency and establishes that autophagy restricts CMV RNA and 2b protein accumulation.
CMV 2b is degraded by autophagy
Quantification of 2b levels revealed strong accumulation in atg5 compared to WT (Figure 3E), suggesting that 2b could be a direct autophagy target and thereby indirectly effect RNA accumulation. In support of this possibility, ectopically expressed 2b was previously shown to be stabilized in tobacco by the potential autophagy inhibitor 3-Methyladenine [19]. As a first approach addressing whether 2b is a target of autophagy, we carried out colocalization of 2b-RFP and GFP-ATG8a in roots of a double transgenic line treated with ConA or DMSO as control (Figure 4A). We found abundant colocalization of 2b-RFP and GFP-ATG8a in vacuoles only after ConA treatment, indeed supporting that 2b is degraded by autophagy. To address this further, we treated transgenic 2b seedlings with ConA and the autophagy-inducing drug AZD8055, both causing expected, albeit minor effects on 2b (Figure 4B and C). Autophagy-dependent accumulation of 2b was even more evident after introgressing the 2b transgene into the atg5 background, leading to increased protein accumulation in atg5 compared to WT (Figure 4D and E). Importantly, transcript levels of the 2b transgene were comparable between WT and atg5 (Figure 4F). We also noticed that the relative decrease in rosette biomass caused by 2b was much higher in atg5 plants compared to WT (Figure 4G), which could be a consequence of higher 2b levels, as 2b was shown to facilitate virulence in a concentration-dependent manner [34].
Figure 4.
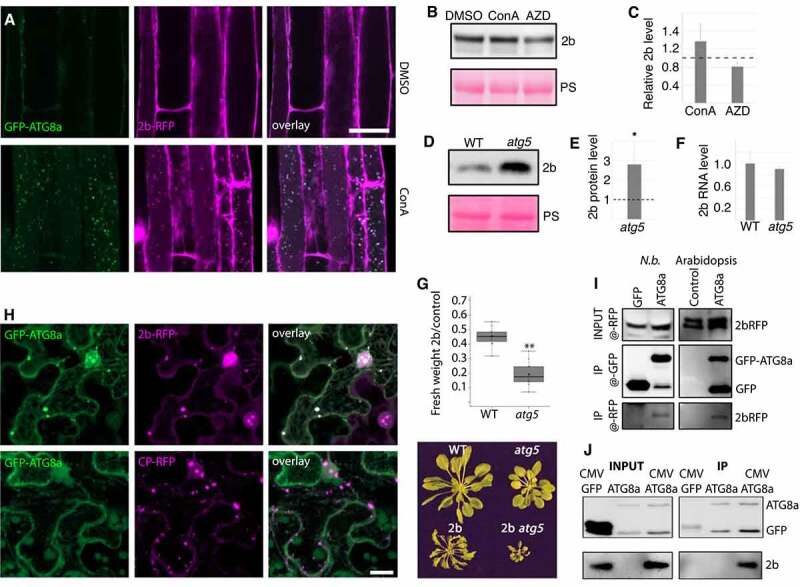
Autophagy degrades 2b within and outside of the infection context. (A) Colocalization analysis of GFP-ATG8a and 2b-RFP in roots of transgenic Arabidopsis after DMSO and ConA treatment for 6 h. (B and C) 2b-RFP transgenic seedlings were treated with DMSO (control), ConA or AZD for 12h followed by western blot detection of 2b using anti-RFP. Ponceau S staining shows loading. Representative blot is shown in (B) and a quantification of 2b levels relative to PS stained Rubisco using ImageJ from three independent experiments with the dashed line indicating DMSO levels (C). (D and E) Accumulation of 2b in transgenic WT and atg5 plants was analyzed by western blotting. Representative blot is shown in (D) with PS-stained Rubisco indicating loading. Quantification of 2b levels using ImageJ from three independent experiments with the dashed line indicating WT levels (E). (F) Expression levels of the 2b transgene in WT and atg5 backgrounds related to (D and E) was determined by RT-qPCR. (n= 4). (G) 2b-dependent virulence measured as relative fresh weight loss caused by transgenic 2b expression compared to non-transgenic plants in WT and atg5 backgrounds. (n= 9). A representative plant image is shown below. (H) Colocalization analysis in N. benthamiana leaves co-expressing 2b-RFP or CP-RFP with GFP-ATG8a. The Z-stack images were acquired 48 h post agroinfiltration. Scale bar: 20 μm. (I) Co-IP (Co-immunoprecipitation) analysis of 2b-RFP with GFP-ATG8a from N. benthamiana (N.b.) and Arabidopsis. Co-expression of GFP was used as control in N. benthamiana and 2b-RFP expression alone in Arabidopsis. Shown is the anti-RFP input signal, as well as anti-GFP and anti-RFP signals from the GFP-based IP samples. (J) Co-IP analysis of 2b with GFP-ATG8a from infected Arabidopsis plants. Non-infected GFP-ATG8a and infected GFP expressing plants were used as control. Shown are the input and IP samples probed with anti-GFP and anti-2b.
Next, we carried out colocalization and immunoprecipitation experiments addressing 2b association with the autophagy marker ATG8a in Nicotiana benthamiana. 2b-RFP, but not the CP-RFP control, colocalized with GFP-ATG8a in larger cytoplasmic structures (Figure 4H). 2b-RFP was also associated with multiple nuclear speckles, including the nucleolus as previously described [35], and caused re-localization of GFP-ATG8a to the same domains (Fig. S2). CP-RFP also localized to the nucleolus but without recruiting GFP-ATG8a. Interestingly, 2b re-localized CP-GFP but not free GFP to the nuclear subcompartments during their co-expression (Figure S2). Notably, we have not observed localization of GFP-ATG8a to nuclear speckles during CMV infection in Arabidopsis and thus limit our present interpretation of these results to support the association between 2b and ATG8a. When 2b-RFP was co-expressed with either GFP or GFP-ATG8a in N. benthamiana followed by GFP-based immunoprecipitation (Figure 4I), we could detect a specific association of 2b with ATG8a. Likewise, we found copurification of 2b-RFP with GFP-ATG8a from transgenic Arabidopsis (Figure 4I), altogether supporting that 2b is also degraded by autophagy outside of an infection context. To further substantiate autophagic degradation of 2b during infection, we performed GFP-based immunoprecipitation of GFP-ATG8a from mock and CMV infected plants and showed that 2b copurified with GFP-ATG8a (Figure 4J). Together, our results provide ample evidence that 2b is a target of autophagic degradation, both within and outside of an infection context.
The virulence protein 2b suppressed SA-activated autophagy during CMV infection
The capacity of SA to induce autophagy [23] prompted us to test whether SA drives autophagy induction during CMV infection. To this end, we crossed the GFP-ATG8a line with NahG, and quantified autophagosomes after ConA treatment with CMV (Figure 5A). In the NahG background, autophagy was only mildly induced by CMV as compared to the GFP-ATG8a control line. This observation was further supported by a free GFP release assay, as CMV infection no longer increased the amount of free GFP in the NahG background (Figure 5B). We therefore concluded that CMV-induced autophagy is mediated by SA.
Figure 5.
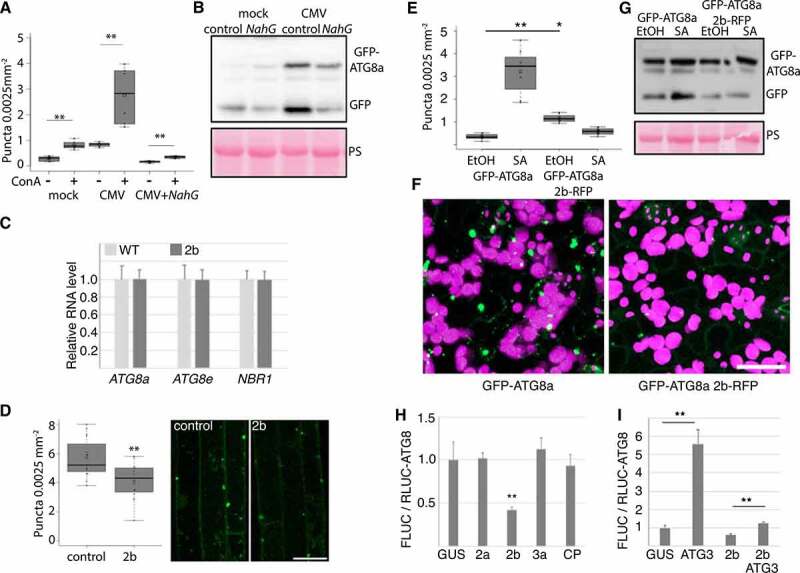
CMV protein 2b dampens SA-induced autophagy. (A) GFP-ATG8a foci were counted from mock and CMV infected GFP-ATG8a and GFP-ATG8a NahG plants after ConA and DMSO treatment using ImageJ (n= 10). (B) Western blot analysis of GFP-ATG8a from mock and infected control and NahG plants. The experiment was repeated twice. (C) Transcript levels of ATG8a, ATG8e and NBR1 were determined by RT-qPCR from 2b transgenic plants shown in (Figure 4G) (n= 4). (D) The number of GFP-ATG8a foci were analyzed in roots of 10 days old seedlings without and with stable expression of 2b after 10 h ConA treatment. Counting was performed using ImageJ on confocal Z-stack projections (n= 15). Representative images are shown with scale bar: 20 μm. (E) Short-day grown 4-week-old GFP-ATG8a and GFP-ATG8a expressing 2b-RFP plants were sprayed with 1 mM SA. GFP-ATG8a foci were quantified 12 h later using confocal imaging and ImageJ software. Control plants were sprayed with corresponding 0.1 % EtOH (n= 9). (F) Representative images of GFP-ATG8a 12 h after SA treatments in control and 2b-RFP plants. Shown is GFP in green and chloroplasts in magenta. Scale bar: 20 μm. (G) SA-induced free GFP release from GFP-ATG8a was analyzed in control and 2b-RFP plants by western blot using anti-GFP. The western blot experiment was repeated twice with similar results. PS staining of the membrane was used to verify loading. (H) RLUC-ATG8a activity was analyzed after co-expression with CMV proteins and GUS control in N. benthamiana leaves at 3 DAI. FLUC was used as an internal control for RLUC activity, and the quantitative data is presented as a ratio of FLUC to RLUC-ATG8a (higher ratio = increased ATG8a degradation). n= 4. (I) As (H), but co-expressed proteins were combinations of 2b and Arabidopsis ATG3 as indicated. Statistical significance (**P < 0.01) was revealed by Student´s t-test.
CMV protein 2b is known to lower SA-mediated virus resistance [28, 29], a mechanism proposed to involve suppression of RDR1-dependent antiviral RNA silencing [36]. 2b also interferes with AGO1 [37], and defects in AGO1 and more generally the microRNA pathway appear to activate autophagy [21]. Thus, we addressed whether the viral 2b effector can modulate autophagy. First, we used a previously published transgenic Arabidopsis line that expresses 2b from the FNY strain [34], resulting in a strong phenotype with reduced growth and morphological abnormalities (Figure 4E). However, the transcript levels of ATG8a, ATG8e, and NBR1 that were induced during infection remained unaltered in the 2b transgenic plants (Figure 5C). We then crossed this line with GFP-ATG8a and quantified the number of foci in ConA treated seedling roots (Figure 5D). When compared to the GFP-ATG8a control, the presence of 2b resulted in a mild but significant decrease of foci.
We reasoned that 2b may exert a more obvious phenotype on SA-induced autophagy. To investigate this, transgenic GFP-ATG8a lines with and without 2b-RFP were sprayed with SA, followed by quantification of GFP-ATG8a (Figure 5E and F). In control plants, SA resulted in the appearance of numerous different sized GFP-ATG8a foci when compared to EtOH treatment, and this response was commonly absent in the 2b-RFP line. Likewise, SA treatment increased free GFP release from GFP-ATG8a in control but not 2b-RFP lines (Figure 5G). Interestingly, the 2b-RFP transgenic plants did not reduce growth upon SA treatment in contrast to WT (Fig. S3), further supporting that 2b interferes with responses to exogenous SA.
Owing to the strong developmental phenotype of transgenic 2b lines with potential pleiotropic effects, we also verified that 2b can suppress autophagy in the transient expression system of N. benthamiana. We used a previously described quantitative assay based on Agrobacterium-mediated transient expression of Renilla luciferase-fused ATG8a (RLUC-ATG8a) together with firefly luciferase (FLUC) as internal control [9]. Co-expression of the viral protein 2b substantially increased RLUC-ATG8a accumulation in relation to FLUC, while the other viral proteins 2a, 3a and CP behaved similar as the GUS (β-glucuronidase) control (Figure 5H), indicating reduced RLUC-ATG8a turnover and hence specific down-regulation of autophagy by 2b. Moreover, the over-expression of ATG3 was previously shown to induce autophagy in N. benthamiana [38] and indeed, ATG3 increased the FLUC to RLUC-ATG8a ratio 5-fold compared to GUS (Fig. 5I). However, while still significant, this increase was efficiently reduced by 2b when co-expressed with ATG3, further supporting the autophagy-suppressing activity of 2b. Taken together, these results support that 2b can suppress SA-induced autophagy.
Autophagy-enhanced CMV resistance likely involves RNA silencing and is uncoupled from disease symptom severity
The 2b protein is essential for CMV suppression of DCL2/Dicer like 2- and DCL4/Dicer like 4-dependent antiviral RNA silencing as shown by restored pathogenicity of 2b-deficient CMV in the dcl2 dcl4 knock-out mutant [36,39]. Furthermore, 2b interacts directly with AGO1 as part of RNA silencing suppression [37]. We therefore reasoned that autophagy could limit CMV RNA accumulation by compromising 2b-dependent suppression of RNA silencing. If this was the case, autophagy-dependent suppression of CMV RNA accumulation should be less pronounced in AT3G03300/dcl2 AT5G20320/dcl4 and AT1G48410/ago1 backgrounds. Indeed, while CMV RNA accumulated to higher levels in both atg7 and dcl2 dcl4 mutants, there was no additive effect in the atg7 dcl2 dcl4 background at 14 DAI (Figure 6A). We obtained similar results for AGO1 as the ago1 atg5 double mutant did not show any additive increase in CMV RNA accumulation compared to atg5 and ago1 single mutants (Figure 6B). The absence of additive effects between autophagy and AGO1/DCL2,4-dependent resistance could support that these pathways are coupled through autophagy targeting of the viral RNA silencing suppressor 2b. However, we are unable to rule out that CMV accumulation was not already capped in single mutants and thus prevented additive accumulation.
Figure 6.
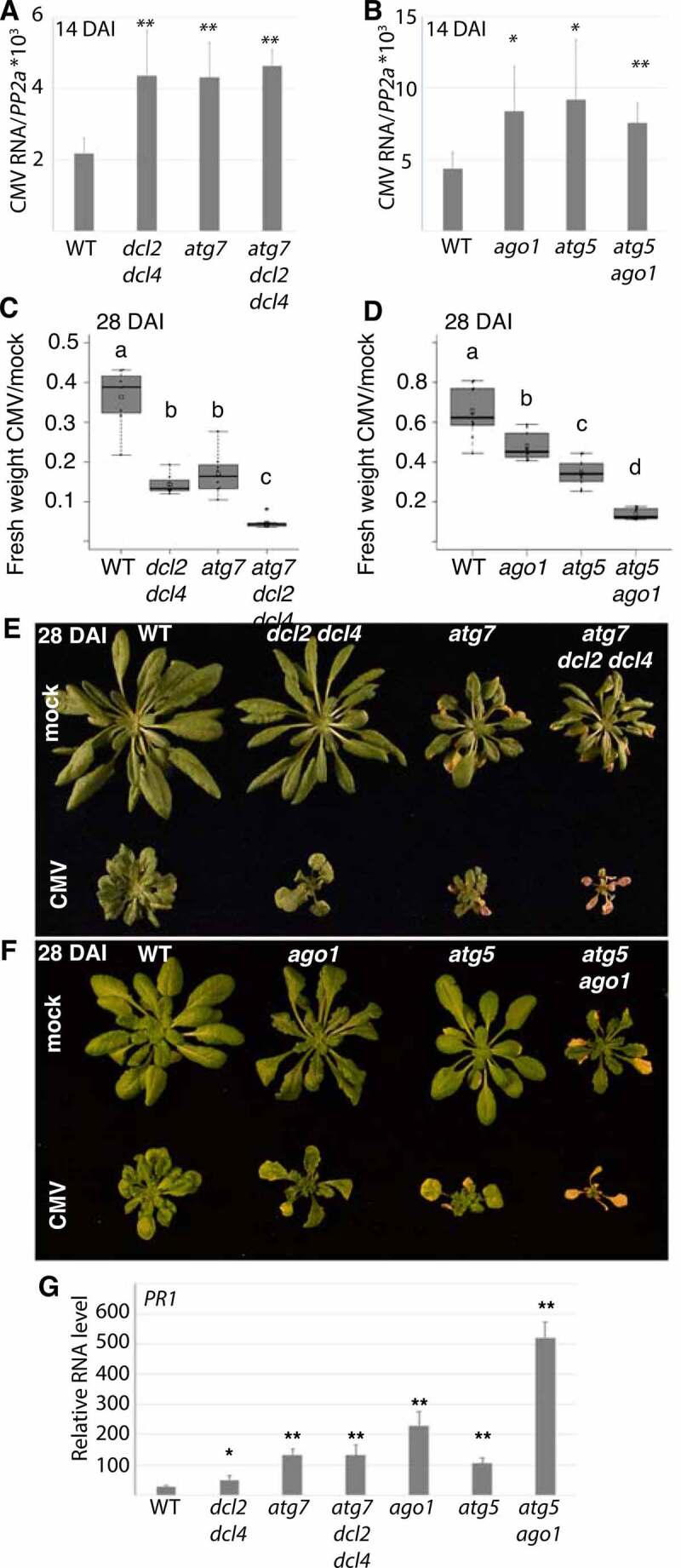
Autophagy-enhanced CMV resistance involves RNA silencing and is uncoupled from disease symptom tolerance. (A) CMV RNA levels relative to PP2A were determined by RT-qPCR at 14 DAI in WT, atg7, dcl2 dcl4 and atg7 dcl2 dcl4 plants (n=4). (B) CMV RNA levels relative to PP2A were determined by RT-qPCR at 14 DAI in WT, atg5, ago1 and atg5 ago1 plants (n= 4). (C) and (D) The relative fresh weight of CMV infected plants compared to mock were determined in the same genotypes as in (A) and (B) at 28 DAI. (n=10). Representative images of mock and CMV infected plants at 28 DAI are shown to the right (E) and (F). (G) PR1 transcript levels were analyzed in the indicated genotypes using RT-qPCR at 18 DAI with CMV (n= 4). Statistical significance (**P < 0.01, *P < 0.05) was revealed by Student´s t-test. Letters indicate genotypes with statistically different levels (ANOVA P < 0.05, Tukey-HSD test).
Despite the apparent non-additive functions of autophagy and RNA silencing in reducing CMV RNA accumulation at an early stage of infection (Figure 6A and B), atg7 dcl2 dcl4 and atg5 ago1 mutants showed strong additive effects on CMV infection-induced biomass loss (Figure 6C and D). Notably, there was no tissue senescence in either WT, dcl2 dcl4, and ago1 plants, while this autophagy-associated phenotype was clearly visible in CMV-infected atg5/atg7 and highly intensified in atg7 dcl2 dcl4 and atg5 ago1 plants (Figure 6E and F). Interestingly, when PR1 levels were analyzed in the infected mutants at 18 DAI before extensive senescence, ago1 and especially atg5 ago1, but not dcl2 dcl4, showed very strong up-regulation (Figure 6G). This indicates that SA responses escalate in atg5 ago1 and could well explain the additive disease phenotype in this mutant. Notably, SA responses have been previously reported to be elevated in ago1 [40] and SA causes premature senescence in autophagy-deficient plants [23]. Because the autophagy and RNA silencing pathways showed additivity in disease symptom severity but not viral RNA accumulation, we propose that autophagy-dependent CMV disease symptomology in RNA silencing mutants is not directly coupled to virus resistance.
Discussion
Autophagy can deliver a wide array of substrates including proteins, RNAs, ribosomes, proteasomes, viral particles, organelles, and aggregates for degradation in the lytic vacuole [2]. Hence, it plays an important role in maintaining plant homeostasis, especially when plants encounter stressful conditions such as viral infections [41]. The complexity of substrates targeted by autophagy underscores the possibility that several distinct autophagic processes operate in parallel upon cellular reprogramming and adaption to new conditions. This is fortified, for instance, by the uncoupling of autophagy-based virus resistance and plant biomass tolerance against TuMV [10], CaMV [11] and as demonstrated by this study, also CMV. The interaction between CMV and autophagy includes resistance and disease intensity regulated through RNA silencing components DCL2/DCL4 and AGO1 as well as the defense hormone SA, all likely coupled to autophagy-based degradation of the major virulence factor 2b. Interestingly, 2b alone reduces plant growth in an autophagy-dependent manner and shows the capacity to dampen SA-activated autophagy. A striking phenotype is the increased SA-dependent disease severity shown by CMV during autophagy deficiency. Together, the connections we have revealed between autophagy and different infection processes emphasize the complexity of this interaction in CMV epidemiology.
Plants reduce disease majorly by tolerance and resistance mechanisms, which are assumed to impose different selective pressures on both pathogens and hosts [42]. In this study, we found that autophagy contributes to the resistance against CMV by reducing virus accumulation. While autophagy-mediated resistance has been established for several animal viruses [16], comparative findings were largely lacking for plant viruses. Only recently, we and others have found that autophagy targets multiple plant viruses, including the positive-stranded RNA viruses TuMV and barley stripe mosaic virus [10,18,43], the negative-stranded RNA virus, rice stripe virus [17], the double-stranded DNA virus CaMV [11] and three single-stranded DNA geminiviruses [12]. These findings established autophagy as a central component of plant immunity against viruses, which is supported further by our current results on CMV. Notably, these studies have revealed a wide mechanistic diversity of autophagy-based virus resistance and viral counter-strategies [41], which ultimately suggests that different plant viruses have co-evolved with the autophagy pathway in an individual manner. We discovered that SA drives autophagy-induction during CMV infection and that 2b alone can suppress SA-induced autophagy. Interfering with SA may represent an indirect but general counter-strategy of plant viruses that reduces autophagy, as a similar observation was reported for P6 of CaMV [44]. In addition to 2b and P6 [28,29,45], also potyviral HCpro can suppress SA responses [46,47], but whether HCpro and other viral effectors can regulate SA-dependent autophagy remains currently unknown. Interestingly, SA is essential for CMV-induced autophagy, and the similar level of resistance and lack of epistasis at earlier infection stage between mutants in SA and autophagy supports that autophagy participates in SA-mediated resistance against CMV.
Despite the profound mechanistic differences in the detailed interaction between individual viruses and autophagy, a common theme arising is the involvement of the RNA silencing pathway in the autophagy-virus interplay [41]. These defense related pathways intersect during CMV infection as we show that autophagy degrades the viral silencing suppressor 2b and that the enhancement of viral RNA accumulation in autophagy- and RNA silencing-deficient plants lack epistasis. Our findings suggest that autophagy-based CMV resistance includes SA and RNA silencing, and we speculate that this is linked to the degradation of the viral silencing suppressor 2b. Notably, 2b is a highly complex virulence factor that among other functions interacts with AGO1/4 to impair slicing and also binds siRNAs (small interfering RNAs) directly [35,37,48,49]. Intriguingly, a very recent report showed that CMV, either lacking 2b completely or carrying a mutated 2b protein compromised in RNA silencing suppression, benefits from autophagy-dependent degradation of the RNA silencing component SGS3 leading to reduced resistance [22]. This finding further underscores the complexity of CMV-autophagy interactions and the likelihood that diverse autophagy mechanisms operate in parallel to influence virus resistances. Similar concepts are present for other viruses as well. Previously we showed that the TuMV silencing suppressor HCpro is degraded by autophagy, also resulting in reduced virus accumulation [10]. Intriguingly, this effect appears to include the degradation of potyvirus-induced structures reminiscent of RNA granules to which also AGO1 localizes [50]. In parallel, the TuMV protein VPg exploits autophagy to degrade SGS3 [20]. Together with the autophagic degradation of the geminiviral satellite ßC1 silencing suppressor [12], autophagy appears to be more generally nested into the antiviral RNA silencing defense with virus-specific adaptations. We also consider the exciting option that autophagy could degrade 2b in complex with siRNA, AGO1, and other components, where 2b would function as a selective autophagy cargo receptor to mediate the degradation of RNA silencing components. Our results, however, do not support that 2b uses this strategy to counteract RNA silencing-dependent resistance, and in general, autophagy seems to support rather than antagonize resistance against plant viruses [10–12,17,18,43].
One striking observation is the escalated severity of CMV disease in autophagy-deficient plants, as also seen for other viruses [41]. For CMV, we show that this phenotype is SA-dependent and correlates with PR1 levels. Defects in RNA silencing alone also increase CMV virulence, but when combined with autophagy deficiency the virulence intensifies to the extent that plants collapse and thus terminate the infection. It is likely that elevated 2b levels contribute to this phenotype as 2b virulence is dose-dependent [34] and the growth of 2b transgenic atg5 is severely reduced compared to 2b transgenic control plants. In the case of AGO1, the escalated disease could well be associated with the enhanced SA responses reported in the ago1 mutant [40] according to the elevated PR1 levels we observed. As 2b is known to directly bind and inhibit AGO1 [37], this interaction could contribute to SA-dependent disease under autophagy deficiency. Theory suggests that under strict vertical transmission, virulence should be negatively correlated with the transmission rate [51–53]. This can be reasoned because viral fitness is tightly linked to the reproductive success of the host [54]. Evidently, optimized virulence benefits virus epidemiology, where virus accumulation and virulence need to be balanced with plant longevity and fecundity to promote both successful horizontal and vertical virus transmission [33,52,55]. Thus, as autophagy and RNA silencing enhance plant longevity and vertical transmission of CMV, the antiviral nature of these pathways in a more holistic and epidemiological context becomes less clear. 2b of CMV subgroup I, including the virulent strains PV0187 (this study) and FNY, but not the mild LS strain of subgroup II, localize to the nucleolus [35,56]. Interestingly, nucleolar 2b appears to promote virulence uncoupled from virus accumulation and this virulence is furthermore attenuated by the RNA silencing pathway [56]. This finding is similar to our observation in combined autophagy and RNA silencing knockout mutants. In this context, it is notable that 2b re-localized ATG8a to the nucleolus in N. benthamiana co-expressions. Functions associated with nuclear and nucleolar localized LC3, the mammalian homolog of ATG8, are still largely undefined [57]. Recently, however, ATG8 was proposed to bind a geminiviral protein in the plant nucleus and target it for autophagic degradation in the cytoplasm [58]. Despite a potential connection between these events, we are unable to conclude whether 2b-dependent virulence involves nucleolar ATG8 functions at this stage, but notably, we have not observed evident ATG8a redistribution into the nucleolus during CMV infection.
In our hypothetical model (Figure 7), we stipulate that the intricate equilibrium between plant health, virus accumulation and virus transmission during CMV infection is determined by the reciprocity between the viral 2b protein, defense hormone SA, plant autophagy and RNA silencing pathway. 2b is a major virulence factor and RNA silencing suppressor of CMV [56] and the strong penalty of super-disease arising from autophagy- and RNA silencing-deficiency highlights the importance for CMV to fine-tune these interactions. One exciting possibility is that CMV recruits the autophagy pathway to degrade 2b in a regulated manner to balance the resistance penalty with long-term plant health. At the plant level, virus infections are prolonged processes, and the absolute amount of virus can continue to increase for several weeks during systemic infection. However, the active cellular infection cycle is assumed to be largely completed within 24 h for most plant viruses, including CMV [59]. At the single cell level, CMV may benefit from dampening the autophagy response using 2b without any long-term tolerance costs. Once the active stage of infection ceases in a cell, the autophagy-mediated clean-up of virulence factors like 2b should promote long-term tolerance and possibly even reduce coinfection competition from weaker viruses that would benefit from high levels of 2b. In line with this, the CP of CMV was recently proposed to destabilize 2b in a self-attenuation mechanism by which the virus achieves long-term disease reduction [60]. Taken together, through our model, we suggest that autophagy balanced by 2b regulates divergent aspects CMV infection, influencing virus accumulation, transmission, and plant disease (Figure 7).
Figure 7.
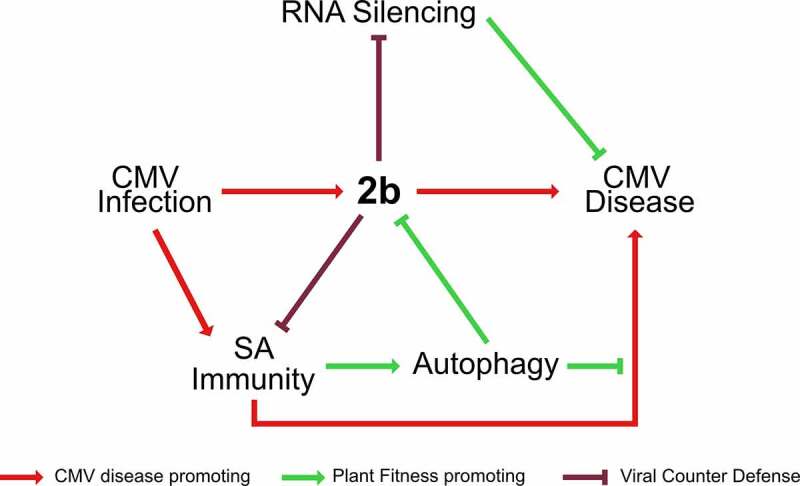
Model for the interplay between autophagy, SA, RNA silencing and 2b in CMV disease. The 2b protein is the major pathogenicity determinant in CMV infection. SA activates autophagy upon CMV infection and mediates 2b degradation. By limiting 2b levels, autophagy relaxes 2b mediated suppression of antiviral RNA silencing and virulence. Vice versa, 2b itself has the capacity to restrict both SA-dependent autophagy and plant growth reduction, predicting complex interactions between SA, autophagy and 2b in CMV disease. Furthermore, SA, autophagy and antiviral RNA silencing pathways all suppress virus accumulation, and the lack of additivity between the pathways suggests the possibility that they interact in the process, potentially through 2b degradation and SA-dependent autophagy. Taking SA-driven disease development under autophagy deficiency together with the prominent synergism of autophagy and RNA-silencing in disease attenuation, we propose that autophagy and RNA silencing-based plant health is not quantitatively coupled to virus accumulation and that the pathways rather operate in a parallel manner to promote survival of infected plants. Virulence evolution and trade-offs are complicated for pathogens that utilize both vertical and horizontal transmission. We consider that CMV has adapted to and benefits from these potential antiviral pathways. Thus, the interplay between the viral 2b protein, SA, plant autophagy and RNA silencing pathways determines the delicate balance between virus accumulation, transmission and plant fitness in CMV disease.
Materials and Methods
Plant material and growth conditions
Wild-type (WT) plant was Arabidopsis thaliana ecotype Columbia (Col-0) (NASC ID:N1093). Mutants atg5, NahG, atg5 NahG, sid2, atg5 sid2, npr1, atg5 npr1, ago1-27, dcl2 dcl4, atg7 dcl2 dcl4, and the GFP-ATG8a transgenic line have been described previously [23,61–64]. The ago1-27 atg5-1 double mutant and NahG ATG8a were generated by crossing. The transgenic line 2b3C was described previously [34], and used for crossing with GFP-ATG8a and the atg5-1 background. A 2b-RFP expressing GFP-ATG8a transgenic line was obtained by transformation of Arabidopsis. Arabidopsis plants were grown on soil for infection experiments under short-day conditions (10 /14 h light/dark cycles) at a light intensity of 150 μE/ m2s in a growth cabinet, and Nicotiana benthamiana plants were cultivated for transient expression assays under long-day conditions (16/8 h light/dark cycles) in a growth room at 150 μE/ m2s, 21 °C, and 70 % relative humidity, respectively.
DNA constructs
2a, 2b, 3a, CP and ATG3 were amplified using cDNA prepared from CMV infected plant total RNA as template and cloned into pENTRY-Topo (Thermo Fisher Scientific, K240020) and further recombined into pGWB660 except ATG3 into pGWB614 [65]. Expression constructs for GUS and GFP-ATG8a were described in [11]. All binary vectors were transformed into Agrobacterium C58C1 GV3101 [66] for transient expression in N. benthamiana or transformation of Arabidopsis by the floral dip method.
CMV inoculation and quantification
The first true leaves of 3-week-old Arabidopsis plants were inoculated mechanically with sap prepared from N. benthamiana plants infected with the CMV strain PV0187 described [67]. Plants were sampled in biological replicates, each containing 3 individual plants from which inoculated leaves were removed. For CMV RNA or plant transcript quantitation, total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, 74904), and on-column DNA digestion was performed with DNase I (Qiagen, 72954). First-strand cDNA was done using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1641). Quantitative RT-PCR analysis (qPCR) was performed with Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Fisher Scientific, K0241) using the CFX ConnectTM Real-Time PCR detection system (BIO-RAD) with gene-specific primers listed in Table S1. Normalization was done using AT1G69960/PP2A.
Drug and SA treatments
Seedlings were treated with 0.5 μM concanamycin A (Santa Cruz Biotechnology, 202111) or 15 μM AZD8055 (Santa Cruz Biotechnology, 364424) in liquid 1/2 MS (Duchefa biochemicals, M0221) for 10 h before analysis. Infected plants were treated at 21 DAI stage by vacuum-infiltration of 0.5 μM concanamycin A or DMSO (Sigma Aldrich, D8418) in liquid 1/2 MS and imaged after 10 h. For SA (Sigma Aldrich, 247588), 21 DAI plants were sprayed with 1 mM SA and 0.1 % EtOH and imaged 15 h later.
Confocal microscopy
Live-cell images were acquired from abaxial leaf epidermal cells using Zeiss LSM 780/800 microscopes. Excitation/detection parameters for GFP was 488 nm/490-552 nm. Confocal images were processed with ZEN (version 2011). Quantitation of GFP-ATG8a labelled puncta was done using ImageJ (version 1.48v). For the quantification of ATG8a labelled puncta, images were stacked using ‘Z-projection’ followed by ‘Gaussian blur’ to negate the background, and then puncta were counted under ‘Find maxima’ with a set threshold.
Immunoblot analysis
Proteins were extracted in 100 mM Tris, pH 7.5 with 2% SDS, boiled for 5 min in Laemmli sample buffer, and cleared by centrifugation at 9,000 g for 5 min. The protein extracts were then separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Amersham, GE Healthcare, 10061-494), blocked with 5% skimmed milk in 1 X PBS (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.2), and incubated with primary antibodies anti-NBR1 [68], anti-2b [35], and anti-GFP (Santa Cruz Biotechnology, sc-9996) using 1:2000 dilution in PBS 0.1% Tween-20 (VWR International, 97062), and secondary horseradish peroxidase-conjugated antibodies (Amersham, GE Healthcare, NA931) 1:10,000 in PBS 0.1% Tween-20. The immunoreaction was developed using the ECL Prime kit (Amersham, GE Healthcare, RPN2232) and detected in a LAS-3000 Luminescent Image Analyzer (Fujifilm, Fuji Photo Film). Quantification of band intensities in western blots and Rubisco large in Ponceau S-stained membranes as loading control was done using ImageJ 1.48v.
Immunoprecipitation
For immunoprecipitation, plant tissue was homogenized in 2 ml buffer (100 mM Tris, pH 8, 150 mM NaCl, 0.5% [v:v] Triton X-100 [Sigma Aldrich, T8787], protease inhibitor cocktail [Roche diagnostics, 11873580001]) per gram of tissue. The lysate was cleared at 4000 x g for 5 min at +4°C, filtered through two layers of Mira cloth (VWR Internationals, 475855-1R), and incubated 1 h with anti-GFP μbeads according to manufacturer’s instruction (Miltenyi Bioteck Norden AB, 130-091-125) for infected Arabidopsis tissue and GFP-agarose beads (Chromotech, GTMA-20) for transiently expressing N. benthamiana and transgenic Arabidopsis tissue. After washing 4 times with buffer, samples were eluted using 2 X Laemmli sample buffer and analyzed by immunoblotting.
Analysis of vertical virus transmission
Plants grown under long-day conditions were infected with CMV and allowed to set seeds. For vertical transmission, 10 pools of 20 seedlings grown for 10 days on ½ MS plates supplemented with 1 % sucrose each were analyzed by RT-PCR per parental plant for the presence of CMV. The frequency was calculated assuming that positive pools had only one infected plant out of 20 and thus provides a theoretical minimal transmission rate. The probability of virus transmission by a single seed was also calculated by the Gibbs and Gower’s formulae [32] ; where p is the probability of virus transmission by a single seed, y is the number of positive samples, n denotes the total number of samples assayed and k represents the number of seedlings taken per sample.
Luciferase-based quantitative autophagy assay
The assay was performed essentially as described previously in [9] using the Dual-Luciferase Reporter Assay System (Promega Biotech AB, E1910) following the manufacturer’s instructions. Briefly, three days post infiltration of N. benthamiana leaves with Agrobacteria carrying expression cassettes for RLUC-ATG8a, FLUC and co-expressed proteins, four leaf discs were harvested per sample in four replicates for lysate preparation. The measurement was performed in an Omega Fluostar plate reader.
Statistical analysis
Data are presented as mean ± SD (standard deviation) and statistical significance was analyzed by two-sided Student´s t-test with p-values <0.05 denoted * and p-values <0.01 denoted **. The number of replicates is given in the respective figure legends (n). Letters indicating groups with statistically different levels was tested by ANOVA P < 0.05 followed by Tukey-HSD test, performed in R-studio v 4.1.0.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge John Carr for providing the CMV PV0187 strain and FNY 2b transgenic lines as well as Thomas Canto for the 2b antibody. We are also grateful acknowledge Kohki Yoshimoto and Céline Masclaux-Daubresse for providing NahG, sid2 and npr1 with atg5 mutants.
Funding Statement
This work was supported by the Knut and Alice Wallenberg Foundation [2019-0062] for A.H.; EU Horizon 2020-MSCA-IF [898053] for N.K.K; Swedishresearchcouncil Formas [22915-000] for D.H; Swedishresearchcouncil Formas [2016-01044] for A.H.
Supplementary Material
Supplemental data for this article can be accessed here.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20(5):521–7. [DOI] [PubMed] [Google Scholar]
- 2.Batoko H, Dagdas Y, Baluska F, et al. Understanding and exploiting autophagy signaling in plants. Essays Biochem. 2017;61(6):675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogov V, Dotsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53(2):167–78. [DOI] [PubMed] [Google Scholar]
- 4.Michaeli S, Galili G, Genschik P, et al. Autophagy in Plants–What’s New on the Menu? Trends Plant Sci. 2016;21(2):134–44. [DOI] [PubMed] [Google Scholar]
- 5.Gatica D, Lahiri V, Klionsky DJ.. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20(3):233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephani M, Dagdas Y.. Plant Selective Autophagy-Still an Uncharted Territory With a Lot of Hidden Gems. J Mol Biol. 2020;432(1):63–79. [DOI] [PubMed] [Google Scholar]
- 7.Soto-Burgos J, Zhuang X, Jiang L, et al. Dynamics of Autophagosome Formation. Plant Physiol. 2018;176(1):219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofius D, Li L, Hafren A, et al. Autophagy as an emerging arena for plant-pathogen interactions. Curr Opin Plant Biol. 2017;38:117–23. [DOI] [PubMed] [Google Scholar]
- 9.Ustun S, Hafren A, Liu Q, et al. Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants. Plant Cell. 2018;30(3):668–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafren A, Ustun S, Hochmuth A, et al. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol. 2018;176(1):649–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafren A, Macia JL, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci U S A. 2017;114(10):E2026–E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haxim Y, Ismayil A, Jia Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagdas YF, Belhaj K, Maqbool A, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paudel DB, Sanfacon H.. Exploring the Diversity of Mechanisms Associated With Plant Tolerance to Virus Infection. Front Plant Sci. 2018;9:1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagan I, Garcia-Arenal F.. Tolerance to Plant Pathogens: Theory and Experimental Evidence. Int J Mol Sci. 2018;19(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong XN, Autophagy Levine B. and Viruses: Adversaries or Allies? Journal of Innate Immunity. 2013;5(5):480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu S, Xu Y, Li C, et al. Rice Stripe Virus Interferes with S-acylation of Remorin and Induces Its Autophagic Degradation to Facilitate Virus Infection. Mol Plant. 2018;11(2):269–87. [DOI] [PubMed] [Google Scholar]
- 18.Yang M, Zhang Y, Xie X, et al. Barley stripe mosaic virus gammab Protein Subverts Autophagy to Promote Viral Infection by Disrupting the ATG7-ATG8 Interaction. Plant Cell. 2018;30(7):1582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahara KS, Masuta C, Yamada S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(25):10113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng X, Wang A.. The Potyvirus Silencing Suppressor Protein VPg Mediates Degradation of SGS3 via Ubiquitination and Autophagy Pathways. J Virol. 2017;91(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derrien B, Baumberger N, Schepetilnikov M, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci U S A. 2012;109(39):15942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong X, Liu SY, Zou JZ, et al. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021:e108050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimoto K, Jikumaru Y, Kamiya Y, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy AM, Zhou T, Carr JP.. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr Opin Virol. 2020;42:8–17. [DOI] [PubMed] [Google Scholar]
- 25.Carr JP, Murphy AM, Tungadi T, et al. Plant defense signals: Players and pawns in plant-virus-vector interactions. Plant Sci. 2019;279:87–95. [DOI] [PubMed] [Google Scholar]
- 26.Mayers CN, Lee KC, Moore CA, et al. Salicylic acid-induced resistance to Cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol Plant Microbe Interact. 2005;18(5):428–34. [DOI] [PubMed] [Google Scholar]
- 27.Lee WS, Fu SF, Verchot-Lubicz J, et al. Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to potato virus X. BMC Plant Biol. 2011;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji LH, Ding SW.. The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol Plant Microbe Interact. 2001;14(6):715–24. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Murphy AM, Lewsey MG, et al. Domains of the cucumber mosaic virus 2b silencing suppressor protein affecting inhibition of salicylic acid-induced resistance and priming of salicylic acid accumulation during infection. J Gen Virol. 2014;95(Pt 6):1408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung T, Phillips AR, Vierstra RD.. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62(3):483–93. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Li X.. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol. 2019;50:29–36. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs AJ, Gower, J.C.. The use of multiple transfer method in plant virus transmission studies- some statistical points arising from the analysis of results. Annals of Applied Biology. 1960;48(1):75–83. [Google Scholar]
- 33.Pagan I, Fraile A, Fernandez-Fueyo E, et al. Arabidopsis thaliana as a model for the study of plant-virus co-evolution. Philos Trans R Soc Lond B Biol Sci. 2010;365(1548):1983–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewsey M, Robertson FC, Canto T, et al. Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 2007;50(2):240–52. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez I, Martinez L, Rakitina DV, et al. Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol Plant Microbe Interact. 2010;23(3):294–303. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Pendon JA, Li F, Li WX, et al. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19(6):2053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Yuan YR, Pei Y, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20(23):3255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han S, Wang Y, Zheng X, et al. Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell. 2015;27(4):1316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebell H, Carr JP.. Effects of dicer-like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by cucumber mosaic virus and a mutant virus lacking the 2b counter-defence protein gene. J Gen Virol. 2009;90(Pt 9):2288–92. [DOI] [PubMed] [Google Scholar]
- 40.Mason GA, Lemus T, Queitsch C.. The Mechanistic Underpinnings of an ago1-Mediated, Environmentally Dependent, and Stochastic Phenotype. Plant Physiol. 2016;170(4):2420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kushwaha NK, Hafren A, Hofius D.. Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation. Mol Plant Pathol. 2019;20(9):1211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy BA, Kirchner JW.. Evolutionary dynamics of pathogen resistance and tolerance. Evolution. 2000;54(1):51–63. [DOI] [PubMed] [Google Scholar]
- 43.Li F, Zhang C, Li Y, et al. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat Commun. 2018;9(1):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zvereva AS, Golyaev V, Turco S, et al. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016. [DOI] [PubMed] [Google Scholar]
- 45.Love AJ, Geri C, Laird J, et al. Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS One. 2012;7(10):e47535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alamillo JM, Saenz P, Garcia JA.. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 2006;48(2):217–27. [DOI] [PubMed] [Google Scholar]
- 47.Poque S, Wu HW, Huang CH, et al. Potyviral Gene-Silencing Suppressor HCPro Interacts with Salicylic Acid (SA)-Binding Protein 3 to Weaken SA-Mediated Defense Responses. Mol Plant Microbe Interact. 2018;31(1):86–100. [DOI] [PubMed] [Google Scholar]
- 48.Zhao JH, Liu XL, Fang YY, et al. CMV2b-Dependent Regulation of Host Defense Pathways in the Context of Viral Infection. Viruses. 2018;10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goto K, Kobori T, Kosaka Y, et al. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48(7):1050–60. [DOI] [PubMed] [Google Scholar]
- 50.Hafren A, Lohmus A, Makinen K.. Formation of Potato Virus A-Induced RNA Granules and Viral Translation Are Interrelated Processes Required for Optimal Virus Accumulation. PLoS Pathog. 2015;11(12):e1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elena SF, Agudelo-Romero P, Carrasco P, et al. Experimental evolution of plant RNA viruses. Heredity (Edinb). 2008;100(5):478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenski RE, May RM.. The evolution of virulence in parasites and pathogens: reconciliation between two competing hypotheses. J Theor Biol. 1994;169(3):253–65. [DOI] [PubMed] [Google Scholar]
- 53.Alizon S, Hurford A, Mideo N, et al. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol. 2009;22(2):245–59. [DOI] [PubMed] [Google Scholar]
- 54.Lipsitch M, Siller S, Nowak MA.. The Evolution of Virulence in Pathogens with Vertical and Horizontal Transmission. Evolution. 1996;50(5):1729–41. [DOI] [PubMed] [Google Scholar]
- 55.Stewart AD, Logsdon JM, Jr., Kelley SE.. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution. 2005;59(4):730–9. [PubMed] [Google Scholar]
- 56.Du Z, Chen A, Chen W, et al. Nuclear-cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA-silencing suppressor. J Virol. 2014;88(10):5228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kraft LJ, Manral P, Dowler J, et al. Nuclear LC3 Associates with Slowly Diffusing Complexes that Survey the Nucleolus. Traffic. 2016;17(4):369–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Zhang M, Zhang C, et al. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020;225(4):1746–61. [DOI] [PubMed] [Google Scholar]
- 59.Gonda T, Symons R.. Cucumber mosaic virus replication in Cowpea protoplasts: Time course of virus, coat protein and RNA synthesis. Journal of General Virology. 1979;45:723–36. [Google Scholar]
- 60.Zhang XP, Liu DS, Yan T, et al. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 2017;13(7):e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137(4):773–83. [DOI] [PubMed] [Google Scholar]
- 62.Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munch D, Teh OK, Malinovsky FG, et al. Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell. 2015;27(2):463–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shinozaki D, Merkulova EA, Naya L, et al. Autophagy Increases Zinc Bioavailability to Avoid Light-Mediated Reactive Oxygen Species Production under Zinc Deficiency. Plant Physiol. 2020;182(3):1284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakagawa T, Kurose T, Hino T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. [DOI] [PubMed] [Google Scholar]
- 66.Koncz C, Schell J.. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204: 383–396. [Google Scholar]
- 67.Groen SC, Jiang S, Murphy AM, et al. Virus Infection of Plants Alters Pollinator Preference: A Payback for Susceptible Hosts? PLoS Pathog. 2016;12(8):e1005790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7(9):993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


