Abstract
Dilated cardiomyopathy (DCM) remains an enigmatic cardiovascular disease (CVD) condition characterized by contractile dysfunction of the myocardium due to dilation of the ventricles. DCM is one of the major forms of CVD contributing to heart failure. Dilation of the left or both ventricles with systolic dysfunction, not explained by known causes, is a hallmark of DCM. Progression of DCM leads to heart failure. Genetic and various other factors greatly contribute to the development of DCM, but the etiology has still remained elusive in a large number of cases. A significant number of studies have been carried out to identify the genetic causes of DCM. These candidate-gene studies revealed that mutations in the genes of the fibrous, cytoskeletal, and sarcomeric proteins of cardiomyocytes result in the development of DCM. However, a significant proportion of DCM patients are idiopathic in nature. In this review, we holistically described the symptoms, causes (in adults and newborns), genetic basis, and mechanistic progression of DCM. Further, we also summarized the state-of-the-art diagnosis, available biomarkers, treatments, and ongoing clinical trials of potential drug regimens. DCM-mediated heart failure is on the rise worldwide including in India. The discovery of biomarkers with a better prognostic value is the need of the hour for better management of DCM-mediated heart failure patients. With the advent of next-generation omics-based technologies, it is now possible to probe systems-level alterations in DCM patients pertaining to the identification of novel proteomic and lipidomic biomarkers. Here, we also highlight the onset of a systems-level study in Indian DCM patients by applying state-of-the-art mass-spectrometry-based “clinical proteomics” and “clinical lipidomics”.
Keywords: dilated cardiomyopathy (DCM), heart failure, gene, biomarkers, mechanism, treatment, proteomics, lipidomics
1. Introduction
Dilated cardiomyopathy (DCM) is characterized as a disorder of the heart muscle. It is distinguished by the widening/dilation of the left ventricle of the heart with left ventricular or biventricular systolic dysfunction [1,2]. Contractile functioning of the left ventricle is highly compromised due to dilation. However, in some cases of DCM, the right ventricle is also dilated. Dilation of the ventricle leads to progressive disease resulting in heart failure. DCM is positioned third on the list of the main causes of heart failure [3,4], and it is most prevalent among all forms of cardiomyopathies in children [5]. Dilation and dysfunction in the contractility of the left ventricle can be caused by several pathological conditions including viral infection, drug abuse, toxins, and autoimmune disorders. However, around 50% of cases of DCM are etiologically unknown [6]. These etiologically unknown cases are termed as idiopathic DCM. In many cases of DCM, the early clinical symptoms are not highly observable and some patients are asymptomatic. The symptoms of DCM can be intermittently observed at the initial phase of DCM for certain patients. These clinical symptoms (Table 1) become more pronounced at the advanced (severe) stage of DCM [7,8,9,10]. Genetically inherited defects are a major cause of DCM. Around 30–35% of idiopathic cardiomyopathies are caused by genetic defects [10]. The very first attempt to assess the prevalence of DCM (36.5 in 100,000) was performed by Codd et al. during 1975–1984 in the Olmsted Country population (Minnesota, USA). Males were found more prone to DCM than females with a prevalence ratio of 3:1 [11]. In Italy, 2 years of data (1987–1989) of postmortem study in the Department of Pathology at Trieste University estimated the prevalence of DCM to be 4.5 in 100,000 subjects per year, and clinical data showed the prevalence of 2.5 in 100,000 subjects. However, combining DCM causing mortality, the total prevalence was found to be 6.95 in 100,000 subjects [12]. In the last 30–40 years, many epidemiological studies have been undertaken on DCM. However comprehensive data about the incidence of DCM globally is not known but the estimate of prevalence is >1 out of every 250–400 persons [13,14].
Table 1.
| Symptoms of DCM in Adults | Symptoms of DCM in Children/Newborns |
|---|---|
| Dyspnea and fatigue | Poor appetite/Problems in feeding |
| Dizziness | Poor growth |
| Syncope | Excessive sweating during feeding |
| Edema | Sweating while doing any activity |
| Palpitations | Palpitations |
| Controlled weight loss/gain | Fast breathing/difficult breathing |
| Excessive Sweating | Edema |
| Abdominal discomfort | |
| Nausea | |
| Anorexia | |
| Cachexia |
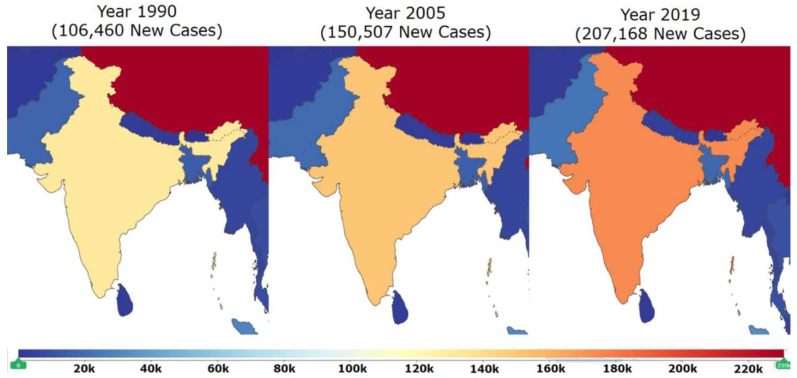
The prevalence of DCM has not been well studied in India compared to many western countries in the world. However, the global burden of disease [15] data shows that there is a drastic increase in the incidence of DCM in India. There were 106,460 new cases of DCM in 1990 in India. In one and a half decades, incidence increased massively and 150,507 new cases were reported in the year 2005. A spike in the incidence is consistent because 207,168 new cases were reported in the year 2019 [15]. This data clearly indicates the massive increase in the incidence of DCM in the past three decades (Figure 1) in India. Although, it is difficult to ascertain whether this increase in the incidence of DCM is highlighted because of the advanced detection systems [16], but the increasing incidence of DCM in India is alarming and has emerged as a major public health problem in the current scenario. There is the utmost need for better early diagnostic, prognostic biomarkers, effective treatments, and optimized preventive strategies to combat this disease. In this comprehensive review of DCM, we summarize the symptoms, causes, mechanistic insights of DCM development, available state-of-the-art diagnosis, and treatments, with emphasis on potential novel therapies and ongoing clinical trials. Further, we also showcase the importance of identifying a newer systems-level biomarker panel for DCM patients in India using next-generation proteomic- and lipidomic-based methods.
Figure 1.
This map of India (modified from Global Burden of Disease; GBD) shows the increase in the incidence of DCM in India in years between 1990 to 2019. There were 106,460 new cases in 1990. Incidents massively increased and 150,507 new cases were reported in 2005. Spike in incidence increased further and 207,168 new cases were reported in 2019. This data clearly indicates that the incidence of DCM is increasing at an alarming rate in India [Adapted with permission from IHME, Seattle, DC, USA “Source: Institute for Health Metrics Evaluation. Used with permission. All rights reserved” [15]. Accessed on 18 April 2021.
2. Causes of DCM
2.1. Causes of DCM in Children and Newborns
2.1.1. Myocarditis
Virus, allergens, toxins and autoimmune responses are responsible for myocardial damage [17]. This damage, caused by inflammation in general, is evidenced by the infiltration of immune cells into the myocardium associated with the degeneration and necrosis of myocytes during myocarditis. Importantly, histological evidence along with a specific threshold of leucocytes, monocytes and T-lymphocytes defines the myocarditis status [17,18,19,20]. Viral myocarditis is a common cause of DCM. Coxsackie B virus infection has a high tendency to lead to DCM. Coxsackie B is categorized under the picornavirus family and enterovirus genus. It also has a close association with echovirus, poliovirus, and rhinovirus. Other viruses are also found to contribute to DCM. Coxsackie (A, B), Herpes simplex, Epstein-Barr virus, Rubeola, Coronavirus, Adenovirus, Cytomegalovirus, Mumps, Vaccinia, Hepatitis B, Influenza (A, B), Varicella-zoster, Rubella, HIV and rabies viruses cause myocarditis that leads to the DCM [21].
2.1.2. Selenium Deficiency
Selenium is one of the trace elements present in animal bodies. Selenium is an important element of selenoproteins which play a crucial role in various endocrine, immune, and reproductive processes. Glutathione peroxidases (GSH-Px) is a selenoprotein that inactivates peroxidase. Selenium is useful in cardiomyocytes for the function of the antioxidant defense mechanism and a deficiency of selenium can lead to DCM [22].
2.1.3. Malformation in Pulmonary Arteriovenous
Pulmonary arteriovenous malformation (PAVM) is a condition of anomalous communication between veins and pulmonary arteries. Congenital pulmonary arteriovenous malformations have a close relation with hereditary hemorrhage telangiectasia. The shunting of blood from the pulmonary artery to veins causes the delivery of low oxygen to the heart. Shunting of blood causes hypoxia which leads to pulmonary hypertension and imparts a high load on the left ventricle resulting in DCM causing a reduced ejection fraction [23]. However, congenital heart diseases should be diagnosed to understand the overlapping contributory phenotypes of the DCM pathology in patients.
2.1.4. Endocardial Fibroelastosis
Endocardial fibroelastosis is caused by the deposition of collagen and fibroelastin fibers in the endocardium. The mitral valve leaflet is thickened, and the left ventricle is dilated in endocardial fibroelastin. Over a period of time, it develops into DCM [24].
2.1.5. Noncompacted Myocardium
Noncompacted myocardium is associated with the regional thickness of the ventricular wall and it can lead to heart failure [25]. Mutations in the protein-encoding genes of tafazzin, lamin, actin, titin and others are involved in non-compact myocardium disease. These gene mutations eventually lead to DCM and heart failure.
2.1.6. Calcium Deficiency
Calcium deficiency causes irreversible DCM in infants. Calcium is essential for myocardial contraction. A deficiency of calcium in infants affects myocardial contractility [26].
2.1.7. Idiopathic DCM (IDCM)
IDCM contributes to 50 to 70% of the total number of cases of DCM in children [5]. Many cases of IDCM are assumed to have genetic causes. In a number of cases, IDCM is reclassified as familial DCM after family investigation and testing. There is no specific observable characteristic difference present between familial DCM and idiopathic DCM [5].
2.1.8. Barth Syndrome
Barth syndrome is a heart muscle syndrome caused by mutation in the tazzafin protein-encoding TAZ (G4.5) gene [27]. It is mostly present in males and it is often present from birth. It is characterized by muscle dilation and skeletal myopathy. In Barth syndrome, the heart muscles fail to perform the movement function properly. Children with Barth syndrome find it hard to move or crawl and they feel very tired even after little amount of effort. In many cases, Barth syndrome goes unnoticed in the early life of a patient. The development of noticeable symptoms varies greatly from person to person [28].
2.1.9. Familial/Genetic Pediatric DCM
Genetic causes are one of the major reasons for the occurrence of DCM. There are similarities in the genetic causes of newborns and adults. Rampersaud et al. [29] studied 41 cases of pediatric DCM. They identified 15 genes associated with DCM pediatric patients. Out of these 15 genes, 9 genes (MYH7, SCN5A, TNNT2, LMNA, MYBPC3, MYH6, TNNC1, TNNI3 and TPM1) are also known to cause DCM in adults. Recently, Khan et al. [30] did a cohort study on 109 pediatric patients and found that TTN (Titin) truncation was the most prevalent cause of genetic DCM in pediatric patients. Along with TTNtv, genetic variations (mutation) were also detected in MYH7 among pediatric DCM patients.
2.2. Causes of DCM in Adults/Adolescents
2.2.1. Familial/Genetic
It is estimated that 30–50% of DCM cases fall under this category [31]. DCM is highly linked with autosomal dominant inheritance, but X-linked and autosomal recessive inheritance has also been found to be associated with DCM. Mutations in the cytoskeletal and sarcomeric genes of cardiomyocytes are considered to be responsible for DCM [31]. Cytoskeletal genes including TTN (titin), DES (desmin), LMNA (lamin A/C), ABLIM1 (actin binding LIM domain protein), ACTN2 (α-actinin-2), NEBL (nebulette), MYPN (myopalladin), SGCD (δ sarcoglycan) and ZASP (Z band alternatively spliced PDZ domain protein) are involved in DCM [32]. Mutations in sarcomeric genes including ACTC1 (cardiac actin alpha), TNNT2, TNNI3, TNNC1 (troponin T2, I3 and C1), MHY7 (β- myosin heavy chain), TPM1 (tropomyosin-1), PLN (phospholamban), MYBPC3 (myosin binding protein C) and SCN5A (Sodium channel protein type 5 subunit alpha) are associated with DCM (Table 2).
Table 2.
Mutations in cytoskeletal and sarcomeric protein-coding genes are majorly associated with DCM [31,32,33,34].
| Cytoskeletal and Sarcomeric Genes | |
|---|---|
| Titin (TTN) | Cardiac actin alpha |
| Desmin (DES) | Troponin T2, I3, C1 (TNNT2, TNNI3, TNNC1) |
| Lamin A/C (LMNA) | β-myosin heavy chain (MHY7) |
| α-actinin-2 (ACTN2) | Tropomyosin-1 (TPM1) |
| Actin binding LIM domain protein (ABLIM1) | Phospholamban (PLN) |
| Nebulette (NEBL) | Myosin binding protein C (MYBPC3) |
| Myopalladin (MYPN) | Sodium channel protein type 5 subunit alpha (SCN5A) |
| Filamin C (FLNC) | BCL2-associated athanogene 3 (BAG3) |
| δ sarcoglycan (SGCD) | |
| Vinculin (VCL) | |
| Z band alternatively spliced PDZ domain protein (ZASP) | |
| Dystrophin gene (DMD) |
2.2.2. Alcohol
A daily consumption of alcohol of more than 35.2 mL in males and 17.6 mL in females can cause alcohol cardiomyopathy [35]. Although it remains debatable that a low consumption of alcohol can be useful in coronary artery disease (CAD), hemorrhagic stroke and heart failure, alcohol abuse increases blood pressure and adversely affects the immune system, which increases the chances of myocarditis [35]. Carbohydrate-deficient transferrin and elevated levels of liver enzyme with cardiomyopathy are diagnostic measures of alcohol-induced cardiomyopathy [35].
2.2.3. Myocarditis
Myocarditis affects both at early age and adult age due to several virus-mediated infections. The mode of action of myocarditis remains the same at all stages of life.
2.2.4. Tachycardiomyopathy
Tachycardiomyopathy, also known as tachycardia-induced cardiomyopathy, is a left ventricle disorder. It is a cardiac condition which can be partially or completely reversed. If tachyarrhythmia is controlled then tachycardiomyopathy can also be reversed to normal stage [36].
2.2.5. Mitochondrial Diseases
Mitochondrial disorders including NADH-coenzyme Q reductase (complex I) deficiency, deficiency of cytochrome C oxidase, lactic acidosis, myoclonic epilepsy with ragged red fibers (MERFF), Kearns–Sayre syndrome, encephalopathy associated with mitochondria, and most importantly carnitine palmitoyl transferase II deficiency, significantly contribute to developing DCM [37].
2.2.6. Cardiomyopathy Associated with Right Ventricular Arrhythmia
Although less prevalent, it is a life-threatening condition entailing sudden death in young and sports persons. Mutation of the desmosomal protein gene leads to this disease which is now considered as genetic myocardial dystrophy [38].
2.2.7. Eosinophilic Myocarditis
Eosinophilic granulomatosis with polyangiitis (EGPA), also known as Churg-Strauss syndrome, is characterized by systemic vasculitis. This vasculitis affects the small and medium sized blood vessels [39]. The effects of this disease in the heart are dangerous and can lead to heart failure [40]. Eosinophilic myocarditis is diagnosed with left ventricle enlargement and decreased systolic function. If eosinophilic myocarditis is caused by Churg-Strauss syndrome then treatment of the Churg-Strauss syndrome can reverse the eosinophilic myocarditis to the normal stage [41].
2.2.8. Toxins
Over-exposure to heavy metal toxins can lead to DCM. Lead (Pb), cobalt (Co) and arsenic (As) over-intoxication can lead to DCM. Iron overload is also found to cause DCM [42]. Under certain genetic and metabolic disorders, the amount of circulating iron exceeds the handling capacity of transferrin. Free, unbound iron is a potent free radical which causes oxidative damage to the cardiomyocytes. Lead is a toxin which attacks the cardiovascular system adversely. Cobalt is useful in the production of red blood cells, but a very high amount of blood cells due to a higher level of cobalt can affect the heart rate by interfering with calcium binding to sarcolemma [42]. Thus, it causes trouble in contractility in the heart. Arsenic is a toxin which directly attacks cardiac muscles. It dilates cardiac capillaries and it induces DCM [42].
2.2.9. Peripartum Cardiomyopathy
It is a life-threatening disease. It is developed during pregnancy in females. It severely affects females during late pregnancy or the puerperium phase. In peripartum DCM, cardiac failure can be observed towards the end of the gestation period to the 5-month postpartum period [43].
2.2.10. Endocrinopathy
A hormonal imbalance in the body can lead to the development of DCM. An imbalance of the thyroid hormones leads to heart disorders. Tachycardia and palpitations are common in patients with thyroid hormone imbalance [44]. The catecholamine secretion from tumors induces cardiac remodeling [45]. A low level of insulin causing persistent hyperglycemia severely compromises cardiac functions leading to DCM or other cardiomyopathies [46].
2.2.11. Nutritional Deficiency
Low levels of thiamine (B6), carnitine deficiency and hypophosphatemia have a correlation with DCM progression. Nutritional deficiency leads to the abnormal functioning of cardiac muscles and long-term nutritional deficiencies can lead to DCM and other disorders [47].
3. Genes Involved in Molecular Mechanisms of DCM
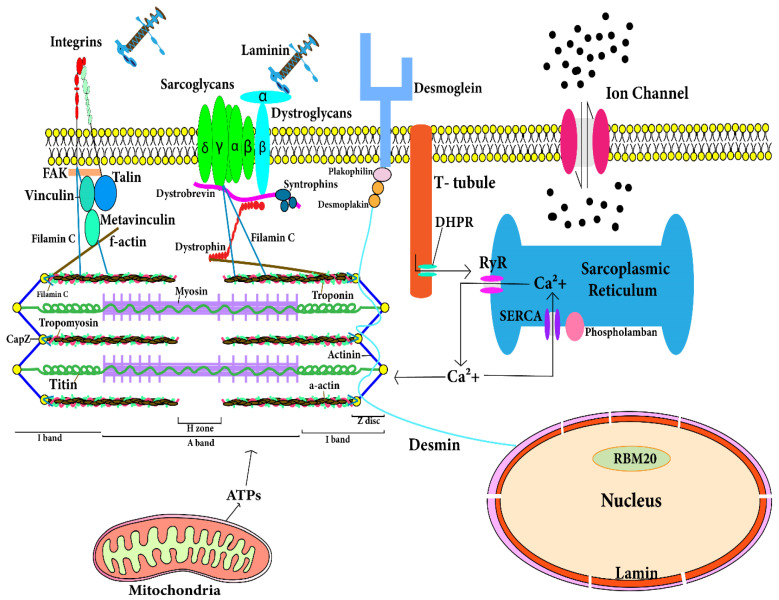
Cardiomyocytes are comprised of fibrous proteins, cytoskeletal proteins, and sarcomeres. Mutations in the following genes encoding the respective proteins lead to the development of DCM. A comprehensive spatial map of such structural proteins is depicted in Figure 2. Following are the genes which significantly contribute to the development of DCM.
Figure 2.
Location of proteins in cardiomyocytes that are responsible for development of dilated cardiomyopathy. The figure depicts the spatial location of the cytosolic and sarcomeric proteins encoded by corresponding genes. Mutations in these genes contribute to development of dilated cardiomyopathy. The T-tubule has a large number of ion channels and transporters. T-tubule regulates the calcium concentration in cardiomyocytes and transmits the action potential into the cardiomyocyte. RyR (ryanodine receptor) in sarcoplasmic reticulum (SR) of cardiomyocyte interacts with T-tubule for action potential transmission and RyR is responsible for release of stored calcium from SR. Calcium and ATPs are utilized by sarcomere in contraction. After contraction, calcium released from sarcomere is stored in SR through SERCA. Sarcolemmal and cytoskeletal proteins maintain architecture and functions of cardiomyocytes. Mutation in any of these protein-encoding genes leads to dilated cardiomyopathy.
3.1. Troponin T Gene (TNNT2)
Troponin T protein of the thin sarcomere filament is encoded by TNNT2 gene. TNNT2 is 17 exons containing 17 kb long genomic DNA. Alternative splicing of this gene produces different isoforms of troponin T. Some 288 amino acids are found in the primary isoform of troponin T and it has two major domains. Tropomyosin interacts with the NH2 terminal domain of troponin T, troponin C and troponin I interact with the COOH terminal domain of troponin T. Deletion (Lys210) in the TNNT2 gene is found in DCM [48]. In DCM, deletion in TNNT2 gene is found on the carboxyl domain of 207–234 residues which communicate with calcium-sensitive troponin C. Thus, this deletion contributes to a calcium imbalance in cardiomyocytes and their misfunction.
3.2. Cardiac Actin Alpha Gene (ACTC1)
Actin, a protein of the thin sarcomere filament is encoded by the ACTC1 gene. Out of 20 genes associated with actin which are present in the human genome, four genes code for smooth muscle actins, skeletal actins and cardiac actins. Actin corresponding to cardiac and skeletal tissues are present in the heart, but skeletal actin has a lesser amount in the adult human [49]. The cardiac actin gene (ACTC1) encodes 375 amino acids which are coded by six exons. The myosin cross-bridge attaches to the NH2 terminal domain of the cardiac actin protein. Further, α-actinin and dystrophin bind with the COOH terminal domain of ACTC1. ACTC1 participates mainly in the contraction of muscles. This gene was found to be involved in autosomal dominant DCM in adults [49]. Two missense mutations are found in the ACTC1 gene present in DCM patients [49]. Mutation in the cardiac actin at the juncture of the Z line is found in DCM. This immobilized end of actin in the Z line is believed to be associated in force transfer to the extra-sarcomeric cytoskeleton via the sarcomere.
3.3. Myosin Heavy Chain—β Gene (MYH7)
The protein β-myosin heavy chain of the thick sarcomere filament is encoded by the MYH7 gene. The myosin protein is present in hexameric form with two heavy chains and four light chains. β-MHC and α-MHC are two isoforms of the myosin heavy chain protein present in cardiac myocytes. The β-myosin heavy chain is predominantly expressed in the atrium and ventricles of embryos or some skeletal muscles. However, in adults, the α-MHC (encoded by MYHC6) predominates. MYHC7 is a 23 kb long DNA. Out of 41, 38 exons make up a 1935 amino acid long protein product. Two missense alterations associated with the MYH7 gene have been found to significantly contribute to the development of DCM [48]. First is the MYHC7 (Ser532Pro) mutation which affects α-helical of the lower domain of the myosin heavy chain. Mutation in this domain disrupts the actin–myosin binding. The second missense mutation is the MYH7 (Phe764Leu) mutation. It affects the hinge region and it adversely affects contraction by changing the polarity of the cross-bridge movement.
3.4. Cardiac Myosin Binding Protein C Gene (MYPBC3)
Cardiac myosin binding protein C (cMBP-C) is a thick filamented protein that is encoded by the MYPBC3 gene. This cMBP-C is the exclusive isoform present in the A band of sarcomere, and it forms 7 to 9 transverse bands with a spacing of 43 nm. The MYPBC3 gene is made up of 24 kb of DNA. The 37 exons of the MYPBC3 gene encode 1274 amino acids. This protein has multiple sites including a phosphorylation region, fibronectin type 3 domain, a cardiac-specific region and intervening titin and myosin binding sites. Missense mutation in this gene is found in DCM patients [50].
3.5. Tropomyosin α Gene (TPM1)
Its protein product can be found in fast skeletal muscle or ventricular myocardium. Fifteen exons are present in TPM1 gene. Five exons are constitutively present in all isoforms and alternating splicing of the other 10 exons gives rise to different isoforms of the protein. Some 288 amino acids are encoded by 10 exons of cardiac α-tropomyosin isoform. α-tropomyosin contains Ca2+ sensitive and Ca2+ insensitive binding sites. Two missense mutations in the TPM1 gene are found in individuals having DCM [51]. These two mutations are believed to be responsible for change in actin interactions and the structural integrity of α-tropomyosin.
3.6. Titin Gene (TTN)
Giant sarcomeric protein titin is encoded by the TTN gene. Titin (3816 kDa) is the largest known polypeptide. Titin has a stiffer portion within the A band and an extensible portion within the I band. The repeating 298 immunoglobulin domain and fibronectin 3 domain contribute 90% of the total mass of titin. The elasticity and organization of sarcomere is maintained by titin. Frameshift and missense mutation in the TTN gene are found in individuals having DCM [52]. Mutations resulting in titin truncation variants (TTNtv) contribute to almost 25% of genetic cause in idiopathic DCM and almost 18% is estimated for sporadic cases [53]. Verdonschot et al. [54] studied 689 DCM patients and found TTNtv were most abundant in the patients with DCM. This study evidenced that the titin truncation contributes the most to familial/genetic DCM.
3.7. Desmin Gene (DES)
Desmin, generally considered a cytoskeletal protein of intermediate filament is encoded by the DES gene. Desmin is present in all muscle tissues. It is believed that it regulates myogenic differentiation and the desmin intermediate filament significantly contributes to force transmission from the sarcomere [55]. The DES gene is made up of 8.4 kb of genomic material. The desmin protein has 470 amino acids encoded by nine exons of the DES gene. Desmin tail domain missense mutation (Ile451Met) disrupts myofibril organization, and it is reported to significantly contribute to the development of DCM [55]. Desmin knockout in mouse models also develops DCM [55].
3.8. δ-Sarcoglycan Gene (SGCD)
δ-sarcoglycan glycoprotein of transmembrane of sarcolemma is encoded by the SGCD gene. Some sarcoglycan proteins (α, β, γ and δ-sarcoglycan) form the sarcoglycan complex, which is a part of a glycoprotein transmembrane complex associated with dystrophin. The SGCD gene is active in striated and smooth muscles. The highest levels of the SGCD gene are expressed in cardiac and skeletal muscle. The SGCD gene is composed of around 100 kb of genomic DNA. δ-sarcoglycan has 290 amino acids encoded by eight exons of the SGCD gene. The δ-sarcoglycan protein has a large extracellular COOH terminal domain, a hydrophobic transmembrane domain, and a small intracellular domain. One missense alteration of the SGCD gene is associated with DCM and early unprecedented death [56]. Two other mutations in the SGCD gene leading to codon deletion, i.e., Lys238 codon deletion, are found in individuals with DCM [56].
3.9. Vinculin and Metavinculin Gene (VCL)
Metavinculin and vinculin are cytoskeletal proteins encoded by alternative splicing of the VCL gene. Metavinculin and vinculin are coexpressed in smooth, skeletal and cardiac muscle. α-Actinin, talin, and γ-actin interact with metavinculin and vinculin at subsarcolemmal costameres and they form a micro-filamentous network. This network links sarcolemma via the cytoskeleton. Metavinculin along with vinculin are seen on the intercalating disc of the adherens junction and they actively participate in cell-to-cell adhesion. Over 75 kb of genomic DNA makes up the structure of the VCL gene. It has 22 exons that make the vinculin protein comprising 1066 amino acids and metavinculin protein comprising 1134 amino acids. Their multimers of four to six molecules have head and tail domains. The head domain interacts with talin and the tail domain interacts with γ-actin and paxillin. High levels of vinculin to 1.2 fold were found in explanted human hearts from individuals having DCM [57]. On the other hand, absence or low levels of metavinculin are also found in individuals having DCM [58].
3.10. Lamin A and Lamin C Gene (LMNA)
Lamin A and lamin C protein of intermediate filament are encoded by the LMNA gene. Lamin proteins are present in nuclear lamina, located on the internal side of nuclear membrane. Lamin A has four [4] transcripts (lamin A, C, Aδ10, and C2) made from alternative splicing of LMNA gene, while lamin B has two transcripts (lamin B1 and B2) made by LMNB1 and LMNB2 genes. A, B2 and C types of lamins are present in skeletal muscle and cardiac muscle. These are believed to contribute to the integrity of nuclear membrane; LMNA gene is made up of 2400 bp of DNA and 12 exons. Lamin A transcript has 664 amino acids while lamin C transcript has 562 amino acids. Lamin A and C proteins have an α-helical dimer rod domain, globular head domain (NH2-terminal) and tail (COOH-terminal) domains. LMNA mutations are prevalent in families of autosomal dominant DCM [59]. A total of 19 mutations in LMNA gene are found in relation with autosomal dominant DCM [59].
3.11. Dystrophin Gene (DMD)
Dystrophin is a massive cytoskeletal protein, which is coded by the DMD gene. It is present in high amounts in skeletal, smooth and cardiac muscle cells and also in low amounts in brain cells [60]. Dystrophin is a large 2.4 mb gene. The 3685 amino acids are encoded by 79 exons of the DMD gene. The DMD gene has different promoters expressed in different tissues. Alternative splicing of the DMD gene give rise to different isoforms of dystrophin protein. Dystrophin is a rod-shaped protein and is around 150 mm long in length. Dystrophin is believed to contribute to maintaining membrane stability, force transmission and intracellular organization. Mutation in the DMD gene is found to contribute to X-linked DCM-associated death within just 1–2 years after diagnosis [61]. Deletion, missense, insertion, inversion and point mutations in the DMD gene result in abnormalities of the dystrophin protein [62].
3.12. Tafazzin Gene (G4.5)
Tafazzin, a group of proteins present in abundance in cardiac and skeletal muscles is encoded by G4.5 gene. The molecular functions of tafazzins have remained elusive. The G4.5 gene is made up of around 1100 bp of genomic DNA. It has 11 exons whose alternative splicing produce 10 different transcripts. Mutations in G4.4 are found to be associated with X-linked Barth syndrome which is characterized by infantile-onset DCM [63]. Missense mutations in exon 8 and exon 10 conserved regions are found in the individuals with X-linked DCM [63].
3.13. Phospholamban Gene (PLN)
Phospholamban, a membrane protein is encoded by the PLN gene. Phospholamban is made up of 52 amino acids. It is a key regulatory protein in cardiac diastolic function and Ca2+ cycling in sarcoplasmic reticulum. Mutations in the PLN gene result in an imbalance of calcium in the cells. Deletion of arginine 14 (PLN-R14Del) in the PLN gene is found in individuals having DCM [64]. Mutation in the PLN gene (R9C) leads to the development of DCM in mice [65].
3.14. Mitochondrial DNA
Mitochondrial inheritance is maternal in nature. Cardiomyocytes contain high numbers of mitochondria for the production of energy and contractile function. Mitochondria has 16.6 kb DNA, which encodes 13 proteins involved in the oxidative phosphorylation. Certain genetic alterations such as deletions or point mutations in mitochondrial DNA lead to mitochondrial abnormality [66]. Mitochondrial abnormality contributes to the development of the fatal infantile form of DCM [67].
3.15. Filamin C (FLNC)
Filamin C is a large 2725 amino acid long protein. It is encoded by the FLNC gene. It plays important role in the function of skeletal and cardiac muscles. Begay et al. [68] found filamin C truncations to be associated with arrhythmogenic dilated cardiomyopathy. Recently Aggarwal et al. [69] discussed that haploinsufficiency (FLNC+/−) can cause dilated cardiomyopathy. Studies carried out on filamin C revealed that mutations in FLNC can lead to cardiomyopathies.
4. Mechanistic Insights for DCM Development
DCM progression is initiated by various genetic and idiopathic factors. Classically, “Frank-Starling law” described the causal relationship between resultant contraction force with cardiac fiber length [70]. Mutations in sarcomeric, non-sarcomeric and cytoskeletal protein components have been attributed mechanically to cardiac contraction reduction during DCM [33]. During the pathological progression of DCM, cardiomyocytes gradually undergo various changes leading to compromised function and morphology leading to reduced systolic performance with dilated ventricles [71]. In DCM, significant death of cardiomyocytes occurs by the process of apoptosis (Figure 3) [72]. Accumulation of collagens and other extracellular matrices (ECM) proteins in the ECM followed by the cardiomyocyte apoptosis leads to myocardial fibrosis [73]. The proliferation of cardiac fibroblasts to myofibroblasts in response to profibrotic stimuli contributes majorly to the fibrosis [74]. These processes lead to the dilation and stiffening of the left ventricle, a definitive hallmark of DCM [75] (Figure 3). The myocardium becomes thinner and the ventricle is stretched, leading to dysfunction in heart [76].
Figure 3.
In DCM, cardiomyocytes undergo apoptosis and proliferation of cardiac fibroblasts to myofibroblasts causes fibrosis in the left ventricle of the heart. These processes lead to left ventricle dilation and wall thinning.
4.1. Signaling Pathways Involved in DCM
Cardiomyocyte death and fibrosis are two major processes that lead to the development of DCM. The following pathways involved in cell death and fibrosis during DCM are described below.
4.1.1. Cell Death Pathways in Cardiomyocytes
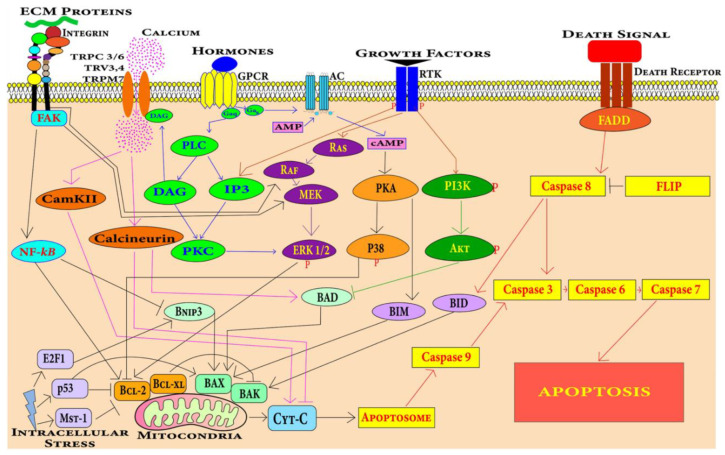
Apoptosis in cardiomyocytes can be induced by various stimuli including hypoxia, mechanical stress, oxidative stress, hormones, growth factors, death ligand including tumor necrosis factor, Fas ligand and anthracycline drugs [77,78]. Myocardium-specific apoptosis in the transgenic mouse model revealed that even low levels of myocyte apoptosis leads to the development of DCM [79]. There are two central pathways involved in apoptosis, i.e., extrinsic pathway and intrinsic pathway. Several key pathways and potential signaling mechanisms during DCM progression have been summarized in Figure 4.
Figure 4.
Cardiomyocytes can undergo apoptosis through extrinsic or intrinsic apoptosis pathways. Death receptors mediate the extrinsic apoptosis pathway and various factors including mechanical stress, oxidative stress, DNA damage, growth factors and hormones induce the intrinsic cardiomyocyte apoptosis pathway.
Extrinsic Pathway
The extrinsic apoptotic pathway involving caspase 8 is found to be associated with cardiomyocyte apoptosis leading to DCM in mice [79]. The extrinsic apoptosis pathway is mediated by a class of receptors called death receptors present on cardiomyocytes. These receptors include TNF-related apoptosis-inducing ligand (TRAIL) receptors, Fas receptors and TNF receptors [80]. These generic pathways become activated during the pathological progression of DCM.
Intrinsic Pathway
The intrinsic apoptosis pathway mediated by mitochondria dysfunction leads to cardiomyocyte apoptosis in DCM [81]. The intrinsic pathway is the integration of stress cues from the internal environment of the cells and can be triggered by different stimuli such as nutrient deficiency, radiation, drug, oxidative stress, irreparable DNA damage, and the misfolding of proteins. Mitochondrial and endoplasmic-reticulum stress has also been found to be involved in the apoptosis of cardiomyocytes in cardiovascular disease conditions [82].
4.1.2. Fibrosis Pathways Associated with DCM
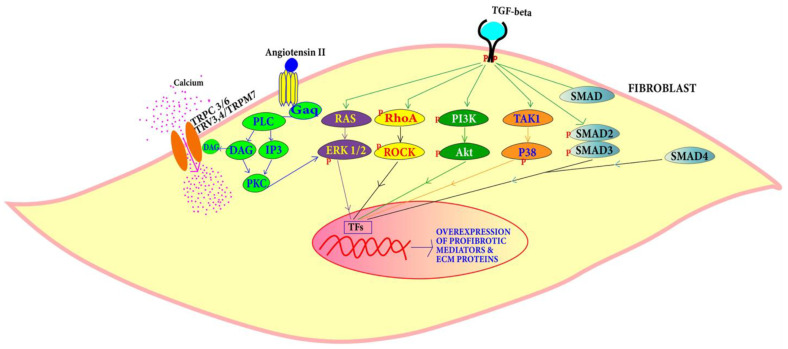
TGF-β and Ang-II pathways are known in the DCM condition to induce the proliferation of cardiac fibroblasts to myofibroblasts leading to fibrosis (Figure 5) [83,84,85,86,87].
Figure 5.
TGF-β can induce cardiac fibroblast proliferation through SMAD-dependent and SMAD-independent pathways, while proliferation through Ang-II is mediated by PLC, DAG, IP3 and PKC.
TGF-β Pathway: TGF-β is a signaling molecule that is involved in a plethora of signaling pathways. TGF-β can change the expression of extracellular matrix components [88]. TGF-β levels are found to be high in DCM patients. The overexpression of TGF-β in mouse heart leads to myocardial fibrosis [83]. Canonically, TGF-β type II binding with its receptor causes TGF-β type I receptor stimulation which activates the SMAD complex. This heterotrimeric complex of SMAD 2, SMAD 3 along with SMAD 4 that is translocated in the nucleus, binds to a section called the SMAD binding elements (SBE) and promotes collagen deposition and fibrosis. SMAD-independent pathways involve PI3K/AKT and MAPK pathways and RHO/RAC1. High TGF-β expression triggers the differentiation of myofibroblasts and the deposition of matrix components associated with hypertrophic cardiomyopathy and DCM [83,84]. An isoform of TGF-β called TGF-β1 promotes cardiac apoptosis and hypertrophy and may be involved in cardiomyopathic pathology [85,86]. TGF-β binding to type II of TGF-β receptor activates TGF-β-activated kinase 1 (TAK1). TAK1 activation leads to the activation of p38 [89]. The action of TGF-β on Ras-Raf-MEK-ERK cascade causes the translocation of extracellular regulated protein kinases (ERK) and various transcription factor phosphorylation [90]. For example, activator protein 1 (AP1) gene is expressed to produce various ECM proteins. Along with regulating actin cytoskeleton, ROCK phosphorylate targets that are important for remodeling include myosin light chain (MLC), myosin-binding subunit (MBS) on MLC phosphatase (MLCP). Upregulation of ROCK leads to cardiac remodeling [91].
4.1.3. Angiotensin Pathway
The renin angiotensin pathway has been revealed to be far more complex than was previously perceived. It is known to be involved in local transduction via the intracrine and paracrine or autocrine pathways as well as acting systemically. Mice develop DCM in response to angiotensin-II [87]. Angiotensin II (Ang II) can bind to two receptors: angiotensin type I (AT1) and angiotensin type II (AT2). The release of Ang II activates G protein-coupled receptors inducing the conversion of guanosine-5′-diphosphate (GTP) to guanosine-5′-triphosphate (GDP) via Gαq that interacts with phospholipase C (PLC). This stimulates diacylglycerol (DAG) and IP3 that activate the protein kinase C (PKC). GPCR activation also triggers Ca2+ release via DAG that is then converted to triacylglycerol (TAG) which controls Ca2+ influx into the fibroblasts from TRPC 3/6 TRV3,4/TRPM7 and regulates cardiac contractile adaptation [92]. PKC induces ERK1/2 leading to the activation of transcription factors that cause fibroblast proliferation into myofibroblasts.
5. Diagnosis of DCM
5.1. Clinical Investigation
Clinical investigation is the front-line diagnosis procedure. Many patients do not manifest diagnostic symptoms until they are at the advanced stage of heart failure [93]. Decreased cardiac output and arrhythmia are general symptoms of DCM but many individuals with DCM are asymptomatic. Family history plays a significant role in DCM [3]. The investigation of family history of disease includes type 2 diabetes, epilepsy, deafness and other metabolic disorders [94].
5.2. Electrocardiography (ECG)
In patients with DCM, the electrocardiograms are not generally normal and patients with extensive fibrosis in the left ventricle are observed with isolated T wave change to the septal Q wave [95]. Delay in atrioventricular conduction and a bundle branch block are also likely to be observed [95]. Due to atrial fibrillation, sinus tachycardia and supraventricular arrhythmia can be observed [96].
5.3. Echocardiography
Echocardiography is most essential and one of the gold-standard techniques in the diagnosis of DCM. The echo diagnostic criteria of DCM includes the left ventricle area >112% which indicates dilation of the left ventricle. A fraction shortening <25% indicates abnormal systolic function. A left ventricle area >117% is a criterion for screening familial DCM (Table 3) [97].
Table 3.
Echocardiographic diagnosis of dilated cardiomyopathy [Adapted with permission from Mathew et al.] [97].
| View | Measurement | Description |
|---|---|---|
| Plax (parasternal long axis) M-mode | LVIDd (left ventricular internal dimension in diastole). LVPWd (left ventricular posterior (inferolateral) wall thickness in diastole). IVSd (interventricular septal thickness diastole). |
Left ventricle cavity size >112% (2 S.D.) based on surface area and age. Left ventricle cavity size >117% is specific criterion in screening. |
| Fraction shortening | FS < 25% | |
| Mitral valve E-septal separation | Normal range—0–5.3 mm. Reduced systolic function is indicated by value above 7 mm. | |
| PLAX (parasternal long axis) color flow Doppler | Mitral regurgitation |
|
| A4C (apical four chamber) PW | Diastolic Dysfunction |
Diastolic dysfunction is present in patients with EF < 45%. |
| A4C 2D | Left ventricle ejection fraction | EF < 45% indicates DCM |
| Sphericity index | Normal > 1.5, decrease in sphericity index value to near 1 indicates DCM [97]. |
5.4. Magnetic Resonance Imaging (MRI)
MRI is useful in determining chamber dimensions, wall thickness and ventricle mass. MRI is also useful in the assessment of flow rate, abnormality in wall motion, edema, fibrosis and hyperemia [98].
5.5. Serological Test for Virus
Serological testing for virus by specific IgM antibody titer test can be useful in detecting viral myocarditis [99].
5.6. Endomyocardial Biopsy
This is the most definitive method for the diagnosis of DCM. An endomyocardial biopsy can diagnose patients who cannot be screened through other methods. Endomyocardial biopsy can detect various myocardial diseases [100]. However, this is an invasive, costly method warranting specialized expertise. Hence, it has limitations for screening in general population.
5.7. Clinical Genetics of DCM
Genetic testing is now widely used in the detection of DCM. Genetic testing is crucial in the case of asymptomatic patients for better management and early intervention of subjects. Family genetic history using pedigree analysis has remained a classical approach in the diagnosis of genetic DCM. Pedigree analysis can help in the diagnosis of mutation in family-specific genes by genetic testing. However, it has remained a challenge to ascertain the specific genes causing DCM. Moreover, more than 100 genes have been reported to be involved in the etiology of DCM [101]. Commercial laboratories are using a gene panel (~60 genes) to test the susceptibility of developing DCM in different populations. Recently, Jordan et al. [101] evaluated the potential of a large gene panel for DCM prediction and summarized 12 definitive (strongly associated, included in Table 2 and shown in Figure 2) genes to be exclusively associated with the DCM pathophysiology, not hypertrophic cardiomyopathy (HCM) or any other cardiomyopathy. The panel of these 12 genes should be evaluated across different populations to be implemented as a standard genetic test to screen for DCM patients [13,34].
5.8. Pathological Tests for DCM Diagnosis
Several pathological tests in isolation or in combination are performed in order to detect the clinical severity of DCM patients (Table 4) [22,47,77,78,102].
Table 4.
| Pathological Tests | Other Tests in Specific Indications |
|---|---|
| Erythrocyte sedimentation rate (ESR) | Coronary angiography |
| Viral serology | Blood content tests—carnitine, pyruvate, lactate, autoantibodies, selenium, acylcarnitine profile and drug screening. |
| Creatine kinase (CK) | Red cell transketolase (beri beri) |
| Liver function tests | Urine |
| Renal function | Enteroviruses test |
| Serum ferritin/iron/transferring | Infective screening (HIV/hepatitis C) |
| Thyroid function tests | Organic acid/amino acids |
5.9. Protein Biomarkers of DCM
There are alterations in the protein content in the heart during the development of DCM. This alteration can be detected by proteomic techniques [103]. These alterations in the protein content indicate the progression of DCM. Various gene mutations work as genetic biomarkers for the diagnosis of DCM, in the same way that proteins are generally used as biomarkers of DCM.
5.9.1. Brain Natriuretic Peptide (BNP)
Brain natriuretic peptide indicates stress on the ventricles. Brain natriuretic peptide levels predict DCM as well as heart failure. BNP levels accurately diagnose DCM and heart failure. Brain natriuretic peptide can also be used to assess the effectiveness of a heart therapy [104].
5.9.2. ST2
ST2 is a cardiac biomarker, which is a member of the interleukin-1 receptor family. ST2 is released by cardiomyocytes in stress conditions. The presence of ST2 indicates tissue fibrosis, cardiac remodeling and abnormality in the heart. ST2 can diagnose a heart abnormality at an earlier stage than the brain natriuretic peptide [105].
5.9.3. Troponins T, I
Troponins T and I are sensitive biomarkers of heart failure. A modest increase in the serum level of troponins T and I predict abnormality in the heart. Most of heart failure patients have elevated levels of high sensitivity troponin in the serum [106].
5.9.4. Procollagen Type III
Collagen also works as heart failure biomarker. Elevated levels of collagen predict abnormality in the heart. Procollagen type III independently predicts progression of heart diseases [107].
5.9.5. Matrix Metalloproteinase (MMP)
Matrix metalloproteinases work as biomarkers for DCM and heart failure. Elevated levels of matrix metalloproteinase predict cardiac dilation and a decrease in ejection fraction. Elevated levels of MMP-2, -7, -8, and -9 are found in children with DCM [108].
5.9.6. Galectin-3
Under stressed conditions, activated macrophages produce galectin-3 protein. Galectin-3 protein predicts abnormality in the heart. Galectin-3 in combination with N-terminal brain natriuretic peptide (proBNP) is the best biomarker for the diagnosis of acute heart failure. It is also useful in assessing the effectiveness of a therapy in individuals with heart disease [109].
6. Treatment Strategies of DCM
There is no treatment available to permanently cure or ameliorate the DCM condition, but several treatment regimens are clinically practiced to restrict the progression of DCM and further reduce the symptoms. The current state-of-the-art therapeutic regimens described below aim to prevent heart failure and thromboembolism.
6.1. Angiotensin-Converting Enzyme (ACE) Inhibitors
In all etiologies of heart failure, the activation of the renin angiotensin aldosterone system (RAAS) is significant. To block this pathway, ACE inhibitors are used. These inhibitors are found to relieve dyspnea and reduce heart disease progression. ACE inhibitors work by preventing the formation of angiotensin II which is a vasoconstrictor that inhibits the hydrolysis of bradykinin, which is a vasodilator [110]. Thus, ACE inhibitors prevent blood load on blood vessels and decrease blood pressure. Commonly used ACE inhibitor drugs are Benazepril, Perindopril, Trandolapril, Zofenopril, and Ramipril. Adverse effects of ACE inhibitors are also observed in some patients [111]. Impairment in renal function is a major side effect, hypotension and cough are also adverse effects of the use of ACE inhibitors [111].
6.2. Diuretics
Diuretic drugs are useful in treating heart disease. Diuretics increase the excretion of water and salts from the body, which lowers the pumping load from the heart. These drugs work by inhibiting the antidiuretic hormone vasopressin and they work to reduce the blood pressure, swelling, and water buildup [112,113]. The side effects of diuretics include increased peeing, tiredness, weakness, muscle cramps, blurred vision, headache, increased sweating, and dehydration.
6.3. Angiotensin II (Ang-II) Receptor Antagonists
These are angiotensin II receptor blocking drugs. These drugs bind to angiotensin II receptors and actively inhibit their function. Thus, arteriolar contraction and sodium retention is prevented by angiotensin II receptor antagonists [114].
6.4. Beta Blockers
Beta-blockers are widely used to control heart disease. Beta-blockers are efficient in temporarily reducing blood pressure, angina, arrhythmia, anxiety, migraine, glaucoma, and overactive thyroid [115]. Beta blockers are useful in preventing heart attack [116]. Adrenaline and noradrenaline hormones are responsible for inducing the fight or flight response in the body in the presence of any danger. These hormones increase oxygen demand, heartbeat, arrhythmia, blood pressure, anxiety, sweating and palpitation in the body [116]. These hormones excessively increase the load on the heart. Beta blockers block the action of these hormones by competitively binding with the beta-adrenergic receptors of endogenous catecholamines epinephrine (adrenaline) and norepinephrine (noradrenaline) hormones. Β1-adrenergic receptors are found in the cells of the heart which are blocked by beta blockers [117].
6.5. Spironolactone
Spironolactones are used to lower blood pressure. Spironolactones are useful in preventing heart failure and in some cases, they are also useful in managing edema. Aldosterone is present in high amounts in DCM patients, retains sodium inside the body, excretes potassium and causes baroreceptor dysfunction [118]. Mineralocorticoid receptors in the distal convoluted tubule are competitively inhibited by spironolactones [118]. This inhibition increases water and sodium excretion and potassium retention [118].
6.6. SGLT2 Inhibitors (SGLT2i)
In the comorbid condition of diabetes mellitus and cardiovascular disease, SGLT2 inhibitors are quite useful. These molecular inhibitors of sodium-glucose-co-transporter 2 reduce the blood glucose level by promoting the urinary excretion of glucose. The unique mode of action of SGLT2 inhibitors results in glycosuria. SGLT2 inhibitors show diuretic and natriuretic actions. These actions help in decreasing blood pressure and plasma volume. A decrease in albuminuria and the glomerular infiltration rate is also associated with SGLT2 inhibitor treatment [119]. SGLT2 are becoming the primary choice for treatment in patients having diabetes with diabetic kidney disease (DKD) and cardiovascular diseases [120]. Empagliflozin, canagliflozin and dapagliflozin are available as SGLT2 inhibitor drugs used in clinical practice.
6.7. Potential Novel Treatments of DCM
6.7.1. Cytokine Antagonists
Degraded myocardium, activated macrophages and T cells secrete tumor necrosis factor α (TNF-α) and other secreted proinflammatory cytokines to promote the progression of cardiomyopathy [121]. The pentoxifylline drug is a xanthine derivative which actively suppresses the production of TNF-α [122]. Endothelin peptides are produced in high amounts in DCM patients and these peptides contribute to vasoconstriction leading to increased blood pressure. Bosentan is an endothelin antagonist which helps in lowering the blood pressure [123].
6.7.2. Anticoagulants
In patients with a history of thromboembolism, high ventricular dilation, and severe systolic dysfunction, anticoagulants are used in treatment [124]. Blood thinner drugs prevent stroke in people with severe heart disease. Warfarin is an anticoagulant drug that prevents blood coagulation by blocking the enzyme vitamin K epoxide reductase.
6.7.3. Natriuretic Peptides
Atrial natriuretic peptides (ANP) are natural vasodilators and diuretics which are secreted by atrial myocytes in response to stretch/stress or other stimulations [125]. These may be useful in DCM. Tolerance against these peptides is gained in a short time so these cannot be administrated for long periods and research is needed to make drugs based on them for use in heart disease.
6.7.4. Stem Cell Therapy
Stem cell therapy is an exciting newer approach studied for the treatment of DCM. Autologous bone marrow cell therapy for DCM in the short term (6 months) and long term (3 years) showed beneficial results [126]. In the future, stem cell therapy could be highly useful in the treatment of DCM.
6.7.5. Clinical Trials
Drugs undergoing clinical trials for DCM in the United States are listed in Table 5.
Table 5.
Description of drugs for dilated cardiomyopathy undergoing clinical trials in the United States [127].
| Drug | Title of Project | Description | Disease or Condition | Location |
|---|---|---|---|---|
| ARRY-371797, (p38 inhibitor) | A rollover study of ARRY-371797 in patients with LMNA-related dilated cardiomyopathy | Assessment of effectiveness of drug ARRY-371797 is being investigated in this clinical trial. | LMNA-related dilated cardiomyopathy | University of Colorado Aurora, Colorado, United States Johns Hopkins University Baltimore, Maryland, United States |
| ARRY-371797 | A study of ARRY-371797 in patients with symptomatic dilated cardiomyopathy due to a lamin A/C gene mutation | This study is a placebo controlled, dose-dependent efficacy assessment of ARRY-371797 drug on LMNA gene mutation dilated cardiomyopathy patients. | Lamin A/C gene mutation dilated cardiomyopathy | Pfizer Investigational Site Birmingham, Alabama, United States. CB Flock Research Corporation Mobile, Alabama, United States, and 64 more. |
| Ivabradine | Pulse reduction on beta-blocker and Ivabradine therapy | Ivabradine improves ejection fraction by reducing heart rate independently from beta-blockade. | Dilated cardiomyopathy ventricular remodeling electrical remodeling |
University of Colorado Anschutz Medical Campus Aurora, Colorado, United States The Ohio State University Wexner Medical Center Columbus, Ohio, United States |
| Ifetroban | Oral Ifetroban in subjects with Duchenne muscular dystrophy (DMD) | X-linked Duchenne muscular dystrophy (DMD) is a fatal genetic disorder. This lacks effective treatment therapy. Ifetroban is assessed in this study for the treatment of DMD. | Duchenne muscular dystrophy cardiomyopathy dilated cardiomyopathy | Mattel Children’s Hospital Los Angeles, California, United States and 3 more. |
6.8. Assisting Devices and Mechanical Support
6.8.1. Partial Left Ventriculectomy (PLV)
PLV is used in the treatment of DCM. It reduces heart wall pressure by resection of a portion of the left ventricle which lowers cardiac volume [128]. PLV is used on patients who are ready to receive a heart transplant but are unable to receive it [129].
6.8.2. Left Ventricular Assist Devices (LVADs)
LVADs are used in end stage heart failure patients. LVADs are used as a bridge to heart transplantation. Advanced LVADs can be used as a replacement of transplant [130].
6.8.3. Multisite Ventricular Pacing
Patients with DCM have abnormal left ventricle function. A delay in conduction of the left ventricle is also associated in the prolongation of conduction of the atrioventricle. The synchrony in the function is restored by dual-chamber pacing [131]. Single ventricular pacing is not considered sufficient, so biventricular pacing is used. Pacing is used in patients having a QRS duration of more than 150 ms [131].
7. Epidemiological Studies in India on DCM
Several candidate-gene-based epidemiological studies have been performed in India. Most importantly, Dhandapani et al., in 2009, published a work highlighting the role of defects (25 bp deletion) in the gene myosin binding protein C in the heart represented as MYBPC3, in a South Asian population with heritable conditions of cardiomyopathies such as DCM or HCM and contributing to heart failure. The sample collection for the cardiomyopathy investigation was carried out in hospitals in Madurai, Trivandrum, Hyderabad, Kozhikode, Chandigarh, and Mumbai [132]. Das et al. and Kothari et al. examined DCM clinically and genetically, focusing on the Indian adult and child population, respectively [133,134,135]. The samples were collected under the “Epidemiology of Cardiomyopathy DCM study” with 80 people out of the cohort being DCM patients. Forty percent were the result of the familial type of DCM with the majority showing autosomal dominant inheritance and 48% were sporadic. Rai et al.’s work on the clinical and genetic profiling of cardiomyopathy (idiopathic restrictive and hypertrophy) investigated sarcomere-associated gene mutation effects [136]. They found the contribution of TNN13 and a novel MYH7 (beta myosin heavy chain) mutation in cardiomyopathy development and a new mutation found to be associated with DCM induction [136]. Another study on MYH7 genetic variations carried out by Tanjore et al. analyzed the mechanism behind DCM-phenotype development. The samples belonged to a hospital in Hyderabad and common SNPs were found in exons 7, 12, 19 and 20 in DCM and HCM conditions. These SNPs when present in homozygous conditions gave rise to DCM [137]. Similarly, in another work, they aimed to reveal ACE indel (I/D) mutation-associated cardiomyopathic development in the Indian population showing reduced LVEF in DCM patients, which pointed towards the influence of ID genotype on cardiomyopathic phenotype [138]. A first case study presented by Jadhav et al. unravels a unique case of Emery-Dreifuss muscular dystrophy (EDMD2) which is autosomal dominant and associated with LMNA mutations appearing in a familial DCM patient [139]. Endothelial nitric oxide synthase gene (NOS3) with three functional polymorphisms resulted in its variable expression. This led to DCM progression due to oxidative stress from the imbalanced production of nitric oxide which causes impairment in the contractile property of cardiomyocytes [140]. The role of the endothelin 1 gene (EDN1) was investigated by Matsa et al. in the manifestation of DCM in 115 DCM patients from a hospital in Hyderabad, using PCR-based followed by single-stranded confirmation polymorphism analysis along with the commercial screening insertion variation (+138 A) in a heterozygotic state leading to a four-fold increased risk of DCM [141]. Troponin T (cTNNT)-related genetic variations were studied by Rani et al. in 147 DCM patients of south-Indian origin. Exon analysis revealed a novel mutation R144W in the tropomyosin binding domain that was responsible for distorting the protein structure and function by changing the residue charge [142]. A paper presented by Deshmukh et al. from Uttar Pradesh, where DCM is sparsely studied, revealed different causes and etiologies of this condition in 100 patients over a four-year period [143]. Similarly, a cross-sectional study over a period of three years was conducted in Gujarat with 180 DCM patients. This descriptive study calculated the etiological factors contributing to DCM such as diabetes, alcohol, nutritional, post-partum, etc. [144]. A significant DCM cross-sectional study from Ranchi by Saha et al. of 30 DCM cases provided prevalence data of DCM [145]. Ushasree et al. explored the genetic heterogeneity and epidemiological factors associated with disease expression in the Indian context [146]. Another rare account of DCM from the north of the country came from Shairgojri et al., providing a comprehensive view of the condition in children. From the analysis of samples collected from a pediatric hospital in Srinagar, a 51.35% DCM pathology was found, arising due to different causes [147]. Recently, a DCM study from Assam provided a comprehensive perspective on the cardiac condition. An observational study characterizing DCM prevalence based on its causes was carried out in North East India. A cadaveric study of DCM patients to measure the anatomical alterations of the cardiac wall in samples from North India, giving significant insight into the morphological changes undergone by the heart muscle, was made by Bose et al. [148]. A study showing the effects of hypertension on DCM patients was performed by Balije et al. that would help in the diagnosis of DCM by evaluating blood pressure response [149]. Ahmad et al. presented a clinical and etiological profile of DCM patients from 55 samples and evaluated the echocardiographic and electrocardiographic profiles which would prove to be highly significant in diagnosis of the condition [150].
Table 6 (supplementary Figure S1) summarizes these independent DCM studies carried out across different states of India. It can be clearly observed that most of these studies are from the candidate genomic perspective. There has been major progress in DCM study in the Indian context and genomics has disclosed major secrets about the said pathology. However, recently it has been shown that approximately only 40% of the DCM cases stem from a genetic defect or mutation [13]. This implies that the genetic variations could explain only a certain number of cases of DCM pathological progression in patients.
Table 6.
Summary of DCM studies performed across different states of India.
| Age Group | Male or Female (%) | Number of Cardiomyopathy Patients | Study Description | Sample Collection | Focus Population | Reference |
|---|---|---|---|---|---|---|
| 41.7 ± 16.5 | 38.70% males | 80 DCM | 40% familial, 48% sporadic. LMNA (c. 639 + G > C) associated novel splice site and MYH7 (c. 2769 > T) rare varient found. | EPOCH-D Study, AIIMS, New Delhi | General [151] | Das et al., 2015 |
| 15–67 | 35.3% males | 61 DCM | 61% DCM patients with MYH7 gene (exon 8–24) mutation with a novel p.Gly377Ser mutation found. | Nehru Hospital, Chandigarh | Asian Indians [136] | Rai et al., 2009 |
| 45.41 ± 14.35 | 54% males | 51 DCM | Influence of D allele in DCM development associated with ACE I/D | PGIMER, Chandigarh, Rajaji Government hospital, Madurai, and, Sri Chitra Tirunal Institute of Medical Sciences and Technology, Trivandrum | General [138] | Rai et al., 2008 |
| Juvenile–Adult | Undefined | 97 DCM | Common exon link of MYH7 gene IN DCM and HCM. Homozygous condition develops DCM. | CARE Hospitals, Mahavir hospitals and Niloufer hospitals, Hyderabad | General [137] | Tanjore et al., 2010 |
| 53 year old | 1 male | DCM Case Study | Familial DCM along with Emery-Dreifuss myopathy associated with mutation in LMNA gene. | Lourdes Heart Institute and Neuro Center, Kochi, Kerala | Kerala [139] | Jadhav et al., 2012 [22] |
| >25 years old | Undefined | 115 DCM | Influence of endothelial NOS3 gene in DCM manifestation by oxidative stress production. | CARE hospitals, Krishna Institute of Medical Sciences (KIMS) and Niloufer hospital for Children, Hyderabad, India | General [140] | Matsa et al., 2013 |
| 33.2± 16.1 | 70% male 30% females | 115 DCM | Screening of EDN1 gene and two rare genetic variations found. Insertion variations (+138 A) corresponding to heterozygotic condition leads to 4-fold increased risk of DCM. | CARE hospitals, Krishna Institute of Medical Sciences (KIMS) and Niloufer Hospital for Children, Hyderabad, India | General [141] | Matsa et al., 2014 |
| NA | NA | 147 DCM | Role of novel mutation R144W in Toponin T binding domain found by overall exon analysis in DCM development. | NA | South Indian [142] | Rani et al., 2014 |
| 2.9 ± 3.07 | 50 males | 80 DCM | Pediatric DCM was explored in this observational study. | General [135] | Kothari et al., 2003 | |
| Above 60 | 66.6% Males 33.4% females | 100 DCM | 4-year study of DCM charecterization based on cause. DCM in peripartum females accounted for 9%, DCM in smokers was 65%, 30% alcoholic DCM. | M.M.C. Muzaffarnagar, U.P, & SIMS, Hapur, UP | Western Uttar Pradesh [143] | Deshmukh et al., 2011 |
| Above 60 | 56.6% males 43.25% females | 30 DCM | Cross-sectional study of DCM over 1 year period. Ischemic DCM 33.3%+ diabetic cardiomyopathy (23.3%) peripartum cardiomyopathy (16.6%) 6.6 Alcoholic and miscellaneous DCM |
Rajendra Institute of Medical Sciences, Ranchi. |
India General [145] | Saha et al., 2018 |
| Above 13 | 43% male, 56.66% females | 180 DCM | 3-year DCM etiology study. Diabetes 13.33% alcohol 23.33% postpartum 15.00% idiopathic 30.00% inflammation 03.33% nutritional 6.66% multifactorial 06.66% | GMERS General Hospital, Gotri, Vadodara. | Indian General [144] | Rana et al., 2015 |
| 6 months–Adult | 62.61% males 37.38% females | 107 DCM | Parental consanguinity 22.42% alcoholic/smoking DCM | KIMS and Niloufer Hospital for children, Hyderabad | Indian General [146] | Ushasree et al., 2009 |
| 1 month–18 | 52.65% males 47.6% female | 19 DCM | Hospital based observational profile of DCM in children. Study period of two and half years, charecterization made on symptoms, gender preponderance parental consanguinity and myocarditis. | Post Graduate Department of Pediatrics Government Medical College, Srinagar | North (Kashmir) [147] | Shairgojri et al., 2021 |
| 18–80 | 61.29% males 38.71% females | 31 DCM | Observational study characterizing DCM on the basis of its causes. Alcoholic DCM 23.33% + viral DCM 03.33% peripartum cardiomyopathy 22.58% idiopathic DCM 41.93% familial DCM 9.6% | Jorhat Medical College and Hospital, Jorhat, Assam | North East India [152] | Sonowal et al., 2014 |
| 15–75 | 18.3% females | 60 DCM | An analytical and observational study showed 35 patients DCM with hypertension and 25 DCM patients without hypertension | Cardiology, GSL Medical College and Hospital, Rajahmundry India | Andhra Pradesh [149] | Balije et al., 2016 |
| 50 ± 15 | 65.45% males 34% females | 55 DCM | 1-year clinical and incidence profiling of idiopathic DCM | JN Medical College, Aligarh | General [150] | Ahmad et al., 2005 |
| All | 61.4% males 38.6% females | 70 DCM | Pilot study to determine different demographic parameters in DCM patients. 27% alcoholic 46% smokers in the cohort | Tertiary Medical College of Eastern India | Eastern India [153] | Paul et al., 2014 |
| 10–70 years | NA | 10 DCM | Measuring the gross morphological changes in heart wall of DCM patients | Institute of Medical Sciences, BHU, Varanasi (U.P) | North Indian [148] | Prasenjit Bose et al., 2017 |
8. Limitations of Candidate-Gene Based Studies: Newer System-Level OMICS Approaches to the Rescue
So far in India, DCM has been looked at from a candidate-gene perspective. Genetic evidence from Indian populations points to the fact that the majority of the descendants of ethnolinguistic groups include a blend of various divergent ancestral natives such as those of North Indian lineage sharing kinship with West Eurasians, and Native Andaman Islanders being ancestral to Southern Indians [154,155]. This admixture of genes and genetic diversity contributes to many variations in genome-reliant studies. Thus next-generation omics-based methods focusing on “proteins” and “lipids” may shed critical insights into the pathological progression of DCM as well as the identification of newer potential biomarker panels with better prognostic value. In recent years, proteomics has emerged to be a highly revelatory and effective approach in cardiovascular disease research [156]. It makes use of high throughput and sensitive techniques such as mass spectrometry for biomarker investigation, pathway elucidation, and post-translation modification studies [88]. Additionally, lipidomics provides a comprehensive view of the metabolic rewiring of individuals progressing towards a disease state [157,158]. It is now time to exploit the collaboration of the latest technological advancements and medical data to broaden our understanding of pathology. This would provide an integrative view instead of a reductionist approach to the disease. This can be achieved by incorporating systems biology techniques in our research that integrate molecular information from different hierarchies to interpret into models that demonstrate complex biological processes [159]. Systems biology and proteomic analysis have given a new platform for biomarker studies and extensive research is being carried out to discover novel biomarkers. These biomolecules would help assess the progression of CVD just by the quantitative analysis of components of plasma. Recently, Izquierdo et al. performed a biomarker study by proteomic analysis using 2D-DIGE and mass spectrometry in DCM patients carrying the LMNA pathological gene mutation [160]. Another interesting study systematically characterized the circulating proteins at different stages of heart failure by employing aptamer-based proteomic profiling [161]. These significant studies will be corroborated with the novel proteomic and lipidomic candidate biomarkers identified in Indian DCM patients.
9. Systems Biology Approach for Better Diagnosis of DCM
Proteomic techniques combined with lipidomic concept application to study heart disease can give us new insights into the biomolecular mechanisms and changes undergone by tissue during disease progression. Our next focus in this context is to probe mass-spectrometry-based “clinical-omics” studies of DCM patients from India. This initiative has been recently funded by the Indian Council of Medical Research (ICMR). We aim to develop a noninvasive diagnostic multimarker panel (proteomic and lipidomic) that would be indicative of DCM status in Indian patients.
Acknowledgments
The Seed grant (IITM/SG/TB/75) from IIT-Mandi and the Indian Council of Medical Research (ICMR) funded “High-Resolution Plasma Proteomic and Lipidomic Analyses For Fibrosis-Related Metabolic Assessment In DCM Patients in India: A Multi-Center Based Study (NCD/Adhoc/40/2020-2021)” to T.B. is acknowledged for this work. V.S. also acknowledges the HTRA fellowship (MHRD, Govt. of India) for the doctoral program.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcdd9060174/s1, Figure S1: Representative map of India highlighting different studies performed on DCM across the country.
Author Contributions
V.S. and S.S. worked on summarizing the recent knowledge generation and state-of-the-art developments in DCM. V.S. and S.S. prepared the figures and wrote the initial drafts of the manuscript. T.B. conceptualized the overall structure of the review and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
Authors declare no conflict of interest.
Funding Statement
This research was funded by [from IIT-Mandi and ICMR] grant number [Seed grant (IITM/SG/TB/75) and (NCD/Adhoc/40/2020-2021)] And The APC was funded by [NCD/Adhoc/40/2020-2021-IITM/ICMR/TB/334].
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elliott P., Andersson B., Arbustini E., Bilinska Z., Cecchi F., Charron P., Dubourg O., Kuhl U., Maisch B., McKenna W.J., et al. Classification of the cardiomyopathies: A position statement from the european society of cardiology working group on myocardial and pericardial diseases. Eur. Hear J. 2007;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 2.Maron B.J., Towbin J.A., Thiene G., Antzelevitch C., Corrado D., Arnett D., Moss A.J., Seidman C.E., Young J.B., American Heart A., et al. Contemporary definitions and classification of the cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 3.Ku L., Feiger J., Taylor M., Mestroni L. Familial Dilated Cardiomyopathy. Circulation. 2003;108:e118–e121. doi: 10.1161/01.CIR.0000097493.70422.50. [DOI] [PubMed] [Google Scholar]
- 4.Costa M.C., Calderon-Dominguez M., Mangas A., Campuzano O., Sarquella-Brugada G., Ramos M., Quezada-Feijoo M., Pinilla J.M.G., Robles-Mezcua A., Pacheco-Cruz G.d.A., et al. Circulating circRNA as biomarkers for dilated cardiomyopathy etiology. Klin. Wochenschr. 2021;99:1711–1725. doi: 10.1007/s00109-021-02119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Towbin J.A., Lowe A.M., Colan S.D., Sleeper L.A., Orav E.J., Clunie S., Messere J., Cox G.F., Lurie P.R., Hsu D., et al. Incidence, causes, and outcomes of dilated cardio-myopathy in children. Jama. 2006;296:1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 6.Hazebroek M., Dennert R., Heymans S. Idiopathic dilated cardiomyopathy: Possible triggers and treatment strategies. Neth. Hear J. 2012;20:332–335. doi: 10.1007/s12471-012-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weintraub R.G., Semsarian C., Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390:400–414. doi: 10.1016/S0140-6736(16)31713-5. [DOI] [PubMed] [Google Scholar]
- 8.Morales A., Cowan J., Dagua J., Hershberger R.E. Family History: An Essential Tool for Cardiovascular Genetic Medicine. Congest. Heart Fail. 2008;14:37–45. doi: 10.1111/j.1751-7133.2008.08201.x. [DOI] [PubMed] [Google Scholar]
- 9.Dilated Cardiomyopathy (DCM)|Pediatric Cardiomyopathy [Internet] [(accessed on 21 April 2021)]. Available online: https://www.cincinnatichildrens.org/service/c/cardiomyopathy/types/dilated-cardiomyopathy.
- 10.Hershberger R.E., Morales A., Siegfried J.D. Clinical and genetic issues in dilated cardiomyopathy: A review for genetics professionals. Genet. Med. 2010;12:655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Codd M.B., Sugrue D.D., Gersh B.J., Melton L.J., 3rd Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation. 1989;80:564–572. doi: 10.1161/01.CIR.80.3.564. [DOI] [PubMed] [Google Scholar]
- 12.Rakar S., Sinagra G., Di Lenarda A., Poletti A., Bussani R., Silvestri F., Camerini F., Heart Muscle Disease Study Group Epidemiology of dilated cardiomyopathy: A prospective post-mortem study of 5252 necropsies. Eur. Heart J. 1997;18:117–123. doi: 10.1093/oxfordjournals.eurheartj.a015092. [DOI] [PubMed] [Google Scholar]
- 13.Hershberger R.E., Hedges D.J., Morales A. Dilated cardiomyopathy: The complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 14.Reichart D., Magnussen C., Zeller T., Blankenberg S. Dilated cardiomyopathy: From epidemiologic to genetic phenotypes. J. Intern. Med. 2019;286:362–372. doi: 10.1111/joim.12944. [DOI] [PubMed] [Google Scholar]
- 15.Institute for Health Metrics and Evaluation Global Burden of Diseases (GBD) Compare. [(accessed on 18 April 2021)]. Available online: https://vizhub.healthdata.org/gbd-compare/
- 16.Heymans S., Eriksson U., Lehtonen J., Cooper L.T. The Quest for New Approaches in Myocarditis and Inflammatory Car-diomyopathy. J. Am. Coll. Cardiol. 2016;68:2348–2364. doi: 10.1016/j.jacc.2016.09.937. [DOI] [PubMed] [Google Scholar]
- 17.Caforio A.L.P., Pankuweit S., Arbustini E., Basso C., Gimeno-Blanes J., Felix S.B., Fu M., Heliö T., Heymans S., Jahns R., et al. Current state of knowledge on aetiology, diagnosis, management, and therapy of myocarditis: A position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2013;34:2636–2648. doi: 10.1093/eurheartj/eht210. [DOI] [PubMed] [Google Scholar]
- 18.Maisch B., Richter A., Sandmöller A., Portig I., Pankuweit S. Inflammatory Dilated Cardiomyopathy (DCMI) Herz. 2005;30:535–544. doi: 10.1007/s00059-005-2730-5. [DOI] [PubMed] [Google Scholar]
- 19.Kindermann I., Kindermann M., Kandolf R., Klingel K., Bültmann B., Müller T., Lindinger A., Boühm M. Predictors of Outcome in Patients with Suspected Myocarditis. Circulation. 2008;118:639–648. doi: 10.1161/CIRCULATIONAHA.108.769489. [DOI] [PubMed] [Google Scholar]
- 20.Mahrholdt H., Goedecke C., Wagner A., Meinhardt G., Athanasiadis A., Vogelsberg H., Fritz P., Klingel K., Kandolf R., Sechtem U. Cardiovascular magnetic resonance assessment of human myocarditis: A comparison to histology and molecular pathology. Circulation. 2004;109:1250–1258. doi: 10.1161/01.CIR.0000118493.13323.81. [DOI] [PubMed] [Google Scholar]
- 21.Kearney M.T., Cotton J., Richardson P.J., Shah A. Viral myocarditis and dilated cardiomyopathy: Mechanisms, manifestations, and management. Postgrad. Med. J. 2001;77:4–10. doi: 10.1136/pmj.77.903.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oropeza-Moe M., Wisløff H., Bernhoft A. Selenium deficiency associated porcine and human cardiomyopathies. J. Trace Elem. Med. Biol. 2015;31:148–156. doi: 10.1016/j.jtemb.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Tirumanisetty P., Abdullah A., Matos M. Pulmonary arteriovenous malformation: A myriad of presentations. Chest. 2018;154:1032A. doi: 10.1016/j.chest.2018.08.930. [DOI] [Google Scholar]
- 24.Seki A., Patel S., Ashraf S., Perens G., Fishbein M.C. Primary endocardial fibroelastosis: An underappreciated cause of car-diomyopathy in children. Cardiovasc. Pathol. 2013;22:345–350. doi: 10.1016/j.carpath.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Yin L. Non-compact cardiomyopathy or ventricular non-compact syndrome? J. Cardiovasc. Ultrasound. 2014;22:165–172. doi: 10.4250/jcu.2014.22.4.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanyal D., Raychaudhuri M. Infants with dilated cardiomyopathy and hypocalcemia. Indian J. Endocrinol. Metab. 2013;17:221–223. doi: 10.4103/2230-8210.119578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferies J.L. Barth syndrome. Am. J. Med. Genet. Part C (Semin. Med. Genet.) 2013;163:198–205. doi: 10.1002/ajmg.c.31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aprikyan A.A., Khuchua Z. Advances in the understanding of Barth syndrome. Br. J. Haematol. 2013;161:330–338. doi: 10.1111/bjh.12271. [DOI] [PubMed] [Google Scholar]
- 29.Rampersaud E., Siegfried J.D., Norton N., Li D., Martin E., Hershberger R.E. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog. Pediatr. Cardiol. 2011;31:39–47. doi: 10.1016/j.ppedcard.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan R.S., Pahl E., Dellefave-Castillo L., Rychlik K., Ing A., Yap K.L., Brew C., Johnston J.R., McNally E.M., Webster G. Genotype and Cardiac Outcomes in Pediatric Dilated Cardiomyopathy. J. Am. Hear. Assoc. 2022;11:e022854. doi: 10.1161/JAHA.121.022854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luisa M., Francesca B., Anita S., Gianfranco S., Taylor M.R. Genetic causes of Dilated Cardiomyopathy. Prog. Pediatr. Cardiol. 2014;37:13–18. doi: 10.1016/j.ppedcard.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arbustini E., Narula N., Dec G.W., Reddy K.S., Greenberg B., Kushwaha S., Marwick T., Pinney S., Bellazzi R., Favalli V., et al. The MOGE(S) classification for a phenotype-genotype nomenclature of cardiomyopathy: Endorsed by the World Heart Federation. J. Am. Coll. Cardiol. 2013;62:2046–2072. doi: 10.1016/j.jacc.2013.08.1644. [DOI] [PubMed] [Google Scholar]
- 33.Schultheiss H.P., Fairweather D., Caforio A.L., Escher F., Hershberger R.E., Lipshultz S.E., Liu P.P., Matsumori A., Mazzanti A., McMurray J., et al. Dilated cardiomyopathy. Nat. Rev. Dis. Primers. 2019;5:32. doi: 10.1038/s41572-019-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hershberger R.E., Cowan J., Jordan E., Kinnamon D.D. The Complex and Diverse Genetic Architecture of Dilated Cardiomyopathy. Circ. Res. 2021;128:1514–1532. doi: 10.1161/CIRCRESAHA.121.318157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maisch B. Alcoholic cardiomyopathy: The result of dosage and individual predisposition. Herz. 2016;41:484–493. doi: 10.1007/s00059-016-4469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohamed H.A. Tachycardia-induced Cardiomyopathy (Tachycardiomyopathy) Libyan J. Med. 2007;2:26–29. doi: 10.3402/ljm.v2i1.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates M.G., Bourke J.P., Giordano C., D’Amati G., Turnbull D.M., Taylor R.W. Cardiac involvement in mitochondrial DNA disease: Clinical spectrum, diagnosis, and management. Eur. Heart J. 2012;33:3023–3033. doi: 10.1093/eurheartj/ehs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basso C., Corrado D., Marcus F.I., Nava A., Thiene G. Arrhythmogenic right ventricular cardiomyopathy. Lancet. 2009;373:1289–1300. doi: 10.1016/S0140-6736(09)60256-7. [DOI] [PubMed] [Google Scholar]
- 39.Bluett R., McDonnell D., O’Dowling C., Vaughan C. Eosinophilic myocarditis as a first presentation of eosinophilic granulomatosis with polyangiitis (Churg-Strauss syndrome) Case Rep. 2017;2017:bcr-2017. doi: 10.1136/bcr-2017-221227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama T., Murai S., Ohte N. Dilated Cardiomyopathy with Eosinophilic Granulomatosis with Polyangiitis in Which Active Myocardial Inflammation Was Only Detected by Endomyocardial Biopsy. Intern. Med. 2018;57:2675–2679. doi: 10.2169/internalmedicine.0330-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M.-X., Yu B.-L., Peng D.-Q., Zhou S.-H. Eosinophilic myocarditis due to Churg–Strauss syndrome mimicking reversible dilated cardiomyopathy. Hear Lung. 2014;43:45–47. doi: 10.1016/j.hrtlng.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Lipshultz S.E., Law Y.M., Asante-Korang A., Austin E.D., Dipchand A.I., Everitt M.D., Hsu D.T., Lin K.Y., Price J.F., Wilkinson J.D., et al. Cardiomyopathy in Children: Clas-sification and Diagnosis: A Scientific Statement from the American Heart Association. Circulation. 2019;140:e9–e68. doi: 10.1161/CIR.0000000000000682. [DOI] [PubMed] [Google Scholar]
- 43.Pearson G.D., Veille J.C., Rahimtoola S., Hsia J., Oakley C.M., Hosenpud J.D., Ansari A., Baughman K.L. Peripartum cardiomyopathy: National Heart, Lung, and Blood Institute and Office of Rare Diseases (National Institutes of Health) workshop recommendations and review. Jama. 2000;283:1183–1188. doi: 10.1001/jama.283.9.1183. [DOI] [PubMed] [Google Scholar]
- 44.Klein I., Danzi S. Thyroid disease and the heart. Circulation. 2007;116:1725–1735. doi: 10.1161/CIRCULATIONAHA.106.678326. [DOI] [PubMed] [Google Scholar]
- 45.Mobine H.R., Baker A.B., Wang L., Wakimoto H., Jacobsen K.C., Seidman C.E., Seidman J.G., Edelman E.R. Pheochromocytoma-induced cardiomyopathy is modulated by the synergistic effects of cell-secreted factors. Circ. Heart Fail. 2009;2:121–128. doi: 10.1161/CIRCHEARTFAILURE.108.813261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradley T.J., Slorach C., Mahmud F.H., Dunger D.B., Deanfield J., Deda L., Elia Y., Har R.L.H., Hui W., Moineddin R., et al. Early changes in cardiovascular structure and function in adolescents with type 1 diabetes. Cardiovasc. Diabetol. 2016;15:31. doi: 10.1186/s12933-016-0351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albakri A. Nutritional deficiency cardiomyopathy: A review and pooled analysis of pathophysiology, diagnosis and clinical management. Res. Rev. Insights. 2019;3:1–19. doi: 10.15761/RRI.1000149. [DOI] [Google Scholar]
- 48.Kamisago M., Sharma S.D., De Palma S.R., Solomon. S., Sharma P., McDonough B., Smoot L., Mullen M.P., Woolf P.K., Wigle E.D., et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. ACC Curr. J. Rev. 2001;10:55. doi: 10.1016/S1062-1458(01)00258-6. [DOI] [PubMed] [Google Scholar]
- 49.Olson T.M., Michels V.V., Thibodeau S.N., Tai Y.-S., Keating M.T. Actin Mutations in Dilated Cardiomyopathy, a Heritable Form of Heart Failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 50.Daehmlow S., Erdmann J., Knueppel T., Gille C., Froemmel C., Hummel M., Hetzer R., Regitz-Zagrosek V. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;298:116–120. doi: 10.1016/S0006-291X(02)02374-4. [DOI] [PubMed] [Google Scholar]
- 51.Olson T.M., Kishimoto N.Y., Whitby F.G., Michels V.V. Mutations that alter the surface charge of alpha-tropomyosin are asso-ciated with dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2001;33:723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 52.Gerull B., Gramlich M., Atherton J., McNabb M., Trombitás K., Sasse-Klaassen S., Seidman J.G., Seidman C., Granzier H., Labeit S., et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 53.Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E., et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verdonschot J.A., Hazebroek M.R., Krapels I.P., Henkens M.T., Raafs A., Wang P., Merken J.J., Claes G.R., Vanhoutte E.K., Wijngaard A.V.D., et al. Implications of Genetic Testing in Dilated Cardiomyopathy. Circ. Genom. Precis. Med. 2020;13:476–487. doi: 10.1161/CIRCGEN.120.003031. [DOI] [PubMed] [Google Scholar]
- 55.Li D., Tapscoft T., Gonzalez O., Burch P.E., Quiñones M.A., Zoghbi W.A., Hill R., Bachinski L.L., Mann D., Roberts R. Desmin Mutation Responsible for Idiopathic Dilated Cardiomyopathy. Circulation. 1999;100:461–464. doi: 10.1161/01.CIR.100.5.461. [DOI] [PubMed] [Google Scholar]
- 56.Tsubata S., Bowles K.R., Vatta M., Zintz C., Titus J., Muhonen L., Bowles N.E., Towbin J.A. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J. Clin. Investig. 2000;106:655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heling A., Zimmermann R., Kostin S., Maeno Y., Hein S., Devaux B., Bauer E., Kloüvekorn W.P., Schlepper M., Schaper W., et al. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ. Res. 2000;86:846–853. doi: 10.1161/01.RES.86.8.846. [DOI] [PubMed] [Google Scholar]
- 58.Olson T.M., Illenberger S., Kishimoto N.Y., Huttelmaier S., Keating M.T., Jockusch B.M. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 59.Fatkin D., Macrae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Jr., Spudich S., De Girolami U., et al. Missense Mutations in the Rod Domain of the Lamin A/C Gene as Causes of Dilated Cardiomyopathy and Conduction System Disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 60.DMD Gene: MedlinePlus Genetics. [(accessed on 21 April 2021)]; Available online: https://medlineplus.gov/genetics/gene/dmd/#references.
- 61.Towbin J.A., Hejtmancik J.F., Brink P., Gelb B., Zhu X.M., Chamberlain J.S., McCabe E.R., Swift M. X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87:1854–1865. doi: 10.1161/01.CIR.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 62.Franz W.-M., Müller M., Muller O.J., Herrmann R., Rothmann T., Cremer M., Cohn R.D., Voit T., Katus A.H. Association of nonsense mutation of dystrophin gene with disruption of sarcoglycan complex in X-linked dilated cardiomyopathy. Lancet. 2000;355:1781–1785. doi: 10.1016/S0140-6736(00)02266-2. [DOI] [PubMed] [Google Scholar]
- 63.Bleyl S.B., Mumford B.R., Thompson V., Carey J.C., Pysher T.J., Chin T.K., Ward K. Neonatal, lethal noncompaction of the left ventricular myocardium is allelic with Barth syndrome. Am. J. Hum. Genet. 1997;61:868–872. doi: 10.1086/514879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haghighi K., Kolokathis F., Gramolini A.O., Waggoner J.R., Pater L., Lynch R.A., Fan G.C., Tsiapras D., Parekh R.R., Dorn G.W., et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc. Natl. Acad. Sci. USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmitt J.P., Kamisago M., Asahi M., Li G.H., Ahmad F., Mende U., Kranias E.G., MacLennan D.H., Seidman J.G., Seidman C.E. Dilated Cardiomyopathy and Heart Failure Caused by a Mutation in Phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 66.Zhang D., Mott J.L., Farrar P., Ryerse J.S., Chang S.-W., Stevens M., Denniger G., Zassenhaus H.P. Mitochondrial DNA mutations activate the mitochondrial apoptotic pathway and cause dilated cardiomyopathy. Cardiovasc. Res. 2003;57:147–157. doi: 10.1016/S0008-6363(02)00695-8. [DOI] [PubMed] [Google Scholar]
- 67.Arbustini E., Diegoli M., Fasani R., Grasso M., Morbini P., Banchieri N., Bellini O., Bello B.D., Pilotto A., Magrini G., et al. Mitochondrial DNA Mutations and Mitochondrial Abnormalities in Dilated Cardiomyopathy. Am. J. Pathol. 1998;153:1501–1510. doi: 10.1016/S0002-9440(10)65738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Begay R., Graw S.L., Sinagra G., Asimaki A., Rowland T.J., Slavov D.B., Gowan K., Jones K.L., Brun F., Merlo M., et al. Filamin C Truncation Mutations Are Associated with Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell–Cell Adhesion Structures. JACC Clin. Electrophysiol. 2018;4:504–514. doi: 10.1016/j.jacep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agarwal R., Paulo J.A., Toepfer C.N., Ewoldt J.K., Sundaram S., Chopra A., Zhang Q., Gorham J., DePalma S.R., Chen C.S., et al. Filamin C Cardiomyopathy Variants Cause Protein and Lysosome Accumulation. Circ. Res. 2021;129:751–766. doi: 10.1161/CIRCRESAHA.120.317076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crocini C., Gotthardt M. Cardiac sarcomere mechanics in health and disease. Biophys. Rev. 2021;13:637–652. doi: 10.1007/s12551-021-00840-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNally E.M., Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ. Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong B.K., Kwon H.M., Byun K.H., Kim D., Choi E.Y., Kang T.S., Kang S., Chun K.J., Jang Y., Kim H.S., et al. Apoptosis in dilated cardiomyopathy. [(accessed on 21 April 2021)];Korean J. Intern. Med. 2000 15:56–64. doi: 10.3904/kjim.2000.15.1.56. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4531747/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agapitos E., Kavantzas N., Nanas J., Margari Z., Bakouris M., Kassis K., Panolaridis A., Davaris P. The myocardial fibrosis in patients with dilated cardiomyopathy: The application of image analysis in the myocardial biopsies. Gen. Diagn. Pathol. 1995;141:305–311. doi: 10.1117/12.238788. [DOI] [PubMed] [Google Scholar]
- 74.Shinde A.V., Frangogiannis N.G. Fibroblasts in myocardial infarction: A role in inflammation and repair. J. Mol. Cell. Cardiol. 2014;70:74–82. doi: 10.1016/j.yjmcc.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin K.Y., Miyamoto S.D., Schumacher K.R. Heart Failure in the Child and Young Adult: From Bench to Bedside. Elsevier; Amsterdam, The Netherlands: 2018. Dilated Cardiomyopathy; pp. 203–213. [Google Scholar]
- 76.DCM-Symptoms and Causes-Mayo Clinic [Internet] [(accessed on 21 April 2021)]. Available online: https://www.mayoclinic.org/diseases-conditions/dilated-cardiomyopathy/symptoms-causes/syc-20353149.
- 77.Twu C., Liu N.Q., Popik W., Bukrinsky M., Sayre J., Roberts J., Rania S., Bramhandam V., Roos K.P., MacLellan W.R., et al. Cardiomyocytes undergo apoptosis in human immunode-ficiency virus cardiomyopathy through mitochondrion and death receptor-controlled pathways. [(accessed on 21 April 2021)];Proc. Natl. Acad. Sci. USA. 2002 99:14386–14391. doi: 10.1073/pnas.212327899. Available online: www.pnas.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGowan J.V., Chung R., Maulik A., Piotrowska I., Walker J.M., Yellon D.M. Anthracycline Chemotherapy and Cardiotoxicity. [(accessed on 21 April 2021)];Cardiovasc. Drugs Ther. 2017 31:63–75. doi: 10.1007/s10557-016-6711-0. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5346598/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wencker D., Chandra M., Nguyen K., Miao W., Garantziotis S., Factor S.M., Shirani J., Armstrong R.C., Kitsis R.N. A mechanistic role for cardiac myocyte apoptosis in heart failure. J. Clin. Investig. 2003;111:1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia P., Liu Y., Cheng Z. Signaling Pathways in Cardiac Myocyte Apoptosis. BioMed Res. Int. 2016;2016:9583268. doi: 10.1155/2016/9583268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramaccini D., Montoya-Uribe V., Aan F.J., Modesti L., Potes Y., Wieckowski M.R., Krga I., Glibetić M., Pinton P., Giorgi C., et al. Mitochondrial Function and Dysfunction in Dilated Cardiomyopathy. Front. Cell Dev. Biol. 2021;8:1800. doi: 10.3389/fcell.2020.624216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitra A., Basak T., Datta K., Naskar S., Sengupta S., Sarkar S. Role of α-crystallin B as a regulatory switch in modulating cardiomyocyte apoptosis by mitochondria or endoplasmic reticulum during cardiac hypertrophy and myocardial infarction. Cell Death Dis. 2013;4:e582. doi: 10.1038/cddis.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobaczewski M., Chen W., Frangogiannis N.G. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011;51:600–606. doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li R.K., Li G., Mickle D.A., Weisel R.D., Merante F., Luss H., Rao V., Christakis G.T., Williams W.G. Overexpression of transforming growth factor-beta1 and insulinlike growth factor-I in patients with idiopathic hypertrophic cardiomyopathy. Circulation. 1997;96:874–881. doi: 10.1161/01.CIR.96.3.874. [DOI] [PubMed] [Google Scholar]
- 85.Schneiders D., Heger J., Best P., Taimor G., Piper H.M. SMAD proteins are involved in apoptosis induction in ventricular cardiomyocytes. Cardiovasc. Res. 2005;67:87–96. doi: 10.1016/j.cardiores.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 86.Huntgeburth M., Tiemann K., Shahverdyan R., Schlüter K.-D., Schreckenberg R., Gross M.-L., Mödersheim S., Caglayan E., Müller-Ehmsen J., Ghanem A., et al. Transforming Growth Factor β1 Oppositely Regulates the Hypertrophic and Contractile Response to β-Adrenergic Stimulation in the Heart. PLoS ONE. 2011;6:e26628. doi: 10.1371/journal.pone.0026628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pellieux C., Foletti A., Peduto G., Aubert J.F., Nussberger J., Beermann F., Brunner H.R., Pedrazzini T. Dilated cardiomyopathy and impaired cardiac hypertrophic response to angiotensin II in mice lacking FGF-2. J. Clin. Investig. 2001;108:1843–1851. doi: 10.1172/JCI13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Merl-Pham J., Basak T., Knüppel L., Ramanujam D., Athanason M., Behr J., Engelhardt S., Eickelberg O., Hauck S.M., Vanacore R., et al. Quantitative proteomic profiling of extracellular matrix and site-specific collagen post-translational modifications in an in vitro model of lung fibrosis. Matrix Biol. Plus. 2019;1:100005. doi: 10.1016/j.mbplus.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W., Zhou G., Hu M.C., Yao Z., Tan T.H. Activation of the hematopoietic progenitor kinase-1 (HPK1)-dependent, stress-activated c-Jun N-terminal kinase (JNK) pathway by transforming growth factor beta (TGF-β)-activated kinase (TAK1), a kinase mediator of TGF beta signal transduction. J. Biol. Chem. 1997;272:22771–22775. doi: 10.1074/jbc.272.36.22771. [DOI] [PubMed] [Google Scholar]
- 90.Courcelles M., Frémin C., Voisin L., Lemieux S., Meloche S., Thibault P. Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions. Mol. Syst. Biol. 2013;9:669. doi: 10.1038/msb.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimizu T., Liao J.K. Rho Kinases and Cardiac Remodeling. Circ. J. Off. J. Jpn. Circ. Soc. 2016;80:1491–1498. doi: 10.1253/circj.CJ-16-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wieder N., Greka A. Calcium, TRPC channels, and regulation of the actin cytoskeleton in podocytes: Towards a future of targeted therapies. Pediatric Nephrol. 2016;31:1047–1054. doi: 10.1007/s00467-015-3224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dilated Cardiomyopathy (DCM)-Cardiomyopathy UK [Internet] [(accessed on 20 April 2021)]. Available online: https://www.cardiomyopathy.org/dilated-cardiomyopathy/intro.
- 94.Duran J., Martinez A., Adler E. Cardiovascular Manifestations of Mitochondrial Disease. Biology. 2019;8:34. doi: 10.3390/biology8020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dilated Cardiomyopathy (DCM) • LITFL • ECG Library Diagnosis [Internet] [(accessed on 20 April 2021)]. Available online: https://litfl.com/dilated-cardiomyopathy-dcm-ecg-library/
- 96.Finocchiaro G., Merlo M., Sheikh N., De Angelis G., Papadakis M., Olivotto I., Rapezzi C., Carr-White G., Sharma S., Mestroni L., et al. The electrocardiogram in the diagnosis and management of patients with dilated cardiomyopathy. Eur. J. Hear Fail. 2020;22:1097–1107. doi: 10.1002/ejhf.1815. [DOI] [PubMed] [Google Scholar]
- 97.Mathew T., Williams L., Navaratnam G., Rana B., Wheeler R., Collins K., Harkness A., Jones R., Knight D., O’Gallagher K., et al. Diagnosis and assessment of dilated cardio-myopathy: A guideline protocol from the British Society of Echocardiography. Echo Res. Pract. 2017;4:G1–G13. doi: 10.1530/ERP-16-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Green J.J., Berger J.S., Kramer C.M., Salerno M. Prognostic Value of Late Gadolinium Enhancement in Clinical Outcomes for Hypertrophic Cardiomyopathy. JACC Cardiovasc. Imaging. 2012;5:370–377. doi: 10.1016/j.jcmg.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 99.Japp A.G., Gulati A., Cook S.A., Cowie M., Prasad S.K. The Diagnosis and Evaluation of Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2016;67:2996–3010. doi: 10.1016/j.jacc.2016.03.590. [DOI] [PubMed] [Google Scholar]
- 100.Wu L.A., Lapeyre A.C., Cooper L.T. Current Role of Endomyocardial Biopsy in the Management of Dilated Cardiomyopathy and Myocarditis. Mayo Clin. Proc. 2001;76:1030–1038. doi: 10.4065/76.10.1030. [DOI] [PubMed] [Google Scholar]
- 101.Jordan E., Peterson L., Ai T., Asatryan B., Bronicki L., Brown E., Celeghin R., Edwards M., Fan J., Ingles J., et al. Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation. 2021;144:7–19. doi: 10.1161/CIRCULATIONAHA.120.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rapezzi C., Arbustini E., Caforio A.L.P., Charron P., Blanes J.G., Heliö T., Linhart A., Mogensen J., Pinto Y., Ristic A., et al. Diagnostic work-up in cardiomyopathies: Bridging the gap between clinical phenotypes and final diagnosis. A position statement from the ESC Working Group on Myocardial and Pericardial Diseases. Eur. Hear J. 2012;34:1448–1458. doi: 10.1093/eurheartj/ehs397. [DOI] [PubMed] [Google Scholar]
- 103.Suresh C.P., Saha A., Kaur M., Kumar R., Dubey N.K., Basak T., Tanwar V.S., Bhardwaj G., Sengupta S., Batra V.V., et al. Differentially expressed urinary biomarkers in children with idiopathic nephrotic syndrome. Clin. Exp. Nephrol. 2015;20:273–283. doi: 10.1007/s10157-015-1162-7. [DOI] [PubMed] [Google Scholar]
- 104.Felker G.M., Hasselblad V., Hernandez A.F., O’Connor C.M. Biomarker-guided therapy in chronic heart failure: A meta-analysis of randomized controlled trials. Am. Hear J. 2009;158:422–430. doi: 10.1016/j.ahj.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 105.Januzzi J.L., Peacock W.F., Maisel A.S., Chae C.U., Jesse R.L., Baggish A.L., O’Donoghue M., Sakhuja R., Chen A.A., van Kimmenade R.R., et al. Measurement of the Interleukin Family Member ST2 in Patients with Acute Dyspnea: Results from the PRIDE (Pro-Brain Natriuretic Peptide Investigation of Dyspnea in the Emergency Department) Study. J. Am. Coll. Cardiol. 2007;50:607–613. doi: 10.1016/j.jacc.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 106.DeFilippi C.R., de Lemos J.A., Christenson R.H., Gottdiener J.S., Kop W.J., Zhan M., Seliger S.L. Association of serial measures of cardiac troponin T using a sensitive assay with incident heart failure and cardiovascular mortality in older adults. Jama. 2010;304:2494–2502. doi: 10.1001/jama.2010.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cicoira M., Rossi A., Bonapace S., Zanolla L., Golia G., Franceschini L., Caruso B., Marino P.N., Zardini P. Independent and additional prognostic value of aminoterminal propeptide of type III procollagen circulating levels in patients with chronic heart failure. J. Card. Fail. 2004;10:403–411. doi: 10.1016/j.cardfail.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 108.Yan A.T., Yan R.T., Spinale F.G., Afzal R., Gunasinghe H.R., Arnold M., Demers C., Mckelvie R.S., Liu P.P. Plasma Matrix Metalloproteinase-9 Level Is Correlated with Left Ventricular Volumes and Ejection Fraction in Patients with Heart Failure. J. Card. Fail. 2006;12:514–519. doi: 10.1016/j.cardfail.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 109.Kapur N.K., Heffernan K.S., Yunis A.A., Parpos P., Kiernan M.S., Sahasrabudhe N.A., Kimmelstiel C.D., Kass D.A., Karas R.H., Mendelsohn M.E. Usefulness of Soluble Endoglin as a Noninvasive Measure of Left Ventricular Filling Pressure in Heart Failure. Am. J. Cardiol. 2010;106:1770–1776. doi: 10.1016/j.amjcard.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Herman L.L., Padala S.A., Annamaraju P., Bashir K. Angiotensin Converting Enzyme Inhibitors (ACEI) StatPearls Publishing; Treasure Island, FL, USA: 2021. [PubMed] [Google Scholar]
- 111.Packer M., Poole-Wilson P.A., Armstrong P.W., Cleland J.G., Horowitz J.D., Massie B.M., Rydén L., Thygesen K., Uretsky B.F. Comparative effects of low and high doses of the angiotensin-converting enzyme inhibitor, lisinopril, on morbidity and mortality in chronic heart failure. ATLAS Study Group. Circulation. 1999;100:2312–2318. doi: 10.1161/01.CIR.100.23.2312. [DOI] [PubMed] [Google Scholar]
- 112.Diuretics [Internet] [(accessed on 19 April 2021)]. Available online: https://www.cvpharmacology.com/diuretic/diuretics.
- 113.Ballew J.R., Fink G.D. Characterization of the antihypertensive effect of a thiazide diuretic in angiotensin II-induced hy-pertension. J. Hypertens. 2001;19:1601–1606. doi: 10.1097/00004872-200109000-00012. [DOI] [PubMed] [Google Scholar]
- 114.Barreras A., Gurk-Turner C. Angiotensin Ii Receptor Blockers. Bayl. Univ. Med Cent. Proc. 2003;16:123–126. doi: 10.1080/08998280.2003.11927893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beta-Blockers for Heart Disease: Benefits, Risks, and More [Internet] [(accessed on 19 April 2021)]. Available online: https://www.healthline.com/health/heart-disease/beta-blockers.
- 116.Böhm M., Maack C. Treatment of heart failure with beta-blockers. Mechanisms and results. Basic Res. Cardiol. 2000;95:I15–I24. doi: 10.1007/s003950070004. [DOI] [PubMed] [Google Scholar]
- 117.James P.A., Oparil S., Carter B.L., Cushman W.C., Dennison-Himmelfarb C., Handler J., Lackland D.T., LeFevre M.L., MacKenzie T.D., Ogedegbe O., et al. 2014 evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the Eighth Joint National Committee (JNC 8) Jama. 2014;311:507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 118.Sica D.A. Pharmacokinetics and Pharmacodynamics of Mineralocorticoid Blocking Agents and their Effects on Potassium Homeostasis. Hear Fail. Rev. 2005;10:23–29. doi: 10.1007/s10741-005-2345-1. [DOI] [PubMed] [Google Scholar]
- 119.Miller E., Shubrook J.H. Sodium glucose co-transporter 2 inhibitors in the treatment of type 2 diabetes mellitus. Am. Coll. Osteopath. Fam. Physicians. 2015;7:10–30. [Google Scholar]
- 120.Zou H., Zhou B., Xu G. SGLT2 inhibitors: A novel choice for the combination therapy in diabetic kidney disease. Cardiovasc. Diabetol. 2017;16:65. doi: 10.1186/s12933-017-0547-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blum A., Miller H. Pathophysiological role of cytokines in congestive heart failure. Annu. Rev. Med. 2001;52:15–27. doi: 10.1146/annurev.med.52.1.15. [DOI] [PubMed] [Google Scholar]
- 122.Hassan I., Dorjay K., Anwar P. Pentoxifylline and its applications in dermatology. Indian Dermatol. Online J. 2014;5:510–516. doi: 10.4103/2229-5178.142528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rubin L.J., Roux S. Bosentan: A dual endothelin receptor antagonist. Expert Opin. Investig. Drugs. 2002;11:991–1002. doi: 10.1517/13543784.11.7.991. [DOI] [PubMed] [Google Scholar]
- 124.Koniaris L.S., Goldhaber S.Z. Anticoagulation in dilated cardiomyopathy. J. Am. Coll. Cardi-Ology. 1998;31:745–748. doi: 10.1016/S0735-1097(98)00003-5. [DOI] [PubMed] [Google Scholar]
- 125.Riegger A.J.G. Endocrinology of the Heart. Springer; Berlin, Heidelberg: 1989. Atrial Natriuretic Peptide in Heart Failure; pp. 85–89. [Google Scholar]
- 126.Seth S., Bhargava B., Narang R., Ray R., Mohanty S., Gulati G., Kumar L., Airan B., Venugopal P. The ABCD (Autologous Bone Marrow Cells in Dilated Cardiomyopathy) Trial. J. Am. Coll. Cardiol. 2010;55:1643–1644. doi: 10.1016/j.jacc.2009.11.070. [DOI] [PubMed] [Google Scholar]
- 127.U.S. National Library of Medicine. Clinical Trials.gov. [(accessed on 17 September 2020)]; Available online: https://www.clinicaltrials.gov/
- 128.Yoshii S., Hosaka S., Suzuki S., Osawa H., Akashi O., Tada Y., Sugiyama H., Hoshiai M., Tan T., Kadono T., et al. Indications for partial left ventriculectomy in pediatric dilated cardiomyopathy. Circ. J. 2002;66:337–340. doi: 10.1253/circj.66.337. [DOI] [PubMed] [Google Scholar]
- 129.Franco-Cereceda A., McCarthy P.M., Blackstone E.H., Hoercher K.J., White J.A., Young J.B., Starling R.C. Partial left ventriculectomy for dilated cardiomyopathy: Is this an alternative to transplantation? J. Thorac. Cardiovasc. Surg. 2001;121:879–893. doi: 10.1067/mtc.2001.113598. [DOI] [PubMed] [Google Scholar]
- 130.Mancini D., Colombo P.C. Left ventricular assist devices: A rapidly evolving alternative to transplant. J. Am. Coll. Cardiol. 2015;65:2542–2555. doi: 10.1016/j.jacc.2015.04.039. [DOI] [PubMed] [Google Scholar]
- 131.Sipahi I., Fang J.C. QRS duration criteria to select patients for cardiac resynchronization therapy: Crt should be reserved for a QRS duration ≥150 ms: Pro. Circ. Arrhythmia Electrophysiol. 2013;6:436–441. doi: 10.1161/CIRCEP.112.000160. [DOI] [PubMed] [Google Scholar]
- 132.Dhandapany P.S., Sadayappan S., Xue Y., Powell G.T., Rani D.S., Nallari P., Rai T.S., Khullar M., Soares P., Bahl A., et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Das S., Biswas A., Kapoor M., Bhargava B., Seth S., Rao V.R. Epidemiology of Cardiomyopathy-A Clinical and Genetic Study of Dilated Cardiomyopathy: The EPOCH-D study. J. Pract. Cardiovasc. Sci. 2015;1:30–34. [Google Scholar]
- 134.Kothari S.S., Dhopeshwarkar R.A., Saxena A., Juneja R. Dilated cardiomyopathy in Indian children. Indian Hear J. 2003;55:147–151. [PubMed] [Google Scholar]
- 135.Dilated Cardiomyopathy in Indian Children-PubMed [Internet] [(accessed on 19 April 2021)]; Available online: https://pubmed.ncbi.nlm.nih.gov/12921329/
- 136.Rai T.S., Ahmad S., Ahluwalia T.S., Ahuja M., Bahl A., Saikia U.N., Singh B., Talwar K.K., Khullar M. Genetic and clinical profile of Indian patients of idiopathic restrictive cardiomyopathy with and without hypertrophy. Mol. Cell. Biochem. 2009;331:187–192. doi: 10.1007/s11010-009-0157-7. [DOI] [PubMed] [Google Scholar]
- 137.Nallari P., Tanjore R., RangaRaju A., Vadapalli S., Remersu S., Narsimhan C. Genetic variations of β-MYH7 in hypertrophic cardiomyopathy and dilated cardiomyopathy. Indian J. Hum. Genet. 2010;16:67–71. doi: 10.4103/0971-6866.69348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perundurai D., Dhandapany P.S., Ahluwalia T.S., Bhardwaj M., Bahl A., Talwar K.K., Nair K., Rathinavel A., Khullar M. ACE I/D polymorphism in Indian patients with hypertrophic cardiomyopathy and dilated cardiomyopathy. Mol. Cell. Biochem. 2007;311:67–72. doi: 10.1007/s11010-007-9695-z. [DOI] [PubMed] [Google Scholar]
- 139.Jadhav K.B., Karpe K.K., Maramattom B.V. An Indian family with an Emery-Dreifuss myopathy and familial dilated cardiomyopathy due to a novel LMNA mutation. Ann. Indian Acad. Neurol. 2012;15:344–346. doi: 10.4103/0972-2327.104355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matsa L.S., Rangaraju A., Vengaldas V., Latifi M., Jahromi H.M., Ananthapur V., Nallari P. Haplotypes of NOS3 Gene Polymorphisms in Dilated Cardiomyopathy. PLoS ONE. 2013;8:e70523. doi: 10.1371/journal.pone.0070523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matsa L.S., Sagurthi S.R., Ananthapur V., Nalla S., Nallari P. Endothelin 1 gene as a modifier in dilated cardiomyopathy. Gene. 2014;548:256–262. doi: 10.1016/j.gene.2014.07.043. [DOI] [PubMed] [Google Scholar]
- 142.Rani D.S., Dhandapany P.S., Nallari P., Narasimhan C., Thangaraj K. A Novel Arginine to Tryptophan (R144W) Mutation in Troponin T (cTnT) Gene in an Indian Multigenerational Family with Dilated Cardiomyopathy (FDCM) PLoS ONE. 2014;9:e101451. doi: 10.1371/journal.pone.0101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deshmukh A., Deshmukh A., Deshmukh G., Garg P.K. A pilot study of dilated cardiomyopathy (DCM) in western Uttar Pradesh, India: A four year review. Med.-Leg. Update. 2011;11:1. [Google Scholar]
- 144.Rana H., Rathod C., Chavda P., Patel S., Deshpande S. Clinical profile of Dilated Cardiomyopathy patients presenting to a tertiary care hospital Clinical profile of Dilated Cardiomyopathy patients presenting to a tertiary care hospital from central Gujarat. Int J Res Med. 2015;4:143–146. [Google Scholar]
- 145.Saha K.K., Kumar A., Sneha K., Kumar P., Mishra A., Tiwari M., Roshan R. A Clinical Study of Dilated Cardiomyopathy with Correlation to Electrocardiography and Echocardiography: A Cross Sectional Study. Int. J. Sci. Study. 2018;5:91–95. Available online: www.ijss-sn.com. [Google Scholar]
- 146.Ushasree B., Shivani V., Venkateshwari A., Jain R.K., Narsimhan C., Nallari P. Epidemiology and genetics of dilated cardio-myopathy in the Indian context. Indian J. Med. Sci. 2009;63:288–296. [PubMed] [Google Scholar]
- 147.Shairgojri A.A., Mohiuddin K., Bashir A., Jan M. Childhood cardiomyopathies: A study in tertiary care hospital of Kashmir. Int. J. Contemp. Pediatr. 2021;8:289–294. doi: 10.18203/2349-3291.ijcp20210115. [DOI] [Google Scholar]
- 148.Histopathological Changes Seen in The Heart Samples of Dcm Patients from North India a Cadaveric Study, IJSR-International Journal of Scientific Research(IJSR), IJSR|World Wide Journals [Internet] [(accessed on 22 April 2021)]. Available online: https://www.worldwidejournals.com/international-journal-of-scientific-research-(IJSR)/article/histopathological-changes-seen-in-the-heart-samples-of-dcm-patients-from-north-india-a-cadaveric-study/MTA4NjE=/
- 149.Bhawani G., Balije S., Kumar A., Murthy K.S.N., Kumari N. Effect of hypertension at presentation on prognosis in patients with dilated cardiomyopathy presenting with normal renal angiogram. Indian J. Med Res. 2016;144:281–287. doi: 10.4103/0971-5916.195045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Clinical E.C.G. EKG, Profile of Patients with DCM [Internet] [(accessed on 22 April 2021)]. Available online: http://iscindia.co.in/Uploads/635343547865542527.PDF.
- 151.Das S., Biswas A., Kapoor M., Seth S., Bhargava B., Rao V. A novel donor site mutation in LMNA gene leading to severe form of Dilated Cardiomyopathy in a proband of a family from Bihar, India. Mol. Cytogenet. 2014;7:P35. doi: 10.1186/1755-8166-7-S1-P35. [DOI] [Google Scholar]
- 152.Sonowal N., Vanamali D.R. Clinical profile of patients with dilated cardiomyopathy in a tertiary care center in north east India. J. Evol. Med. Dent. Sci. 2014;3:8378–8386. doi: 10.14260/jemds/2014/3056. [DOI] [Google Scholar]
- 153.Paul R., Nandi S., Sinha P.K. Epidemiological study of dilated cardiomyopathy from eastern India with special reference to left atrial size. Int. J. Med. Res. Heal Sci. 2014;3:639. doi: 10.5958/2319-5886.2014.00410.X. [DOI] [Google Scholar]
- 154.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moorjani P., Thangaraj K., Patterson N., Lipson M., Loh P.-R., Govindaraj P., Berger B., Reich D., Singh L. Genetic Evidence for Recent Population Mixture in India. Am. J. Hum. Genet. 2013;93:422–438. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Basak T., Varshney S., Akhtar S., Sengupta S. Understanding different facets of cardiovascular diseases based on model sys-tems to human studies: A proteomic and metabolomic perspective. J. Proteom. 2015;127:50–60. doi: 10.1016/j.jprot.2015.04.027. Elsevier. [DOI] [PubMed] [Google Scholar]
- 157.Basak T., Varshney S., Hamid Z., Ghosh S., Seth S., Sengupta S. Identification of metabolic markers in coronary artery disease using an untargeted LC-MS based metabolomic approach. J. Proteom. 2015;127:169–177. doi: 10.1016/j.jprot.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 158.Zhou L., Wang Z., Hu C., Zhang C., Kovatcheva-Datchary P., Yu D., Liu S., Ren F., Wang X., Li Y., et al. Integrated Metabolomics and Lipidomics Analyses Reveal Metabolic Reprogramming in Human Glioma with IDH1 Mutation. J. Proteome Res. 2018;18:960–969. doi: 10.1021/acs.jproteome.8b00663. [DOI] [PubMed] [Google Scholar]
- 159.Louridas G.E., Lourida K.G. Systems Biology and Biomechanical Model of Heart Failure. Curr. Cardiol. Rev. 2012;8:220–230. doi: 10.2174/157340312803217238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Izquierdo I., Rosa I., Bravo S.B., Guitián E., Pérez-Serra A., Campuzano O., Brugada R., Mangas A., García Á., Toro R. Proteomic identification of putative biomarkers for early detection of sudden cardiac death in a family with a LMNA gene mutation causing dilated cardiomyopathy. J. Proteom. 2016;148:75–84. doi: 10.1016/j.jprot.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 161.Egerstedt A., Berntsson J., Smith M.L., Gidlöf O., Nilsson R., Benson M., Wells Q.S., Celik S., Lejonberg C., Farrell L., et al. Profiling of the plasma proteome across different stages of human heart failure. Nat. Commun. 2019;10:5830. doi: 10.1038/s41467-019-13306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.