ABSTRACT
In selective macroautophagy/autophagy, cargo receptors are recruited to the forming autophagosome by interacting with Atg8 (autophagy-related 8)-family proteins and facilitate the selective sequestration of specific cargoes for autophagic degradation. In addition, Atg8 interacts with a number of adaptors essential for autophagosome biogenesis, including ATG and non-ATG proteins. The majority of these adaptors and receptors are characterized by an Atg8-family interacting motif (AIM) for binding to Atg8. However, the molecular basis for the interaction mode between ATG8 and regulators or cargo receptors in plants remains largely unclear. In this study, we unveiled an atypical interaction mode for Arabidopsis ATG8f with a plant unique adaptor protein, SH3P2 (SH3 domain-containing protein 2), but not with the other two SH3 proteins. By structure analysis of the unbound form of ATG8f, we identified the unique conformational changes in ATG8f upon binding to the AIM sequence of a plant known autophagic receptor, NBR1. To compare the binding affinity of SH3P2-ATG8f with that of ATG8f-NBR1, we performed a gel filtration assay to show that ubiquitin-associated domain of NBR1 outcompetes the SH3 domain of SH3P2 for ATG8f interaction. Biochemical and cellular analysis revealed that distinct interfaces were employed by ATG8f to interact with NBR1 and SH3P2. Further subcellular analysis showed that the AIM-like motif of SH3P2 is essential for its recruitment to the phagophore membrane but is dispensable for its trafficking in endocytosis. Taken together, our study provides an insightful structural basis for the ATG8 binding specificity toward a plant-specific autophagic adaptor and a conserved autophagic receptor.
Abbreviations: ATG, autophagy-related; AIM, Atg8-family interacting motif; BAR, Bin-Amphiphysin-Rvs; BFA, brefeldin A; BTH, benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester; CCV, clathrin-coated-vesicle; CLC2, clathrin light chain 2; Conc A, concanamycin A; ER, endoplasmic reticulum; LDS, LIR docking site; MAP1LC3/LC3, microtubule associated protein 1 light chain 3; LIR, LC3-interacting region; PE, phosphatidylethanolamine; SH3P2, SH3 domain containing protein 2; SH3, Src-Homology-3; UBA, ubiquitin-associated; UIM, ubiquitin-interacting motif.
KEYWORDS: Arabidopsis ATG8, ATG8 interacting motif, NBR1, selective autophagy, SH3P2
Introduction
Derived from the Greek word meaning “self-eating”, autophagy acts as a clearing-up process by breaking down damaged or unwanted proteins/cellular structures, thereby balancing cellular homeostasis in almost all eukaryotes [1]. Macroautophagy (hereafter as autophagy) requires the formation of a double-membrane vesicle known as an autophagosome, and is actively induced under nutrient-limiting conditions or pathogen infections [2,3]. Phagophores, the precursors to autophagosomes, either randomly sequester bulk cytoplasm cargo in a nonselective manner, or target specific cargo molecules selectively, whereby the latter requires selective autophagy receptors for cargo recognition and recruitment. In recent years, a growing number of studies on plants have shown that selective autophagy is under tight control to maintain cellular homeostasis for plant development [4–16].
At the heart of the autophagy process is the Atg8 protein (also known as MAP1LC3/LC3 [microtubule associated protein 1 light chain 3] in mammals) [17,18], which decorates the outer and inner membranes of the phagophore and autophagosome being covalently conjugated to phosphatidylethanolamine (PE) [19]. Subsequently, the lipidated Atg8 (Atg8–PE) proteins further recruit numbers of receptors and adaptors to regulate the initiation, expansion, and maturation of the phagophore, and, ultimately, fusion of the autophagosome with the vacuole [20–23]. Atg8–PE thus provides a docking platform for autophagy adaptors to facilitate autophagosome formation, or for receptor recognition to selectively recruit the cargos to the nascent phagophore membrane, respectively. A number of autophagy adaptors and receptors have been shown to interact with Atg8/LC3 via the Atg8-family interacting motif (AIM)/LC3-interacting region (LIR) [20,24]. Emerging structural studies in yeast and mammals have shed novel light on the regulatory mechanism of Atg8-receptor/adaptor interactions [25–29]. The canonical AIM contains a consensus sequence xxx[W/F/Y]xx[L/I/V]xxx, where “x” stands for any residue. In the current model, the aromatic residue [W/F/Y] and the hydrophobic residue [L/I/V] in AIM bind to the two hydrophobic pockets within Atg8 protein, whereas the negatively charged residues interact with the positively charged residues surrounding the two hydrophobic pockets in Atg8 to stabilize the interaction [30].
In Arabidopsis plants, there is a great expansion of the ATG8 family (ATG8a-i), which has been suggested to function in specific interactions with different plant autophagic adaptors or receptors [31]. NBR1, a selective autophagy receptor mainly responsible for recognizing ubiquitinated protein aggregates, represents one of the best-studied AIM-containing proteins in Arabidopsis [12,32–36]. The AIM sequence in the ubiquitin-associated (UBA) domain of NBR1 (657–667 aa: GVSEWDPILEE), has been shown to be indispensable for ATG8 binding [32]. Of note, based on the domain organization, it is suggested that Arabidopsis NBR1 might be a hybrid of mammalian autophagic receptors SQSTM1/p62 and NBR1 [32]. Nevertheless, the molecular basis for the binding specificity between NBR1 and ATG8 during plant selective autophagy remains unclear, especially at the structural level. To date, the only available plant ATG8 structure was obtained from a potato ATG8CL in its bound form with a canonical AIM peptide (PDB: 5L83) (WEIV) of PexRD54 [37]. In this structure, the Trp and Val residues of the AIM motif bind canonically to the W- and L-sites, respectively, of ATG8CL.
In addition to the autophagy receptors, several ATG proteins, including Atg1, Atg3, Atg4, Atg7, and Atg13, have been shown to directly interact with Atg8 or LC3 in mediating autophagosome formation in yeast, mammals or plants [38–44]. Particularly, structural analysis has provided novel insights into their hierarchy order for the execution of autophagosome assembly spatially or temporally. In a general view, regulators for autophagosome formation are initially recruited for phagophore assembly and expansion, while cargo is subsequently recognized via Atg8 and encapsulated within the autophagosome. Despite recent advances unveiling Atg8 binding specificity by matching different autophagic regulators or cargo receptors via the AIMs, how the specificity is executed spatially and temporally remains unclear, particularly the balance between the autophagic adaptors and cargo receptors toward Atg8 to facilitate autophagosome formation or cargo sequestration.
Our previous study showed that a plant unique adaptor protein named SH3P2 (SH3 domain-containing protein 2, AT4G34660), binds to ATG8 via its C-terminal Src-Homology-3 (SH3) domain [45,46]. Of note, in the Arabidopsis genome, there are three isoforms of SH3Ps, namely SH3P1, SH3P2, SH3P3, all of which contain a conserved N-terminal BAR (Bin-Amphiphysin-Rvs) domain and C-terminal SH3 domain (Figure 1A), to function in endocytosis [47]. However, the molecular element for the binding specificity between SH3P2 and ATG8 interaction remains uncharacterized, and nothing is known about the function of SH3P2-ATG8 interaction in Arabidopsis. In regarding the membrane deformation feature of the BAR domain and its ability to recruit different membrane remodeling machinery, it is plausible that the transition for SH3P2 binding to ATG8 might serve as a plant unique mechanism for membrane scaffold assembly on the phagophore membrane. Subsequently, it might facilitate membrane remodeling, or provide a docking site for building further contacts with other membrane compartments. Several recent studies have demonstrated a crucial role for SH3P2 in the clathrin-coated vesicle (CCV)-mediated endocytic pathway and cell plate formation, in which SH3P2 is associated with several CCV related regulators, including clathrin, auxilin-like protein and ESCRT machinery [47–51]. Our previous study also showed that SH3P2 interacts with a plant-specific ESCRT subunit FREE1 to function in the maturation of autophagosome [50]. Therefore, SH3P2 might function as a multi-tasking regulator in various membrane remodeling processes.
Figure 1.
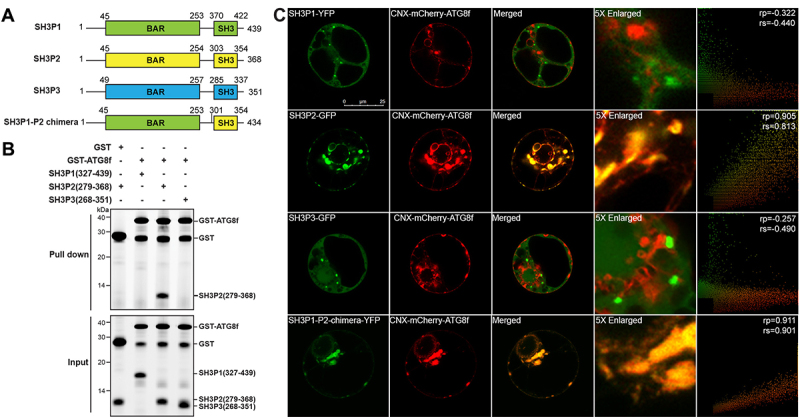
SH3P2, but not SH3P1 and SH3P3, binds to Arabidopsis ATG8f. (A) Domain organization of Arabidopsis SH3Ps proteins. SH3P1, SH3P2 and SH3P3 all contain an N-terminal BAR domain and a C-terminal SH3 domain. (B) GST affinity-isolation assay. 1 µM SH3 domain protein of SH3P1, SH3P2 and SH3P3 was mixed with 1 µM GST-ATG8f and incubated with glutathione resin (upper panel) respectively. The bound protein was eluted with 10 mM glutathione, and then pre-stained by Instant-Bands and analyzed by SDS-PAGE (lower panel). Our results suggested that GST-ATG8f only interacted with the SH3 domain of SH3P2 but not with the SH3 domains of SH3P1 and SH3P3. (C) SH3Ps-GFP/YFP, or SH3P1-P2-chimera-YFP, and CNX-mCherry-ATG8f were transiently co-expressed in Arabidopsis protoplasts. Confocal analysis showed that SH3P2, but not SH3P1 and SH3P3, colocalized with CNX-mCherry-ATG8f. Similar results were obtained from three different independent experiments. The right column shows the scatterplot images obtained from ImageJ with the PSC colocalization plug-in. The linear Pearson correlation coefficient (rp) and the nonlinear Spearman correlation coefficient (rs) indicate the extent of colocalization with the value of +1.0 for complete colocalization.
Here, we have investigated the molecular mechanism underlying the specificity for SH3P2-ATG8 interaction in Arabidopsis. As a first step to characterize the determinants for the SH3P2-ATG8 interaction, we compared the specificity of the SH3 domain from SH3P2 with the other two SH3 homologs, SH3P1 and SH3P3. Surprisingly, by mapping the sequence in SH3P2 for binding to ATG8, we identified an AIM-like motif essential for ATG8 interaction, which however is highly conserved among SH3P1 to SH3P3. Next, we solved the solution structure of the unbound form of Arabidopsis ATG8f. With a combination of chemical shift perturbations and mutagenesis analysis, we have identified distinct interaction residues in ATG8f for SH3P2 and the well-known plant receptor NBR1. To gain more insights into the SH3P2-ATG8 interaction at the molecular level, we compared the binding mode for SH3P2 and NBR1 toward ATG8f. Interestingly, by gel filtration analysis, we found that the SH3 domain of SH3P2 is outcompeted by the UBA domain of NBR1 for ATG8f interaction. Further in-vitro and in-vivo analysis unveiled an essential role of two residues in ATG8f unique for ATG8f-NBR1 interaction, but dispensable for ATG8f-SH3P2 interaction. In addition, we demonstrated that the AIM-like motif of SH3P2 is essential for the recruitment of SH3P2 to the phagophore membrane in Arabidopsis transgenic plants, while its trafficking in the endocytic pathway seems not compromised. Taken together, our study provides new mechanistic insights into the structural basis in the binding specificity and plasticity of plant ATG8 for a plant unique autophagic adaptor and a conserved autophagic receptor.
Results
SH3P2 contains an AIM-like motif and atypically interacts with Arabidopsis ATG8
To test the specificity in ATG8-SH3P2 interaction, we first examined the interactions between SH3P2 and the nine Arabidopsis ATG8 isoforms (ATG8a-i). Using yeast-two-hybrid assay, coimmunoprecipitation (CoIP) and GST affinity-isolation analysis, we found that all ATG8 isoforms interact with SH3P2 (Figure S1). In our hands, protein samples of ATG8f exist as a monomer in solution without the problem of aggregation, and, hence, were used in further structural and biochemical studies of ATG8 in this work. Next, we tested the binding specificity between ATG8f and SH3Ps by expressing ATG8f, and the SH3 domains of SH3Ps recombinantly in E. coli. We showed by in-vitro GST affinity-isolation assay that only the SH3 domain of SH3P2 (SH3P2-SH3), but not that of SH3P1 and SH3P3, binds to ATG8f (Figure 1B). To further evaluate the specificity for ATG8f interaction in-vivo, we established a recruitment assay by co-expressing an ER-localized ATG8f (CNX-mCherry-ATG8f) and SH3Ps (Figure 1C). In this assay, a construct by fusing mCherry-tagged ATG8f to the transmembrane and C-terminal domain of calnexin, which anchors the fusion protein to the ER membrane [12,52], was co-expressed with GFP/YFP-tagged SH3Ps. We observed that only SH3P2-GFP, but not SH3P1-YFP or SH3P3-GFP, was specifically rerouted and colocalized with CNX-mCherry-ATG8f (Figure 1C). On the other hand, a chimeric construct, SH3P1-P2, which is composed of the N-terminal BAR domain of SH3P1 (1–366 aa) and the C-terminal SH3 domain of SH3P2 (301–368 aa), was also rerouted by and colocalized with CNX-mCherry-ATG8f (Figure 1C).
Interestingly, it has been suggested that SH3P2 binds to ATG8 via the AIM motif [53], but there are three candidate motifs (310YHGV313, 325YVVV328, 350YGYI353) predicted in the SH3 domain of SH3P2 (Figure 2A). To test whether these putative AIM motifs of SH3P2 are responsible for ATG8f interaction, we performed an in-vitro peptide affinity-isolation assay. Our results revealed that ATG8f only displayed a strong interaction with the peptide sequence containing the 325YVVV328 motif, but not with the other two AIM-like motifs, 310YHGV313 and 350YGYI353 (Figure 2B). Next, we performed a yeast-two-hybrid assay, showing that mutation in 325YVVV328 disturbed the interaction between SH3P2 and ATG8f, while no obvious change was observed when mutation in 350YGYI353 (Figure 2C). Consistently, immunoprecipitation assay by the GFP trap assay showed that SH3P2Y325A,V328A mutation also suppressed the association between SH3P2 and ATG8f, when YFP-ATG8f and HA-tagged SH3P2 wild type or SH3P2Y325A,V328A were transiently co-expressed in the Arabidopsis protoplast cells (Figure 2D). To further test whether the 325YVVV328 motif is crucial for ATG8 binding in-vivo, we carried out the recruitment assay using CNX-mCherry-ATG8f. As shown in Figure 2E, the mutation in 325YVVV328 significantly prevented recruitment of SH3P2 by CNX-mCherry-ATG8f, whereas the mutation in 350YGYI353 did not affect the colocalization of SH3P2 with ATG8f (Figure 2E). Consistent results were also obtained when tested using another ATG8 isoform ATG8e, which sharing 83% sequence identity with ATG8f (Figure S2).
Figure 2.
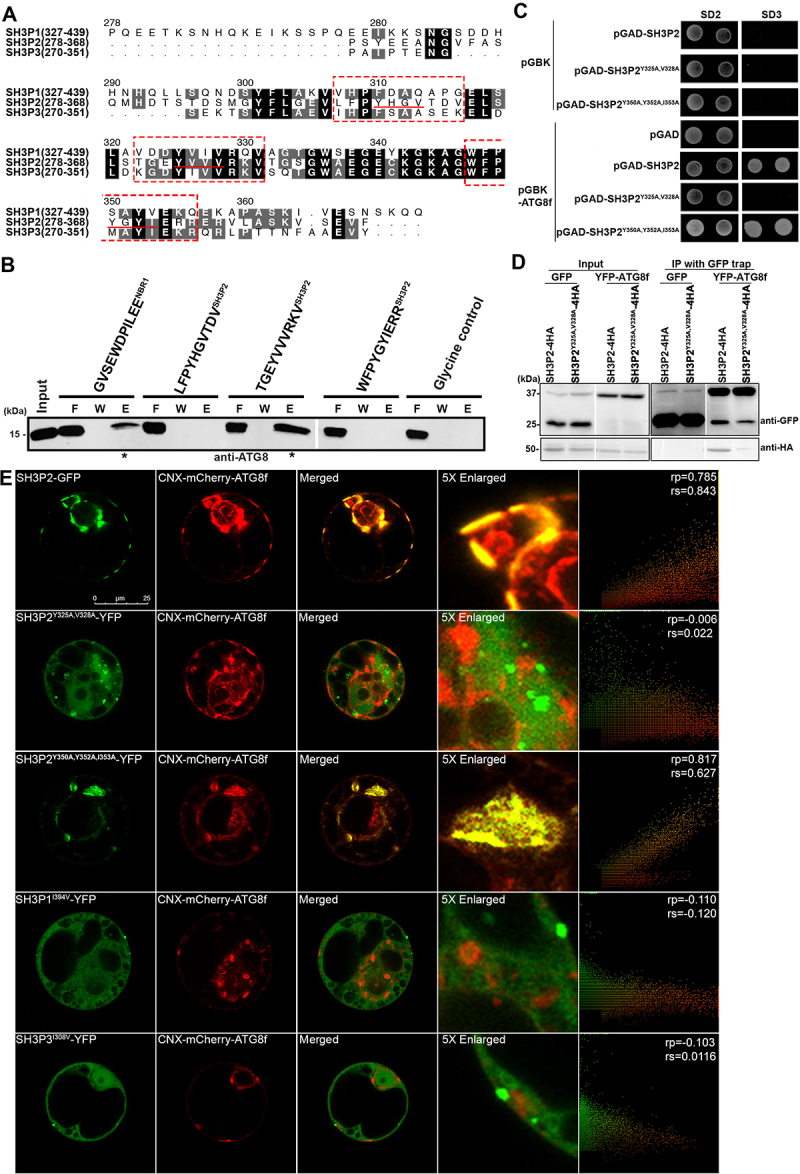
SH3P2 contains an AIM-like motif for interaction with ATG8f. (A)The sequences of the SH3 domain of SH3Ps protein in Arabidopsis were aligned by the Clustal-Omega algorithm [92] implemented in the program SEAVIEW [93]. Three AIM-like sequences (underlined) were identified in SH3P2. Residues were numbered according to the Arabidopsis SH3P2 sequence. (B) The purified ATG8f protein was incubated with NHS-activated Sepharose resins coupled with peptide sequences containing the putative AIM-like motifs of SH3P2. The flow-through (F), wash (W) and elution (E) fractions were analyzed by western blot using ATG8 antibodies. Our affinity-isolation assay showed that ATG8f interacted with the peptide sequence containing the 325YVVV328 AIM motif, but not with the 310YHGV313 and 350YGYI353 motifs. (C) Yeast-two-hybrid assay showed mutation in 325YVVV328, but not in 350YGYI353, compromised SH3P2-ATG8f interaction. (D) Immunoprecipitation assay showed that mutation in 325YVVV328 affects ATG8-SH3P2 interaction. Cell lysate from Arabidopsis PSBD protoplasts transiently co-expressed GFP or YFP-ATG8f with SH3P2-4HA and SH3P2Y325A,V328A-4HA respectively for 12 h were subjected to GFP trap assay. The resulting immunoprecipitation (IP) and cell lysate were analyzed by immunoblotting (IB) using anti-HA or anti-GFP antibodies. (E) Wild-type and mutants of SH3P2-YFP/GFP, SH3P1I394V-YFP, SH3P3I308V-YFP, and CNX-mCherry-ATG8f were transiently co-expressed in Arabidopsis protoplast respectively. The recruitment assay showed that wild-type SH3P2 and SH3P2Y350A, Y352A, I353A, but not SH3P2Y325A,V328A, was relocalized to overlap with CNX-mCherry-ATG8f. Neither SH3P1I394V-YFP, nor SH3P3I308V-YFP were recruited by CNX-mCherry-ATG8f. Similar results were obtained from three different independent experiments. The right column shows the scatterplot images obtained from ImageJ with the PSC colocalization plug-in. The linear Pearson correlation coefficient (rp) and the nonlinear Spearman correlation coefficient (rs) indicate the extent of colocalization with the value of +1.0 for complete colocalization.
We are intrigued by the observation that the AIM-like motif of SH3P2 (325–328 aa: YVVV) is also highly conserved in SH3P1 (392–395 aa: YVIV) and SH3P3 (307–310 aa: YIVV) (Figure 2A), yet only SH3P2 could interact with ATG8f. To further examine whether this motif is sufficient for ATG8 interaction, the 392YVIV395 motif of SH3P1 and 307YIVV310 motif of SH3P3 were substituted into “YVVV” as SH3P2. Our results revealed that neither of the resulting mutants, SH3P1I394V and SH3P3I308V, was rerouted by and colocalized with CNX-mCherry-ATG8f (Figure 2E), suggesting that the 325YVVV328 AIM-like motif of SH3P2 is not the sole determinant for the binding specificity of SH3 domain toward ATG8f.
Conformational changes around the ligand binding site in ATG8f upon binding with the NBR1 AIM peptide
To gain more insights into the interaction mode between SH3P2 and ATG8f, we took a further step to examine the conformational changes in plant ATG8f upon ligand binding. Firstly, we solved the solution structure of Arabidopsis ATG8f in ligand-free form (Figure 3A). Statistics of the ten best structures are summarized in Table 1 . The overall structure of ATG8f contains a four-stranded ß-sheet sandwiched by N-terminal helix-1/2 and C-terminal helix-3/4 (Figure 3B), resembling the folding topology of ATG8 homologs found in other species. The structure of Arabidopsis ATG8f in its apo form was aligned to the structure of ATG8CL in complex with the Pexrd54 AIM peptide (DWEIV) [37]. Significant conformational changes were detected on several residues (e.g., I22, Y26, R29, K47, R68) around the ligand binding sites of ATG8f (Figure 3C), which are highly conserved among all nine Arabidopsis ATG8 isoforms. It has been well established that Arabidopsis NBR1 also binds ATG8 through the AIM motif [32]. Consistent with previous work, we showed by in-vitro peptide affinity-isolation assay that ATG8f could interact with the NBR1 AIM peptide (657–667 aa: GVSEWDPILEE) (Figure S3A). Next, we performed a chemical shift perturbation experiment to identify residues in ATG8f that may participate in binding with the NBR1 AIM peptide. Backbone amide 1H and 15N chemical shifts of ATG8f in its free and bound forms were assigned and compared (Figure S4A,B).
Figure 3.
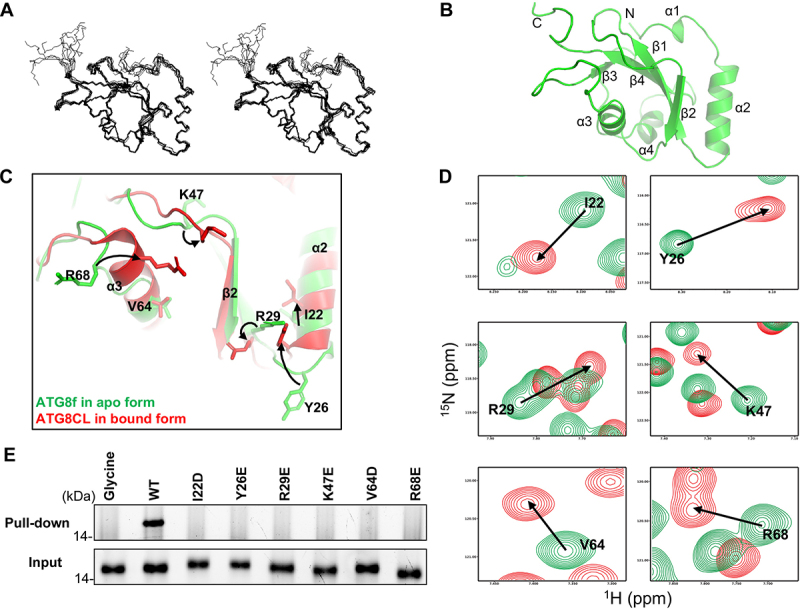
Binding of the NBR1 AIM peptide induces conformational changes around the ligand binding sites in ATG8f. (A) Solution structure of ATG8f. Stereodiagram of an ensemble of the 10 best structures showing the best-fit superposition of the backbone atoms. (B) Ribbon representation of the representative ATG8f structure. N- and C- termini and secondary structure elements were labeled. (C) Structure of Arabidopsis ATG8f in apo form (green) was compared to that of Irish potato ATG8CL in complex with the AIM peptide (red) of PexRD54 (PDB code: 5L83). Upon binding of the cognate AIM peptide, I22, Y26, R29, K47, R68 with conformation changes were evident in regions around the ligand binding sites of ATG8. (D) 1H-15N correlation spectra of ATG8f in the absence (green contours) and in the presence (red contours) of the NBR1 AIM peptide (657–667 aa: GVSEWDPILEE) were compared. Significant chemical shift perturbations were found for residues (I22, Y26, R29, K47, V64, R68) around the ligand binding sites of ATG8f, suggesting binding of the NBR1 AIM peptide induced changes in the chemical environment around these residues. (E) The role of residues with large chemical shift perturbations was tested by mutagenesis and affinity-isolation assay. 1 μM wild-type or variants of ATG8f was mixed with NHS-activated Sepharose resins coupled with the NBR1 AIM peptide, and the bound proteins were stained by Instant-Bands and analyzed by SDS-PAGE. Our results showed that all substitutions weaken the interaction between ATG8f and the NBR1 AIM peptide, suggesting these residues are important in the interaction. Wild-type ATG8f loaded to the NHS-activated Sepharose coupled with glycine was included as a negative control.
Table 1.
NMR and refinement statistics for the 10 structures of ATG8f with lowest energy and no restraint violation
| Distance and dihedral restraints | |
| Total number of NOE | 1312 |
| unambiguous | 1141 |
| intra-residue (|i-j|=0) | 570 |
| sequential (|i-j|=1) | 213 |
| medium-range (1<|i-j|<5) | 130 |
| long-range (|i-j| >4) | 228 |
| ambiguous | 171 |
| Number of hydrogen bond restraints | 58 |
| Number of dihedral angle restraints | 173 |
| Structure statistics | |
| Violations | |
| Distance restraints (Å) | 0.057 ± 0.003 |
| Dihedral restraints (o) | 0.71 ± 0.05 |
| No. of dihedral angles with violation > 5o | 0 |
| No. of distant restraints with violation > 0.5Å | 0 |
| Deviation from idealized geometry | |
| Bond length (Å) | 0.0037 ± 0.0002 |
| Bond angle (o) | 0.59 ± 0.02 |
| Improper (o) | 0.54 ±0.01 |
| Average pairwise r.m.s deviation (Å) * | |
| Backbone | 0.487 Å |
| Heavy | 1.041 Å |
| Average Ramachandran plot statistics (%) | |
| Most favored | 64 |
| Additional allowed | 28 |
| Generously allowed | 8 |
| Disallowed | 0 |
* r.m.s.d of residue 10-117 was reported.
To further understand how ATG8f interacts with the AIM motif of NBR1, we modeled the structure of Arabidopsis ATG8f in complex with the NBR1 AIM peptide by homology modeling [54], using potato ATG8CL in complex with the Pexrd54 AIM peptide as the template (Figure S4C). Our model predicted that I22, Y26 and V64 on ATG8f form hydrophobic interactions with W661 and I664 of NBR1, while R29, K47 and R68 on ATG8f form salt bridges with E666, E660, D662 of NBR1 (Figure S4C, right). The sequences of Arabidopsis ATG8f and potato ATG8CL are highly homologous to each other, sharing 84% sequence identity (Figure S4D). Moreover, the AIM motif of NBR1 (WDPI) is similar to that of PexRD54 (WEIV). Our model of ATG8f-NBR1 AIM complex is consistent with the chemical shift perturbation experiment, where significant chemical shift perturbations were found for residues (I22, Y26, R29, K47, V64, R68) around the ligand binding sites of ATG8f (Figure 3D). Next, we performed site-directed mutagenesis to further test our model. By peptide affinity-isolation assay using variants of ATG8f (I22D, Y26E, R29E, K47E, V64D, R68E), we found that these ATG8f residues are all involved in interaction with the NBR1 AIM peptide (Figure 3E).
Differential binding affinities of ATG8f toward NBR1-UBA and SH3P2-SH3
Based on the above observation, we next tested whether the ATG8f-NBR1 interaction mode is similar to that of ATG8f-SH3P2. We showed that ATG8f interacted with the UBA domain (619–704 aa) of NBR1 (NBR1-UBA) containing the AIM motif by GST affinity-isolation assay (Figure S3B). We therefore carried out a gel filtration assay to compare the binding affinities of ATG8f toward NBR1-UBA and SH3P2-SH3. When ATG8f was mixed with NBR1-UBA in 1:1 molar ratio and loaded to a gel filtration column, they formed an ATG8f-NBR-UBA complex that eluted at a volume of ~11.3 ml (Figure 4Ai). When ATG8f was mixed with the SH3 domain of SH3P2 (SH3P2-SH3), they formed an ATG8f-SH3P2-SH3 complex that eluted at a volume of ~12.3 ml (Figure 4Aii). Surprisingly, we found that ATG8f exhibits differential binding affinities toward NBR1 and SH3P2 respectively. When ATG8f, SH3P2-SH3 and NBR1-UBA were mixed at 2:1:1 ratio, both ATG8f-NBR1-UBA and ATG8f-SH3P2-SH3 complex were detected (Figure 4Aiii). In contrast, when three proteins were mixed at 1:1:1 ratio, the majority of ATG8f formed a complex with NBR1-UBA and the excess SH3P2-SH3 was eluted as a free form at a volume of ~18.5 ml, suggesting that NBR1-UBA might outcompete SH3P2-SH3 to favor the formation of ATG8f-NBR1-UBA complex (Figure 4Aiv).
To further verify this observation in-vivo, we performed a coimmunoprecipitation assay by transiently co-expressing an increasing amount of 5HA-NBR1 at 0:1:3 ratio, together with SH3P2-5Flag and YFP-ATG8f in Arabidopsis protoplasts using the GFP trap method. Consistent with the in-vitro data, less SH3P2-5Flag was immunoprecipitated with YFP-ATG8f when a higher amount of 5HA-NBR1 was expressed (Figure 4B).
Figure 4.
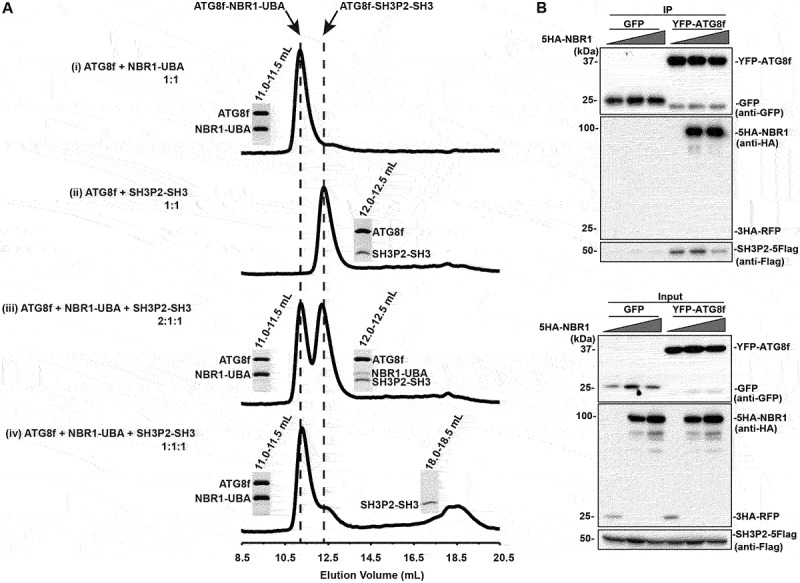
NBR1-UBA outcompetes SH3P2-SH3 for ATG8 binding. (A) ATG8f was mixed with NBR1-UBA and/or SH3P2-SH3 and injected into a Superdex 75 10/300 gel filtration column pre-equilibrated in 1X PBS, 5 mM DTT. The fractions corresponding to the peaks were resolved by SDS–PAGE followed by Coomassie Brilliant Blue staining. (i) When 40 µM ATG8f was mixed with 40 µM NBR1-UBA in 1:1 molar ratio, they formed an ATG8f-NBR1-UBA complex that eluted at ~11.2 ml. (ii) When 40 µM ATG8f was mixed with 40 µM SH3P2-SH3 in 1:1 molar ratio, they formed an ATG8f-SH3P2-SH3 complex that eluted at ~12.3 ml. (iii) When 80 µM ATG8f was mixed with 40 µM NBR1-UBA and SH3P2-SH3 in 2:1:1 molar ratio, both peaks at ~11.2 and ~12.3 ml were present, suggesting both ATG8f-NBR1-UBA and ATG8f-SH3P2-SH3 were formed. (iv) When 40 µM ATG8f was mixed with 40 µM NBR1-UBA and SH3P2-SH3 in 1:1:1 molar ratio, majority of ATG8f formed a complex with NBR1-UBA. On the other hand, a peak at ~18.5 ml representing the free form of SH3P2-SH3 was observed. (B) Increasing amount of 5HA-NBR1 (0:1:3) was transiently co-expressed with SH3P2-5Flag and YFP-ATG8f or GFP in Arabidopsis protoplasts for 12 h, followed by the GFP trap assay. 3HA-RFP was used as a control. The resulting immunoprecipitation and cell lysate were analyzed by immunoblotting using anti-Flag, anti-HA or anti-GFP antibodies respectively.
ATG8f interaction with SH3P2-SH3 differs from that with NBR1-UBA
That NBR1-UBA has a stronger binding affinity for ATG8f binding when compared with SH3P2-SH3, prompts us to further explore whether their interactions with ATG8f rely on distinct interfaces on ATG8f. We first examined whether substitutions (I22D, Y26E, R29E, K47E, V64D, R68E) of ATG8f that break the interaction with the NBR1 AIM peptide will affect the interaction with the SH3 domain of SH3P2 (SH3P2-SH3) by GST affinity-isolation assay. We found that substitutions of I22D, V64D and R68E on ATG8f also disturbed the interaction with SH3P2-SH3, implying that these residues of ATG8f might participate in ATG8f binding with either NBR1-UBA or SH3P2-SH3 (Figure S5).
Intriguingly, substitutions of other sites on ATG8f, including Y26E, R29E and K47E, did not interfere with its binding to SH3P2 (Figure S5). To further examine the possible function of these residues, we created a double variant of ATG8f (Y26E R29E) to compare its interaction with SH3P2 and NBR1 respectively. Our results showed that the Y26E R29E substitutions significantly broke the interaction with NBR1-UBA, but retained the interaction with SH3P2-SH3 in the GST affinity-isolation assay (Figure 5A). Similarly, when transiently expressed in Arabidopsis cells, the YFP-ATG8f (Y26E R29E) variant was coimmunoprecipitated with SH3P2, but not with NBR1 (Figure 5B). A previous study showed that mutation in the LDS (LIR docking site) by substitution with Y50A L51A in Arabidopsis ATG8a abolished the LDS binding affinity [53]. As this hydrophobic pocket is highly conserved in other ATG8 isoforms, we thus created another double variant of ATG8f (Y50A L51A) as an LDS null mutant control to compare with the residues we identified. Consistent with previous reports, we showed Y50A L51A substitutions significantly weakened the interaction with both NBR1 and SH3P2 in the GST affinity-isolation assay (Figure 5A), as well as in the co-immunoprecipitation experiment (Figure 5B).
Figure 5.
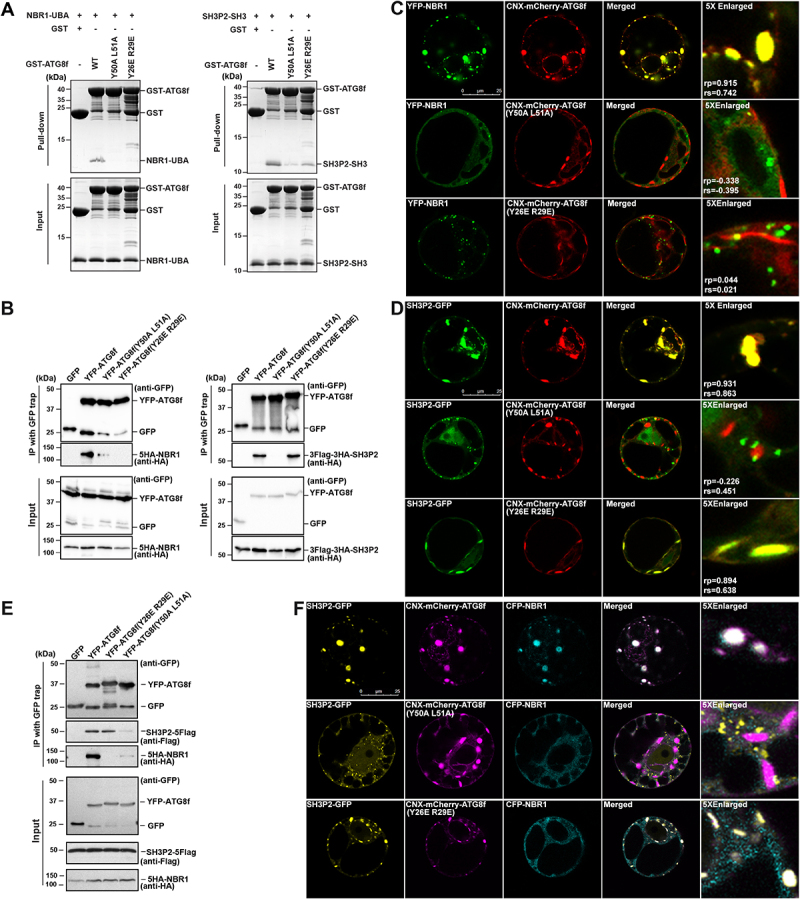
Distinct interaction surfaces in ATG8 for binding with SH3P2-SH3 and NBR1-UBA. (A) 10 μM wild-type or variants of GST-ATG8f was mixed with 10 μM SH3P2-SH3 (left panel) or NBR1-UBA (right panel) respectively, and incubated with glutathione resins in 1X PBS, 5 mM DTT. The bound proteins were eluted with glutathione and analyzed on SDS-PGAE with Coomassie Brilliant Blue staining. (B) YFP-ATG8f or its variants was co-expressed with HA-tagged NBR1 or HA-tagged SH3P2 in Arabidopsis protoplasts respectively. Lysates were immunoprecipitated by GFP-Trap method and detected by anti-HA or anti-GFP antibodies. (C) YFP-NBR1 was transiently co-expressed with wild-type or mutants of CNX-mCherry-ATG8f in Arabidopsis protoplasts. YFP-NBR1 was overlapped with CNX-mCherry-ATG8f, but not with the other two variants (Y26E R29E and Y50A L51A), suggesting these mutations are essential for the interaction between NBR1 and ATG8f in Arabidopsis protoplasts. Similar results were obtained from three different independent experiments. The linear Pearson correlation coefficient (rp) and the nonlinear Spearman correlation coefficient (rs) indicate the extent of colocalization with the value of +1.0 for complete colocalization. (D) SH3P2-GFP was transiently co-expressed with wild-type or mutants CNX-mCherry-ATG8f in Arabidopsis protoplast. SH3P2-GFP was recruited by wild-type or the Y26E R29E variant of CNX-mCherry-ATG8f, but not by the Y50A L51A variant. Similar results were obtained from three different independent experiments. The linear Pearson correlation coefficient (rp) and the nonlinear Spearman correlation coefficient (rs) indicate the extent of colocalization with the value of +1.0 for complete colocalization. (E) YFP-ATG8f, its variants (Y26E R29E, Y50A L51A) and GFP were co-expressed together with 5HA-NBR1 and SH3P2-5Flag in Arabidopsis protoplasts for 12 h respectively, and were subjected to GFP trap assay. The resulting immunoprecipitation (IP) and cell lysate were analyzed by immunoblotting (IB) using anti-Flag, anti-HA or anti-GFP antibodies. (F) SH3P2-GFP, CFP-NBR1 were transiently co-expressed together with wild-type or mutants CNX-mCherry-ATG8f in Arabidopsis protoplast. SH3P2-GFP was recruited by wild-type and the Y26E R29E variant of CNX-mCherry-ATG8f, but not by the Y50A L51A variant. In contrast, both Y26E R29E and Y50A L51A variants compromised the recruitment of CFP-NBR1.
To further confirm the above observation, we performed the recruitment assay to test whether these variants of ATG8f would redirect NBR1 or SH3P2 to the ER as the wild type ATG8f (Figure 5C,D). We found that SH3P2-GFP, but not YFP-NBR1, was rerouted by and colocalized with the Y26E R29E variant of CNX-mCherry-ATG8f in Arabidopsis protoplast cells (Figure 5C,D, bottom panel). However, both SH3P2 and NBR1 failed to be recruited by the Y50A L51A variant of CNX-mCherry-ATG8f (Figure 5C,D, middle panel). In addition, we also performed a coimmunoprecipitation assay by transient expression of YFP-ATG8f or its variants, together with 5HA-NBR1 and SH3P2-5Flag in Arabidopsis cells. We found that SH3P2-5Flag but not 5HA-NBR1 was coimmunoprecipitated with the Y26E R29E variant of YFP-ATG8f, whereas their associations with the Y50A L51A variant were both strongly diminished (Figure 5E). Consistent results were also obtained in confocal recruitment assays when CFP-NBR1, SH3P2-GFP and CNX-mCherry-ATG8f wild type or its variants were co-expressed in Arabidopsis protoplasts (Figure 5F).
The AIM-like motif of SH3P2 is essential for SH3P2 recruitment to the phagophore membrane
The mechanism for SH3P2 recruitment to the phagophore membrane yet remains unclear. Since ATG8 is anchored to the phagophore membrane, the ATG8-SH3P2 interaction may serve as a strategy for recruiting SH3P2 to phagophore to induce membrane remodeling. To test this hypothesis, we first generated transgenic Arabidopsis plants expressing GFP-tagged SH3P2Y325A,V328A. Consistently with previous finding [45], benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH, an autophagy inducer) and concanamycin A (Conc A, a vacuolar proton pump inhibitor) cotreatment could induce the accumulation of puncta in both cytosol and the vacuole in SH3P2-GFP transgenic plants (Figure S6A). In contrast, the number of SH3P2Y325A,V328A-GFP foci was significantly reduced in SH3P2Y325A,V328A-GFP transgenic plants upon the same treatment (Figure S6B). Instead, delivery of SH3P2-GFP into the vacuole was suppressed in the autophagy deficiency mutant atg5 background (Figure S6C).
To further examine whether ATG8-interaction is required for SH3P2 translocation to the phagophore membrane during autophagy in plants, we created double transgenic plants expressing SH3P2-GFP or SH3P2Y325A,V328A-GFP with the autophagosome marker mCherry-ATG8e [45]. When the SH3P2-GFP/mCherry-ATG8e double transgenic plants were treated with BTH to induce autophagy, SH3P2-GFP formed puncta or ring-like structures that were overlapped with mCherry-ATG8e (Figure 6A). However, neither obvious foci nor ring-like structures labeled by SH3P2Y325A,V328A-GFP were detected to overlap with mCherry-ATG8e in the SH3P2Y325A,V328A-GFP/mCherry-ATG8e double transgenic plants (Figure 6B). When the double transgenic plants were treated with both BTH and Conc A, numerous autophagic bodies labeled by both SH3P2-GFP and mCherry-ATG8e were clearly observed (Figure 6C, upper panel). Conversely, the number of autophagic bodies labeled by SH3P2Y325A,V328A-GFP was dramatically decreased, but a pronounced accumulation of autophagic bodies labeled by mCherry-ATG8e was still detected (Figure 6C, lower panel). Next, we conducted an ATG8 lipidation assay to further examine whether the autophagic activity is altered in the SH3P2Y325A,V328A-GFP transgenic plants (Figure 6D). Immunoblotting analysis with the ATG8 antibody showed that ATG8–PE adducts were also accumulated in SH3P2Y325A,V328A-GFP plants after BTH induction, similar to that in the wild type, whereas no ATG8–PE was detected in the atg5 mutant plants.
Figure 6.
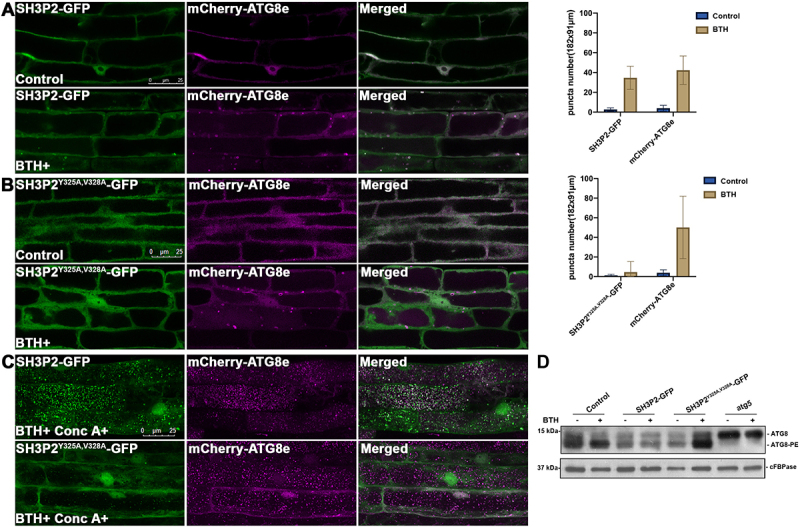
The AIM-like motif is essential for SH3P2 recruitment to the phagophore membrane upon autophagic induction. (A) 4-d-old seedlings were transferred to medium with or without BTH for 6 h respectively. Upon BTH treatment, SH3P2-GFP was redistributed to puncta and ring-like structures that overlapped with mCherry-ATG8e in transgenic plants expressing both SH3P2-GFP and mCherry-ATG8e. Quantification of the puncta labeled by SH3P2 or mCherry-ATG8e were obtained from more than 10 individual seedlings (error bars ±SD). (B) Upon BTH treatment, no co-localization of SH3P2-GFP-labeled structures with mCherry-ATG8e was observed in transgenic plants expressing SH3P2Y325A,V328A-GFP and mCherry-ATG8e, suggesting that the SH3P2Y325A,V328A mutation impaired recruitment of SH3P2-GFP to autophagosomes. Quantification of the puncta labeled by SH3P2Y325A,V328A-GFP or mCherry-ATG8e were obtained from more than 10 individual seedlings (error bars ±SD). (C) SH3P2-GFP, and SH3P2Y325A,V328A-GFP seedlings were incubated in medium with/without BTH and Conc A treatment for 6 h respectively. Autophagic bodies upon BTH and Conc A treatment labeled by GFP signals were significantly downregulated in SH3P2Y325A,V328A-GFP transgenic plants. (D) Immunoblot detection of the ATG8 lipidation level in wild-type, SH3P2, SH3P2Y325A,V328A-GFP and atg5 plants. 5-d-old wild type, SH3P2-GFP, SH3P2Y325A,V328A-GFP and atg5 seedlings were incubated in medium with/without BTH treatment for 6 h respectively. Membrane fractions were subjected to immunoblot analysis with ATG8 antibodies. Immunoblotting with cFBPase antibodies was used as a loading control.
The AIM-like motif of SH3P2 is dispensable for SH3P2 trafficking in the endocytic pathway
Of note, we also found that SH3P2 interacts with the CCV-related regulator Auxilin2 (also known as Auxilin-like 2) [51,55] via the SH3 domain in coimmunoprecipitation, yeast-two-hybrid and recruitment assays (Figure S7A-C). Since both Auxilin2 and ATG8 binds to the SH3 domain of SH3P2, we next tested whether the AIM-like motif of SH3P2 plays a role in binding to Auxilin2. Using yeast-two-hybrid assay, we showed that the interaction between SH3P2 and Auxilin2 was not altered by the SH3P2Y325A,V328A mutation (Figure 7A). Moreover, coimmunoprecipitation analysis showed that SH3P2Y325A,V328A was coimmunoprecipitated with Auxilin2 (Figure 7B). Consistently, SH3P2Y325A,V328A-RFP was colocalized with Auxilin2-YFP when transiently expressed in Arabidopsis cells (Figure 7C). Furthermore, the recruitment assay showed that only a portion of SH3P2-CFP was recruited by and colocalized with CNX-RFP-ATG8e, whereas the majority of SH3P2-CFP signals still overlapped with Auxilin2-YFP (Figure S7D).
Figure 7.
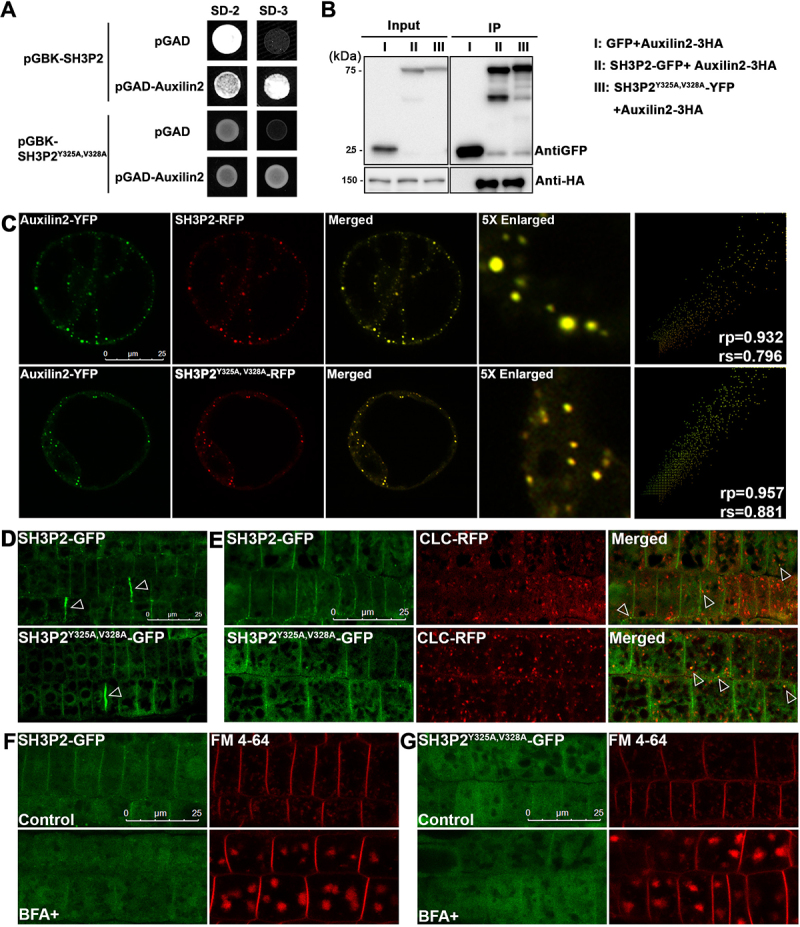
The AIM-like motif of SH3P2 is dispensable for its trafficking in endocytosis. (A) Yeast-two-hybrid assay for analyzing the interaction between SH3P2 and Auxilin2. (B) Immunoprecipitation assay showed Auxilin2-3HA is associated with both SH3P2-GFP and SH3P2Y325A,V328A-YFP. Auxilin2-3HA was co-expressed with SH3P2-GFP and SH3P2Y325A,V328A-YFP in Arabidopsis protoplasts respectively. Lysates were immunoprecipitated by GFP-Trap and detected by anti-GFP or anti-HA antibodies. (C) Subcellular analysis showed that Auxilin2-YFP is colocalized with both SH3P2-RFP and SH3P2Y325A,V328A-RFP in Arabidopsis protoplasts. Similar results were obtained from three different independent experiments. The right column shows the scatterplot images obtained from ImageJ with the PSC colocalization plug-in. The linear Pearson correlation coefficient (rp) and the nonlinear Spearman correlation coefficient (rs) indicate the extent of colocalization with the value of +1.0 for complete colocalization. (D) Both SH3P2-GFP and SH3P2Y325A,V328A-GFP signals were mainly detected in the cytosol and on the plasma membrane, as well as the cell pate forming sites (arrows) in Arabidopsis root cells. (E) Both SH3P2-GFP and SH3P2Y325A,V328A-GFP were associated with the CCV (clathrin-coated vesicle) marker CLC2-RFP in transgenic plants. (F-G) A treatment with FM 4–64 dye or cotreatment with brefeldin A (BFA) showed that uptake of FM 4–64 and overaccumulation of FM 4–64 dye-labeled BFA-bodies were similar in SH3P2-GFP and SH3P2Y325A,V328A-GFP transgenic plants. Consistent results were obtained from three independent experiments.
To further test whether the SH3P2Y325A,V328A mutation will affect the function of SH3P2 in endocytosis, we examined the expression patterns of SH3P2Y325A,V328A-GFP transgenic plants in comparison with that in SH3P2-GFP transgenic plants (Figure 7D). Similar to the wild type SH3P2-GFP, SH3P2Y325A,V328A-GFP displayed a plasma membrane pattern and was recruited to the cell plate forming site (Figure 7D). Moreover, we observed that some puncta labeled by SH3P2Y325A,V328A-GFP were overlapped with the CCV marker CLC2 (clathrin light chain 2)-RFP in double transgenic plants expressing both SH3P2Y325A,V328A-GFP and CLC2-RFP, resembling those observed in the SH3P2-GFP/CLC2-RFP double transgenic plants (Figure 7E). To further analyze the possible effect of SH3P2Y325A,V328A-GFP during endocytosis, we treated the plants with an endocytic dye, FM 4–64, which is taken up into the plant cell from the PM via endocytosis to pass through the trans-Golgi network/early endosome and finally into the vacuole [51]. Brefeldin A (BFA) treatment will interfere with the trafficking from the trans-Golgi network/early endosome to form large “BFA bodies”, but not the endocytic process [56]. Therefore, a combination of BFA and FM 4–64 cotreatment to observe the FM 4–64 labeled “BFA bodies” may inform the endocytosis rate of FM 4–64 from the PM [51]. As shown in Figure 7F,G, FM 4–64 labeled puncta or similar “BFA bodies” were detected in both the SH3P2-GFP and SH3P2Y325A,V328A-GFP transgenic plants, implying that there was no significant change in the rate of endocytosis.
Discussion
Arabidopsis ATG8 interacts canonically with NBR1 but atypically with SH3P2
Here we have reported on the structure of an Arabidopsis ATG8 isoform, ATG8f, in its unbound form and showed how it interacts with the AIM motif of NBR1 (WDPI) by chemical shift perturbation and mutagenesis experiments (Figure 3). Structural comparison suggests that binding of the NBR1 AIM peptide induces conformational changes around the ligand binding site in ATG8f (Figure 3C). In particular, Tyr26 located on the loop between helix-2 and strand-1 swings toward the W-site, and Arg29 is expected to form a charge-charge interaction with Glu667 of the NBR1 AIM peptide (Figure S4). Chemical shift perturbations and mutagenesis results are consistent with the observed conformational changes, showing a canonical mode of interaction between ATG8f and NBR1 (Figures 3 and S4). For example, significant chemical shift changes were observed for Tyr26 and Arg29, whereas substitutions of Tyr26 and Arg29 by Glu (Y26E and R29E) disturbed interaction between ATG8f and NBR1 (Figures 3 and S4).
In contrast, our results revealed that Arabidopsis ATG8 interacts atypically with SH3P2. First, the other two SH3 proteins, SH3P1 and SH3P3 both contain conserved AIM-like motif as SH3P2, but neither interacts with ATG8f (Figures 1B,C and 2A). Second, SH3P1 or SH3P3 with substitution of the SH3P2 AIM-like motif (325YVVV328) was not recruited by CNX-mCherry-ATG8f, but SH3P1-SH3P2 chimera did, suggesting other determinant(s) exists within the SH3 domain of SH3P2 for ATG8f interaction (Figures 1C and 2E). Third, distinct residues on ATG8f (Y26E R29E) for ATG8f and NBR1-UBA interaction dependent on a typical AIM motif have been identified, which however do not compromise SH3P2-ATG8 interaction (Figures. 3E and 5, and S5).
Decision of cargo sequestration and autophagosome formation: a competition mode for ATG8 binding specificity and plasticity between cargo receptor and autophagic adaptor in Arabidopsis
Our results suggest that the typical cargo receptor NBR1 is likely to outcompete SH3P2 via binding to an additional distinct interface on ATG8f in a dosage-dependent manner (Figures 4 and 5). In response to the drastic accumulation of the autophagic substrates upon stress, it is plausible that the competition for ATG8 by the cargo receptors from other ATG8-binding proteins would facilitate cargo sequestration. We identified that the conserved residues in the ATG8 hydrophobic pocket are essential for binding to both NBR1 and SH3P2. Furthermore, our data showed that when there were mutations in the conserved ATG8 docking pocket (Y50A L51A), the NBR1- and SH3P2-ATG8 interactions were both disturbed (Figure 5). In addition to the conserved hydrophobic pocket, we found the ATG8f-NBR1 interaction is significantly weakened when ATG8f carries additional mutations (Y26E R29E) (Figure 5). One possible explanation is that this distinct binding interface would further strengthen the ATG8f-NBR1 interaction, which might serve as a double-check to ensure that NBR1 is sensed by ATG8 and subsequently outcompetes other binding adaptor proteins to release the binding pocket. Another possibility is that when accumulating NBR1 (or substrates) proteins bind to multiple ATG8 interfaces, it might induce a conformational change on ATG8, which subsequently releases SH3P2 from the ATG8 binding pocket.
Because the receptors bind to the autophagic substrates, such receptor-dosage-dependent competition action by the autophagic receptor is likely to serve as a possible fine-tuning mechanism between autophagosome formation and cargo recognition. By doing so, ATG8 plasticity can be achieved in a spatial and temporal manner by switching its binding affinity from autophagic adaptors like SH3P2 to cargo receptors like NBR1. The weaker interaction between ATG8 and SH3P2 is probably important for the creation of a transient regulatory complex for autophagosome formation. However, the conjugation of the ATG8 proteins onto the phagophore membrane is an energy-dependent process [57]. Therefore, when the phagophore has been initiated and receptor-cargo proteins are accumulated, the ability of the receptors to outcompete adaptors probably enables the receptors to be recognized by the existing ATG8 in a “cost-effective” manner, while the adaptor might be released back into the cytosol to assist nascent autophagosome formation. Nevertheless, future efforts are required to investigate whether such a “switch-off’ mechanism functions in plants to balance autophagosome formation and cargo sequestration.
Up till now, only a few non-ATG proteins carrying the canonical AIM motif have been experimentally verified in plants [7–9,32,58–60]. Whether other plant cargo receptors use a similar strategy to compete with the ATG proteins or non-ATG adaptors for ATG8 binding remains unknown. Recently, several studies in plants unveiled that noncanonical AIM motifs have also been adopted to mediate selective autophagy. For example, ubiquitin-interacting motif (UIM) has been reported to mediate the binding between ubiquitin binding protein RPN10 and ATG8 for proteasome degradation [53]. Another study also demonstrated that a reticulophagy receptor c53 contains a shuffle AIM for ATG8 binding [61]. Of note, both UIM and shuffle AIM have also been identified in mammals. In addition, other forms of AIMs have also been reported in various systems, including linear, helical, 3D AIM [62]. Whether these noncanonical AIM motifs also employ a competition action toward ATG8 to outcompete the canonical AIMs would be another interesting question to explore in the future.
ATG8-SH3 domain interaction: a divergence of SH3 domains for plant autophagic membrane scaffold assembly?
Significantly, we found among the three Arabidopsis SH3Ps, only SH3P2 translocates to the phagophore membrane and binds to ATG8 in response to autophagic stimuli in Arabidopsis. We have mapped the minimal SH3P2 binding region for ATG8 from residues 322 to 331 and identified an “AIM-like” sequence (Figure 2B). However, it seems that such an “AIM-like” sequence is highly conserved in both SH3P1 and SH3P3, raising another question that why neither SH3P1 nor SH3P3 interacts with ATG8 as SH3P2 (Figure 2A). Replacing the SH3P2 AIM-like sequence into SH3P1 or SH3P3 did not lead to recruitment of SH3P1 or SH3P3 by ATG8 (Figure 2E). Together with our SH3P1-P2 chimera data (Figure 1C), our results suggest that the divergence of the SH3 domain for binding specificity toward ATG8, might require a coordination of the AIM-like motif and additional unknown element(s) specific to the SH3 domain of SH3P2.
Conversely, it is possible that a yet unknown mechanism within the SH3 domain may safeguard SH3P2 to prevent its binding to ATG8 under normal conditions. Posttranslational modification might play an essential role in regulating the binding strength and specificity to ATG8, such as phosphorylation and ubiquitination. In mammals, the counterpart of SH3P2 belongs to the endophilin protein family, with the consensus BAR domain and SH3 domain, which however does not interact with LC3. Interestingly, it has also been reported that endophilin-A is phosphorylated by the kinase LRRK2, thus leading to an activity switch in membrane remodeling during autophagy and endocytosis [63]. A recent study showed that an E3-ubiquitin ligase FBXO32/atrogin-1 interacts with endophilin-A to regulate its ubiquitination level to participate in autophagosome formation [64]. Whether phosphorylation or ubiquitination level influences SH3P2 binding specificity/affinity for ATG8 during autophagy needs further investigation. Clearly, knowledge of the structural flexibility during autophagy should provide us a better understanding of how the divergence of plant SH3 domain and ATG8 binding specificity is achieved spatially and temporally, as well as its contribution to plant physiological function.
Methods and materials
Plasmid construction
The GFP/YFP/RFP/mCherry/CFP fusion constructs used for transient expression in protoplasts were created by cloning the PCR-amplified cDNA into the pBI221 [45] backbone by restriction digestion, gateway method or by Q5 mutagenesis kit (New England Biolabs, E0554). To generate 221-UBQ-CNX-RFP-ATG8e, the CNX-RFP fragment was PCR-amplified from 221-CNX-RFP [45] and inserted into 221-UBQ-YFP-ATG8e [45] by restriction digestion. To generate 221-UBQ-CNX-mCherry-ATG8f and its variants, CNX-mCherry was firstly PCR-amplified by fusion PCR, and inserted into 221-UBQ-YFP-ATG8e by restriction digestion to create 221-UBQ-CNX-mCherry, followed by the insertion of ATG8f or its various point mutations. The primers for various corresponding constructs are listed in Table S1. All constructs were confirmed by DNA sequencing.
Protein expression and purification
Arabidopsis ATG8f was inserted between the NdeI and BamHI site of the pET-3a expression vector (Novagen, 69,418–3) under the control of the T7 promoter. The expression construct of tag-free ATG8f was transformed into E. coli C41(DE3) strain [65] for protein expression and was induced at OD600 of 0.5–0.7 with 0.4 mM isopropyl beta-D-1-thiogalactopyranoside (IPTG; Gold Biotechnology, I2481 C) for 16 h at 16°C. The cells were harvested and resuspended in lysis buffer (20 mM sodium phosphate, 5 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF; USB Corporation, 20,203], pH 7.4) and lysed by sonication. The lysate was centrifuged at 21,000 g for 45 min, the supernatant was loaded on a 5 mL HiTrap SP HP (GE Healthcare, 17–1152-01) cation exchange chromatography column pre-equilibrated with 20 mM sodium phosphate, 5 mM DTT, pH 7.4 and proteins were eluted at 3 mL/min with a 300 mL linear gradient of 0–500 mM NaCl. Fractions containing ATG8f were pooled, concentrated using a Vivacell 100 with a molecular mass cutoff of 5000 Da (Sartorius, VC1011) to a final volume of 5 mL or 10 mL. Then protein was loaded onto a preparative grade HiLoad 26/60 Superdex 75 gel filtration column (GE Healthcare, 17–1044-01) equilibrated with 1X phosphate-buffered saline (PBS), 5 mM DTT, pH 7.4. Fractions containing the purified ATG8f were concentrated to 4.8 mg/mL, and stored at −80°C. Mutants of tag-free ATG8f protein used for peptide affinity-isolation assay were expressed and purified using the same method.
GST-tagged ATG8a-i were cloned by subcloning ATG8a-i into the pGEX-4T-2 (GE Healthcare, 28–9545-50) or pGEX-6P-1 (GE Healthcare, 28–9546-48) vector using restriction enzyme digestion. Mutants of ATG8f used in the GST affinity-isolation assay were obtained by Q5 mutagenesis kit using GST-ATG8f as the template. The plasmid was transformed into E. coli C41 (DE3) strain for protein production. The cells were induced by 0.4 mM IPTG at OD 0.6 to 0.8 and were harvested after 16 h growth at 16°C. Cells were lysed in lysis buffer (1X PBS, 5 mM DTT, pH 7.4) by sonication. Cell debris was removed by centrifuge at 21,000 g for 45 min. The supernatant was filtered by a 0.45-μm filter before being applied to a 5 mL HiTrap GST FF column (GE Healthcare, 17–5131-02). Protein was eluted by 10 mM reduced glutathione in 1X PBS, 5 mM DTT. Mutants of GST-ATG8f were purified as wild type. Proteins were concentrated to 2 mg/ml, and stored at −80°C.
The SH3 domains of SH3P1, SH3P2, and SH3P3 were cloned by Q5 mutagenesis kit from His-MBP-pp-SH3Ps full-length constructs. Primers used to generate these constructs were listed in Table S1. The obtained His-MBP-pp-SH3P1 (327–439), His-MBP-pp-SH3P2 (279–368) and His-MBP-pp-SH3P3 (268–351) constructs were transformed into E. coli BL21 (DE3) pLysS strains (Novagen, 69,451) for protein production. Protein was expressed by IPTG induction at a final concentration of 0.4 mM for 16 h at 16°C. Cells were collected by centrifugation at 9000 g for 6 min and lysed by the binding buffer (20 mM Tris, 150 mM NaCl, 1 mM Tris [2-carboxyethyl] phosphine hydrochloride [TCEP; Gold Biotechnology, TCEP25], 20 mM imidazole [Sigma, I202], pH 7.4) with 1 mM PMSF. Cell debris was removed by centrifugation at 21,000 g for 45 min. The supernatant was loaded onto a 5 mL HiTrap Ni-chelating column (GE Healthcare, 17–0921-04) pre-equilibrated with binding buffer. The column was washed with the binding buffer for 10 column volume before being eluted with the elution buffer (20 mM Tris, 150 mM NaCl, 1 mM TCEP, 300 mM imidazole, pH 7.4). The eluted proteins were concentrated to 1 mg/mL before being loaded onto a preparative grade HiLoad 26/60 Superdex 200 gel filtration column (GE Healthcare, 17–1043-01) equilibrated with 1XPBS, 5 mM DTT, pH 7.4. Fractions corresponding to His-MBP-pp-SH3Ps-SH3 monomer were pooled and concentrated to no more than 2 mg/mL. Then the protein was cleaved using PreScission protease (GE Healthcare, 27–0843-01) by incubation overnight (1:1000 in molar ratio) at 4°C. The cleaved mixture was loaded onto a preparative grade HiLoad 26/60 Superdex 75 gel filtration column (GE Healthcare, 17–1044-01) equilibrated with 1X PBS, 5 mM DTT, pH 7.4 buffer to remove the His-MBP tag. Fractions corresponding to SH3Ps-SH3 were collected and concentrated to around 1 mg/mL and stored at −80°C.
NBR1-UBA was expressed as a His-tagged protein. The coding sequence corresponding to 619–704 aa of NBR1 was amplified and inserted between the NcoI and BamHI site of the pET-3a vector. E. coli BL21 plysS (DE3) strain containing the plasmid was induced at OD600 of 0.6–0.8 with 0.4 mM IPTG for 16–20 h at 16°C. Cells were harvested and lysed by binding buffer (20 mM Tris, 150 mM NaCl, 1 mM TCEP, 20 mM imidazole, pH 8.0) with 1 mM PMSF. Cell debris was removed by centrifugation and the supernatant was applied to a 5 mL HiTrap Ni-chelating column pre-equilibrated with binding buffer. After washing with 10 column volume of binding buffer, the bound protein was eluted with elution buffer (20 mM Tris, 150 mM NaCl, 1 mM TCEP, 300 mM imidazole, pH 8.0).
GST affinity-isolation assay
To determine whether the single mutations on GST-ATG8f affect its interaction with SH3P2 (279–368), GST-ATG8f (0.5 μM) and its variants were mixed with SH3P2 (279–368) (0.5 μM) in 400 μL binding buffer (1X PBS, 5 mM DTT, pH 7.4). Then, the mixture was added to prewashed glutathione 4B agarose resins (Thermo Fisher Scientific, 16,101) and incubated at 4°C overnight with gentle rotation. The resins were washed three times with the binding buffer before it was eluted by 100 μL reduced glutathione (10 mM) in the binding buffer. Samples pretreated with Instant-Bands (EZBiolab, PFS001P) were analyzed in 16.5% tricine gel and visualized by an LED blue transilluminator.
For the GST affinity-isolation assay between GST-ATG8f double mutants with SH3P2-SH3 and NBR1-UBA, GST-ATG8f wild type or its mutants (10 μM) was mixed with SH3P2-SH3 or NBR1-UBA (10 μM) in 400 μL binding buffer (1X PBS, 5 mM DTT, pH 7.4) respectively, followed by incubating with the prewashed glutathione 4B agarose resins at 4°C for 2 h with gentle rotation. The resins were washed three times with the binding buffer, and then eluted with 100 μL reduced glutathione (10 mM) in binding buffer. Bound proteins were analyzed by SDS-PAGE and stained by Coomassie Brilliant Blue.
To determine the specificity between SH3P2-SH3 and ATG8 isoforms, GST-ATG8a to ATG8i (1 μM) was mixed with SH3P2-SH3 (1 μM) in 500 μL binding buffer (1X PBS, 5 mM DTT, pH 7.4) respectively. Then the protein mixture was added into prewashed glutathione 4B agarose resins and incubated at 4°C for 2 h with gentle rotation. The resins were washed three times with the binding buffer, and then eluted with 100 μL reduced glutathione (10 mM) in the binding buffer. Samples pretreated with Instant-Bands were analyzed in 16.5% tricine gel and visualized by an LED blue transilluminator.
NMR and chemical shift perturbation
For labeled protein samples, cells transformed with the pET-3a-ATG8f plasmid were first grown in King Broth (6 g/L Na2HPO4, 3 g/L KH2PO4, pH 7.0, 5 g/L NaCl, 1 g/L NH4Cl, 20 g/L tryptone [Formedium Ltd, TRP04], 5 g/L yeast extract [Formedium, YEA03], 1 mM MgSO4, 40% glucose) to OD ~ 0.4, the cells were collected and resuspend in M9 minimal medium (12.8 g/L Na2HPO4, 3 g/L KH2PO4, pH 7.0, 0.5 g/L NaCl, 2 mM MgSO4, 1x vitamin mix [4.1 µM biotin, 7.2 µM choline chloride, 4.2 µM calcium pantothenate, 2.3 µM folic acid, 11.1 µM myo-inositol, 8.2 µM nicotinamide, 4.9 µM pyridoxal HCl, 0.3 µM riboflavin, 3.0 µM thiamine HCl], 0.25x metal mix [12.5 µM FeCl3, 5 µM CaCl2, 2.5 µM MnCl2, 2.5 µM ZnSO4, 0.5 µM CoCl2, 0.5 µM CuCl2, 0.5 µM NiCl2]) containing 4 g/L 13C glucose, 1 g/L 15N ammonium chloride [66]. The culture was then incubated at 25°C for 2 h and induced with 0.4 mM IPTG for 16 h at 16°C.
All NMR experiments were carried out using 0.3 mM ATG8f containing 5% (v:v) D2O (Cambridge Isotope, DLM-4-99.8–1000) at 298 K using a Bruker Avance 700 MHz spectrometer. The sequential backbone assignments were obtained from four triple-resonance experiments: HNCACB [67,68], CBCA(CO)NH [69], HNCA [70,71] and HN(CO)CA [70,71]. The side chain resonances were assigned by seven spectra: TOCSY-HSQC [72,73], NOESY-HSQC [73,74], HCCH-COSY [75], HCCH-TOCSY [75,76], H(CC)(CO)NH [77], (H)CC(CO)NH [77], HBHA(CO)NH [68,78]. Stereospecific assignments for the methyl groups of valine and leucine were obtained using a 10% 13C-labeled sample [79]. Inter-proton distance restraint information was obtained from the following NOESY-type experiments: 3D 1H,15N-NOESY-HSQC [73,80], 3D 1H, 13C-NOESY-HSQC [74], 4D 1H,13C-HSQC-NOESY-1H,13C-HSQC [81,82]. All multidimensional NMR spectra were processed with NMRpipe [83] and analyzed by NMRViewJ [84]. Dihedral angle restraints were derived from the TALOS program [85]. Hydrogen bond restraints, derived from hydrogen-deuterium exchange experiments, were included for secondary structure elements. Structural calculation was performed using ARIA 2.3 [86] and CNS 1.2 [87,88] with an initial set of manually assigned nuclear Overhauser effects (NOEs). ARIA-assigned NOEs were checked manually and were included in subsequent rounds of calculation iteratively. Structural models and all the figures were visualized and prepared by MOLMOL or PyMOL. The coordinates for the structure have been deposited in the Protein Databank (http://www.rcsb.org) with the PDB ID code 7DHT.
Chemical perturbation experiments were performed with a 0.3 mM 15N-labeled ATG8f titrating with three times molar excess of the NBR1 AIM peptide. The chemical shift changes in ATG8f were monitored in 1H-15N HSQC spectra. The composite chemical shift changes (∆σ) were calculated using the equation where ∆N and ∆HN are the chemical shift changes of the backbone nitrogen and amide proton respectively.
Peptide affinity-isolation assay
Peptide was coupled to NHS-activated Sepharose resin (GE Healthcare, 17,090,601) according to the manufacturer’s instruction. Peptide (10 μmol) was coupled to 500 μL resin slurry and finally stored in 2 mL 20% ethanol. Resin slurry (20 μL) was used for each peptide affinity-isolation assay. Peptide-coupled resin was equilibrated with the binding buffer (1XPBS [137 mM NaCl, 2.7 mM KCl, 18.9 mM Na2HPO4, 1.8 mM KH2PO4], 5 mM DTT, pH 7.4) before prey protein was added. Peptide-coupled resin and 1 μM protein were mixed and incubated at 4°C for 2 h with gentle rotation. The resin was washed three times by binding buffer before being eluted by 10-fold peptide in binding buffer. Protein loaded to the NHS-activated Sepharose coupled with glycine was included as a negative control.
Analytical gel filtration
Protein mixture (100 μL) of ATG8f (40 µM) with SH3P2-SH3 (40 µM) in 1:1 molar ratio, ATG8f (40 µM) with NBR1-UBA (40 µM) in 1:1 molar ratio, and protein samples of ATG8f (80 µM) with SH3P2-SH3 (40 µM) and NBR1-UBA (40 µM) in 2:1:1 molar ratio, ATG8f (40 µM) with SH3P2-SH3 (40 µM) and NBR1-UBA (40 µM) in 1:1:1 molar ratio were injected into Superdex 75 10/300 GL column (GE Healthcare, 17,517,401) pre-equilibrated with 1XPBS, 5 mM DTT, pH 7.4. The protein samples were incubated for 5 min before being loaded into the column. Fractions were collected and analyzed on SDS-PAGE with Coomassie Brilliant Blue staining.
Yeast-two-hybrid
Yeast-two-hybrid analysis was performed using the MatchMaker GAL4 Two-Hybrid System (Clontech, 630,489) according to the manufacturer’s instructions. The cDNAs were cloned into the pGBKT7 and pGADT7 vectors (Clontech, 630,442 and 630,443). Pairs of pGBKT7 and pGADT7 vectors were co-transformed into the yeast cells. Diploids were selected on SD medium lacking tryptophan and leucine (SD -Trp -Leu), while the selection of yeast cells expressing interacting proteins was made on SD medium lacking histidine, tryptophan and leucine (SD3 -His -Trp -Leu) or (SD4 -Ade -His -Leu -Trp). The experiments (yeast transformation and selection) were repeated at least 3 times independently.
Protein extraction and immunoprecipitation
Protein extraction and immunoprecipitation were performed as described previously [89]. Total cell lysates were prepared in lysis buffer (10 mM Tris/HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 5% glycerol, 0.2% NP-40 [Sigma, I8896] containing complete protease inhibitor Cocktail [Roche, 5,056,489,001]) and were then incubated with GFP-Trap magnetic beads (ChromoTek, gtma) for 4–6 h at 4°C. Samples were washed 5 times in the wash buffer (10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA with complete protease inhibitor Cocktail) and then eluted by boiling in 2X SDS sample buffer. Then, samples were separated by SDS-PAGE and analyzed by western blot using appropriate antibodies. Rabbit ATG8 (AS142769), and cFBPase (AS04043) antibodies were purchased from Agrisera. Rabbit HA (ab9110) and GFP (ab290) were purchased from Abcam.
Transient expression in protoplasts and confocal imaging
Transient expression in Arabidopsis PSBD protoplasts was carried out essentially as described previously [89]. Confocal fluorescence images were acquired 16–18 h after transformation using a Leica SP8 confocal microscope (Leica, Wetzlar, Germany) with a 63× water lens. CFP, GFP/YFP and RFP/mCherry were excited by 408-nm, 488-nm or 561-nm laser, respectively. Images were processed using Adobe Photoshop software (http://www.adobe.com) as previously described [45]. For each experiment or construct, more than 30 individual cells or individual 10 plants were observed for confocal imaging that represented >75% of the samples showing similar expression levels and patterns.
ATG8 lipidation assay
ATG8 lipidation assay was carried out essentially as described previously [89]. 4 or 5-d-old seedlings were transferred in liquid MS with methanol (1:100) as control or 100 μM BTH for 6 h, then extracted in lysis buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1X Complete Protease Inhibitor Cocktail. The total cell extracts were centrifuged at 500 g for 5 min at 4°C. The supernatant was further centrifuged at 100,000 g for 45 min and the membrane pellet was washed 2 times with lysis buffer, then solubilized in an equal volume of lysis buffer with additional 1% Triton X-100 (Sigma, X100). Protein samples were subjected to SDS-PAGE in the presence of 6 M urea and analyzed by immunoblotting with ATG8 antibodies.
Plant materials
Arabidopsis thaliana Columbia-0 seeds were surface sterilized and sown on plates with Murashige and Skoog (MS) salts (Sigma, M5519) plus 0.8% agar. The seeded plates were kept at 4°C for 3 days before being moved to the growth chamber. The plates were incubated at 22°C under a long-day (LD 16 h light and 8 h dark) photoperiod. Plants exposed to LD conditions were transferred to the soil after 2 weeks. 121-UBQ-SH3P2Y325A,V328A-GFP transgenic plants were generated using the flora dip method as previously described [90]. For autophagic induction, 4 or 5-d-old seedlings were transferred in liquid MS with methanol (1:100) as control, or 100 μM BTH (Sigma, 32,820) at least for 5 h prior to observation or as indicated. Conc A (Santa Cruz Biotechnology, sc-202,111 C) was used at a concentration of 0.5 μM. FM 4–64 uptake experiments were performed by incubation with 2 μM FM 4–64 (Invitrogen, T3166). 10 μg/mL BFA (Sigma, B6542) was applied in the medium and incubated for 1 h before observation.
Statistical analyses
The colocalization of two fluorescent signals was quantified using the PSC colocalization plug-in in the ImageJ program and results were presented by scatterplot images as previously described [91]. Quantifications of the puncta were also performed with ImageJ, and statistical results were analyzed and presented using GraphPad Prism 8.
Supplementary Material
Acknowledgments
We thanks to Professor Daniël Van Damme (Ghent University) for sharing the CLC2-RFP seeds.
Funding Statement
This work was supported by Research Grants Council of Hong Kong [14177217, 24108820, N_CUHK405/20, G-CUHK404/18, C4002-17G, C4033-19E, R4005-18F and AoE/M-05/12]; the National Natural Science Foundation of China [91854201]; CUHK Research Committee.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Liu Y, Bassham DC.. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. [DOI] [PubMed] [Google Scholar]
- [2].Soto-Burgos J, Zhuang X, Jiang L, et al. Dynamics of autophagosome formation. Plant Physiol. 2018;176(1):219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhuang X, Chung KP, Luo M, et al. Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci. 2018;23(8):677–692. [DOI] [PubMed] [Google Scholar]
- [4].Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–What’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. [DOI] [PubMed] [Google Scholar]
- [5].Floyd BE, Morriss SC, MacIntosh GC, et al. What to eat: evidence for selective autophagy in plants. J Integr Plant Biol. 2012;54(11):907–920 [DOI] [PubMed] [Google Scholar]
- [6].Michaeli S, Clavel M, Lechner E, et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci U S A. 2019;116(45):22872–22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vanhee C, Zapotoczny G, Masquelier D, et al. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23(2):785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nolan TM, Brennan B, Yang M, et al. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017;41(1):33–46 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang X, Ding X, Marshall RS, et al. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. Elife, 2020; 9: e51918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu ML, Yao MC.. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot Cell. 2012;11(4):494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hu S, Ye H, Cui Y, et al. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J Integr Plant Biol. 2020;62(2):181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ji C, Zhou J, Guo R, et al. AtNBR1 is a selective autophagic receptor for AtExo70E2 in Arabidopsis. Plant Physiol. 2020;184(2):777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bao Y, Song W-M, Wang P, et al. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020;117(13):7482–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stephani M, Picchianti L, Gajic A, et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 2020;9. DOI: 10.7554/eLife.58396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haxim Y, Ismayil A, Jia Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017;6. DOI: 10.7554/eLife.23897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y, Yu B, Zhao J, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013;25(4):1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nakatogawa H, Ichimura Y, Ohsumi Y, et al. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. [DOI] [PubMed] [Google Scholar]
- [18].Xie Z, Nair U, Klionsky DJ, et al. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu Z-Q, Ni T, Hong B, et al. Dual roles of Atg8− PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Birgisdottir ÅB, Lamark T, Johansen T, et al. The LIR motif–crucial for selective autophagy. J Cell Sci. 2013;126(15):3237–3247. [DOI] [PubMed] [Google Scholar]
- [21].Kriegenburg F, Ungermann C, Reggiori F, et al. Coordination of autophagosome–lysosome fusion by ATG8 family members. Curr Biol. 2018;28(8):R512–R518. [DOI] [PubMed] [Google Scholar]
- [22].Alemu EA, Lamark T, Torgersen KM, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287(47):39275–39290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Birgisdottir ÅB, Mouilleron S, Bhujabal Z, et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15(8):1333–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci, 2014; 127(1): 3–9 [DOI] [PubMed] [Google Scholar]
- [25].Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13(12):1211–1218. [DOI] [PubMed] [Google Scholar]
- [26].Ichimura Y, Kumanomidou T, Sou Y-S, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283(33):22847–22857. [DOI] [PubMed] [Google Scholar]
- [27].Cheng X, Wang Y, Gong Y, et al. Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins. Autophagy. 2016;12(8):1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu F, Watanabe Y, Guo X-Y, et al. Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol Cell. 2015;60(6):914–929. [DOI] [PubMed] [Google Scholar]
- [29].Rogov VV, Stolz A, Ravichandran AC, et al. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep. 2017;18(8):1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432(1):80–103. [DOI] [PubMed] [Google Scholar]
- [31].Kellner R, De La Concepcion JC, Maqbool A, et al. ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 2017;22(3):204–214. [DOI] [PubMed] [Google Scholar]
- [32].Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7(9):993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jung H, Lee HN, Marshall RS, et al. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J Exp Bot. 2020;71(1):73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tarnowski L, Rodriguez MC, Brzywczy J, et al. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci Rep. 2020;10(1):7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thirumalaikumar VP, Gorka M, Schulz K, et al. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy. 2020:1–16. DOI: 10.1080/15548627.2020.1820778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maqbool A, Hughes RK, Dagdas YF, et al. Structural basis of host autophagy-related protein 8 (ATG8) binding by the irish potato famine pathogen effector protein PexRD54. J Biol Chem. 2016;291(38):20270–20282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nakatogawa H, Ohbayashi S, Sakoh-Nakatogawa M, et al. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J Biol Chem. 2012;287(34):28503–28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kraft C, Kijanska M, Kalie E, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin‐like protein Atg8 regulates autophagy. EMBO J. 2012;31(18):3691–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hain AU, Weltzer RR, Hammond H, et al. Structural characterization and inhibition of the plasmodium Atg8–Atg3 interaction. J Struct Biol. 2012;180(3):551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hong SB, Kim B-W, Lee K-E, et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol. 2011;18(12):1323. [DOI] [PubMed] [Google Scholar]
- [42].Noda NN, Satoo K, Fujioka Y, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44(3):462–475. [DOI] [PubMed] [Google Scholar]
- [43].Satoo K, Noda NN, Kumeta H, et al. The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28(9):1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Suzuki H, Tabata K, Morita E, et al. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure. 2014;22(1):47–58. [DOI] [PubMed] [Google Scholar]
- [45].Zhuang X, Wang H, Lam SK, et al. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25(11):4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhuang X, Jiang L. Autophagosome biogenesis in plants: roles of SH3P2. Autophagy. 2014;10(4):704–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lam BCH, Sage TL, Bianchi F, et al. Role of SH3 domain-containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell. 2001;13(11):2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ahn G, Kim H, Kim DH, et al. SH3 domain-containing protein 2 plays a crucial role at the step of membrane tubulation during cell plate formation. Plant Cell. 2017;29(6):1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nagel MK, Kalinowska K, Vogel K, et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci U S A. 2017;114(34):E7197–E7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gao C, Zhuang X, Cui Y, et al. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci U S A. 2015;112(6):1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Adamowski M, Narasimhan M, Kania U, et al. A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell. 2018;30(3):700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Niemes S, Labs M, Scheuring D, et al. Sorting of plant vacuolar proteins is initiated in the ER. Plant J. 2010;62(4):601–614. [DOI] [PubMed] [Google Scholar]
- [53].Marshall RS, Hua Z, Mali S, et al. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177(3):766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nim YS, Sun S, Wong KB. Using Homology Modeling to Understand the Structural Basis of Specific Interaction of a Plant-Specific AtSar1a-AtSec23a Pair Involved in Protein ER Export. Methods Mol Biol. 2017; 1662:59–73 [DOI] [PubMed] [Google Scholar]
- [55].Ortiz-Morea FA, Savatin DV, Dejonghe W, et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci U S A. 2016;113(39):11028–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lam SK, Cai Y, Tse YC, et al. BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J. 2009;60(5):865–881. [DOI] [PubMed] [Google Scholar]
- [57].Martens S, Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou J, Wang Z, Wang X, et al. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14(3):487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. :agdas YF, Belhaj K, Maqbool A, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife. 2016; 5:e10856.. doi: 10.7554/eLife.10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Honig A, Avin-Wittenberg T, Ufaz S, et al. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of arabidopsis plants to carbon starvation. Plant Cell. 2012;24(1):288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stephani M, Picchianti L, Gajic A, et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 2020; 9: e58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wesch N, Kirkin V, Rogov VV, et al. Atg8-family proteins—Structural features and molecular interactions in autophagy and beyond. Cells. 2020;9(9):2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Soukup SF, Kuenen S, Vanhauwaert R, et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 2016; 92(4):829-844. [DOI] [PubMed] [Google Scholar]
- [64].Murdoch JD, Rostosky C, Gowrisankaran S, et al. Endophilin-A deficiency induces the Foxo3a-Fbxo32 network in the brain and causes dysregulation of autophagy and the ubiquitin-proteasome system. Cell Rep. 2016;17(4):1071–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260(3):289–298. [DOI] [PubMed] [Google Scholar]
- [66].Murray V, Huang Y, Chen J, et al. A novel bacterial expression method with optimized parameters for very high yield production of triple-labeled proteins. Methods Mol Biol. 2012; 831:1–18 [DOI] [PubMed] [Google Scholar]
- [67].Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-and beta-carbon resonances in proteins. J Magn Reson Ser B. 1993;101(2):201–205. [Google Scholar]
- [68].Muhandiram D, Kay LE, Haxim Y, et al. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson Ser B. 1994;103(3):203–216. [Google Scholar]
- [69].Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114(16):6291–6293. [Google Scholar]
- [70].Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson (1969). 1992;96(2):432–440. [Google Scholar]
- [71].Kay LE, Xu GY, Yamazaki T, et al. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson Ser A. 1994;109(1):129–133. [Google Scholar]
- [72].Schleucher J, Schwendinger M, Sattler M, et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. 1994;4(2):301–306. [DOI] [PubMed] [Google Scholar]
- [73].Marion D, Driscoll PC, Kay LE, et al. Overcoming the overlap problem in the assignment of proton NMR spectra of larger proteins by use of three-dimensional heteronuclear proton-nitrogen-15 Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1. beta. Biochemistry. 1989;28(15):6150–6156 [DOI] [PubMed] [Google Scholar]
- [74].Muhandiram D, Farrow NA, Xu GY, et al. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O. J Magn Reson Ser B. 1993;102(3):317–321. [Google Scholar]
- [75].Kay LE, Xu GY, Singer AU, et al. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples for proteins. J Magn Reson Ser B. 1993;101(3):333–337. [Google Scholar]
- [76].Bax A, Clore GM, Gronenborn AM, et al. 1H 1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J Magn Reson (1969). 1990;88(2):425–431. [Google Scholar]
- [77].Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J Magn Reson Ser B. 1993;101(1):114–119 [Google Scholar]
- [78].Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13 C/15 N-enriched proteins. J Biomol NMR. 1993;3(2):185–204. [DOI] [PubMed] [Google Scholar]
- [79].Szyperski T, Neri D, Leiting B, et al. Support of 1 H NMR assignments in proteins by biosynthetically directed fractional 13 C-labeling. J Biomol NMR. 1992;2(4):323–334. [DOI] [PubMed] [Google Scholar]
- [80].Wider G, Neri D, Otting G, et al. A heteronuclear three-dimensional NMR experiment for measurements of small heteronuclear coupling constants in biological macromolecules. J Magn Reson. 1989;85(2):426–431 [Google Scholar]
- [81].Palmer III AG, Cavanagh J, Wright PE, et al. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson (1969). 1991;93(1):151–170. [Google Scholar]
- [82].Kay L, Keifer P, Saarinen T, et al. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114(26):10663–10665. [Google Scholar]
- [83].Delaglio F, Grzesiek S, Vuister G, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. [DOI] [PubMed] [Google Scholar]
- [84].Johnson BA, Blevins RA. NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. [DOI] [PubMed] [Google Scholar]
- [85].Cornilescu G, Delaglio F, Bax A, et al. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. [DOI] [PubMed] [Google Scholar]
- [86].Linge JP, O'Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90 [DOI] [PubMed] [Google Scholar]
- [87].Brünger AT, Adams PD, Clore GM, et al.Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D: Biol Crystallogr. 1998;54(5):905–921 [DOI] [PubMed] [Google Scholar]
- [88].Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728. [DOI] [PubMed] [Google Scholar]
- [89].Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114(3):E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhang X, Henriques R, Lin SS, et al. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1(2):641–646 [DOI] [PubMed] [Google Scholar]
- [91].French AP, Mills S, Swarup R, et al. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3(4):619–628 [DOI] [PubMed] [Google Scholar]
- [92].Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


