Figure 1.
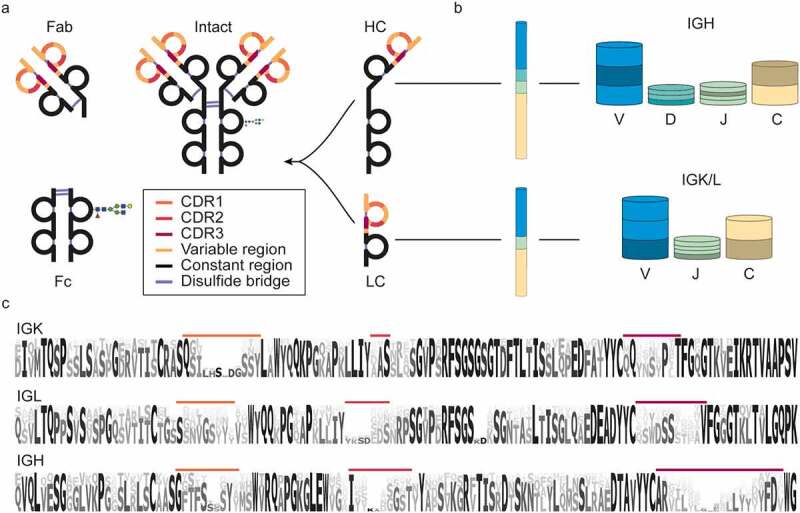
Nomenclature, structure, and diversity of IgG1 antibodies. (a) Nomenclature and protein fragments of an IgG1 molecule. The antigen-binding domain, containing light and heavy chain (LC and HC respectively) variable regions, is termed Fab (or Fab2 when dimerized). The constant part of the heavy chain carrying an N-glycosylation site is called Fc. Other IgG subclasses vary in their heavy chain constant region (Fc) and disulfide patterns. (b) The diversity in antibodies originates primarily from the V, D, J, and C-allele (each annotated with a distinct color) recombination process. In this process, each of many individual V, D, J, and C-alleles can recombine with any of the other gene segments, yielding thousands of possible combinations, in particular for the heavy chain, which incorporates the most diverse D region. (c) Sequence logo created by the alignment of in silico generated sequences of Ig kappa (IGK) and lambda (IGL) light chains and Ig heavy chain (IGH) from the international ImMunoGeneTics (IMGT) information system database.20,21 Even though the displayed sequences are part of the variable domain, large stretches of these sequences, also known as the framework regions (FRs), are relatively conserved, compared to the hypervariable complementarity determining regions (CDRs), colored in accordance with (a).
