Figure 2.
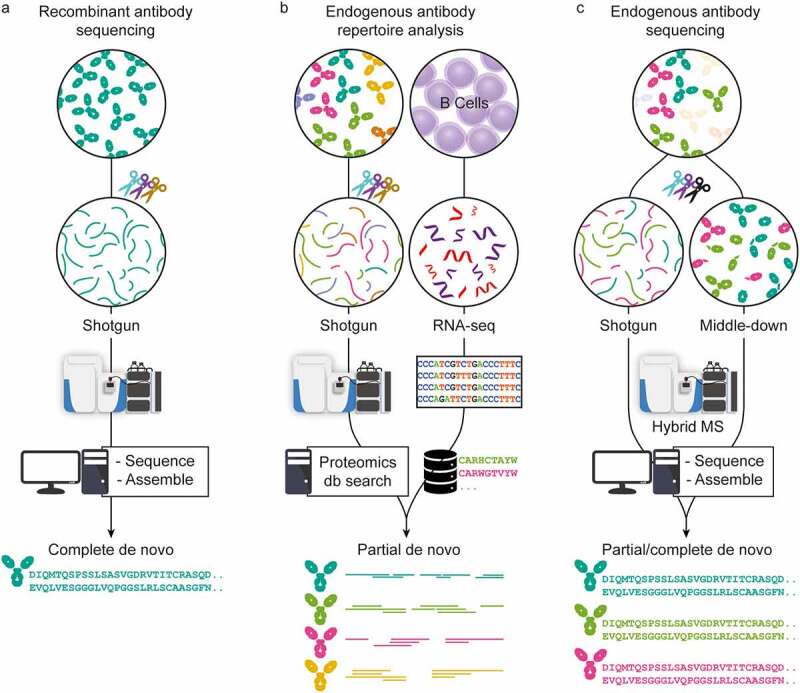
Three approaches in MS-based antibody sequencing. (a) Recombinant antibody sequencing generally starts with abundant highly purified mAbs, which can be fully sequenced through BU MS, where hundreds of peptides are generated by digesting the mAb with one or several proteases, providing multiple overlapping short sequence reads. After liquid chromatography-mass spectrometry (LC-MS) measurement, the spectra can be processed by several different de novo sequencing software solutions and assembled into full-length mAb sequences.16,17 (b) In repertoire analysis, a sequence database is generated through B-cell sequencing, and MS-data is obtained through BU MS experiments. After generation of the personalized database, a high throughput of the LC-MS is possible.44 While not strictly de novo since only hits from the sequence database are identified, it is a powerful tool for antibody repertoire analysis. (c) Endogenous antibody sequencing cannot rely on BU MS alone, as direct sequencing of endogenous humoral antibodies is hampered by inherent challenges and complexity. Emerging MD and TD MS techniques provide clone-specific sequence information highly complementary to traditional sequencing. Integrating BU MS and MD/TD MS makes it possible to achieve full-length coverage of antibody sequences.31
