Abstract
We sequenced the complete mitochondrial genomes of one Iolania perkinsi (Kirkaldy 1902) and one Oliarus cf. filicicola (Kirkaldy 1909) planthopper (Hemiptera: Fulgoroidea: Cixiidae) from Volcano Village, located on the eastern flank of Mauna Loa. The I. perkinsi complete mitogenome is 14,949 bp in length and contains 13 protein-coding genes, 22 tRNAs and 2 rRNAs. The O. cf. filicicola nearly complete mitogenome is 15,196 bp in length due to an expanded (but incomplete) control region, and contains all of the typical metazoan genes: 13 protein-coding genes, 22 tRNAs and two rRNAs. Relative to the inferred ancestral gene order of insect mitochondrial genomes, no rearrangements were identified in either species. In addition to the shorter control region in I. perkinsi, the differences between the two mitogenomes consist of longer cox1, cox2, cob, nad1, nad5, nad6 genes but shorter nad4 gene in I. perkinsi relative to O. cf. filicicola. These mitogenomes represent the first sequences from species within the Hawaiian Cixiidae. The sequence data here will provide a foundation for continued studies of speciation patterns and dynamics of evolutionary radiation across Hawaiian planthoppers.
Keywords: Planthopper, mitogenomes, Cixiidae, Iolania, Oliarus
Within the family Cixiidae, there have been only two genera – Iolania and Oliarus – that have colonized the Hawaiian archipelago. While these independent colonizations have led to radiations across the island chain, the degree of speciation differs drastically between them. Although in Oliarus ∼58 species have been described, with radiations of multiple species on each Hawaiian island (Zimmerman 1948), in Iolania only six endemic species are recognized with each Hawaiian island generally harboring only a single-island endemic (Hoch 2005). These drastic differences in speciation patterns lead to questions regarding what is driving the observed differences, and more specifically why the Iolania lineage is so species poor. To begin to build genomic resources to address these questions, here we present the first mitogenome sequences for Hawaiian Cixiidae species - one complete mitogenome from Iolania perkinsi Kirkaldy 1902, and a second nearly complete mitogenome from Oliarus cf. filicicola Kirkaldy, 1909. While both of these species are widely distributed on Hawaiʻi Island and generally associated with rainforest habitats (Zimmerman 1948; Hoch 2005), there are ∼13 species of Oliarus described from the island while I. perkinsi is a single island endemic. These mitogenomes serves as an important step toward understanding the distinct patterns of diversification within each genus across the Hawaiian archipelago.
Samples of Iolania perkinsi and Oliarus cf. filicicola were collected from Volcano, Hawaiʻi [19.44128 − 155.23535] in 2019 under Hawaiʻi State Permit #I1063. For each species, total genomic DNA was extracted from an entire individual using a Qiagen DNEasy blood and tissue kit. The resulting DNA vouchers were deposited in the Hawaiʻi Cave Arthropod Collection located in the Porter Lab (Megan Porter, mlporter@hawaii.edu) at the University of Hawaiʻi at Mānoa (accession number HI01086 and HI01081 for I. perkinsi and O. cf. filicicola, respectively). Genomic libraries were prepared using NEB Next® Ultra II DNA Library Prep Kit with average insert sizes of approximately 500 bp, and were sequenced using Illumina Technology at NovoGene Corporation (New Jersey, USA). The generated reads were filtered, adapters were trimmed, and reads were assembled using NOVOplasty v4.3.1 (Dierckxsens et al. 2016). A complete mitogenome for I. perkinsi (GenBank: MZ748292) and a nearly complete mitogenome for O. cf. filicicola (GenBank: MZ748293) were generated and genes were annotated with the MITOS2 server (Bernt et al. 2013).
The complete mitogenome of I. perkinsi was 14,949 bp in length with a GC content of 21.3% and contained 13 protein-coding, 22 tRNAs, and 2 rRNA (12S and 16S) genes. The nearly complete mitogenome of O. cf. filicicola was 15,196 bp in length with a GC content of 21.0%, and also contained the same 37 genes. Gene content and gene order for both genomes is consistent with the predicted ancestral insect mitochondrial genome (Cameron 2014). Nucleotide composition in both genomes was AT-biased, with GC content in other fulgoroid mitogenomes ranging from 19.4 to 25.4%. Despite being nearly complete, the O. cf. filicicola mitogenome was larger than the complete I. perkinsi mitogenome because of an expansion in the incomplete control region. In addition to this difference in the control region, there were also differences in gene length between the two mitogenomes, with I. perkinsi having longer cox1, cox2, cob, nad1, nad5, and nad6 genes, and O. cf. filicicola having a longer nad4 gene. Finally, I. perkinsi had a complete stop codon (TAA) at the end of the atp-6 gene, whereas the O. cf. filicicola atp6 gene is most likely completed by polyadenylation of the transcript.
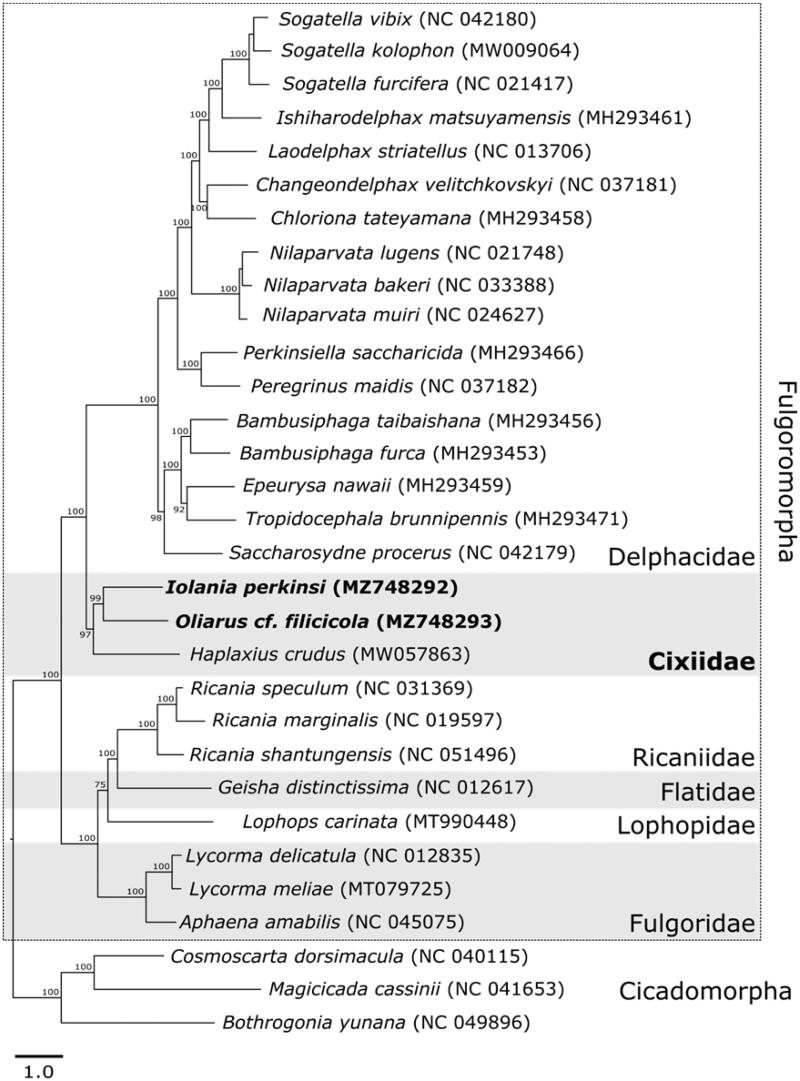
Representative species from the infraorder Flugoromorpha with complete mitochondrial genome sequences were obtained from GenBank for phylogenetic analysis; the mitogenome sequences from three Cicadomorpha representatives were used as outgroup taxa (Figure 1). All 13 protein-coding genes were extracted and aligned using Geneious® v10.2.6 with MAFFT v7.450 (Katoh and Standley 2013) to infer the phylogenetic placement of I. perkinsi and O. cf. filicicola. We used PartitionFinder2 (Stamatakis 2006; Lanfear et al. 2014, 2017) to identify the best partitioning scheme and nucleotide substitution models. We estimated a maximum-likelihood phylogeny using RAxML (Stamatakis 2006) based on the partitioned dataset with 1000 bootstrap replicates using the GTRGAMMAI substitution model for each partition. The phylogenetic tree suggests Iolania and Oliarus are sister; however, there is limited taxonomic representation to infer phylogenetic relationships within the family (Figure 1). There is phylogenetic support for Cixiidae being sister to the family Delphacidae (Figure 1).
Figure 1.
Maximum Likelihood phylogeny for Iolania perkinsi and Oliarus cf. filicicola based on 13 mitochondrial protein-coding genes generated using RAxML with a GTRGAMMAI substitution model. Numbers along branches indicate bootstrap support percentages. GeneBank accession numbers are given after species names, sequences added from this study are in bold. Members from both infraorders of suborder Auchenorrhyncha are included: three cicadas (infraorder: Cicadomorpha) were used as an outgroup, and family name is provided for planthoppers (infraorder: Fulgoromorpha).
As the first sequenced Cixiidae from Hawaiʻi, the I. perkinsi and O. cf. filicicola mitogenomes provide a foundation for continued studies of genetic divergence among island endemics. This is also the first step toward understanding the species-poor radiation patterns of Iolania species relative to the more speciose Oliarus group across the Hawaiian archipelago.
Author contributions
RAC and MLP contributed to the conception and design; RAC and MS contributed to analysis and interpretation of the data; RAC, MLP, and MS contributed to the drafting of the paper, revising it critically for intellectual content, and the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Funding Statement
This work was supported by the HICAVEs research group; the Cave Conservancy Foundation under [Grant HI1819]; the National Science Foundation under [grant DEB1556819] and DEB2204670; and XSEDE [MCB200211]. This is publication #XX from the School of Life Sciences, University of Hawaiʻi at Mānoa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ748292 and MZ748293. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA772291, SAMN22374426 and SAMN22374427, and SRX13212179 and SRX13212180 respectively.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):1015–319. [DOI] [PubMed] [Google Scholar]
- Cameron S. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch H. 2005. Systematics and evolution of Iolania (Hemiptera: Fulgoromorpha: Cixiidae) from Hawai'i. Systemat Entomol. 31(2):302–320. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A.. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Zimmerman EC. 1948. Homoptera: Auchenorrhyncha. In: Insects of Hawaii. Vol. 4. Honolulu: University of Hawai'i Press; p. 1–268. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ748292 and MZ748293. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA772291, SAMN22374426 and SAMN22374427, and SRX13212179 and SRX13212180 respectively.