Abstract
HIV-1 pol gene sequences were analyzed from 77 HIV-1 positive children infected perinatally and exhibiting virological failure (VF). Viral subtyping, phylogenetic analysis, and genotypic drug resistance analysis were carried out on samples collected before start of anti retroviral treatment (ART) (baseline, BL), and at 12 months post-ART initiation (M12). Subtype C was found to be most predominant, seen in 75 of the 77 (97.4%) children. The level of pretreatment drug resistance (PDR) was 14% among these children. At BL, K103N (5), E138A/G (4), and M184V (3) were the most common mutations. At M12 the prevalence of any resistance-associated mutation (RAM) (acquired drug resistance/ADR) was 81.8% (63/77). Dual class resistance mutations were seen in 64% (49/77) of children. M184V/I, K103N/S, and Y181C were the most commonly occurring mutations, seen in 76%, 51%, and 36% children. RAMs to the second-generation non-nucleoside reverse transcriptase inhibitors (NNRTI), etravirine (ETR) and rilpivirine (RPV), were seen in 40.2% (31/77) and 48.05% (37/77) of the children, respectively. Our findings reveal similar prevalence rates of PDR and ADR in children with VF as reported in other studies. Occurrence of ETR and RPV resistance associated mutations (RAMs) is of concern and highlights the need for timely switch of regimens guided by genotypic resistance testing in perinatally infected children from India.
Keywords: vertical transmission, HIV-1 pol gene, HIV-1 drug resistance, South India, virological failure, HIV-1 subtype C
According to UNICEF reports, children belonging to the age group 0–19 years constituted 2.78 million of the total 38 million individuals living with HIV in 2020 and there were 880 new infections and 310 deaths among children each day. The total number of new infections in children with HIV in the age group between 0 and 9 years was 160,000 (100,000–240,000), taking the total to 1.03 million (730,000–1.29 million) in children of this age group. Despite these grim statistics, a slow decline of ∼34% and a rapid decline of ∼52% in new HIV cases in the adolescent age group (10–19 years) and pediatric group (0–9 years), respectively, has been observed since 2010, owing to the prevention of mother-to-child transmission (PMTCT) strategies.1
As per the 2016 National AIDS Control Organization (NACO) data, the estimated new HIV infections in children that occurred between 1998 and 2015 in India were 200,000.2 In the year 2017 alone, out of the total 88,000 new HIV-infected cases, 34,000 infections occurred among women and 3,700 infections were reported among children aged between 0 and 14 years.3 During the past decade there has been rapid scale up of prevention of parent-to-child transmission (PPTCT) services in the country. As on September 2019, there were >30,000 HIV counseling and testing centers in the country offering PPTCT services. In addition, the quality of life in people living with HIV has been largely improved by expansion of the National anti retroviral treatment (ART) program.
According to the Ministry of Health and Family Welfare's annual report (2019–2020), 118.4 lakh pregnant women were screened for HIV during the year 2019–2020 (up to September), with 8,949 HIV-positive cases reported (5,906 new cases and 3,043 known cases); 85% (7,570) of these individuals were on lifelong antiretroviral therapy. Around 6,165 HIV-exposed live births were reported during the same time span, with 5,358 (87%) babies receiving antiretroviral drug (ARV) prophylaxis.4
Despite these efforts and successes, the emergence and spread of drug-resistant viral variants pose a challenge to the country's long-term success and viability of combination ART. There are very limited data on virological failure (VF) and drug resistance among perinatally infected children from India. This study aimed to characterize the HIV-1 pol gene sequences in a group of perinatally infected children exhibiting VF after 12 months on cART.
Molecular analysis of the HIV-1 pol gene sequences was performed using established phylogenetic and bioinformatics methods. The study used stored plasma samples from perinatally infected children who participated in the HIV-Associated Lipodystrophy Syndrome (HALS) study conducted at the National Institute for Research in Tuberculosis in Chennai and St. John's Medical College Hospital in Bangalore, India, between 2010 and 2015.5
This study reported immunological failure among 11% (28/261) and VF among 29% (94/328) of children after 12 months of first-line ART.5 Of the 94 children with VF, partial HIV-1 pol gene sequences were available for 77 children at two time points—(1) pre-ART initiation/baseline (BL) and (2) 12 months post-ART initiation (M12). Viral subtyping, phylogenetic analysis (PA) and genotypic drug resistance analysis were carried out in this subset of 77 children at BL and M12. In brief, viral RNA extraction was carried out from plasma using the Qiagen viral RNA MiniAmp kit (Qiagen, Valencia, CA).
In the initial step, the sample was centrifuged briefly at 14,000 RPM to concentrate the virus. All the remaining steps were carried out as per manufacturer's recommendations. The HIV-1 pol region comprising the first 297 nucleotides of the protease (PR) gene and first 260 codons of the reverse transcriptase (RT) gene were amplified and sequenced bidirectionally using the primers described in Table 1. Seqscape software v2.6 (Applied Biosystems) was used for assembling the sequences with the reference genome and for mutation detection analysis. All samples and sequences were coded using unique identifiers as part of data anonymization.
Table 1.
List of Primers Used for the Amplification and Sequencing of HIV-1 Pol Gene
| Primer ID | Sequence 5′→3′ | Direction/comments | Target region |
|---|---|---|---|
| 5′ Prot1 | TAA TTT TTT AGG GAA GAT CTG GCC TTC C | Sense/outer primers | PR |
| 3′ Prot1 | GCA AAT ACT GGA GTA TTG TAT GGA TTT TCA GG | Antisense/outer primers | PR |
| 5′ Prot2 | TCA GAG CAG ACC AGA GCC AAC AGC CCC A | Sense/inner and sequencing primers | PR |
| 3′ Prot2 | AAT GCT TTT ATT TTT TCT TCT GTC AAT GGC | Antisense/inner and sequencing primers | PR |
| MJ3 | AGT AGG ACC TAC ACC TGT CA | Sense/outer primers | RT |
| MJ4 | CTG TTA GTG CTT TGG TTC CTC T | Antisense/outer primers | RT |
| A35 | TTG GTT GCA CTT TAA ATT TTC CCA TTA GTC CTA TT | Sense/inner and sequencing primers | RT |
| NE135 | CCT ACT AAC TTC TGT ATG TCA TTG ACA GTC CAG CT | Antisense/inner and sequencing primers | RT |
PR, protease; RT, reverse transcriptase.
After sequence assembly and editing, individual sequence files were exported in FASTA format and the sequences were aligned with the Gene cutter tool available in the HIV Los Alamos database. The PR and RT regions of the individual sequences were extracted using the gene cutter protein extraction option. For subtyping of the isolates, the extracted PR and RT sequences were analyzed separately using COMET (https://comet.lih.lu/),6 REGA version 3 online subtyping tool (www.bioafrica.net/rega-genotype/html/subtypinghiv.html),7 and Stanford University HIV Drug Resistance Database (http://hivdb.stanford).8 PA was carried out using the IQ tree web server (CIBIV, Austria).
The model finder implemented in the server9 identified maximum likelihood (ML) as the best fit model for the sequence data, and the aligned-trimmed PR and RT sequences were submitted separately for PA with standard bootstrap values (100 replicates). All HIV-1 group M subtype sequences were downloaded from the Los Alamos database (accessed on December 2020) for phylogenetic tree construction. Resistance mutations were interpreted using the genotypic resistance interpretation algorithm of the Stanford HIV database program, V7.0.10 Among the total pool of 116 PR sequences and 149 RT sequences available for the study, 62 and 72 belonged to BL, and 54 and 77 belonged to M12 time point for PR and RT, respectively.
Consistent with the epidemiology of the HIV epidemic in India, we found a predominance of HIV-1 subtype C (75 out of 77; 97.4%) among the study participants. The remaining two children (2.59%) harbored subtype A virus. The overall concordance between the three bioinformatics tools used for subtype prediction was good; in case of discrepancy, subtype was assigned based on the majority prediction.
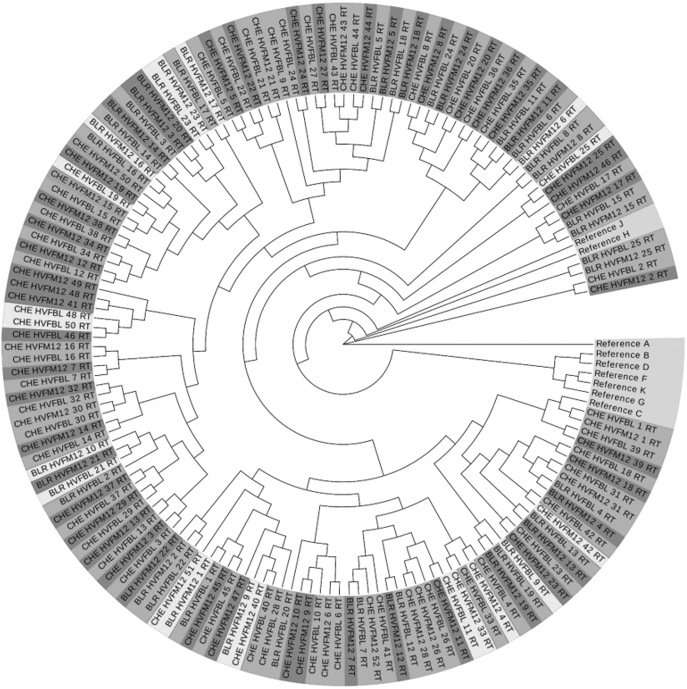
The ML phylogenetic tree constructed using the IQ-tree server based on the trimmed and aligned PR and RT sequences showed that all the subtype C study isolates clustered tightly with the Indian subtype C reference sequence. Tree topology showed a subtype specific pattern. However, visual clustering of viral isolates with similar drug resistance patterns such as resistant variants to single, dual, or triple class of ARVs was not seen. Clustering was apparent in the sequence data sets derived from same participant at different time points (Fig. 1).
FIG. 1.
Molecular phylogenetic analysis of the reverse transcriptase sequences of the 77 vertically transmitted viruses (during suppressive ART and at the time of viral rebound) with HIV-1 group-M subtype-specific reference sequences downloaded from the Los Alamos database. The phylogenetic tree was constructed from nucleotide alignments using maximum likelihood method based on the TPM3+F+I+G4 using the model finder algorithm implemented in the IQ tree server. Number of bootstrap replications was set at 100. ART, anti retroviral treatment.
Recent findings indicate that inadequate suppression of the virus and accumulation of drug resistance mutations (DRMs) are linked to the increasing trend in the prevalence of pretreatment HIV drug resistance in developing and underdeveloped countries.11,12 Children who receive ARVs for preventive therapy in the PMTCT program are particularly vulnerable to development of drug resistance. Efavirenz (EFV)-based treatment was the most widely utilized ART regimen in children over the age of 3 years in India during the study period. More recent guidelines, however, recommend a dolutegravir (DTG)-based regimen along with two nucleoside reverse transcriptase inhibitors (NRTIs) in children aged 6 years and older.
In this study, we analyzed 62 available PR and 72 RT sequences of HIV-infected children sampled before initiation of ART for estimating the level of PDR. Retrospective DRM testing at BL revealed that 10 out of 72 children had sequences carrying DRMs resulting in a PDR level of 14% in the study population; 7 (9.7%) children had non-nucleoside reverse transcriptase inhibitors (NNRTI) resistance, and 2 (2.77%) had NRTI resistance. One child had dual class resistance. The most common NNRTI mutations were K103N,5 E138A/G,4 and K101E1 found in study participants with any NNRTI resistance. M184V was present in 4% (3/72) of the children.
The rates of PDR observed in our study was comparable with that reported in Ugandan HIV-infected children (16.9%)13 and Ethiopian children (14%).14 Another study reported 20% PDR in an urban cohort of Ugandan children15 who did not exhibit VF.
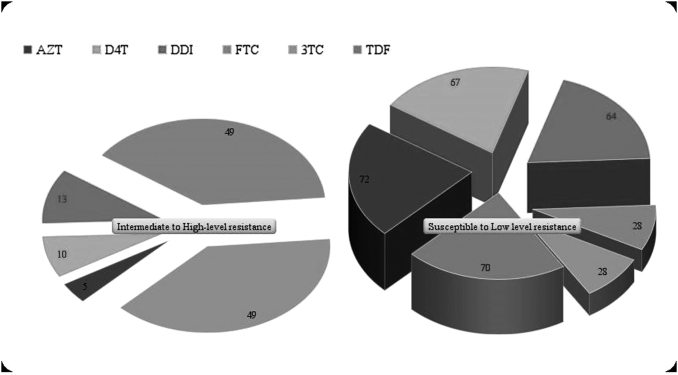
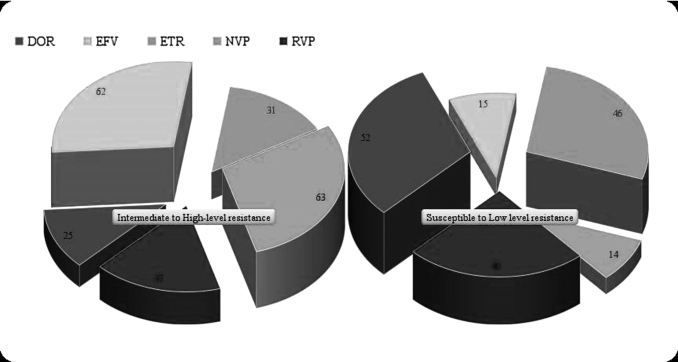
We also determined the level of acquired drug resistance (ADR) in our cohort. Figures 2–4 depict predicted drug resistance/susceptibility of the children to individual NNRTI and NRTI drugs. At M12, 18.2% (14/77) of children with VF had no DRM. The remaining 81.8% (62/77) children had RAMs or acquired drug resistance (ADR), which is similar to the findings in other such studies.13,16 Dual class resistance mutations were seen in 64% (49/77) of the children, and triple class resistance mutations were seen in 2 of the 77 children. The most common NRTI mutation among the children with DRMs was M184V/I, which was present in 76% (48/63) of the children, followed by K65R, D67N (9.5%), and K70R (6%). K103N/S (51%), Y181C (36%), G190A/S (22%), and V106A/M (8%) were the major NNRTI DRMs observed in the 63 children with any RAMs.
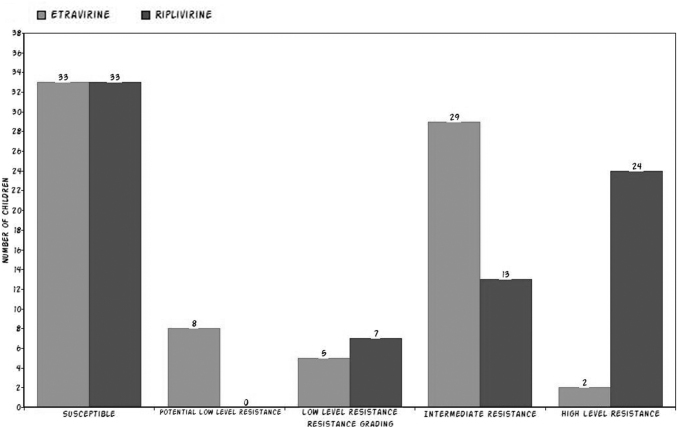
FIG. 2.
ETR and RPV drug susceptibility profile in virologically failed South Indian children. ETR, etravirine; RPV, rilpivirine.
FIG. 3.
Genotypic prediction of NRTI drug resistance/susceptibility profile in virologically failed South Indian children. AZT, zidovudine; d4T, stavudine; DDI, didanosine; FTC, emtricitabine; 3TC, lamivudine; NRTI, nucleoside reverse transcriptase inhibitor; TDF, tenofovir.
FIG. 4.
Genotypic prediction of NNRTI drug resistance/susceptibility profile in virologically failed South Indian children. DOR, doravirine; EFV, efavirenz; NNRTI, non-nucleoside reverse transcriptase inhibitors; NVP, nevirapine.
We also analyzed whether these children had developed any resistance to drugs such as etravirine (ETR) and rilpivirine (RPV) that are considered as second-generation NNRTI drugs. Even though these children are naive to these newer drugs the analysis revealed that 40.2% (31/77) and 48.05% (37/77) children had intermediate to high-level resistance to ETR and RPV, respectively. Another study from South India by Saravanan et al.16 found an even higher prevalence of RPV RAMs in 65% of 97 children. The relatively lower proportion of RPV resistance in our study might be attributed to several cohort characteristics and other factors that are not available and analyzed in the study.
To summarize, we retrospectively analyzed the partial HIV-1 pol gene sequence of 77 paired plasma specimens obtained at baseline and after 1 year of treatment initiation from perinatally infected children who experienced VF at the end of 1 year of treatment with antiretroviral medication. We found that levels of PDR and ADR in children with VF were similar to that reported in other such studies.13–15 The significant finding was, however, the resistance mutations related to ETR and RPV drugs seen in these children, although these drugs are not part of the standard treatment regimen in India. The occurrence of RAMs to the second-generation NNRTIs, ETR and RPV, in this cohort of children who were never exposed to these drugs raises concern and highlights the need for timely switch of regimens guided by genotypic resistance testing to avoid accumulation of DRMs.
Current pediatric treatment guidelines issued by the World Health Organization (WHO) recommend DTG in combination with two NRTIs.17 Although DTG has a high genetic barrier to drug resistance, there is evidence to suggest that the presence of NNRTI resistance before treatment could compromise the efficacy of DTG-based regimens in this population.18,19 Whether the high level of dual class resistance associated mutations (64%) observed in this study compromises the effectiveness of second-line ART and/or DTG-based regimens needs to be studied. Our findings highlight the importance of adequate virological monitoring and genotypic resistance testing to ensure timely switching of regimens in children failing ART in our settings.
Sequence Data
The nucleotide sequences of the pol gene analyzed in this study are available in GenBank under accession numbers OM293757–OM293901.
Acknowledgments
The authors wish to acknowledge the Indian Council of Medical Research (ICMR) and the children who participated in the study.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Research reported in this publication was supported by the intramural funding of ICMR-National Institute for Research in Tuberculosis.
References
- 1. Global and Regional Trends—UNICEF DATA: Monitoring the situation of children and women. Available at https://data.unicef.org/topic/hivaids/global-regional-trends/. accessed November 10, 2021.
- 2. NACO “Annual report 2015–2016” [pdf] Available at http://naco.gov.in/sites/default/files/Annual%20Report%202015-16_NACO.pdf (2015), accessed November 12, 2021.
- 3. UNAIDS “AIDS Data 2018,” p.140–141.[pdf] Available at https://www.unaids.org/sites/default/files/media_asset/unaids-data-2018_en.pdf (2018), accessed November 12, 2021.
- 4. Available at https://main.mohfw.gov.in/sites/default/files/Annual%20Report%202019-2020%20English.pdf, accessed November 12, 2021.
- 5. Chandrasekaran P, Shet A, Srinivasan R, et al. : Long-term virological outcome in children receiving first-line antiretroviral therapy. AIDS Res Ther 2018;15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP: COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014;42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pineda-Peña AC, Faria NR, Imbrechts S, et al. : Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013;19:337–348. [DOI] [PubMed] [Google Scholar]
- 8. Liu TF, Shafer RW: Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006;42:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ: IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. HIVdb Program: Genotypic Resistance Interpretation Algorithm. Available at http://sierra2.stanford.edu/sierra/servlet/JSierra, accessed February 22, 2021.
- 11. Gupta RK, Gregson J, Parkin N, et al. : HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: A systematic review and meta-regression analysis. Lancet Infect Dis 2018;18:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nuttall J, Pillay V: Antiretroviral resistance patterns in children with HIV infection. Curr Infect Dis Rep 2019;21:7. [DOI] [PubMed] [Google Scholar]
- 13. Kityo C, Boerma RS, Sigaloff KCE, et al. : Pretreatment HIV drug resistance results in virological failure and accumulation of additional resistance mutations in Ugandan children. J Antimicrob Chemother 2017;72:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tadesse BT, Tsai O, Chala A, et al. : Prevalence and correlates of pre-treatment HIV drug resistance among HIV-infected children in Ethiopia. Viruses 2019;11:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soeria-Atmadja S, Amuge P, Nanzigu S, et al. : Pretreatment HIV drug resistance predicts accumulation of new mutations in ART-naïve Ugandan children. Acta Paediatr 2020;109:2706–2716. [DOI] [PubMed] [Google Scholar]
- 16. Saravanan S, Kausalya B, Gomathi S, et al. : Etravirine and rilpivirine drug resistance among HIV-1 subtype c infected children failing non-nucleoside reverse transcriptase inhibitor-based regimens in South India. AIDS Res Hum Retroviruses 2017;33:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization: Update of recommendations on first and second-line antitretroviralregimens. Available at https://apps.who.int/iris/bitstream/handle/10665/325892/WHO-CDS-HIV-19.15-eng.pdf?ua=1, Accessed March 10, 2021.
- 18. Rhee SY, Grant PM, Tzou PL, et al. : A systematic review of the genetic mechanisms of dolutegravir resistance. J Antimicrob Chemother 2019;74:3135–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siedner MJ, Moorhouse MA, Simmons B, et al. : Reduced efficacy of HIV-1 integrase inhibitors in patients with drug resistance mutations in reverse transcriptase. Nat Commun 2020;11:5922. [DOI] [PMC free article] [PubMed] [Google Scholar]