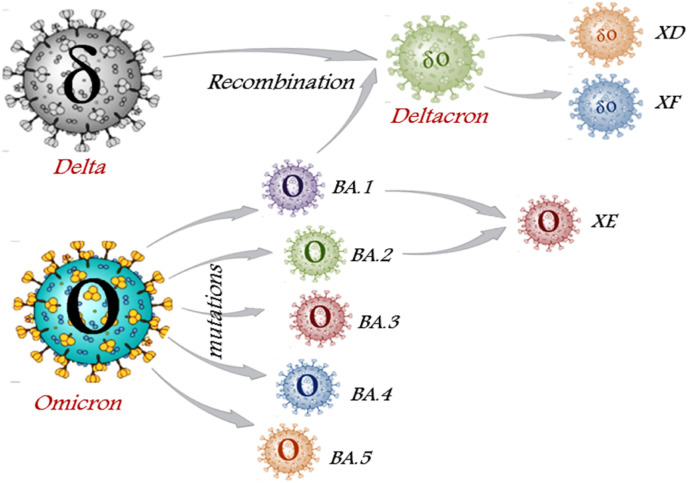
The continuous evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since 2019 created numerous variants like Alpha, Beta, Gamma, Delta, Theta, Lambda, Mu, and finally, Omicron which emerged in November 2021. Viruses are known to create mutations by accumulating errors during viral replications, resulting in emergence of new variants. The Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE) designated Delta (B.1.617.2) and Omicron (B.1.1.529) variants as ‘currently circulating Variants of Concern’ (VOCs) due to their increased transmissibility and virulence in clinical disease presentation [1]. Further computational analysis of the Omicron variant shows distinct sublineages namely BA.1 (B.1.1.529.1), BA.2 (B.1.1.529.2), BA.3 (B.1.1.529.3), BA.4, and BA.5 (Fig. 1 ), all were detected in South Africa indicating their fast evolutionary divergence.
Fig. 1.
Schematic representation of Omicron sublineages and recombinant hybrids: Omicron variant mutates to different sublineages i.e. BA.1, BA.2, BA.3, BA.4, and BA.5. XE subvariant is the hybrid of BA.1 and BA.2. The Deltacron variants i.e. XD and XF is the inter-VOC hybrids formed due to recombination between Omicron BA.1 and Delta variant.
The World Health Organization (WHO) prioritizes monitoring of BA.2.12.1, BA.2.9.1, BA.2.11, BA.2.13, BA.4, and BA.5 sublineages that may pose a threat to global health and require attention and categorize them under VOC lineages under monitoring (VOC-LUM). A large number of genomic mutations as 50 in BA.1, 51 in BA.2, and 43 in BA.3 is the most worrisome attribute of Omicron which may be a matter of concern for public health. As of May 19, 2022, Omicron extended to 187 countries, and around 3.7 million sequences have been submitted in GISAID with a maximum from the United Kingdom (UK, 1,152,917), followed by the United States of America (USA, 1,003,940) [2]. Accounting for >99.9% of total submitted viral genomic sequences, Omicron is now dominating around the world. Additionally, the emergence of Deltacron hybrids also worsened the current pandemic situation.
1. Recombinant subvariants
During the current COVID-19 pandemic, the circulation of several variants facilitates co-infection through the recombination of genetic material from two different strains infecting the same cell, originating new hybrid subvariants such as Deltacron (XD and XF) and XE [3] (Fig. 1). XD and XF variants are the outcome of the Delta and Omicron combination which initially emerged from Europe. ‘X’ denotes the exchange of genetic material between two distinct lineages.
2. The original omicron variant (B.1.1.529.1)
Omicron, first discovered in Botswana, South Africa on November 11, 2021, and designated as a VOC by TAG-VE on November 26, 2021, is now circulating globally as a dominant variant (BA.1.1 to BA.1.22 sublineages) with more than 50 mutations predominantly in the UK (43%), and USA (22%) and is also the main culprit responsible for the third wave of COVID-19 in India in January 2022 [4].
Characterized by 37 RBD mutations, BA.1 lineage preferably infect and replicates in the upper respiratory tract which may account for the increased transmissibility, higher re-infection risk (5.4-fold), and lower severity of illness than the Delta variant [5]. Complimenting this, another study showed a higher infection rate of Omicron (70 fold) in the bronchus and 10 times lower in the lung in comparison to the Delta variant. The same has been validated in the mice and hamster studies, concluding faster transmission of Omicron between humans with lower disease severity [6]. The mutation at 452R/Q is hypothesized to confer intrinsic transmission advantages (immune escape) and may drive further host adaptation and antigenic drift for the evolution of new potentially impactful sub-lineages of Omicron in the future years.
BA.2 (B.1.1.529.2) also known as stealth Omicron carries 31 mutations on the S protein, and is circulating dominantly in more than 122 countries, primarily in the UK (39%), Denmark (15%), and Germany (13%) quickly replacing the Delta and Omicron BA.1 variant [4]. Of the 16 amino acid substitutions in the BA.2's RBD, 12 are common with BA.1. However, the four different substitutions at S371F, T376A, D405 N, and R408S in addition to unique mutations at T19I, L24S, P25del, P26del, A27S, and V213G, are advantageous and confer BA.2 with a better balance of an increased growth rate, better transmissibility, and effective escape immunity from antibodies [7]. Rapid identification is still cumbersome owing to S gene target failure (SGTF)at 69/70del, therefore sequencing is the only confirmatory for BA.2 lineage.
The BA.2.12 and BA.2.12.1 sublineages are estimated to have a 23–27% growth advantage above the original BA.2 variant. In symptomatic BA.2 infection, reduced (13%) vaccine efficacy was recorded for at least 25 weeks following two doses of the vaccine. However, with a third or booster dose, a further 70% enhancement has been recorded at two weeks [5]. Re-infection potential in recovered and vaccinated persons has yet to be determined. BA.3 (B.1.1.529.3), a less known variant has approximately 400 cases reported from at least 19 countries [4].
3. Omicron's BA.4 and BA.5 sub-lineages
Tulio de Oliveira from Stellenbosch University, South Africa discovered the BA.4 and BA.5 subvariants, which are associated with the 5th COVID-19 wave in South Africa and are also responsible for the fresh wave in the USA and Europe. As of May 12, 2022, the European CDC reclassified these sublineages as VOC [4]. The majority of the BA.4 sequences were reported from South Africa (60%), the USA (9%), the UK (7%), and 22 different countries. However, BA.5 is distributed in 19 countries with the highest cases reported from South Africa (31%), Germany (19%), Portugal (18%), and a few BA.5 sequences from China, and France. As of May 21, 2022, India reported one case each of BA.4 and BA.5 subvariants, who had no travel history, however the second BA.5 case had traveled to South Africa [8]. The initial studies suggested a highly infectious nature of the BA.4 and BA.5 sub-lineages appear possibly due to amino-acid (a.a.) substitutions at L452R, F486V, and R493Q, in the Spike RBD. According to a study based on neutralization escape, the unvaccinated group showed an eightfold decrease in antibody production whereas in the vaccinated group 5-fold higher neutralization capacity was observed when exposed to BA.4 and BA.5 in comparison to BA.1 [9].
4. Deltacron (XD): A recombinant variant
While mining the SARS-CoV-2 variants sequences in the GISAID database, Scott Nguyen observed unique recombination of the omicron head (spike) struck onto the body of the Delta variant. Deltacron is an optimized recombination product comprising the best traits from Omicron and Delta variants co-existing in the same cell, synergistically facilitating the mechanism of immune evasion and increasing infectiousness. Formally known as BA.1xAY.4/Deltacron, is thought to have been circulating since its first documentation in France in early January 2022. Later Denmark, Belgium, Netherlands, Germany, U.S., France, and Brazil are witnessing a sudden surge in similar cases. Considering a variant with a potential for future risk, it is designated as VUM on March 9, 2022 [1].
Other hybrid variants designated as XQ, XG, XJ, and XK are identified from the UK, Denmark, Finland, and Belgium respectively. XF is another recombinant sub-lineage of Delta and BA.1 lineage which is confined to the UK with 38 reported since mid-Feb 2022 whereas only 49 cases of XD have been reported worldwide [3,10].
5. Novel omicron XE subvariant
The WHO is continuously tracking and monitoring the emergence of recombinant XE variants which are now scattered globally in countries like the UK, Japan, Canada, and China and are linked with international travel. XE subvariant is a product of recombination between NSP1-6 of original BA.1 and other parts of the more infectious BA.2 strain within a single host [11]. On March 25, 2022, the UKHSA designated this recombinant as ‘XE variant’ which was first detected in the UK through gene sequencing on January 19, 2022 [10]. In the UK alone, more than 637 XE cases have been reported suggesting community transmission. So far, India reported only two cases, one each from Maharashtra and Gujarat [8]. Although the cases are low, the non-effectiveness of the BNT162b2 vaccine against XE reported in two fully vaccinated cases is a concern that needs to be revisited [12].
6. Variants evolutionary theories
The SARS-CoV-2 evolutionary trajectory shows increased transmission during the 1st wave owing to D614G substitution. As immunity gradually builds up, a counteractive mechanism of immune evasion develops in Beta and Gamma variants with E484K mutation [13]. Further delta variant emerges incorporating dual attributes of increased replication fitness and enhanced escape immunity followed by the emergence of omicron with a high basic reproduction number (Ro) and significantly decreased virulence. The reason how current subvariants acquire such unique mutations specifically in the S region could be explained by Dr. Andrew Rambaut from the University of Edinburgh who suggested a theory of intrahost evolution of SARS-CoV-2, especially in immunocompromised HIV-SARS-CoV-2 coinfected persons with reduced CD4+T cells and non-neutralizing antibodies [14]. Another theory stated the origin of Omicron variants with mutations adapting to mink (F486L) which were transmitted back to humans as seen in Denmark. And why viral evolution, possibly to attain better fitness to infect the host more successfully with survival advantages of immune evasion and resistance to neutralizing antibodies. As none of the theories suffice the origin, collectively all are contributing to our understanding of the origin of subvariants but, still, it remains a mystery.
7. New variants, old vaccines: Are they still effective?
Originally designed anti-COVID-19 vaccines are found to be less effective for recently mutating emerging omicron variants, therefore tweaking and re-designing vaccines is an urgent need. Global administration of >11 billion doses of approved COVID-19 vaccine, could only able to vaccinate <65% of the world's population with at least one dose. Although mRNA vaccines offer a good degree of protection against the novel strains, protection wanes within months of a third dose [15]. Also, anti-BA.1 antibody appears to protect against BA.2 infection but for the short term. Relaxation in people's behavior, waning immunity from vaccines, and past infections could be the possible reasons currently helping to fuel surges around the globe. So, vaccination will remain an effective measure to curtail the spread of infection, in addition to maintaining physical distance and wearing masks.
In conclusion, the Omicron, recombinant XE, and Deltacron variants have now spread to all six continents. The WHO is also continuously monitoring the different omicron sublineages for characteristic differences imparting varied transmission and levels of disease severity. Recombination may create highly pathogenic variants with better evolutionary survival fitness, especially in low-vaccinated countries which can lead to different waves of COVID infection. The emergence of a new subvariant of SARS-CoV-2 is being monitored by gene sequencing, particularly in countries where active surveillance does not exist. Therefore, uniform global vaccine campaigns are required to boost the vaccination rate, especially in high-risk groups (unvaccinated adults or children) to tackle the emerging new variants. With much advancement in understanding and handling the global pandemic, there is still speculation that more impactful sub-lineages of this variant can be witnessed in near future.
Provenance and peer review
Not commissioned, internally peer-reviewed.
Data statement
The data and information in this correspondence article is freely available in public domain. Moreover, available data is not sensitive and not of a confidential nature.
Author contributions
Pryanka Thakur: Data curation, Writing-Original Draft, Writing-review & editing. Vikram Thakur: Conceptualization, Data curation, Visualization, Writing-Original Draft, Writing-review & editing. Pradeep Kumar: Supervision, Writing-review & editing. Sanjay Kumar Singh Patel: Supervision, Writing-review & editing. All authors reviewed and approved the final version of the manuscript for publication.
Guarantor
Dr. Vikram Thakur: Research Associate, Department of Virology, Postgraduate Institute of Medical Education and Research (PGIMER) Chandigarh 160012, India; Email: vik5atif@gmail.com.
Dr. Sanjay Kumar Singh Patel: Assistant Professor, Department of Chemical Engineering, Konkuk University, Seoul 05029, Republic of Korea; Email: sanjaykspatel@gmail.com.
Sources of funding for your research
No specific grants from the institute or any public, commercial or not-for-profit sectors received to carry out this study.
Ethical approval
As no animal or human subjects are involved in this study, therefore not require any ethical approval.
Research registration Unique Identifying number (UIN)
-
1.
Name of the registry: Not applicable
-
2.
Unique Identifying number or registration ID: Not applicable
-
3.
Hyperlink to your specific registration (must be publicly accessible and will be checked): Not applicable
Declaration of competing interest
None.
Acknowledgment
This work was supported by the KU Research Professor Program of Konkuk University.
References
- 1.Tracking SARS-CoV-2 variants https://www.who.int/activities/tracking-SARS-CoV-2-variants [Internet]. [cited 2022 Jun 10]. Available from:
- 2.GISAID - Initiative https://www.gisaid.org/ [Internet]. [cited 2022 Jun 10]. Available from:
- 3.Basky G., Vogel L.X.E. XD & XF: what to know about the Omicron hybrid variants. CMAJ (Can. Med. Assoc. J.) 2022 May 9 doi: 10.1503/cmaj.1095998. https://www.cmaj.ca/content/194/18/E654 [Internet] [cited 2022 Jun 10];194(18):E654–5. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cov-Lineages https://cov-lineages.org/ [Internet]. [cited 2022 Jun 10]. Available from:
- 5.Mahase E. Covid-19: what do we know about “long covid”? BMJ. 2022 doi: 10.1136/bmj.m2815. https://www.bmj.com/content/370/bmj.m2815 [Internet]. 2020 Jul 14 [cited 2022 Jun 10];370. Available from: [DOI] [PubMed] [Google Scholar]
- 6.Telenti A., Arvin A., Corey L., Corti D., Diamond M.S., García-Sastre A., et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nat. 2021 doi: 10.1038/s41586-021-03792-w. https://www.nature.com/articles/s41586-021-03792-w [Internet]. 2021 Jul 8 [cited 2022 Jun 10];596(7873):495–504. Available from: [DOI] [PubMed] [Google Scholar]
- 7.Dhawan M., Priyanka, Choudhary O.P. Emergence of Omicron sub-variant BA.2: is it a matter of concern amid the COVID-19 pandemic? Int. J. Surg. 2022 Mar 1 doi: 10.1016/j.ijsu.2022.106581. [Internet] [cited 2022 Jun 10];99:106581. Available from:/pmc/articles/PMC8860472/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.INSACOG Depart. Biotechnol. 2022 https://dbtindia.gov.in/insacog [Internet]. [cited 2022 Jun 10]. Available from: [Google Scholar]
- 9.Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., et al. 2022 May 1. Omicron Sub-lineages BA.4/BA.5 Escape BA.1 Infection Elicited Neutralizing Immunity.https://europepmc.org/article/ppr/ppr488046 [cited 2022 Jun 10]; Available from: [Google Scholar]
- 10.Investigation of SARS-CoV-2 variants: technical briefings - GOV.UK. https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings [Internet]. [cited 2022 Jun 10]. Available from:
- 11.Rahimi F., Talebi Bezmin Abadi A. Detection of the XE subvariant of SARS-CoV-2: a perspective. Int. J. Surg. 2022 May 1 doi: 10.1016/j.ijsu.2022.106642. https://pubmed.ncbi.nlm.nih.gov/35490952/ [Internet] [cited 2022 Jun 10];101. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gjorgjievska M., Mehandziska S., Stajkovska A., Pecioska-Dokuzovska S., Dimovska A., Durmish I., et al. Case report: omicron BA.2 subvariant of SARS-CoV-2 outcompetes BA.1 in two Co-infection cases. Front. Genet. 2022 Apr 12;13:821. doi: 10.3389/fgene.2022.892682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du P, Gao GF, Wang Q. The mysterious origins of the Omicron variant of SARS-CoV-2. [cited 2022 Jun 9]; Available from: 10.1016/j.xinn.2022.100206. [DOI] [PMC free article] [PubMed]
- 14.Karim F., Moosa M., Gosnell B., Cele S., Giandhari J., Pillay S., et al. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. 2021 Jun 4 https://www.medrxiv.org/content/10.1101/2021.06.03.21258228v1 [Internet] [cited 2022 Jun 10];2021.06.03.21258228. Available from: [Google Scholar]
- 15.Sidik S.M. Vaccines protect against infection from Omicron subvariant — but not for long. Nature. 2022 Mar 18 doi: 10.1038/d41586-022-00775-3. Epub ahead of print. PMID: 35304887. [DOI] [PubMed] [Google Scholar]