Abstract
Background
In Covid-19, profound systemic inflammatory responses are accompanied by both metabolic risk factors for severity and, separately, metabolic mechanisms have been shown to underly disease progression. It is unknown whether this reflects similar situations in sepsis or is a unique characteristic of Covid-19.
Aims
Define the immunometabolic signature of Covid-19.
Methods
65 patients with Covid-19,19 patients with sepsis and 14 healthy controls were recruited and sampled for plasma, serum and peripheral blood mononuclear cells (PBMCs) through 10 days of critical illness. Metabotyping was performed using the Biocrates p180 kit and multiplex cytokine profiling undertaken. PBMCs underwent phenotyping by flow cytometry. Immune and metabolic readouts were integrated and underwent pathway analysis.
Results
Phopsphatidylcholines (PC) are reduced in Covid-19 but greater than in sepsis. Compared to controls, tryptophan is reduced in Covid-19 and inversely correlated with the severity of the disease and IFN-ɣ concentrations, conversely the kyneurine and kyneurine/tryptophan ratio increased in the most severe cases. These metabolic changes were consistent through 2 pandemic waves in our centre. PD-L1 expression in CD8+ T cells, Tregs and CD14+ monocytes was increased in Covid-19 compared to controls.
Conclusions
In our cohort, Covid-19 is associated with monocytopenia, increased CD14+ and Treg PD-L1 expression correlating with IFN-ɣ plasma concentration and disease severity (SOFA score). The latter is also associated with metabolic derangements of Tryptophan, LPC 16:0 and PCs. Lipid metabolism, in particular phosphatidylcholines and lysophosphatidylcolines, seems strictly linked to immune response in Covid-19. Our results support the hypothesis that IFN-ɣ -PD-L1 axis might be involved in the cytokine release syndrome typical of severe Covid-19 and the phenomenon persisted through multiple pandemic waves despite use of immunomodulation.
Keywords: Immunometabolism, Covid-19, Immune checkpoint, Tryptophan, Phosphatidylcholine
Immunometabolism; Covid-19; immune checkpoint; tryptophan; phosphatidylcholine.
1. Introduction
SARS-CoV-2 is a positive-sense, single-stranded RNA virus, which first emerged in December 2019 from Wuhan, China and is responsible for the current pandemic [1]. Currently more than 470 million people have been affected by Covid-19 and almost 6 million people died because of this disease.
There has been significant interest in the relationship between metabolism and immune responses in Covid-19. Patients living with metabolic diseases such as diabetes and obesity have higher mortality from of Covid-19 and are more prone to more a severe illness [2]. More widely viral disease in patients with underlying metabolic syndromes appears to modify immune activity with cholesterol, carbohydrate and lipid metabolism being implicated in viral clearance or persistence [3]. Therefore several metabolic pathways, including lipid, amine and carbohydrate, may potentiate severe disease and provide novel therapeutic options to improve current treatment [4, 5, 6].
The importance of lipid metabolism in relation to Covid-19 has been highlighted in the most recent literature [7, 8] where the action of cholesterol on membrane entry of SARS CoV2 suggests a wider role for HDL. Lipid metabolism is a pivotal immune modulator in several immune related diseases including liver failure where the action on innate immunity of LDL, phosphocholines and eicosanoids indicates a wide variety of lipid centric immunometabolic cellular responses [9, 10].
Other metabolic pathways, such as nicotinate and nicotinamide metabolism, tryptophan metabolism, and citrate cycle (TCA cycle) also appear to be altered in severe Covid-19 patients [11]. Moreover, metabolic derangement was noted also in patients in the recovery phase, even in those who go on to be PCR test negative [12].
Cytokine responses are primarily driven by viral encounter with alveolar macrophages and subsequent T cell activation [13]. However, modulation of these responses by metabolic risk factors or metabolism has been described [11].
A Covid-19 peripheral blood immune signature [13], including proinflammatory cytokines (Interleukin (IL)-8, IL-6 and IL-10), and CD8+ T cells co-expressing exhaustion-associated markers has recently been reported. An animal model mimicking the cytokine storm in Covid-19 treating with neutralizing antibodies against TNF-α and IFN-ɣ protected mice from mortality during SARS-CoV-2 infection [14].
As yet it has not been possible to compare the effects of therapy on metabolic responses to Covid-19 across the multiple waves of the pandemic as novel therapeutic agents rapidly enter clinical practice. Therefore, the aim of this study was to characterise a Covid-19 related immunometabolic profile that could provide insight in the pathophysiology of the disease and help to predict outcome in patients affected over multiple waves of Covid-19.
2. Material and methods
2.1. Study population
Between April 2020 and February 2021, consecutive patients admitted to King's College Hospital with positive Covid-19 polymerase chain reaction (PCR) were screened and approached for recruitment as part of the Immunometabolism in Sepsis, Inflammation and Liver Failure Syndromes (IMET study (Research Ethics Committee No.: 19/NW/0750, IRAS No.: 244089) within 24 h of admission. Pregnancy, disseminated malignancy, pre-existing immunosuppressive states, including drugs and human immunodeficiency virus (HIV) infection, and chronic granulomatous diseases were exclusion criteria. Healthy controls (HC), and patients with sepsis from other aetiologies were recruited in the same period with the same exclusion criteria and used as controls. Sepsis was defined as per Sepsis 3 criteria [15]. Full blood count, coagulation parameters, liver and renal function tests, lactate, and clinical variables were entered prospectively into a database. Physiological data was captured and the Sequential Organ Failure Assessment (SOFA) score was calculated to assess severity [16].
2.2. Plasma and peripheral blood mononuclear cell isolation and phenotyping
Blood was drawn, form arterial line or peripheral venipuncture, into lithium heparin Vacutainers (BD, Franklin Lakes, NJ) on day 1, 3, 7 and 10 of admission. Laboratory processing started within 20 min from sampling. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Paque Plus (GE Healthcare, UK), as previously described [10]. Monocytes and T cells phenotype was determined by flow cytometry of PBMCs using monoclonal antibodies showed in Table 1. T-regs were defined as CD4+, CD25+, CD-127- cells (Figure 1) [17].
Table 1.
Antibodies used for flow cytometry.
| Primary antibodies | Host | Anti | Clone | Conjugated fluorophore | Catalogue number | Producer | Application |
|---|---|---|---|---|---|---|---|
| CD14 | Mouse | Human | M5E2 | PeCy7 | 557742 | BD | Flowcytometry |
| CD16 | Mouse | Human | 3G8 | APC-H7 | 560195 | BD | Flowcytometry |
| CD163 | Mouse | Human | GHI/61 | PE | 556018 | Invitrogen | Flowcytometry |
| CCR2 | Mouse | Human | K03602 | AlexaFluor 488 | 357226 | Biolegend | Flowcytometry |
| HLA-DR | Mouse | Human | LN3 | PerCp-Cy 5.5 | 45-9956-42 | Invitrogen | Flowcytometry |
| MerTK | Mouse | Human | 125518 | APC | FAB8912A | R&D System | Flowcytometry |
| CD155 | Mouse | Human | SKII.4 | BV421 | 337632 | Biolegend | Flowcytometry |
| PD-L1 | Mouse | Human | 29E.2A3 | BV 605 | 329724 | Biolegend | Flowcytometry |
| PD-1 | Mouse | Human | EH12.1 | BV786 | 563789 | BD | Flowcytometry |
| CD3 | Mouse | Human | SK7 | eFluor 450 | 48-0036-42 | eBioscience | Flowcytometry |
| Tim-3 | Mouse | Human | F38-2E2 | BV711 | 345024 | Biolegend | Flowcytometry |
| TIGIT | Mouse | Human | MBSA43 | FITC | 11-9500-42 | eBioscience | Flowcytometry |
| CTLA-4 | Mouse | Human | 14D3 | PE | 12-1529-42 | eBioscience | Flowcytometry |
| CD25 | Mouse | Human | M-A251 | PE-CF594 | 562403 | BD | Flowcytometry |
| CD127 | Mouse | Human | eBioRDR5 | PE-Cy7 | 25-1278-42 | Invitrogen | Flowcytometry |
| CD8 | Mouse | Human | RPA-T8 | APC | 17-0088-42 | Invitrogen | Flowcytometry |
| CD4 | Mouse | Human | SK3 | APC-Cy7 | 344616 | Biolegend | Flowcytometry |
Figure 1.
A) Gating strategy for CD14+ cells. B) PD-L1 expression in CD14+ cells of healthy controls, Covid -19 positive and septic patients. Results are presented as both Mean Fluorescence Intensity and % of CD14+ cells. C) Gating strategy for CD3+ cells, CD4+, CD8+ and CD25+ CD127-cells (Tregs). D) PD-L1 expression in CD3+ cells (CD4+, CD8+ and Tregs) of healthy controls, Covid -19 positive and septic patients. Results are presented as both Mean Fluorescence Intensity and % of CD3+ cells.
Results are expressed as percentage (%) and/or mean fluorescence intensity (MFI). Acquisition was performed on a BD LSRFortessa™ cell analyzer (BD Biosciences). Flow cytometry data analysis was performed in FlowJo™ v10 (Becton Dickinson & Company).
2.3. Cytokine analysis
Interleukin (IL)-1β, IL-4, IL-6, IL-8, IL-10, IL-12p70, INF(Interferon)-γ and TNF (Tumour Necrosis Factor)-α plasma concentrations were quantified using the Simple Plex™ Ella (Ella™) (ProteinSimple). Samples were analysed by the Contract Research Laboratory (Viapath), King's College Hospital.
2.4. Enzyme linked immuno-absorbent assay (ELISA) techniques
ELISA kits for Autotaxin (ATX/ENPP2) (R&D Systems, Minneapolis, USA), Phospholipases (PLA) 1 and 2 (Biomatik, USA) and Osteopontin (OPN, Biotechne, Minneapolis, USA) were used and plasma concentrations quantified according to the manufacturer's instructions.
2.5. ATX - Quantikine®ELISA human ENPP-2/autotaxin immunoassay from R&D systems
Heparin plasma samples were diluted 20-fold. 100 μL of Assay Diluent RD1-34 was added to each well of a precoated microplate together with 50 μL of standard or sample. After 2 h incubation at room temperature on a horizontal orbital microplate shaker (0.12″ orbit) set at 500 ± 50 rpm, plates were washed three times (25- fold wash buffer concentrate reconstituted). 200 μL of Human ENPP-2 Conjugate were added to each well and after 2 h incubation at room temperature on the shaker, plates were washed three times. Then, 200 μL of Substrate Solution per well were added and plates were incubated for 30 min at room temperature, protected from light. The reaction was stopped with 50 μL of Stop Solution per well. Optical density was assessed soon after, using a FLUOstar® Omega microplate reader (BMG Labtech Ltd, UK) set to 450 nm with 540 nm wavelength correction. Average of the duplicate readings for each standard, control, and sample were analysed after subtraction of the average zero standard optical density. A third order polynomial standard curve was used for quantification the data.
2.6. PLA1 - human PLA1(Phospholipase A1) ELISA kit from Elabscience biotechnology inc
Heparin plasma samples were 50-fold diluted. 100 μL Of standard or sample were added to each well of a precoated plate and incubated for 90 min at 37°. After samples removal, without washing, 100 μL of Biontinylated detection Ab working solution were added to each well and incubated at 37° for 1 h. Plates were then washed three times (25- fold wash buffer concentrate reconstituted). 100 μL Of HRP Conjugate working solution were added to each well and incubated at 37° for 30 min, then plates were washed three times. 90 μL Of Substrate Reagent were added to each well and plates were incubated at 37° for 15 min protected from light. The reaction was stopped with 50 μL of Stop Solution per well. Optical density was assessed soon after, using a FLUOstar® Omega microplate reader (BMG Labtech Ltd, UK) set to 450 nm. Average of the duplicate readings for each standard and sample were analysed after subtraction of the average zero standard optical density. A third order polynomial standard curve was used for quantification.
2.7. PLA2 - human phospholipidase A2, PLA2 ELISA kit from biomatik
Heparin plasma samples were 10-fold diluted. 100 μL of standard or sample were added to each well of a precoated plate and incubated for 2 h at 37°. After samples removal, without washing, 100 μL of Biontin-Antibody (1x) were added to each well and incubated at 37° for 1 h. Plates were then washed three times (25-fold wash buffer concentrate reconstituted). 100 μL of HRP- avidin (1x) solution were added to each well and incubated at 37° for 1 h, then plates were washed three times. 90 μL of TMB Substrate were added to each well and plates were incubated at 37° for 20 min protected from light. The reaction was stopped with 50 μL of Stop Solution per well. Optical density was assessed soon after, using a FLUOstar® Omega microplate reader (BMG Labtech Ltd, UK) set to 450 nm with 540 nm wavelength correction. Average of the duplicate readings for each standard and sample were analysed after subtraction of the average zero standard optical density. A third order polynomial standard curve was used for quantification.
2.8. Human osteopontin (OPN) ELISA kit from bio-techne
Heparin plasma samples were 300-fold diluted. 100 μL of standard or sample were added to each well of a plate previously coated with Mouse Anti-Human Osteopontin Capture Antibody and kept overnight at room temperature. After 2 h incubation at room temperature on a horizontal orbital microplate shaker (0.12″ orbit) set at 500 ± 50 rpm, plates were washed three times (wash buffer: 0.05% Tween® 20 in PBS). 100 μL Of Biotinylated Goat Anti-Human Osteopontin Detection Antibody was added to each well and after 2 h incubation at room temperature on the shaker, plates were washed three times. Then, 100 μL of Streptavidin-HRP were added and incubated for 20 min avoiding light. After washing, 100 μL Of Substrate Solution (10mg O-phenylenediamine dihydrochioride in 25 ml of 0.05M phosphate citrate buffer) per well was added and plates were incubated for 20 min at room temperature, protected from light. The reaction was stopped with 50 μL of Stop Solution (2N H2SO4) per well. Optical density was assessed soon after, using a FLUOstar® Omega microplate reader (BMG Labtech Ltd, UK) set to 490 nm. The mean of the duplicate readings for each standard, control, and sample were analysed after subtraction of the average zero standard optical density. A third order polynomial standard curve was used for quantification.
2.9. Ultra-performance liquid chromatography-mass spectrometry (UPLC®-MS/MS) analysis
Sample preparation and data acquisition for UPLC-MS/MS analysis of lipids from plasma was performed using the AbsoluteIDQ p180 kit BIOCRATES (BIOCRATES Life Sciences AG, Innsbruck, Austria) and a Waters Xevo TQS Micro instrument. Amino acids and biogenic amines were determined in LC-MS mode, acylcarnitines, phospholipids (lyso-phosphatidylcholines with acyl residue at CXX:X, phosphatidylcholine with diacyl residue sum CXX:X (PC aa), and phosphatidylcholine with acyl-alkyl residue sum CXX:X (PC ae)), sphingomyelins, and the sum of hexoses were analysed using flow injection analysis (FIA). Heparin plasma samples were prepared according to the manufacturers protocols.
Briefly, 10 μL of internal standard followed by 10 μL of sample (plasma after centrifugation at 4 °C for 5 min at 2750 x g), calibrator or quality controls (QC), were transferred onto the filter located in the wells of the upper 96-well plate and dried for 30 min under a nitrogen stream. Thereafter, 50 μL of a 5% phenylisothiocyanate (PITC, Sigma-Aldrich, UK) solution (in ethanol:water:pyridine, 1:1:1 (v/v)) was added to derivatize amino acids and biogenic amines. After 20 min incubation, the filter spots were dried again for 60 min before the metabolites were extracted using 5 mM ammonium acetate in methanol (300 μL). After shaking (450 rpm, 30 min), the eluate was collected into the lower 96-well plate by centrifugation (500 x g, 2 min).
150 μL of extract were transferred to an empty 96-deep-well plate and diluted with 150 μL of water, and the plate sealed. After shaking (600 rpm, 2 min) the plate was transferred for analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
For flow injection analysis (FIA)-mass spectrometry, 20 μL of the original, undiluted extract were transferred to a separate 96-deep-well plate, and diluted with 380 μL FIA mobile phase, sealed and shaken (600 rpm, 2 min). A Xevo Acquity TQ-S micro (Waters Corp., MA, US) instrument at King's College Hospital was used for both LC and FIA analysis of the samples. Separation (LC) was achieved using a BEH-C18 UPLC column (75 mm × 2.1 mm i. d., Waters Corp., MA, US), with all mobile phases and instrument settings according to the Biocrates protocol. The ratio of analyte to internal standard was used for quantification purposes.
2.10. Statistical analysis
Assuming that a 180 metabolites panel would generate at least one metabolite with a significant difference between patients with Covid-19 and healthy controls then, taking false discovery rate correction into account, a metabolite difference of 30μM with standard deviation of 15μM in patients with Covid-19 and controls, and a case/control ratio of 4, then 40 patients with Covid-19 and 10 healthy controls would be required. This assumes a power of 90% and alpha of 0.0001 and is based on pilot data for the difference in LPC16:0 between patients with COVID19 and healthy controls in an exploratory analysis.
For UPLC-MS/MS data, principal components analysis (PCA) was performed to visualise any inherent clustering and identify outliers (SIMCA v 16.0, Sartorius Stedim, Goettingen, Germany). Partial least squares (PLS) and orthogonal PLS (OPLS), S-plot loadings (and Variable Importance in Projection (VIP) were used to determine the metabolites contributing to class separation. Enrichment analysis was performed with MetaboAnalyst 5.0 (https://www.metaboanalyst.ca).
T-test, one-way ANOVA/Kruskal-Wallis, Pearson/Spearman's correlations, chi-square and AUROC were calculated with GraphPad Prism v 8.2.1 (GraphPad Software, CA, USA) and SPSS v25 (IBM, USA).
3. Results
3.1. Patient cohort characteristics
Sixty-seven patients with positive Covid-19 PCR were recruited for this study. Among them 3 were on a medical ward and 64 were admitted to intensive care units. Two patients were subsequently excluded from the analysis, since they did not show features of pneumonia and were respectively admitted to the hospital with acute liver failure and acute pancreatitis.
A total of 98 subjects were included in this study (65 patients with Covid-19 pneumonia, 19 patients affected by sepsis and 14 healthy controls). HC were significantly younger compared to patients (p < 0.001), no difference was found between the two patient groups in terms of age. No difference was found in sex distribution and severity of disease between Covid-19 and septic patients (SOFA score) (p = 0.0576). Monocyte count was significantly reduced in Covid-19 compared to sepsis. Characteristic of the study population is showed in Table 2.
Table 2.
Characteristic of population.
| COVID (n = 65) | SEPSIS (n = 19) | HC (n = 14) | p | |
|---|---|---|---|---|
| Female n (%) | 32 (46%) | 6 (30%) | 8 (57%) | |
| Age | 57.3 ± 13.2 | 65 ± 13 | 34 ± 7.6 | <0.0001 |
| SOFA score | 6.8 ± 4.4 | 4.38 ± 3.5 | ns | |
| Mortality 28 days | 17 (24%) | 3 (16%) | ns | |
| Mortality 90 days | 18 (26 %) | 3 (16%) | ns | |
| CRP | 137.4 ± 113.7 | 186.9 ± 131 | ns | |
| WBC | 11.8 ± 6.1 | 14.2 ± 10.9 | ns | |
| Neut | 9.9 ± 5.7 | 12.2 ± 10.1 | ns | |
| Lymph | 1.2 ± 0.7 | 0.99 ± 0.44 | ns | |
| Mono | 0.48 ± 0.29 | 0.76 ± 0.57 | 0.0087 | |
| BMI | 31.4 ± 8.1 | 31.2 ± 11 | ns | |
| Diabetes n (%) | 24 (34%) | |||
| Hypertension n (%) | 33 (47%) | |||
| BMI>30 | 30 (43%) | |||
| BMI>35 | 17 (24%) |
3.2. Phenotyping
Monocyte expression of CD155 was reduced in sepsis compared to Covid-19 and PD-L1 was increased compared to HC. In Covid-19, percentage of CD3+ was reduced compared to controls and PD-L1 was increased in CD4, CD8 and Tregs, reaching the significance in the last two populations (Figure 1).
3.3. Cytokine response other than IFN-ɣ is similar in Covid-19 and septic patients.
Osteopontin (OPN) (Figure 2C) and inflammatory cytokines, namely IL-6, IL-8, IL1β, TNFα, and IL-10 were increased in Covid-19 compared to controls, without any difference with other sepsis (Figure 2A). IFN-ɣ, instead, was increased in Covid-19 compared to both HC and sepsis, and this was dynamically reduced during the hospital stay from admission to day 10 (Figure 2B). Moreover, a correlation analysis (Figure 2D) demonstrated significant positive correlation between IFN-ɣ, SOFA score and monocyte PD-L1 expression.
Figure 2.
A) Pro inflammatory Cytokines are increased in sepsis and Covid-19 compared to healthy controls (HC). Interferon (IFN)-ɣ, Interleukin (IL)-10, IL-1 beta, IL-6, Tumor Necrosis Factor (TNF) alpha and IL-8. B) Interferon (IFN)-ɣ is the only proinflammatory cytokine that showed progressive decrease on sequential samples (day 1,3,7,10) in COVID-19 patients. Median and 95% CI. C) Osteopontin (OPN) is increased in septic patients both with radiological and clinical feature of COVID-19 (Supected = susp) and with positive PCR (Covid). D) Correlation Matrix showing Interferon-gamma is directly correlated to C-reacting protein (CRP), Sofa score, osteopontin and PD-L1 expression in Monocytes.
3.4. UPLC-MS/MS analysis - BIOCRATES p180 assay
3.4.1. Covid-19 patients demonstate a plasma metabotype distinct from patients with sepsis from other causes
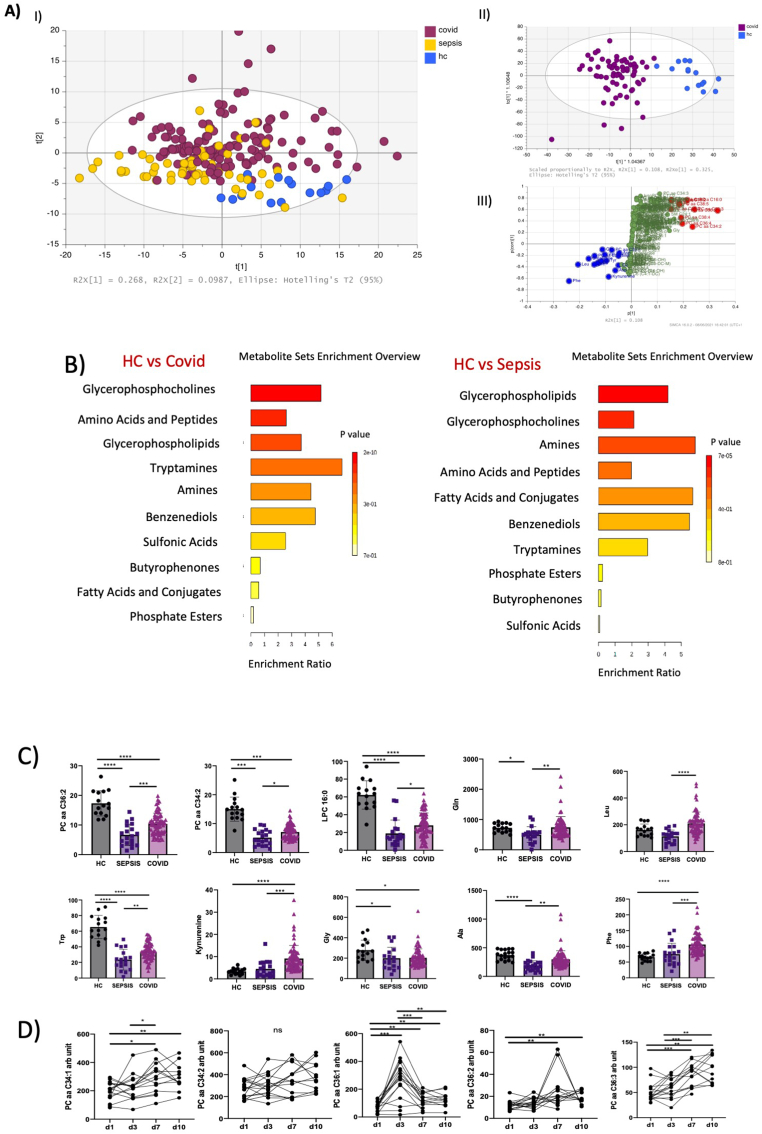
Covid-19 plasma had a metabotype distinct from healthy controls, and patients with sepsis together with patients initially suspected to have Covid-19 but with PCR negative. This was demonstrated by a 3 component PCA model on plasma with R2 of 0.445 and Q2 of 0.394. Assessing baseline samples alone confirmed similar behaviour with R2 of 0.452 and Q2 of 0.365. OPLSDA analysis of 3 groups (Covid-19, sepsis and healthy controls) also gave clear discrimination with a model with 3 + 3+0 components having a R2 of 0.52, Q2 of 0.489 and CV-ANOVA p value of 0 (Figure 3A).
Figure 3.
A) Multivariate analysis including 180 metabolites (Biocrates). I) PCA analysis. Covid-19 positive patients have distinct features compared to septic patients and healthy controls. II) OPLS-DA analysis including Covid -19 positive patients versus healthy control. III) S-plot identifying the panel of metabolites responsible for the greatest variance between Covid-19 positive patients and healthy controls. Covid-19 n = 65, Sepsis n = 19, healthy control n = 14. B) Enrichment Analysis showing the main classes of metabolites. Data analysed with Metaboanalyst, a publicly available platform dedicated for metabolomics data analysis, including several libraries containing about 9,000 metabolite sets from human studies. C) Univariariate analysis comparing Covid-19 positive, septic patients, and healthy controls. Phosphatidylcholines (PCs,) Lysophosphatidylcholine (LPC) 16.0, Tryptophan (Trp) are reduced in sepsis more than in Covid 19, comparing to HC. Glutamine (Gln), Leucine (Leu), Kynurenine and phenylalanine (Phe) are increased in Covid-19 patients. D) Phosphatidylcholines increase progressively during hospital admission (Day 1,3,7,10).
The major classes involved in differentiating between Covid-19 and HC were glycerophosphocholines, amino-acids, glycerophospholypids and tryptamines. Instead comparing Covid-19 with sepsis, the most discriminatory metabolites were glycerophospholipids, amines, glycerophosphocholines, amino-acids and fatty acids (Figure 3B).
We identified a panel of metabolites able to discriminate Covid-19 from HC and sepsis. In Covid-19 patients, glutamine (Gln), leucine (Leu), phenylalanine (Phe) and kyneurine (Kyn) were increased, instead tryptophan (Trp), lysophophatidylcholine (LPC)16:0 as well as several phosphatidylcholines (PC) including PC aa 34:1, 34:2, 36:1,36:2, 36:3 were reduced in Covid-19 compared to HC (Figure 3C). PCs were reduced also in sepsis from other aetiologies.
PC aa C34:1, 34.2, 36:1,36:2, 36:3 also dynamically increase during the hospital stay in Covid-19 positive patients (Figure 3D).
A PLS model using SOFA as Y-variable detected a similar panel of metabolites being Gln, PC aa 32:1, PC aa 34:1, PC aa 34:2, PC aa 36:1, PC aa 36:2, LPC 16:0, kyneurin and tryptophan among those with the highest VIP (Figure 4A).
Figure 4.
A) I) PLS analysis in Covid-19 positive patients using Sequential Organ Failure Assessment (SOFA) score as Y variable. II) PCA analysis failed to identify a panel of metabolites able to predict mortality at 90 days in patients affected by Covid-19. B) PCs are directly correlated with SOFA score, LPC is inversely correlated with the severity of the disease. C) Autotaxin catalyse the conversion of lysophosphatidylcholine (LPC) into Lysophosphatidylcholinic Acid (LPA) and is homogeneously expressed in healthy controls, Covid-19 positive patients (at admission Day 1 and one week after admission, Day 7) and septic patients. Excluding a Covid-19 specific role of the enzyme. Lysophosphatidylcholine (LPC) 16.0 is increased at one week after admission in Covid-19 positive patients. Both PLA1 and PLA 2 expression, measured by ELISA, is increased in Covid-19 compared to controls. D) Kyneurin (Kyn)/Tryptophan (Trp) ratio was significantly increased in Covid-19 and septic patients compared to controls. Trp is significantly reduced with the increased severity of the disease expressed by SOFA score, conversely Kyneurine is increased.
In patients affected by Covid-19, PCs increased according to the severity of the disease SOFA score (Figure 4B), however we did not observe this trend in septic patients. Similar results were found for the Kyn/Trp ratio (Figure 4D).
3.5. Metabotypes do not predict subsequent clinical outcomes
In our cohort, 90-day mortality was 26%. No models satisfactorily detected a metabolic panel predictive for mortality (Figure 4A). Univariate analysis confirmed no difference in the studied metabolites between dead and alive patients (Figure 5A).
Figure 5.
A) Univariate analysis comparing Phosphatidylcholines (PCs), Lysophosphatisylcholine (LPC)16.0, Glutamine (Gln), Tryptophan (Trp), Kyneurine (Kyn) in patients recovered from Covid-19 (alive) and patients dead at 90 days post admission. No one of the metabolites identified was significantly different between the two groups. B) No difference was found in phosphatidylchlines (PCs), Tryptophan (Trp), Kyneurine and Glutamine (Gln) between patients on steroids vs patients managed without steroid therapy. C) Correlation matrix showing numbers indicate Spearman r and intensity of colour indicate the strength of correlations. White blood cell count (WBC), Neutrophils count (Neutr), Lymphocyte count (Lymph), Monocytes coutn (Mono), C-reactive protein (CRP), Sequential Organ failure assessment score (SOFA), Interlerukin (IL) 10, 6 and 8, Tumour Necrosis factor (TNF) alpha, Glutamine (Gln), Tryptophan (Trp) Lysophosphatidylcholine (LPC) 16, Phosphatidylcholine (PC).
A negative correlation was found between LPC 16:0 levels and severity of Covid-19 expressed by SOFA score with an increase at day 7 of admission.
Moreover, chi-square analysis failed to show an association between mortality and high BMI (both >30 and >35) and as well as diabetes.
In an age-adjusted model, BMI, hypertension and presence of diabetes were not associated with 90-day mortality.
When patients were divided according to the two UK “waves”, namely those recruited before (n = 15) and after (n = 50) September 2020, PCA failed to show any difference in the metabolites panel. The same negative result was found when patients were divided according to steroids therapy in two groups, respectively 19 patients who did not receive steroids and 46 patients who received steroids at the time of recruitment (Figure 5B).
3.5.1. PLA 1 and 2 are hyperexpressed in Covid-19 while autotaxin has not difference compared to controls
Given the clear signal of LPC depletion and PC increase, we explored glycerophosphocholines metabolism further. ELISA assays for enzymes involved in phospholipid metbolism showed an increase in PLA1 and PLA2 in Covid-19 patients as compared to HC, but not in autotaxin, the enzyme responsible for LPC conversion to lysophosphatydilcholinic acid (LPA) (Figure 4C). PC 32:1 and 34:1 were directly correlated with proinflammatory cytokines and osteopontin while LPC 16:0 was inversely correlated with them (Figure 5C). Therefore, modulation of PCs concentration may be substrate (choline, phosphate) or lysophospholipase dependent rather than modulation by downstream metabolism.
4. Discussion
Our results suggest an immunometabolic signature in Covid-19 linking monocyte and T-cells PD-L1 expression, IFN-ɣ and systemic metabolism. This underpins disordered immunometabolism in patients hospitalised for severe Covid-19 infection.
Our data extends the findings of previous metabolomics studies in Covid-19. Other authors reported the importance of metabolites involved in arginine metabolism, including glutamine, argininate, asymmetric dimethylarginine (ADMA)and symmetric dimethylarginine (SDMA) that have been found significantly decreased in the sera of non-severe Covid-19 patients [18]. Lipid metabolism was also explored and arachidonic and oleic acids were recently identified as potential biomarkers for Covid-19 infection [19].
We have identified multiple pathways potentially interacting with host immunity. In our multivariate analysis assessing severity of disease (SOFA score), LPC 16:0 was one of the metabolites most significantly reduced with increasing severity, similarly of our findings in immune dysfunction in acute-on-chronic liver failure [10]. Other authors showed the downregulation of glycerophospholipids and the upregulation of lysophospholipids, arachidonic acid, and oleic acid, suggesting a strong involvement of PLA2 in the pathogenesis and progression of Covid-19 [19]. We examined this pathway further and we excluded an increased involvement of ATX in LPC downregulation, confirming both PLA1 and PLA2 hyperexpression in Covid-19 (Figure 4C).
LPC16:0, glutamine and tryptophan were inversely correlated to the profinflammatory cytokines (IL-8, IL-6, TNFa) and osteopontin, with the opposite trend seen in PC aa 32:1 and PC aa 34:1.
These findings are in line with previous studies reporting links between cytokine increase and the alteration of metabolic processes in Covid-19 patients. Increased inflammatory response leads to a loss of key circulating nutrients (in particular lipids and amino acids) [20] and increased mitochondrial dysfunction in immune cells with increased rate of glycolysis and utilisation of glucose as the main substrate for energy production [21, 22].
The proinflammatory profile of Covid-19 infection has been extensively explored [23]. In particular, IL-6 was found associated with adverse clinical outcomes [24], however, and interestingly, its levels were lower in Covid-19 patients than in bacterial sepsis [25].
Authors reported a Covid-19 immuno profile characterised by low absolute T lymphocyte count, with a more severe decrease in critical care patients, and markedly higher percentages of PD-1+CD8+ and CD4+ T cells as marker of exhaustion [26]. In our study PD-L1 was increased in both monocytes and T cells correlating with a reduction in tryptophan and the increase of IFN-ɣ, suggesting pivotal links in immunometabolism.
Immune check points (PD-1,Tim3, TIGIT, CTLA-4) have been involved in several viral infections other than in cancer [27]. T cells from Covid-19 patients have increasing PD-1 and Tim-3 expression as patients progressed from prodromal to overtly symptomatic stages [26]. In a recent study, all subsets of monocytes have been found to express increased levels of PD-1 and PD-L1 in patients affected by Covid-19 [28]. Moreover, in our data monocyte PD-L1 expression is correlated with plasma IFN-ɣ levels (Fig. 2D), a key component of the innate antiviral response [29]. This mirrors what has been observed in cancer where PD-L1 has an essential role in tumor cells escape from antitumor immunity-, and IFN-ɣ cells treatment is proven to upregulate the JAK2/STAT1/IRF1 axis and PD-L1 [30, 31].
Tryptophan metabolism is significantly altered in Covid-19 [20], with decrease in tryptophan (inversely proportional to IL-6 concentration and SOFA score). This is mirrored by an increase in kynurenine (kyn) and Kyn/Trp ratio in Covid-19 patients compared to both HC and Sepsis. This finding is confirmed by other authors who found increased metabolites of kynurenate, kynurenine, and 8-methoxykynurenate in Covid-19 patients [18], although others failed to demonstrate an association [32].
The kynurenine pathway increases nicotinamide adenine dinucleotide (NAD+), the cofactor in many cellular redox reactions, from tryptophan and it can contribute to the switch for macrophage effector responses [18].
Moreover, tryptophan catabolism has been identified as an important suppressor of antitumor immune responses [33]. Depletion of tryptophan, a fundamental factor for T-cell metabolism, is one of the main mechanisms involved in resistance to immunotherapy leading to T-cell anergy and apoptosis. Indoleamine-2,3-dioxygenase (IDO) catalyzes the conversion of tryptophan into kynurenine, inducing an immunosuppressive microenvironment in cancers. IDO activity is involved in peripheral immune tolerance because it can promote the inhibition of T-cell proliferation induced by trp deprivation [33].
IDO is regulated by IFN-ɣ and in our results, Tryptophan is inversely related to IFN-ɣ [34]. The ratio of kynurenine/tryptophan is used as a surrogate method to determine the enzymatic activity of IDO and was increased in Covid-19 patients comparing to the control [35].
Choline metabolism is also interesting since its derivatives were also reported downregulated in Covid-19 patients [18]. Compared to healthy controls, PCs were downregulated in septic patients; while in Covid-19 patients, the reduction was less pronounced, and they increased with the severity of the disease during their admission stay.
Polarisation of macrophages in response to pathogens requires increased absorption of choline for PCs formation, thereby promoting cytokine secretion [36].
Our group recently showed that RNAemia is associated with higher 28-day intensive care mortality [37]. All positive-strand RNA viruses share similar strategies for genomic replication. They proliferate reorganizing host's cellular membranes to assemble viral replication complexes [38]. Cellular membranes are mainly composed of phospholipids and in particular, PCs constitutes ∼50% of total phospholipids. PCs synthesis is significantly enhanced during several viral infections (Dengue virus, poliovirus and hepatitis C virus) in order to promote significantly enhanced accumulation of PCs content at the viral replication sites [38].
Glutamine was increased in Covid-19 patients compared to septic patients, without difference with HC. It is the most abundant free amino acid in human blood, that can be utilized as a major respiratory substrate by glutaminase-expressing tissues including liver, kidney, intestine, lymphocytes and monocytes [39]. Reduced levels of glutamine may therefore have deleterious effects on monocyte function [39].
During sepsis, there is an increase in glutamine synthetase activity in skeletal muscle, depleting this amino acid, indicating accelerated uptake by other organs, in particular the liver [40]. This has led to its use as nutritional supplement in critically ill patients, leading to reduced inflammatory cytokines production and increased heat shock protein expression [41]. In animal models, glutamine shows enhanced protective immunity to Herpes simplex virus (HSV)-1 mucosal infection [42] and may enhance the IFNɣ –associated immune response and reduce the rate of reactivation of latent virus infection [43].
A recent interventional study showed that adding enteral L-glutamine to the normal nutrition in the early period of Covid-19 infection may reduce length of hospital stay and reduce the risk of ICU admission [44].
Metabolic syndrome is recognised as risk factor for Covid-19 related death. Several metanalyses showed that diabetes mellitus doubles the risk of dying from Covid-19 [45, 46]. Elevated plasma glucose, indeed, induces viral replication and proinflammatory cytokine expression [47, 48]. In our smaller cohort of patients, only age was associated with increased mortality, while diabetes, hypertension and BMI>30 were not significantly associated.
In this study, we did not identify a metabolic predictive panel of mortality in patients with Covid-19. The findings of this manuscript are limited by the small sample size. Another limitation of our study is that several patients, even if sampled within 24h since admission, had already received a first dose of immunomodulating or antiviral therapeutics as per our protocol or as part of interventional studies protocol (including steroids, anakinra, tocilizumab, barticinib, ravulizumab, baricitinib, remdesevir). Unfortunately, the number of patients receiving each treatment is too small to draw conclusions on the metabolic effect of novel therapeutics. Moreover, the mortality in our cohort is lower compared to the international data, which may be due to our large pre-existing critical care bed numbers and high levels of involvement in interventional research studies of agents subsequently found to be associated with reduced mortality [49].
5. Conclusions
In our cohort, Covid-19 is associated with monocytopenia, increased CD14+ and Treg PD-L1 expression correlating with IFN-ɣ plasma concentration and disease severity (SOFA score). The latter is also associated with metabolic derangements of tryptophan, LPC 16:0 and PCs. Lipid metabolism, in particular phosphatidylcholines and lysophosphatidylcolines, seems strictly linked to immune response in Covid-19. Our results support the hypothesis that IFN-ɣ -PD-L1 axis might be involved in the cytokine release syndrome typical of severe Covid-19 and the phenomenon persisted through multiple pandemic waves despite use of immunomodulation.
Declarations
Author contribution statement
Francesca M. Trovato, PhD: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Salma Mujib; Ellen Jerome; Anna Cavazza & Salvatore Napoli: Performed the experiments.
Phillip Morgan; James Luxton & Tracey Mare: Performed the experiments; Contributed reagents, materials, analysis tools or data.
John Smith; Maria Theresa Depante & Kevin O'Reilly: Contributed reagents, materials, analysis tools or data.
Mark JW. McPhail: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
MM is grateful to the NIHR Biomedical Research Council at Guys and St Thomas Hospital for salary and infrastructure support.
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare the following conflict of interests: Francesca Trovato is editor in Heliyon.
Additional information
No additional information is available for this paper.
Acknowledgements
MM is grateful to the NIHR Biomedical Research Council at Guys and St Thomas Hospital for salary and infrastructure support.
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Xu J., Wang Y., Hou H., Feng H., Yang H. Obesity and COVID-19 mortality. Metabolism. 2021:154820. doi: 10.1016/j.metabol.2021.154820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su A.I., Pezacki J.P., Wodicka L., et al. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 2002;99(24):15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spinner C.D., Gottlieb R.L., Criner G.J., et al. Effect of remdesivir vs standard care on clinical status at 11 Days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324(11):1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Group R.C., Horby P., Lim W.S., et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group R.C. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397(10285):1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocar E., Rezen T., Rozman D. Cholesterol, lipoproteins, and COVID-19: basic concepts and clinical applications. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866(2):158849. doi: 10.1016/j.bbalip.2020.158849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawler N.G., Gray N., Kimhofer T., et al. Systemic perturbations in amine and kynurenine metabolism associated with acute SARS-CoV-2 infection and inflammatory cytokine responses. J. Proteome Res. 2021;20(5):2796–2811. doi: 10.1021/acs.jproteome.1c00052. [DOI] [PubMed] [Google Scholar]
- 9.Cas M.D., Roda G., Li F., Secundo F. Functional lipids in autoimmune inflammatory diseases. Int. J. Mol. Sci. 2020;21(9) doi: 10.3390/ijms21093074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trovato F.M., Zia R., Napoli S., et al. Dysregulation of the lysophosphatidylcholine/autotaxin/lysophosphatidic acid Axis in acute-on-chronic liver failure is associated with mortality and systemic inflammation by lysophosphatidic acid-dependent monocyte activation. Hepatology. 2021;74(2):907–925. doi: 10.1002/hep.31738. [DOI] [PubMed] [Google Scholar]
- 11.Xiao N., Nie M., Pang H., et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021;12(1):1618. doi: 10.1038/s41467-021-21907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodge S., Nitschke P., Kimhofer T., et al. NMR spectroscopic windows on the systemic effects of SARS-CoV-2 infection on plasma lipoproteins and metabolites in relation to circulating cytokines. J. Proteome Res. 2021;20(2):1382–1396. doi: 10.1021/acs.jproteome.0c00876. [DOI] [PubMed] [Google Scholar]
- 13.Laing A.G., Lorenc A., Del Molino Del Barrio I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 14.Karki R., Sharma B.R., Tuladhar S., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184(1):149–68 e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer M., Deutschman C.S., Seymour C.W., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent J.L., Moreno R., Takala J., et al. The SOFA (Sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the European society of intensive care medicine. Intensive Care Med. 1996;22(7):707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 17.Santegoets S.J., Dijkgraaf E.M., Battaglia A., et al. Monitoring regulatory T cells in clinical samples: consensus on an essential marker set and gating strategy for regulatory T cell analysis by flow cytometry. Cancer Immunol. Immunother. 2015;64(10):1271–1286. doi: 10.1007/s00262-015-1729-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen B., Yi X., Sun Y., et al. Proteomic and metabolomic Characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72 e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barberis E., Timo S., Amede E., et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas T., Stefanoni D., Reisz J.A., et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14) doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y., Chen D., Yuan D., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6):1479–1495 e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajaz S., McPhail M.J., Singh K.K., et al. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Physiol. Cell Physiol. 2021;320(1):C57–C65. doi: 10.1152/ajpcell.00426.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhar S.K., K V, Damodar S., Gujar S., Das M. IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: results from meta-analysis and regression. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coomes E.A., Haghbayan H. Interleukin-6 in Covid-19: a systematic review and meta-analysis. Rev. Med. Virol. 2020;30(6):1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchings S.D., Watchorn J., Trovato F., et al. Microcirculatory, endothelial and inflammatory responses in critically ill patients with COVID-19 are distinct from those seen in septic shock: a case control study. Shock. 2020 doi: 10.1097/SHK.0000000000001672. [DOI] [PubMed] [Google Scholar]
- 26.Diao B., Wang C., Tan Y., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai H., Liu G., Zhong J., et al. Immune checkpoints in viral infections. Viruses. 2020;12(9) doi: 10.3390/v12091051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibellini L., De Biasi S., Paolini A., et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Mol. Med. 2020;12(12) doi: 10.15252/emmm.202013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaath H., Alajez N.M. Identification of PBMC-based molecular signature associational with COVID-19 disease severity. Heliyon. 2021;7(5) doi: 10.1016/j.heliyon.2021.e06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Diaz A., Shin D.S., Moreno B.H., et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19(6):1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimura K., Teh J.L., Okayama H., et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109(1):43–53. doi: 10.1111/cas.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi D., Yan R., Lv L., et al. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118:154739. doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botticelli A., Mezi S., Pomati G., et al. Tryptophan catabolism as immune mechanism of primary resistance to anti-PD-1. Front. Immunol. 2020;11:1243. doi: 10.3389/fimmu.2020.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badawy A.A. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res. 2017;10 doi: 10.1177/1178646917691938. 1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H., Gong J., Liu Y. Indoleamine 2, 3-dioxygenase regulation of immune response (Review) Mol. Med. Rep. 2018;17(4):4867–4873. doi: 10.3892/mmr.2018.8537. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Lopez E., Zhong Z., Stubelius A., et al. Choline uptake and metabolism modulate macrophage IL-1 beta and IL-18 production. Cell Metabol. 2019;29(6):1350–13562 e7. doi: 10.1016/j.cmet.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutmann C., Takov K., Burnap S.A., et al. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat. Commun. 2021;12(1):3406. doi: 10.1038/s41467-021-23494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Zhang Z., Chukkapalli V., et al. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc. Natl. Acad. Sci. U. S. A. 2016;113(8):E1064–E1073. doi: 10.1073/pnas.1519730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zellner M., Gerner C., Munk Eliasen M., et al. Glutamine starvation of monocytes inhibits the ubiquitin-proteasome proteolytic pathway. Biochim. Biophys. Acta. 2003;1638(2):138–148. doi: 10.1016/s0925-4439(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 40.Karinch A.M., Pan M., Lin C.M., Strange R., Souba W.W. Glutamine metabolism in sepsis and infection. J. Nutr. 2001;131(9 Suppl):2535S–2538S. doi: 10.1093/jn/131.9.2535S. discussion 50S-1S. [DOI] [PubMed] [Google Scholar]
- 41.Kim H. Glutamine as an immunonutrient. Yonsei Med. J. 2011;52(6):892–897. doi: 10.3349/ymj.2011.52.6.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uyangaa E., Lee H.K., Eo S.K. Glutamine and leucine provide enhanced protective immunity against mucosal infection with herpes simplex virus type 1. Immune Netw. 2012;12(5):196–206. doi: 10.4110/in.2012.12.5.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K., Hoshino Y., Dowdell K., et al. Glutamine supplementation suppresses herpes simplex virus reactivation. J. Clin. Invest. 2017;127(7):2626–2630. doi: 10.1172/JCI88990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cengiz M., Borku Uysal B., Ikitimur H., et al. Effect of oral l-Glutamine supplementation on Covid-19 treatment. Clin. Nutr. Exp. 2020;33:24–31. doi: 10.1016/j.yclnex.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varikasuvu S.R., Dutt N., Thangappazham B., Varshney S. Diabetes and COVID-19: a pooled analysis related to disease severity and mortality. Prim Care Diabetes. 2021;15(1):24–27. doi: 10.1016/j.pcd.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aggarwal G., Lippi G., Lavie C.J., Henry B.M., Sanchis-Gomar F. Diabetes mellitus association with coronavirus disease 2019 (COVID-19) severity and mortality: a pooled analysis. J. Diabetes. 2020;12(11):851–855. doi: 10.1111/1753-0407.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Codo A.C., Davanzo G.G., Monteiro L.B., et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1 alpha/Glycolysis-Dependent Axis. Cell Metabol. 2020;32(3):437–446 e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Y., Lv Y., Zha W., Zhou N., Hong X. Association of body mass index (BMI) with critical COVID-19 and in-hospital mortality: a dose-response meta-analysis. Metabolism. 2021;117:154373. doi: 10.1016/j.metabol.2020.154373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vlachos S., Wong A., Metaxa V., et al. Hospital mortality and resource implications of hospitalisation with COVID-19 in london, UK: a prospective cohort study. Crit. Care Res. Pract. 2021;2021:8832660. doi: 10.1155/2021/8832660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.