Abstract
Cells of a newly isolated environmental strain of Candida humicola accumulated 10-fold more polyphosphate (polyP), during active growth, when grown in complete glucose-mineral salts medium at pH 5.5 than when grown at pH 7.5. Neither phosphate starvation, nutrient limitation, nor anaerobiosis was required to induce polyP formation. An increase in intracellular polyP was accompanied by a 4.5-fold increase in phosphate uptake from the medium and sixfold-higher levels of cellular polyphosphate kinase activity. This novel accumulation of polyP by C. humicola G-1 in response to acid pH provides further evidence as to the importance of polyP in the physiological adaptation of microbial cells during growth and development and in their response to environmental stresses.
Inorganic polyphosphate (polyP) is a linear polymer of phosphate residues linked together by high-energy phosphoanhydride bonds (34). PolyP, ranging in length from 3 to greater than 1,000 orthophosphate residues, has been detected in almost all organisms studied, including bacteria, yeasts, fungi, plants, and animals (12, 35, 36). Under optimal conditions polyP may amount to 10 to 20% of the cellular dry weight and thus greatly exceed the phosphate requirement of the cell, suggesting metabolic roles other than simply as a phosphate reserve (42). Microbial polyP synthesis is primarily carried out by the enzyme polyphosphate kinase (PPK) (32), which catalyzes the reversible transfer of the gamma phosphate from ATP to polyP (1, 21, 29, 53). PolyP hydrolysis is mediated by exopolyphosphatases (PPX) (2, 60), endopolyphosphatases (38), and specific kinases (59).
The exact physiological function of polyP remains uncertain, although various biological functions have been demonstrated, including as a reservoir of energy and phosphate (32), a chelator of divalent cations (30), a capsule material (54), and a “channelling” agent in the phenomenon of bacterial transformation (32, 48). Moreover, extensive accumulation of polyP has been detected in Escherichia coli in response to osmotic stress or nutritional stress imposed by either nitrogen, amino acid, or phosphate limitation (3, 46). Similarly, Pseudomonas aeruginosa mucoid strain 8830 accumulates intracellular polyP particularly during stationary phase and in response to phosphate and amino acid limitations (3, 31), while both the unicellular alga Dunaliella salina and Saccharomyces cerevisiae utilize their intracellular polyP reserves to provide a pH-stat mechanism to counterbalance alkaline stress (4, 11, 42, 43). PolyP accumulation has also been observed upon exposure of the freshwater sponge Ephydatia muelleri to various organic pollutants (27). Of greatest economical significance is the accumulation of polyP by some Acinetobacter spp. and other undefined organisms when they are exposed to alternating anaerobic/aerobic cycles; this phenomenon is the basis of the biotechnologically important wastewater treatment process designated Enhanced Biological Phosphate Removal (reviewed by VanLoosdrecht et al. [56] and Mino et al. [41]). PolyP may therefore play an important role in the physiological adaptation of microbial cells during growth and development and in their response to nutritional and environmental stresses (3, 32, 39, 45, 46, 61).
In this paper we describe the intracellular accumulation of polyP by the yeast Candida humicola G-1 through increased PPK activity when it is grown under acid conditions; no significant polyP accumulation occurs in cells grown at pH 7.5.
MATERIALS AND METHODS
Enrichment and growth conditions.
C. humicola G-1 was isolated by enrichment culture during a program of study to examine the potential for polyP accumulation by microorganisms when they are subjected to a range of environmental stresses. Enrichment under acid conditions was carried out at pH 5.5 using a basal mineral salts medium which contained the following (per liter): KCl, 0.2 g; MgSO4 · 7H2O, 0.2 g; CaCl2 · 2H2O, 1.0 mg; (NH4)2SO4, 0.4 g; ferric ammonium citrate, 1.0 mg; BME essential amino acids solution (Sigma), 20 ml; KH2PO4, 35 mg; phosphate-free yeast extract (58), 0.05 g; and 1 ml each of trace element solution (33) and vitamin solution (40). Filter-sterilized (pore size, 0.22 μm) glucose (0.65 g/liter) was added as a carbon source. The pH of the medium was adjusted to 5.5 by the addition of 50 mM phthalate–NaOH buffer; 50 mM Tris-HCl buffer was used for media at pH 7.5.
Enrichment cultures were inoculated with a 0.5% (vol/vol) inoculum from an activated sludge plant (Dunmurry, United Kingdom). Cultures were incubated at 30°C on an orbital shaker at 100 rpm. Growth was measured by determining the increase in optical density at 650 nm (OD650) with PU 8200 UV/Vis spectrophotometer (Pye-Unicam Ltd., Cambridge, United Kingdom), while phosphate removal from the culture supernatant was monitored by the method of Fiske and SubbaRow (19). Glucose metabolism was assayed with the Trinder Glucose Determination Kit (Sigma). The presence of intracellular polyP inclusions was determined by light microscopy after the cells were stained by the method of Neisser (25).
Growth, phosphate removal from culture supernatant, and intracellular polyP were scored by comparison with control cultures grown in complete medium at pH 7.5. After three serial transfers, a yeast isolate, designated G-1, capable of growth and intracellular polyP accumulation in response to acid stress, was purified on complete mineral salts medium (pH 5.5) solidified by the addition of 1.2% Purified Agar (Oxoid). It was identified at the Regional Mycology Laboratory (Leeds, United Kingdom) as a strain of C. humicola, a yeast commonly isolated from soil (57). In all experiments C. humicola G-1 was routinely precultured on basal mineral salts medium supplemented with glucose (0.65 g/liter) at pH 7.5. Under such conditions no intracellular polyP accumulation occurred; this ensured a uniform, polyP-free inoculum.
Extraction of polyP.
Total intracellular polyP was assayed by a modification of the methods of Streichan and Schön (52), Rao et al. (47), and Poindexter and Eley (44). Cells of C. humicola G-1, grown at either pH 5.5 or 7.5 as previously described, were harvested by centrifugation at 10,000 × g for 15 min at 4°C and washed twice in 1.5 M NaCl containing 0.01 M EDTA and 1 mM NaF. The cell pellet was resuspended in 1.5 ml of wash buffer and sonicated, on ice, for 10 30-s periods with 2-min intervals at 16 kHz. The resulting homogenate was centrifuged at 25,000 × g for 60 s at 4°C to remove cell debris. To determine total intracellular polyP, 100 μl of concentrated HCl was added to 0.5 ml of cell extract and heated at 100°C for 45 min; the phosphate liberated was assayed by the method of Fiske and SubbaRow (19). The polyP concentrations were expressed in milligrams of phosphate per milligram of cellular protein and are given as means of triplicates. An unhydrolyzed sample was used as a control to determine the background level of Pi. Protein concentration was determined by the method of Bradford (9) using bovine serum albumin as the standard.
Cell lysis and enzyme assays.
C. humicola G-1 was harvested and sonicated as described earlier. Sonicated cell homogenate was centrifuged at 25,000 × g and 4°C for 30 min, and the resultant supernatant or crude extract was stored at 4°C for not more than 24 h. PPK activity was measured by the ATP-regenerating method of Robinson and Wood, which spectrophotometrically assays polyP synthesis (50). This method was chosen in preference to assaying the reverse reaction (production of ATP from polyP using [32P]polyP) since the validity of the latter approach has been questioned due to interference caused by the enzyme diadenosine-5′,5′"-P1-P4-tetraphosphate α,β-phosphorylase in the assay procedure (7). Employment of ATP regeneration is necessary due to the inhibitory nature of ADP (55). PPK assays were carried out by using sulfate-free enzymes and without the addition of the basic protein polylysine; no increase in enzyme activity was observed upon addition of polylysine (0.5 mg/ml). Background levels of both ATP hydrolysis and phosphoenolpyruvate hydrolysis by cell extracts were determined, and PPK activities were corrected accordingly. PPX was assayed by the method of Bonting et al. (6).
Analysis of polyP by gel electrophoresis.
PolyP samples were prepared for gel electrophoresis by the method of Robinson et al. (49). Polyacrylamide gel electrophoresis was performed with Tris-borate buffer (pH 8.3) using precast 15% Tris–borate–EDTA–urea gels on a Novex X cell 11 Mini-cell system (Novex, San Diego, Calif.) for 75 min at 10 mA. Gels were stained with toluidine blue (0.05%) in 25% methanol. The estimated size range of the polyP was determined by comparison to polyP standards (chain lengths, 25, 45, and 75 P molecules; Sigma).
RESULTS
Accumulation of polyP.
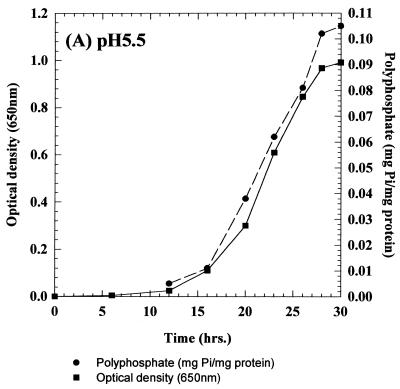
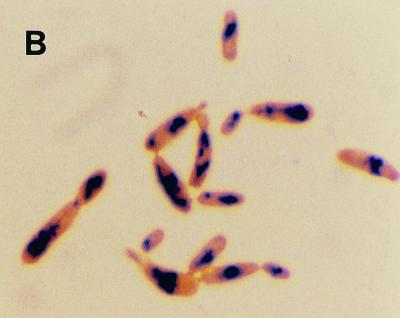
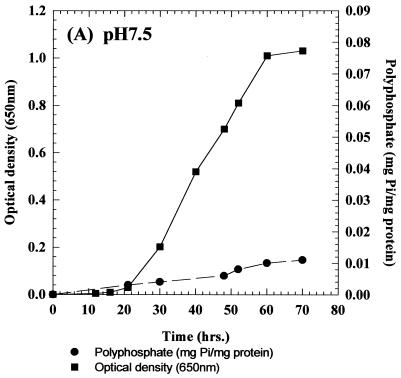
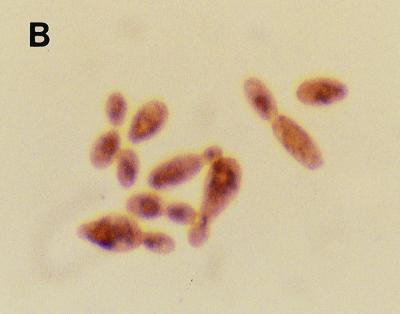
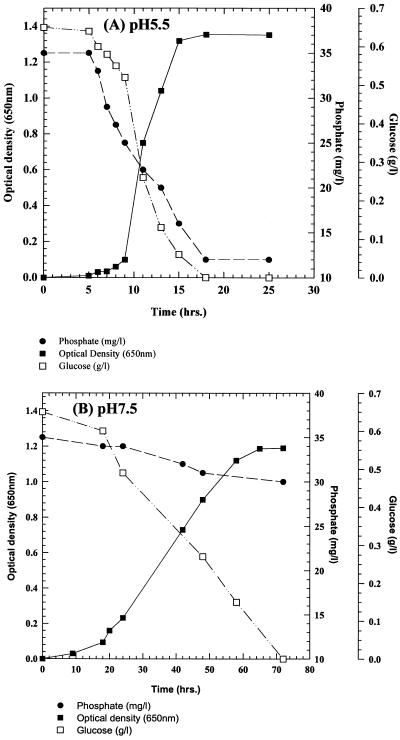
The growth and intracellular accumulation of polyP by C. humicola G-1 grown on basal mineral salts medium containing 0.65 g of glucose per liter at pH 5.5 and 7.5 are shown in Fig. 1A and 2A, respectively. During growth at pH 7.5 the maximum polyP concentration was 0.01 mg of Pi/mg of total cellular protein (Fig. 2A). In contrast, C. humicola G-1 grown at pH 5.5 produced a marked accumulation of intracellular polyP (Fig. 1A), reaching a stationary-phase maximum of 0.105 mg of Pi/mg of total cellular protein, representing some 5.7% of the cellular dry weight. Neisser staining confirmed the presence of intracellular polyP granules in cells grown at pH 5.5 (Fig. 1B); no polyP granules were observed in pH 7.5-grown cells (Fig. 2B). The presence of polyP granules was also confirmed by using the 31P nuclear magnetic resonance method of Rao et al. (47) for polyP detection in intact cells. Despite the increased intracellular accumulation of polyP under acid conditions, stationary-phase cell yields at the two pH values were virtually identical (OD650 values of 0.991 and 1.03 [Fig. 1A and 2A]). No polyP hydrolysis occurred after resuspension of polyP-rich cells, grown at pH 5.5, in complete growth medium at pH 7.5 (data not shown).
FIG. 1.
(A) Growth and intracellular polyP accumulation by C. humicola G-1 at pH 5.5. (B) Stained cells of C. humicola G-1 grown at pH 5.5, showing polyP granules stained blue-black with Neisser stain. Magnification, ×1,000.
FIG. 2.
(A) Growth and intracellular polyP accumulation by C. humicola G-1 at pH 7.5. (B) Stained cells of C. humicola G-1 grown at pH 7.5, showing the absence of polyP granules. Magnification, ×1,000.
Gel electrophoretic analysis of polyP.
Analysis of the polyP that accumulated during growth of C. humicola G-1 at pH 5.5 by gel electrophoresis gives an indication of the mechanism of polyP biosynthesis. PolyP extracted from cells at a culture OD650 of 0.2 consisted solely of low-molecular-weight polyP equivalent to an approximate chain length of 45 residues (results not shown). In contrast, cells harvested at a culture OD650 of 0.8 contained high-molecular-weight polyP containing up to 150 residues in addition to the shorter-chain polyP. This is in accord with the results reported by Schuddemat et al. (51) for polyP synthesis in the yeasts S. cerevisiae and Kluyceromyces marxianus, during which polyP was formed progressively as chains of increasing length.
Phosphate uptake by C. humicola G-1.
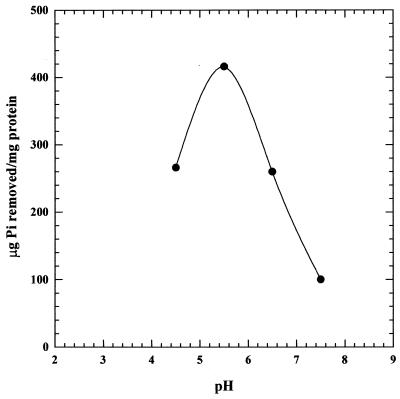
Associated with intracellular polyP production by C. humicola G-1 under acid conditions was the enhanced uptake of exogenous inorganic phosphate from the medium during growth compared to uptake at pH 7.5. C. humicola G-1, grown at pH 5.5 in a medium containing glucose (0.65 g/liter) as carbon source, removed 22 mg of inorganic phosphate per liter (Fig. 3A), compared with the 5 mg/liter removed at pH 7.5 (Fig. 3B). Complete glucose mineralization was observed in each case (Fig. 3). No growth or phosphate uptake occurred in control cultures lacking glucose. Phosphate removal, as determined by the concentration of phosphate remaining in the growth medium upon completion of growth at different pH values, was optimal at pH 5.0 to 5.5 (Fig. 4). At pH values on either side of the optimum the amount of inorganic phosphate taken up by the cells decreased, although at each pH tested both the final cell yields and the total culture protein concentrations (determined by the method of Binks et al. [5]) were identical.
FIG. 3.
(A) Growth, glucose mineralization, and phosphate removal by C. humicola G-1 at pH 5.5. (B) Growth, glucose mineralization, and phosphate removal by C. humicola G-1 at pH 7.5.
FIG. 4.
Phosphate uptake by C. humicola G-1 at pH 4.5 to 7.5.
PPK and polyphosphatase activities.
The activities of the enzymes PPK and polyphosphatase were compared during growth of C. humicola G-1 at pH 5.5 and 7.5 (Table 1). At pH 7.5 both enzymes displayed similar levels of activity. Growth of the organism at pH 5.5 however resulted in a sixfold increase in PPK activity while polyphosphatase activity remained unaltered (Table 1).
TABLE 1.
Activities of PPK and polyphosphatase in C. humicola G-1 grown at either pH 5.5 or 7.5
Enzyme activities are means of triplicate assays from four separate cell extract preparations; for each preparation cells were harvested at a culture OD650 of 0.600 to 0.700.
PPK activities are expressed as units per milligram of total protein in the cell extract; 1 U of activity is defined as the amount of enzyme needed to generate 1 μmol of ATP from polyP and ADP in 1 min at 20°C.
Polyphosphatase activities are expressed as units per milligram of total protein in the cell extract; 1 U of activity is defined as the amount of enzyme that liberated 1 μmol of phosphate from polyP in 1 min at 20°C.
DISCUSSION
Accumulation of polyP has been demonstrated for a wide variety of microorganisms, although the conditions favoring this accumulation differ markedly (12, 37, 59). In general, polyP concentrations are low during exponential growth but may increase either when the stationary phase begins or when growth is arrested due to nutrient imbalance (20). Addition of phosphate to cells previously subjected to phosphate starvation also induces rapid accumulation of polyP: this phenomenon is known as “polyphosphate overplus” (reviewed by Dawes and Senior [12]). Intracellular polyP reserves have often been assumed to mainly provide a reservoir of energy convertible to ATP (32). However, recent studies have demonstrated that even at highly elevated concentrations of polyP, the ATP supply could be sustained for only a matter of seconds, suggesting other metabolic roles (46), such as a role in the survival of microbial cells during stationary phase and a role in the adaptation of cells to environmental stresses (3, 32, 39, 45, 46, 61).
In this paper we describe the intracellular accumulation of polyP by the yeast C. humicola G-1 during growth at acid pH. Neither nutrient limitation, phosphate starvation, nor prior exposure to anaerobiosis is required to induce polyP synthesis. The intracellular polyP concentration increased rapidly during active growth (Fig. 1A); by contrast, polyP accumulation by S. cerevisiae at acid pH was found to be minimal during the maximal growth phase, increasing only during stationary phase (28). PolyP levels were 10-fold greater in cells grown at pH 5.5 than in cells grown at pH 7.5 (Fig. 1A and 2A). The differences are presumably mediated by regulation of either the expression or the activity of the enzymes involved in polyP metabolism (7). Both PPX and PPK have previously been detected in yeasts, although the latter has yet to be purified (36). In cells grown at pH 7.5 the activities of PPX and PPK were approximately equal (Table 1), possibly reflecting the lack of significant polyP accumulation at this pH (Fig. 2). At pH 5.5 PPK activity increased sixfold, while PPX activity remained unaltered (Table 1). This elevation in PPK activity may account for the increased intracellular accumulation of polyP at pH 5.5 (Fig. 1).
The exact physiological role of polyP during growth of C. humicola G-1 at pH 5.5 is at present unknown. Interestingly, no attenuation of growth yield occurred at pH 5.5 in media containing lower phosphate concentrations (5 to 15 mg/liter) (results not shown), suggesting that polyP may not be necessary for survival of cells in the acid environment, although a role in the sequestering of H+ ions cannot be ruled out (26). Such a function could be envisaged given the polyanionic nature of polyP and its known ability to act as an intracellular cation trap (18, 35), which facilitates both intracellular buffering and pH homeostasis. A pH-homeostatic function for polyP has been demonstrated in the unicellular alga D. salina and in S. cerevisiae (grown under conditions necessary to accumulate polyP), in which intracellular polyP levels rapidly decrease in response to the alkalization of the external environment. Hydrolysis of intracellular polyP restores the cytosolic pH by yielding H+ ions (4, 11, 42, 43). The lack of polyP accumulation by C. humicola G-1 at pH 7.5 is not due to an analogous mechanism; no polyP hydrolysis occurs upon resuspension in pH 7.5 medium of cells containing high levels of polyP (data not shown). C. humicola G-1 exhibits a reduced growth rate at pH 7.5 compared to that at pH 5.5 (Fig. 3B). The decrease might suggest the existence of suboptimal growth conditions for C. humicola G-1. It could be envisaged that under such conditions polyP formation is inhibited. It is, however, well documented that yeast cytosolic pH is maintained near neutrality, even if the external pH is modulated between pH 3 and 7.5 (13, 14, 24). The formation of polyP during growth at pH 5.5 might alternatively provide a mechanism to regulate intracellular phosphate levels (26). An acid pH optimum for phosphate transport has been observed in both S. cerevisiae and Rhodotorula rubra (8, 10), while in the present study C. humicola G-1 accumulated phosphate optimally at pH 5.5 (Fig. 4). Growth at an external pH close to the phosphate transport optimum pH may thus result in increased phosphate uptake and elevation of intracellular phosphate concentrations. Such internal phosphate level increases may be countered through the formation of polyP (26).
Regulation of polyP accumulation by C. humicola G-1 grown under acid conditions differs from previous studies involving E. coli and its response to amino acid starvation. Under such nutritional stress E. coli polyP levels may increase 1,000-fold while the cell-free activities of both PPK and PPX remain unaltered (39); polyP accumulation is achieved by selective inhibition of PPX in vivo by either guanosine tetraphosphate or guanosine pentaphosphate generated in response to amino acid starvation (3, 39, 46). This is in contrast to polyP accumulation by C. humicola G-1, which appears to involve the stimulation of PPK activity (Table 1). Such a model for polyP accumulation has been proposed for E. coli in its response to other stress conditions, such as osmotic stress (3). Under these conditions the regulation of polyP accumulation is under the control of additional stress-induced proteins, such as the sigma factor RpoS. RpoS may act, in concert with other regulatory signals, to either inhibit PPX or stimulate PPK (3). An analogous regulatory system could be envisaged within C. humicola G-1 whereby induction of stress proteins in response to either an acid environment or high internal phosphate levels leads to increased PPK activity and hence polyP accumulation. Interestingly, no such response to acid stress occurs in E. coli and no polyP was accumulated under acidic conditions even though RpoS was shown to be present (3).
The intracellular accumulation of polyP observed by us during the exponential growth phase of C. humicola G-1 in response to acid pH is not wholly without precedent. Duguid et al. (15–17) demonstrated that exponentially growing Klebsiella aerogenes produced large quantities of intracellular polyP when it was grown under acid conditions (pH 4.0 to 5.0) but none at neutral pH values. More recently, the tree physiologists Gerlitz et al. (22, 23) have shown that polyP accumulation by the ectomycorrhizal fungus Suillus bovinus, growing in association with the roots of Scotch pine, is maximal at pH 5.5 and some 35% greater than that at pH 7.5. These results are consistent with our findings and may suggest a more widespread acid response mechanism involving polyP accumulation.
Our observations provide further evidence as to the importance of polyP in microbial physiology. Additional investigations are required to identify the exact role played by the increased rate of phosphate uptake and the accumulated intracellular polyP in the response of C. humicola G-1 to external pH.
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom (grant 81E11490) and the Queen's University Environmental Science and Technology Research Centre (QUESTOR).
We thank Noel Duffy for his excellent technical assistance and James Booth (Division of Cell Biology, Toronto Hospital for Sick Children), Robin Rowbury (Department of Biology, University College London), and particularly James Duguid (formerly of the University of Edinburgh) for their helpful discussions on polyP metabolism.
REFERENCES
- 1.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli—isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 2.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli—the enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 3.Ault-Riche D, Fraley C D, Tzeng C M, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bental M, Pick U, Avron M, Degani H. Polyphosphate metabolism in the alga Dunaliella salina studied by P31-NMR. Biochim Biophys Acta. 1991;1092:21–28. doi: 10.1016/0167-4889(91)90173-u. [DOI] [PubMed] [Google Scholar]
- 5.Binks P R, French C E, Nicklin S, Bruce N C. Degradation of pentaerythritol tetranitrate by Enterobacter cloacae PB2. Appl Environ Microbiol. 1996;62:1214–1219. doi: 10.1128/aem.62.4.1214-1219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonting C F C, Vanveen H W, Taverne A, Kortstee G J J, Zehnder A J B. Regulation of polyphosphate metabolism in Acinetobacter strain 210a grown in carbon-limited and phosphate-limited continuous cultures. Arch Microbiol. 1992;158:139–144. [Google Scholar]
- 7.Booth J W, Guidotti G. An alleged yeast polyphosphate kinase is actually diadenosine-5′,5′"-P1,P4-tetraphosphate α,β-phosphorylase. J Biol Chem. 1995;270:19377–19382. doi: 10.1074/jbc.270.33.19377. [DOI] [PubMed] [Google Scholar]
- 8.Borst-Pauwels G W F H, Peters P H J. Effect of the medium pH and the cell pH upon the kinetical parameters of phosphate uptake by yeast. Biochim Biophys Acta. 1977;466:488–495. doi: 10.1016/0005-2736(77)90341-8. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Button D K, Dunker S S, Morse M L. Continuous culture of Rhodotorula rubra: kinetics of phosphate-arsenate uptake, inhibition, and phosphate-limited growth. J Bacteriol. 1973;113:599–611. doi: 10.1128/jb.113.2.599-611.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro C D, Meehan A J, Koretsky A P, Domach M M. In situ 31P nuclear magnetic resonance for observation of polyphosphate and catabolite responses of chemostat-cultivated Saccharomyces cerevisiae after alkalinization. Appl Environ Microbiol. 1995;61:4448–4453. doi: 10.1128/aem.61.12.4448-4453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawes E A, Senior P J. Energy reserve polymers in microorganisms. Adv Microb Physiol. 1973;10:178–203. doi: 10.1016/s0065-2911(08)60088-0. [DOI] [PubMed] [Google Scholar]
- 13.De la Pena P, Barros F, Gascon S, Ramos S, Lazo P S. The electrochemical proton gradient of Saccharomyces cerevisiae—the role of potassium. Eur J Biochem. 1982;123:447–453. doi: 10.1111/j.1432-1033.1982.tb19788.x. [DOI] [PubMed] [Google Scholar]
- 14.den Hollander J A, Ugurbil K, Brown T R, Shulman R G. P31 nuclear magnetic resonance studies of the effect of oxygen upon glycolysis in yeast. Biochemistry. 1981;20:5871–5880. doi: 10.1021/bi00523a034. [DOI] [PubMed] [Google Scholar]
- 15.Duguid J P. The influence of cultural conditions on the morphology of Bacterium aerogenes with reference to nuclear bodies and capsule size. J Pathol Bacteriol. 1948;60:265–274. doi: 10.1002/path.1700600214. [DOI] [PubMed] [Google Scholar]
- 16.Duguid J P, Smith I R, Wilkinson J F. Volutin production in Bacterium aerogenes due to development of an acid reaction. J Pathol Bacteriol. 1954;67:289–300. doi: 10.1002/path.1700670202. [DOI] [PubMed] [Google Scholar]
- 17.Duguid J P, Wilkinson J F. Volutin formation in Klebsiella aerogenes. Proc R Physical Soc. 1956;15:24–27. [Google Scholar]
- 18.Dürr M, Urech K, Boller T, Wiemken A, Schwencke J, Nagy M. Sequestration of arginine by polyphosphate in vacuoles of yeast (Saccharomyces cerevisiae) Arch Microbiol. 1979;121:169–175. [Google Scholar]
- 19.Fiske C H, SubbaRow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- 20.Fuhs G W, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb Ecol. 1975;2:119–138. doi: 10.1007/BF02010434. [DOI] [PubMed] [Google Scholar]
- 21.Geißdörfer W, Ratajczak A, Hillen W. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlitz T G M, Gerlitz A. Phosphate uptake and polyphosphate metabolism of mycorrhizal and nonmycorrhizal roots of pine and of Suillus bovinus at varying external pH measured by in vivo 31P-NMR. Mycorrhiza. 1997;7:101–106. [Google Scholar]
- 23.Gerlitz T G M, Werk W B. Investigations on phosphate-uptake and polyphosphate metabolism by mycorrhized and nonmycorrhized roots of beech and pine as investigated by in-vivo 31P-NMR. Mycorrhiza. 1994;4:207–214. [Google Scholar]
- 24.Greenfield N J, Hussain M, Lenard J. Effects of growth state and amines on cytoplasmic and vacuolar pH, phosphate and polyphosphate levels in Saccharomyces cerevisiae: a 31P-nuclear magnetic resonance study. Biochim Biophys Acta. 1987;926:205–214. doi: 10.1016/0304-4165(87)90205-4. [DOI] [PubMed] [Google Scholar]
- 25.Gurr E. The rational use of dyes in biology. London, United Kingdom: Hill; 1965. p. 216. [Google Scholar]
- 26.Harold F M. Inorganic polyphosphate in biology: structure, metabolism, and function. Bacteriol Rev. 1966;30:772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imsiecke G, Munkner J, Lorenz B, Bachinski N, Muller W E G, Schroder H C. Inorganic polyphosphates in the developing freshwater sponge Ephydatia muelleri: effect of stress by polluted waters. Environ Toxicol Chem. 1996;15:1329–1334. [Google Scholar]
- 28.Katchman B J, Fetty W O. Phosphorus metabolism in growing cultures of Saccharomyces cerevisiae. J Bacteriol. 1955;69:607–615. doi: 10.1128/jb.69.6.607-615.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato J, Yamamoto T, Yamada K, Ohtake H. Cloning, sequence and characterisation of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene. 1993;137:242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- 30.Keasling J D, Hupf G A. Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl Environ Microbiol. 1996;62:743–746. doi: 10.1128/aem.62.2.743-746.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H Y, Schlictman D, Shankar S, Xie Z, Chakrabarty A M, Kornberg A. Aliginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary-phase survival and synthesis of RNA/DNA precursors. Mol Microbiol. 1998;27:717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieg N R. Enrichment and isolation. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 112–142. [Google Scholar]
- 34.Kulaev I S. The biochemistry of inorganic polyphosphates. J. New York, N.Y: Wiley and Sons Inc.; 1979. [Google Scholar]
- 35.Kulaev I S, Vagabov V M. Polyphosphate metabolism in microorganisms. Adv Microbiol. 1983;15:731–738. doi: 10.1016/s0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 36.Kulaev I S, Vagabov V M, Kulakovskaya T V. New aspects of inorganic polyphosphate metabolism and function. J Biosci Bioeng. 1999;88:111–129. doi: 10.1016/s1389-1723(99)80189-3. [DOI] [PubMed] [Google Scholar]
- 37.Kumble K D, Kornberg A. Inorganic polyphosphate in mammalian cells and tissues. J Biol Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- 38.Kumble K D, Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J Biol Chem. 1996;271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 40.McGrath J W, Wisdom G B, McMullan G, Larkin M J, Quinn J P. The purification and properties of phosphonoacetate hydrolase, a novel carbon-phosphorus bond-cleaving enzyme from Pseudomonas fluorescens 23F. Eur. J. Biochem. . 1995;234:225–230. doi: 10.1111/j.1432-1033.1995.225_c.x. [DOI] [PubMed] [Google Scholar]
- 41.Mino T, VanLoosdrecht M C M, Heijnen J J. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 1998;32:3193–3207. [Google Scholar]
- 42.Pick U, Bental M, Chitlaru E, Weiss M. Polyphosphate-hydrolysis—a protective mechanism against alkaline stress. FEBS Lett. 1990;274:15–18. doi: 10.1016/0014-5793(90)81318-i. [DOI] [PubMed] [Google Scholar]
- 43.Pick U, Weiss M. Polyphosphate hydrolysis within acidic vacuoles in response to amine-induced alkaline stress in the halotolerant alga Dunaliella-salina. Plant Physiol. 1991;97:1234–1240. doi: 10.1104/pp.97.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poindexter J S, Eley E F. Combined procedures for assays of poly-β-hydroxybutyric acid and inorganic polyphosphate. J Microbiol Methods. 1983;1:1–17. [Google Scholar]
- 45.Rao N N, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao N N, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao N N, Roberts M F, Torriani A. Amount and chain length of polyphosphates in Escherichia coli depend on cell growth conditions. J Bacteriol. 1985;162:242–247. doi: 10.1128/jb.162.1.242-247.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reusch R N, Sadoff H L. Putative structure and functions of a poly-β-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc Natl Acad Sci USA. 1988;85:4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson N A, Clark J E, Wood H G. Polyphosphate kinase from Propionibacterium shermanii: demonstration that polyphosphates are primers and determination of the size of the synthesized polyphosphate. J Biol Chem. 1987;262:5216–5222. [PubMed] [Google Scholar]
- 50.Robinson N A, Wood H G. Polyphosphate kinase from Propionibacterium shermanii—demonstration that the synthesis and utilization of polyphosphate is by a processive mechanism. J Biol Chem. 1986;261:4481–4485. [PubMed] [Google Scholar]
- 51.Schuddemat J, Deboo R, Vanleeuwen C C M, Vandenbroek P J A, Vansteveninck J. Polyphosphate synthesis in yeast. Biochim Biophys Acta. 1989;1010:191–198. doi: 10.1016/0167-4889(89)90160-2. [DOI] [PubMed] [Google Scholar]
- 52.Streichan M, Schon G. Periplasmic and intracytoplasmic polyphosphate and easily washable phosphate in pure cultures of sewage bacteria. Water Res. 1991;25:9–13. [Google Scholar]
- 53.Tinsley C R, Gotschlich E C. Cloning and characterization of the meningococcal polyphosphate kinase gene: production of polyphosphate synthesis mutants. Infect Immun. 1995;63:1624–1630. doi: 10.1128/iai.63.5.1624-1630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tinsley C R, Manjula B N, Gotschlich E C. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect Immun. 1993;61:3703–3710. doi: 10.1128/iai.61.9.3703-3710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trelstad P L, Purdhani P, Geißdörfer W, Hillen W, Keasling J D. Polyphosphate kinase of Acinetobacter sp. strain ADP1: purification and characterization of the enzyme and its role during changes in extracellular phosphate levels. Appl Environ Microbiol. 1999;65:3780–3786. doi: 10.1128/aem.65.9.3780-3786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VanLoosdrecht M C M, Hooijmans C M, Brdjanovic D, Heijnen J J. Biological phosphate removal processes. Appl Microbiol Biotechnol. 1997;48:289–296. [Google Scholar]
- 57.Van Uden N, Buckley H. The life of yeasts. Cambridge, Mass: Harvard University Press; 1978. pp. 219–220. [Google Scholar]
- 58.Weimberg R, Orton W L. Repressible acid phosphomonoesterase and constitutive pyrophosphatase of Saccharomyces mellis. J Bacteriol. 1963;86:805–813. doi: 10.1128/jb.86.4.805-813.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood H G, Clark J E. Biological aspects of inorganic polyphosphates. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 60.Wurst H, Kornberg A. A soluble exopolyphosphatase of Saccharomyces cerevisiae—purification and characterization. J Biol Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- 61.Zago A, Chugani S, Chakrabarty A M. Cloning and characterization of polyphosphate kinase and exopolyphosphatase genes from Pseudomonas aeruginosa 8830. Appl Environ Microbiol. 1999;65:2065–2071. doi: 10.1128/aem.65.5.2065-2071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]