Abstract
Temozolomide (TMZ) monotherapy is known to be insufficient for resistant/relapsed glioblastoma (GBM), thus seeking a sensitization agent for TMZ is necessary. It was found that regorafenib may improve the overall survival of relapsed GBM patients. We aimed to discover whether regorafenib can enhance the anti-GBM effects of TMZ, and elucidate underlying mechanism. Our analysis of The Cancer Genome Atlas database revealed that the increased expression of CXCR4 is linked to poor survival of GBM patients. Additionally, TMZ treatment may trigger CXCR4/CXCL12 axis of GBM. We used two GBM cell lines, two primary GBM cells, and animal model to identify underlying mechanism and treatment efficacy of regorafenib combined with TMZ by cytotoxicity, apoptosis, reporter gene and invasion/migration assays, chemokine array, Western blotting, MRI, microarray, and immunohistochemistry. We observed that the chemokine CXCL-12 and its receptor CXCR4 regulate the resistance to TMZ, whereas the inhibition of CXCL-12/CXCR4 signaling sensitizes GBM cells to TMZ. The TMZ-induced CXCL-12/CXCR4 signaling, phosphor-extracellular signal-regulated kinases 1 and 2 (ERK1/2) and nuclear factor kappa light chain enhancer of activated B cells (NF-κB), and NF-κB-related proteins can effectively diminish when combining with regorafenib. Regorafenib significantly enhanced the TMZ-induced extrinsic/intrinsic apoptotic pathways, and facilitated the suppression of invasion and migration potential in GBM. Orthotopic tumor experiments demonstrated tumor size reduction and prolonged survival in combination group even with half-dose of TMZ. Our findings provide promising evidence that regorafenib may sensitize GBM to TMZ treatment through inhibition of the CXCL12/CXCR4/ERK/NF-κB signaling.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-022-01194-y.
Keywords: Glioblastoma, Temozolomide, Regorafenib, NF-κB, CXCR4/CXCL12
Introduction
Glioblastoma (GBM), also known as “grade IV glioma”, exhibits poor patient survival due to rapid disease progression [1, 2]. Temozolomide (TMZ), an oral chemotherapeutic agent, has been approved for the treatment of high-grade gliomas (astrocytoma and GBM) and found to improve the survival of GBM patients [3]. In addition, the combination of TMZ with radiotherapy significantly prolonged the median overall survival of patients from 12.1 months (radiotherapy alone) to 14.6 months and increased the two-year survival rate from 10.4% (radiotherapy alone) to 26.5% for patients with newly diagnosed GBM [4, 5].
TMZ, which is an alkylating agent, was found to promote apoptosis by inducing deoxyribonucleic acid [6] lesion O6-methylguanine in GBM [7]. Several key proteins such as O6-alkylguanine DNA alkyltransferase (MGMT), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), chemokine receptor CXCR-4, and its ligand CXCL12 have been shown to plays role on TMZ-treated GBM cells [8, 9]. For instance, The CXCL12/CXCR4 pathway was also found to correlate with the recurrence of GBM after radiotherapy treatment combined with TMZ [10]. Therefore, the anticancer activity of TMZ is limited by the emergence of resistance and insensitivity to TMZ, resulting in GBM treatment failure [11, 12]. Consequently, the development of complementary agents which may sensitize GBM cells to TMZ is an urgent necessity for GBM patients.
Sorafenib, the oral multi-kinase inhibitor, diminishes tumor progression by targeting several oncogenic kinases and receptor tyrosine kinases such as Raf, vascular endothelial growth factor receptor 2 (VEGFR2), platelet-derived growth factor receptor (PDGFR), FLT3, and c-Kit [6, 13]. Sorafenib is approved for treatment of advanced renal cell carcinoma (RCC), hepatocellular carcinoma (HCC), and differentiated thyroid carcinoma (DTC) [14]. Many clinical studies demonstrated the efficacy and toxicity of sorafenib combined with TMZ in patients with GBM. The combination of sorafenib and TMZ had limited activity with acceptable toxicity in patients with relapsed GBM [15, 16].
Regorafenib is an oral multi-kinase inhibitor which has a chemical structure similar to sorafenib which possesses potent activity against the receptors of different oncogenic and tyrosine kinases [17, 18]. Regorafenib is used to treat advanced gastrointestinal stromal tumors, metastatic colorectal cancer, and HCC after the failure of other treatments [19, 20]. Regorafenib was found to significantly improve the overall survival of relapsed GBM patients compared to lomustine, which is an alkylating agent that is used as a positive treatment control in phases 2 and 3 clinical trials in patients with relapsed GBM [21]. Herrlinger et al. showed that the combination of lomustine and TMZ resulted in significant survival benefits for GBM patients compared to the standard TMZ treatment [22]. Furthermore, regorafenib showed potential to suppress CXCL12/CXCR4-mediated resistance on gastric cancer [23].
The knowledge whether regorafenib enhances the therapeutic efficacy of TMZ is still ambiguous. Therefore, the major purposes of this study are to investigate in vitro and in vivo the anticancer efficacy of regorafenib–TMZ combined GBM treatment and explore the underlying therapeutic mechanisms.
Materials and Methods
Cell Culture
U-87 MG cells was purchased from ATCC and GBM8401 cells was kindly provided from professor Tsung-I Hsu. Two patient–derived glioblastoma cells used in this study were obtained from Taipei Medical University (provided by professor Szu-Yi Chou) [24], and successfully transformed into stable cell lines BP-3 and BP-5. Their IRB Protocol approval no.: 201006011 and 201,805,040. Cells were maintained in minimum essential medium eagle medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and 1% sodium pyruvate; Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and 2 mM L-glutamine; and RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and 2 mM L-glutamine (Thermo Fisher Scientific). The atmosphere of incubator was maintained at 37 °C and contained 5% CO2. All cell lines were cultured for less than 3 months for every batch of cells, and checked for mycoplasma after every freeze/thaw cycle, using the Mycoplasma Detection Kit (Lonza Group Ltd, Switzerland).
The Cancer Genome Atlas Expression Profiling and Survival Analysis
The mRNA expression levels and overall survival of glioma patients were determined using GEPIA (http://gepia.cancer-pku.cn), an online analysis software based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, using [log2FC] = 1 and p = 0.01 as the cutoff criteria (Tang et al., 2017). The CXCR4 mRNA expression levels are presented as boxplots (N = 1095). The overall survival difference between glioma patients with either low or high expression levels of CXCR4 was also calculated (N = 676). The dot line presented on figure was represented as 95% confidence interval.
Chemokine Protein Array Immunoassays
To assess the role of chemokine proteins after single agent and combination treatment, the changes of chemokine levels in U-87 MG cells were analyzed. Briefly, U-87 MG cells (1 × 105 cells/well) were cultured in 6-well plates overnight. Cells were then treated with regorafenib (10 μM), TMZ (120 μM), or combination of both for 24 h. Subsequently, proteins from cells were extracted and stored at −80 °C. The production of chemokines was measured by the commercially available kit Human Chemokine Array C1 [AAH-CHE-1] (RayBiotech Life, Norcross, GA, USA) according to the instructions of the manufacturer. The levels of chemokine from human primary culture cells were calculated according to corresponding standards by Bio-Rad Image Lab software.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl-Tetrazolium Bromide Assay (MTT Assay) and Combination Index
Three thousand cells were seeded in 96-well plates. After 1 day of incubation, cells were treated with regorafenib (0–30 μM), TMZ (0–160 μM), 50 ng/ml CXCR4 recombinant protein, 100 ng/ml CXCL12 recombinant protein, 30 μM AMD 3100 (CXCR4 inhibitor), LIT-927 (CXCL12 neutraligand), or combination of both for 24 h. The stock solution of MTT was prepared in PBS at a concentration of 5 mg/ml. Aliquots of 100 μl of MTT were added to every well and incubated for 4 h at 37 °C, then replaced by aliquots of 100 μl of DMSO. The absorbance values of MTT were measured by Thermo Multiskan™ GO Microplate Spectrophotometer at 570 nm. We used the Chou-Talalay method to calculate the CI-isobologram index using the CompuSyn software (ComboSyn) [25, 26]. The definition of CI values was listed as follows: CI = 1, additive effect; CI < 1, synergistic effect; CI > 1, antagonistic effect.
NF-κB Plasmid Transfection, Stable Clone Selection
U-87 MG cells were seeded into 10-cm dishes. The confluence of cells was allowed to reach 80% at the day of transfection. The vector of NF-κB-luciferrase-2 was obtained from Promega (Madison, WI, USA). Transfection process was performed according to the protocol provided by the manufacturer of jetPEI™ kit (Polyplus-transfection®). Ten microliter of jetPEI™ solution (mixed with 240-μl NaCL) was added into 250-μl DNA solution (containing 2 μg pNF-κB/luc2 or pCMV-luc2) to form DNA complexes, and kept at room temperature for 20 min. Media of the culture dishes were refreshed then the DNA complexes were added. After 24 h of incubation, 200-μg/ml Hygromycin B was added for stable clone selection. The stable clones grown from single cells were prepared for selection using the IVIS200 Imaging System (Xenogen) and named as U-87/NF-κB-luc2 or U-87/luc2 [27]. The siRNA (50 μM siRNA) transfection on U-87 MG and BP-5 cells was followed by Lipofectamine™ 3000 transfection reagent commercial protocol (Thermo Fisher Scientific) [28].
NF-κB Luciferase Reporter Assay In Vitro and In Vivo
Three thousand U-87/NF-κB-luc2 cells were seeded into 96-well plates. After 1 day, cells were treated with regorafenib (0–30 μM), TMZ (0–160 μM), QNZ (0.5 μM), PD98059 (10 μM), LY294002 (10 μM), PD98059 (10 μM), SP600125 (10 μM), SB203580 (10 μM), AMD3100 (10 μM) or various drug combinations. After 24 h, 500 μM D-luciferin (Promega) was added into each well and incubated for 5 min before the IVIS 200 scanning. For the animal experiments, 150-mg/kg D-luciferin was intraperitoneally injected into the tested mice. Signals emitted from cells and mice were received by the IVIS200 Imaging System and quantified by Living Image software (Xenogen). The signal intensities emitted from each well were corrected according to the cell viability obtained by MTT assay and normalized using the 0.1% DMSO-treated control. The acquisition time of signals from mice was 1 s. Regions of interest (ROIs) were drawn around the tumors and quantified with the Living Image software as photons/s/cm2/sr [27].
Clonogenic Formation Assay
Cells (1 × 105) were seeded into 6-well plates and treated with regorafenib (10 μM), TMZ (120 μM), AMD3100 (30 μM), or combination for 24 h. Then, different cells number of each group was seeded for 14 days’ growth. Seeded number was listed as followed, 1000 cells (CTRL), 2000 cells (regorafenib, TMZ, AMD3100), and 3000 cells (combination). Fixing and staining procedure were described in previous study [29]. Percent of cells per well were calculated by image J version 1.50 (National Institutes of Health, Bethesda, MD, USA).
Western Blotting
1 × 106 cells were seeded in 10-cm dishes and incubated overnight at the specified conditions. Treatment with regorafenib (10 μM), TMZ (120 μM), 50 ng/ml CXCR4 recombinant protein, 100 ng/ml CXCL12 recombinant protein, 30 μM AMD 3100 (CXCR4 inhibitor), LIT-927 (CXCL12 neutraligand), or combination of both was followed for 24 h. Total protein from cells was extracted with lysis buffer (50 mM Tris–HCl pH 8.0, 120 mM NaCl, 0.5% NP-40, and 1 mM phenylmethanesulfonyl fluoride). Cytosol extraction kit was used to extract the cytosolic fraction of cells following the instructions provided by the manufacturer. Expression of ERK (Thr202/Tyr204), ERK, MMP9, VEGF, CyclinD1, CXCR4, CXCL12, c-FLIP, MCL-1, XIAP, MGMT and cleaved caspase-3 were determined with Western blotting procedures [30]. The protein bands were visualized and quantified using the ChemiDoc MP Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Activation of Caspase-3 and Caspase-8
Fifty thousand cells were seeded in 6-well plates and incubated overnight before treatment. After treatment with regorafenib (10 μM), TMZ (120 μM) or combination of both, the staining procedures of caspase-3 and caspase-8 were performed according to the instructions provided by the manufacturer (CaspGLOW). Briefly, cells were separately stained with fluorescein isothiocyanate-Asp(OCH3)-Glu(OCH3)-Val-Asp(OCH3)-fluoromethyl ketone (FITC-DEVD-FMK; caspase-3 specific) or sulforhodamine-Ile-Glu-Thr-Asp-fluoromethyl ketone (Red-IETD-FMK; caspase-8 specific) and incubated at 37 °C for 30 min [31]. After washing twice, cells were collected in BD Falcon™ tubes and prepared for flow cytometry analysis using (FACSCalibur) device.
Detecting the Loss of Mitochondria Membrane Potential
5 × 105 cells were seeded in 6-well plates, incubated overnight, and followed by treatment with regorafenib (10 μM), TMZ (120 μM), or combination of both. The loss of mitochondrial membrane potential during apoptosis was measured by DIOC6 (3,3′-dihexyloxacarbocyanine Iodide) staining assay using flow cytometry as previously described [19]. Briefly, cells were stained with 4 μM DIOC6 at 500-μl PBS, and the ΔΨm was quantitatively measured and analyzed by a FACSCalibur flow cytometer. All experiments were repeated three times.
Analyzing the Population of SubG1 Phase
5 × 105 cells were seeded in 6-well plates, incubated overnight, and followed by treatment with regorafenib (10 μM), TMZ (120 μM), or combination of both. Briefly, cells were prepared 1 day before treatment, then stained with PI/RNase (cat: 550,625, BD Biosciences) at 37 °C for 1 h [19]. Finally, the cell cycle was quantitatively measured and analyzed by a FACSCalibur flow cytometer. All experiments were repeated three times.
ELISA
1 × 105 cells were seeded in 24-well plates, incubated overnight, and followed by treatment with regorafenib (10 μM), TMZ (120 μM), or combination of both for 24 h. Fresh medium without serum was then replaced and incubate with cells for another 48 h. Supernatant was finally collected for human CXCR4 (Chemokine C-X-C-Motif Receptor 4) ELISA (Elabscience, Houston, TX, USA) and human CXCL12/SDF-1 ELISA (DuoSet®, R&D Systems, Inc., Minneapolis, MN, USA) analysis.
Comet Assay
U-87 MG cells were seeded in 6 wells (1 × 105 cells) treated with regorafenib (10 μM), TMZ (120 μM), QNZ (0.5 μM), or combination for 24 h. The comet assay protocol was followed by Singh et al. and described in our previous studies [32, 33]. DNA fragment results were then imaged by fluorescence microscope (Nikon ECLIPSE Ti-U, Minato City, Tokyo, Japan). Quantification analysis was performed by image J version 1.50 (National Institutes of Health) using the OpenComet v1.3.1 tool box.
Migration and Invasion Assay
Cells were seeded in 6-cm plates with a density of 6 × 105 cells, incubated overnight, and followed by regorafenib (10 μM), TMZ (120 μM), or combination treatment. After treatment, viable cells were harvested and seeded onto 8-μm pore size transwell inserts with or without Matrigel coating for invasion or migration experiments, respectively, and incubated for 24 h. Transwell membranes were fixed with fixation buffer (3:1 methanol and acetic acid) and stained with 0.5% crystal violet. Images of the migrated and invaded cells were acquired by a light Nikon ECLIPSE Ti-U microscope at × 100. Three microscope photographed images were quantified by ImageJ software version 1.50 [34].
Animal Experiments
The animal studies were conducted in accordance with the Animal Use Protocol approved by the Animal Care and Use Committee at Taipei Medical University (approval number: LAC-2017–0248). Six-week-old male BALB/cAnN.Cg-Foxn1nu/CrlNarl nude mice (20–25 g) were used for intracerebral implantations of 1 × 105 U-87/NF-κB-luc2 cells. Animals were anesthetized with 1–2% isoflurane, 1-mm burr hole was drilled into the skull of every animal, the anchor point was set at bregma, and cells were injected at a position of approximately 1 mm posterior, 3 mm lateral, and 3 mm deep of the bregma [35]. The syringe was held still for 15 min after injection, injection hole was sealed with bone wax, and skin was closed with sterilized sutures. The body weights were recorded, and tumor volume measurements were conducted once a week by MRI and BLI until the end of the study. Mice were randomly separated into four groups: CTRL (DMSO 0.1%), temozolomide alone (50-mg/kg TMZ dissolved in 100-μl PBS), regorafenib alone (10-mg/kg regorafenib dissolved in 100-μl PBS), and combination groups (10-mg/kg regorafenib combined with 25-mg/kg TMZ). Mice experiments were performed twice with a total of 17 mice for each group. The experimental groups were divided into survival analysis (N = 7) and image analysis (N = 10) groups. For the survival analysis, mice were killed if they reach a profoundly ill status according to the IACUC regulations, and were scored as a “death” in the survival analysis. For the image analysis, mice were all sacrificed on day 22 for further (Immunohistochemical) IHC and Hematoxylin and eosin (H&E) staining. Mice tumor size was measured by customized MATLAB scripts using T2-RARE images.
RNA Extraction and Microarray Assays
Total RNA of mice tumor tissues (N = 2) was extracted using mirVana miRNA Isolation kit (Invitrogen) according to the manufacturer’s instructions. Quantity and quality of the extracted RNA were evaluated with the RNA 6000 Nano LabChip on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Five hundred nanograms of total RNA were amplified and labeled with the Agilent Quick Amp Labeling Kit according to the manufacturer’s instructions. Signal intensities were quantified from the scanned images by using “Feature Extraction Software version 9.1” (Agilent Technologies). The signal intensities were corrected for the background intensities with the normexp method, then quantile normalized across arrays with limma software [36]. To filter out the non-informative probes, we removed the positive and negative control probes as well as the probes whose overall variances of intensity values were less than the median of the variances [37]. The reported microarray data was deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo).
In Vivo MRI
Mice were kept under anesthesia using 1.2% isoflurane during the MRI scans, which were performed using a Brucker 7 T PharmaScan scanner. Physiological conditions including heart rate, arterial pulse extension, oxygen saturation, and rectal temperature were continually monitored and kept within normal ranges throughout the scan time. A volume coil was used for RF excitation, and a circular surface coil was used for signal receiving. Contrast enhanced T1-, T2-, and dynamic-susceptibility-contrast (DSC) enhanced perfusion-weighted images were acquired every week [38]. MRI data were processed using customized MATLAB scripts (The Mathworks Inc., Natick, MA). Sequences of MR scanning were described as follow.
T1 and T2 Weighted Image
Following initial localization scans, a rapid acquisition with relaxation enhancement (RARE) sequence with TR/TE = 2650/40 ms, FOV = 2.56 × 2.56 cm, matrix size = 192 × 192, 12 slices, and slice thickness of 1 mm was performed to acquire anatomical images for registration. Tumor volumes were obtained from the T2 weighted images (T2-RARE). Subsequent postcontrast axial, coronal, and sagittal T1-weighted imaging was performed after the contrast administration (as CET1).
Dynamic-Susceptibility-Contrast Enhanced Perfusion-Weighted Imaging
To obtain the noteworthy information of angiogenesis in brain tumors, the DSC method (2D echo-plannar imaging pulse sequence, TR/TE = 1000/12 ms, matrix = 96 × 96, flip angle = 90°, 16 slices with slice thickness = 0.5 mm) with contrast injection with 150 dynamic time points were employed. K2 maps were derived according to the method described in detail by Boxerman et al. (Boxerman et al., 2006). This model allows mathematic correction for the T1 leakage effects in cerebral blood volume measurements. Mice tumor sizes and MR K2 map were manually drawn and Image J software (version 1.490, National Institutes of Health, Bethesda, MD, USA). MRI data were processed using customized MATLAB scripts (The Mathworks Inc., Natick, MA).
H&E Staining and Immunohistochemistry Staining
Mice were sacrificed and intracardially perfused with 4% paraformaldehyde prior to brain removal. Formalin-fixed and paraffin-embedded tissues and tumor from mice were subjected to H&E or IHC staining. All staining procedures were performed according to routine protocols [39]. In brief, sections of paraffin embedded tumor tissue on the glass slides obtained from each group was de-paraffinized in xylene, and rehydrated with decreasing concentrations of ethanol. The slides were incubated in 3% H2O2 for 10 min. After washing, the slides will be blocked with 5% normal goat serum for 5 min in a tight container, followed by incubation with primary antibody in a dilution of 1:100–500 at 4 °C overnight [39, 40]. Slides from three different tumors were imaged using a TissueFAXS TissueGnostics Axio Observer Z1 microscope (TissueGnostics GmbH) equipped with a color charge-coupled device using 100 × magnification [40]. Nine views from three different images were quantified by Image J software.
Statistics
All data is presented as mean values ± standard deviation. Differences between the treatment and non-treatment groups were analyzed by one-way ANOVA on Microsoft Excel version 2017 (Redmond, Washington, USA). A P-value of less than 0.05 was defined as significant difference. Determination of the significance in difference of the survival outcomes between the CTRL and combination groups was assessed by Kaplan–Meier survival curve comparisons. The P-values were derived from the log-rank (Mantel-Cox) tests.
Results
Regulation of CXCL12/CXCR4 May Affect the Toxicity of TMZ in GBM
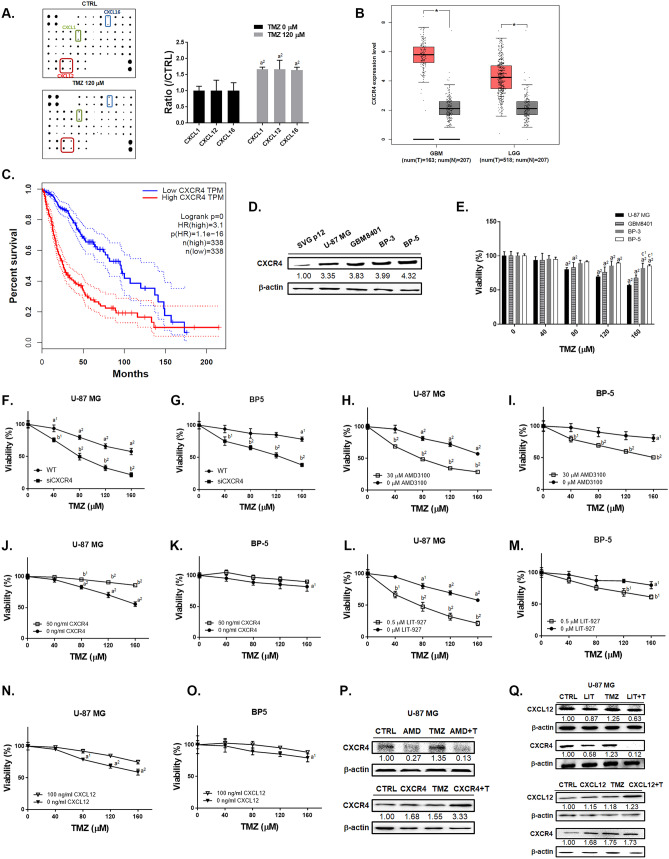
To identify the expression level of chemokine ligands after TMZ treatment, we performed chemokine array immunoassays on U-87 MG cells. The top 3 induction of chemokine ligands by TMZ treatment were CXCL1, CXCL12 (SDF-1-alpha and SDF-1-beta) and CXCL16 (Fig. 1A and Supplementary Table 1). GBM and low grade glioma tumor tissues from TCGA database showed greater expression levels of CXCR4 as compared to normal tissues (Fig. 1B). In Fig. 1C, the overall survival results showed that patients with low expression levels of CXCR4 have longer survival than those who have high expression levels. Four GBM cells showed higher endogenous expression level of CXCR4 as compared to astroglia SVG p12 (Fig. 1D). The cytotoxicity of TMZ was dose dependently increased in four GBM cells (Fig. 1E). However, BP-3 and BP-5 cells with higher endogenous CXCR4 expression were found to be relatively insensitive to TMZ as compare to U-87 MG and GBM8401 cells. To confirm the role of CXCL12/CXCR4 on TMZ-induced toxicity, we transfected siCXCR4 or treated with CXCR4 recombinant protein, CXCL12 recombinant protein, AMD3100 (CXCR4 inhibitor), and LIT-927 (CXCL12 neutraligand) and combined with TMZ in U-87 MG and BP-5 cells. The cytotoxicity of TMZ was sensitized by transfected U-87 MG and BP-5 cells (Supplementary Fig. 1) with siCXCR4 (Fig. 1F–G). Moreover, both U-87 MG and BP-5 cells found the toxicity effect after combining TMZ with AMD3100 (CXCR4 inhibitor) and LIT-927 (neutraligand of CXCL12) (Fig. 1H, I, L, M). Even the highest CXCR4 endogenous BP-5 cells have showed sensitivity to TMZ-induced cytotoxicity after CXCR4 and CXCL12 inhibition. However, the toxicity effect of TMZ was countervailing by additional added CXCR4 and CXCL12 recombinant proteins in U-87 MG and BP-5 cells (Fig. 1J, K, N, and O). In U-87 MG cells, the CXCR4 protein expression that induced by TMZ was diminished by AMD3100 and LIT-927 (Fig. 1P–Q). In the contrary, TMZ combined with CXCR4 and CXCL12 recombinant protein may greatly induce the protein expression of CXCR4 and CXCL12 (Fig. 1P–Q). In sum, the expression of CXCR4 and CXCL12 may insensitive GBM to TMZ; whereas, the inhibition of CXCR4 and CXCL12 may sensitize GBM to TMZ-induced toxicity even with higher endogenous CXCR4 expression.
Fig. 1.
Synergistic cytotoxicity was observed from the combination of regorafenib and TMZ. A Green, blue and red frames point out the expression of CXCL-1, CXCL-12, and CXCL-16 on chemokine protein array membrane after 120 μM TMZ treatment for 24 h in U87 cells. B Box plots of CXCR4 mRNA expression level from GBM and LGG patient’s normal (gray) and tumor (pink) samples (*p < 0.05 vs. normal tissue). C Overall survival of glioma patients of either high or low mRNA expression levels of CXCR4 (dot line represented as 95% confidence interval). D The endogens CXCR4 protein expression levels of SVG p12 and four GBM cells. E The viability of U-87 MG and BP-5 after TMZ alone, TMZ combined F–G siCXCR4, H–I w/o AMD3100, J–K w/o CXCR4 recombinant protein, L–M w/o LIT-927, or N–O w/o CXCL12 recombinant protein are displayed. P–Q Western blotting results of CXCR4 and CXCL12 after different treatments are displayed. Relative full blot images are presented in Supplementary Fig. 10. (a1 p < 0.05, a2 p < 0.01 vs. 0 μM TMZ; b1 p < 0.05, b2 p < 0.01 vs. 0 μM AMD3100, 50 ng/ml CXCR4, 0.5 μM LIT-927 or 100 ng/ml CXCL12; c2 p < 0.01 vs. U-87 MG cells). Abbreviations: LGG, low grade glioma; GBM, GBM; n, normal; T, tumor; TPM, trans per million; AMD, 30 μM AMD3100; LIT, 0.5 μM LIT-927; + T, plus TMZ
Inhibition of CXCL-12/CXCR4 Signaling by Regorafenib May Enhance the Cytotoxicity of TMZ
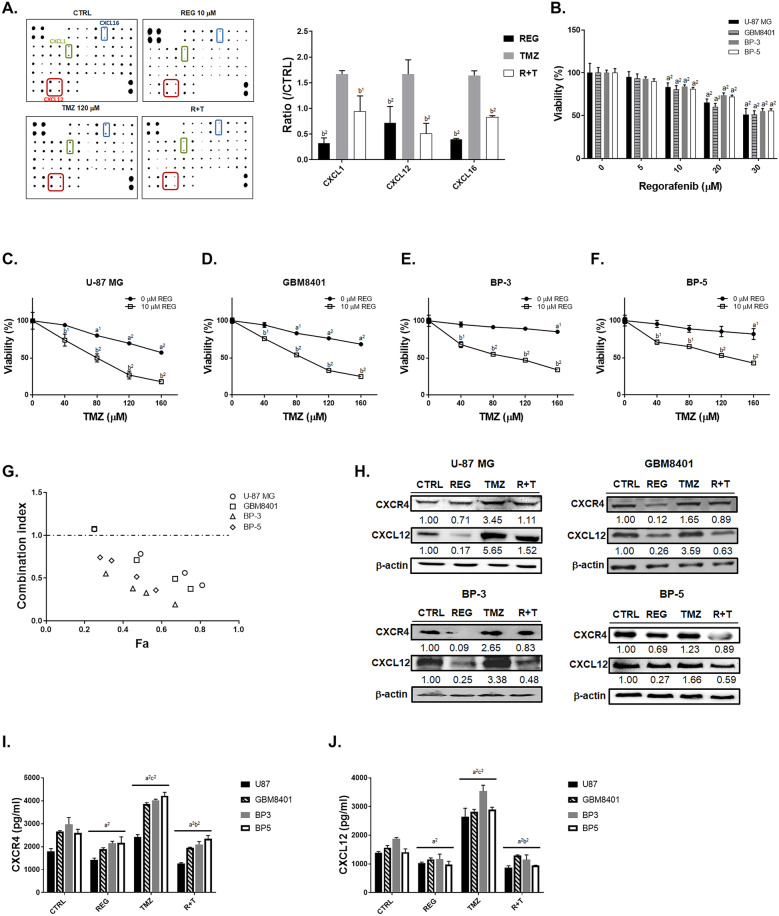
The inactivation of CXCL12 and CXCR4 may associate with TMZ sensitization. In Fig. 2A, regorafenib may diminish TMZ-induced chemokine ligands expression. Among the chemokine ligands that were overexpressed by TMZ treatment, CXCL12 was the one that declined significantly in expression due to the combination with regorafenib, compared to CXCL1 and CXCL-16 (Supplementary Table 2, same CTRL blot and TMZ treated blot on Fig. 1A were used). The cytotoxicity was also induced by regorafenib treatment in four GBM cells (Fig. 2B). The cytotoxicity of TMZ was markedly increased by regorafenib GBM cells compared to monotherapy (Fig. 2C–F). Combination of another multi-kinase inhibitor sorafenib with TMZ was also performed and which indicated poor benefit than combined with regorafenib (Supplementary Fig. 2 and Supplementary Table 3). The combination of 120 μM TMZ and 10 μM regorafenib showed synergistic cytotoxic effect (CI = 1, additive effect; CI < 1, synergistic effect; CI > 1, antagonistic effect) against all cell lines (Fig. 2G and Supplementary Table 4). As elucidated from the CI values, regorafenib may also effectively triggered the cytotoxicity against the BP-3 and BP-5 cell lines, which are relatively resistant to TMZ as compared to other cell lines. Colony formation was also decreased by regorafenib or CXCR4 inhibitor combined with TMZ (Supplementary Fig. 3). We found that CXCR4 and CCXL12 protein level were upregulated in the four GBM cells after TMZ treatment; on the contrary, expression of them was diminished after combination treatment of TMZ with regorafenib (Fig. 2H). In ELISA results, the expression level of CXCR4 and CXCL12 that induced by TMZ may also decrease by regorafenib treatment (Fig. 2I–J). These results suggested that the suppression of CXCR4 and CXCL12 by regorafenib may sensitize GBM cells to TMZ treatment.
Fig. 2.
TMZ-induced expression of CXCR4/CXCL-12 was reduced by combination with regorafenib. A Chemokine protein array membrane and quantification results of CXCL-1, CXCL-12, and CXCL-16 after 10 μM regorafenib, 120 μM TMZ, and combination treatment are displayed. B The MTT viability results of regorafenib treatment in four GBM cells. The viability of C U-87 MG, D GBM-8401, E BP-3, and F BP-5 cells after regorafenib combined with TMZ are displayed. G Combination index calculated by COMPUSYN software is displayed. Symbols represent CI values derived from actual data points (Fa, fraction affected). CI = 1, additive; CI > 1, antagonistic; CI < 1, synergistic. H Western blot images of CXCL-12 and CXCR-4 in four GBM cells are displayed. Relative full blot images are presented in Supplementary Fig. 11. I–J ELISA results of CXCR4 and CXCL12 of four GBM cells are displayed. (a1 p < 0.05, a2 p < 0.01 vs. 0 μM TMZ or CTRL; b1 p < 0.05, b2 p < 0.01 vs. 120 μM TMZ or 0 μM REG). Abbreviations: CTRL, control; REG, regorafenib; R + T, regorafenib plus TMZ
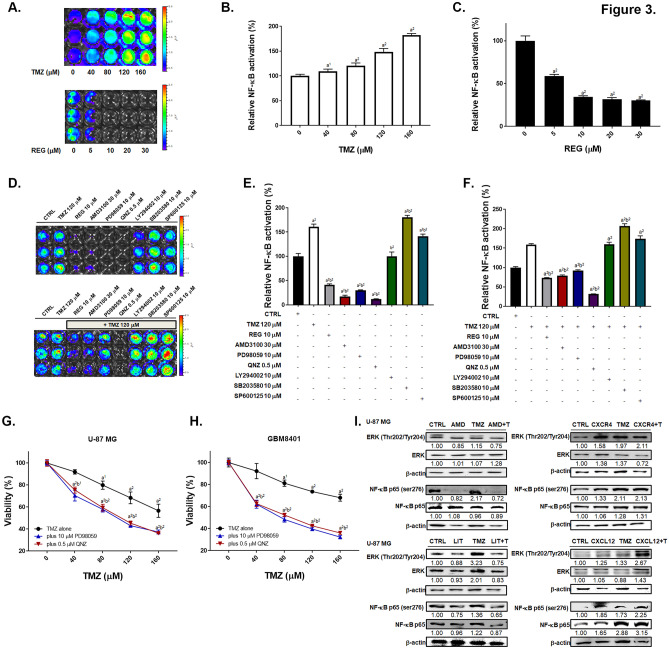
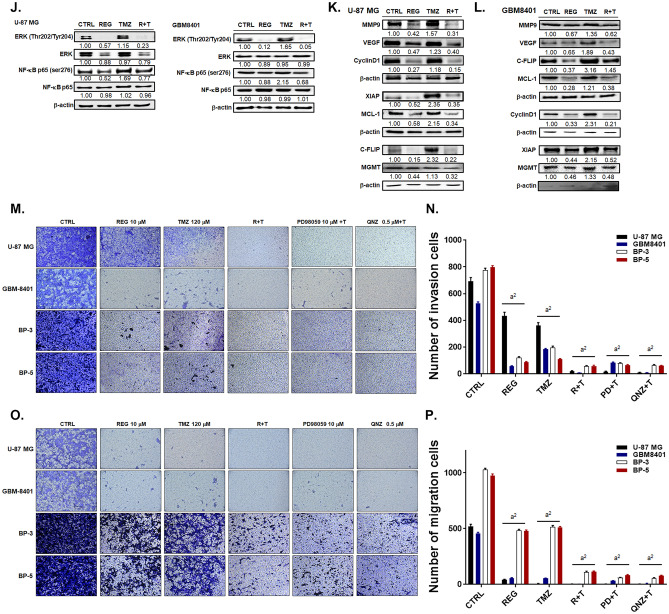
Regorafenib Effectively Suppresses the TMZ-Induced NF-κB Activation and Enhances the TMZ-Induced Inhibition of Invasion/Migration of GBM Cells via the ERK/NF-κB Signaling Pathway
TMZ dose dependently enhanced the activation NF-κB in U-87/NF-κB-luc2 cells, whereas regorafenib inhibited the activation of NF-κB dose dependently as showed in Fig. 3A–C. TMZ treatment may not activate CMV-driven luc2 expression in U87/luc2 cells, which was represented as negative control (Supplementary Fig. 4). To evaluate the key factors of NF-κB upstream signaling, we administrated different inhibitors, including CXCR4 inhibitor (AMD3100), NF-κB inhibitor (QNZ), JNK inhibitor (SP600125), AKT inhibitor (LY294002), ERK inhibitor (PD98059), and P38 inhibitor (SB203580) to the U-87/NF-κB-luc2 cells. We found that AMD3100, QNZ and PD98059 significantly suppressed the activation of NF-κB (Fig. 3D–F) and also diminished TMZ-induced NF-κB activation. Then, we also proved that inhibition of ERK and NF-κB may sensitize GBM to TMZ (Fig. 3G–H). Also, we found that the TMZ-induced phosphorylation of ERK and NF-κB were effectively suppressed by AMD3100 or LIT-927 treatment in U-87 MG cells, but induced by CXCR4 or CXCL12 recombinant proteins treatment (Fig. 3I). Furthermore, TMZ-triggered phosphorylation of both ERK and NF-κB was markedly reduced by regorafenib treatment (Fig. 3J) in U-87 MG and GBM8401 cells. We then examined the alteration of NF-κB–related proteins which play pivotal roles in angiogenesis, proliferation, and anti-apoptosis, due to the combination treatment of TMZ with regorafenib. Regorafenib reversed the protein expression levels of MMP9, VEGF, CyclinD1, XIAP, c-FLIP, and MCL-1 which were induced by TMZ treatment in U-87 MG and GBM-8401 cells (Fig. 3K–L). Furthermore, the expression of MGMT, which is a mediator of the DNA damage/repair process, and key regulator of acquired TMZ resistance, was increased by TMZ treatment and inhibited by combining with regorafenib in U-87 MG and GBM8401 cells (Fig. 3K–L). As illustrated in Supplementary Fig. 5, four GBM cells we used were all represented with methylation pattern, thus the methylation status may not be the major factor that affect regorafinb-induced toxicity on our GBM cells. Finally, we performed the matrigel-coated and non-coated transwell assays. As showed in Fig. 3M-N, invaded cells were markedly reduced by combined treatment of regorafenib with TMZ as compare to single treatment. Similarly, the greatest inhibition of migrated cells was observed for the combination treatment (Fig. 3O–P). Treatment with the ERK inhibitor (PD98059) and NF-κB inhibitor (QNZ) showed similar invasion and migration suppression effects on four GBM cells. These results indicated that regorafenib may not only suppress TMZ-induced ERK/NF-κB activity and its downstream proteins expression, but also enhance TMZ-induced invasion/migration inhibition potential.
Fig. 3.
The ERK/NF-κB signaling pathway and tumor invasion/migration capabilities were inhibited by the combination treatment of TMZ and regorafenib. A U-87/NF-κB-luc2 cells were treated with TMZ or regorafenib for 24 h and the activation of NF-κB were monitored by IVIS200 Imaging System. B–C Quantification result of the NF-κB activation. D U-87/NF-κB-luc2 cells treated with different inhibitors alone or combination with TMZ for 24 h and the E–F quantification result of NF-κB activation. The cytotoxic effect of PD98059 plus TMZ or QNZ plus TMZ on G U87 MG cells, and H GBM-8401 cells. Protein expression level of ERK (Thr202/Tyr204) and NF-κB p65 (Ser276) in U87 MG cells after I combined with 30 μM AMD3100, 0.5 μM LIT-927, 50 ng/ml CXCR4, or 100 ng/ml CXCL12 with TMZ or J regorafenib combined with 120 μM TMZ. Relative full blot images are presented in Supplementary Fig. 12. K-L Protein expression of migration, proliferation and anti-apoptosis related genes after regorafenib, TMZ or combination treatment in U-87 MG and GBM-8401 cells. Micrographs of crystal violet stained membranes of M invasion and O migration assays and N, P its quantification results after different treatments are displayed. (a1 p < 0.05, a2 p < 0.01 vs. 0 μM TMZ, 0 μM REG or CTRL; b1 p < 0.05, b2 p < 0.01 vs. 120 μM TMZ)
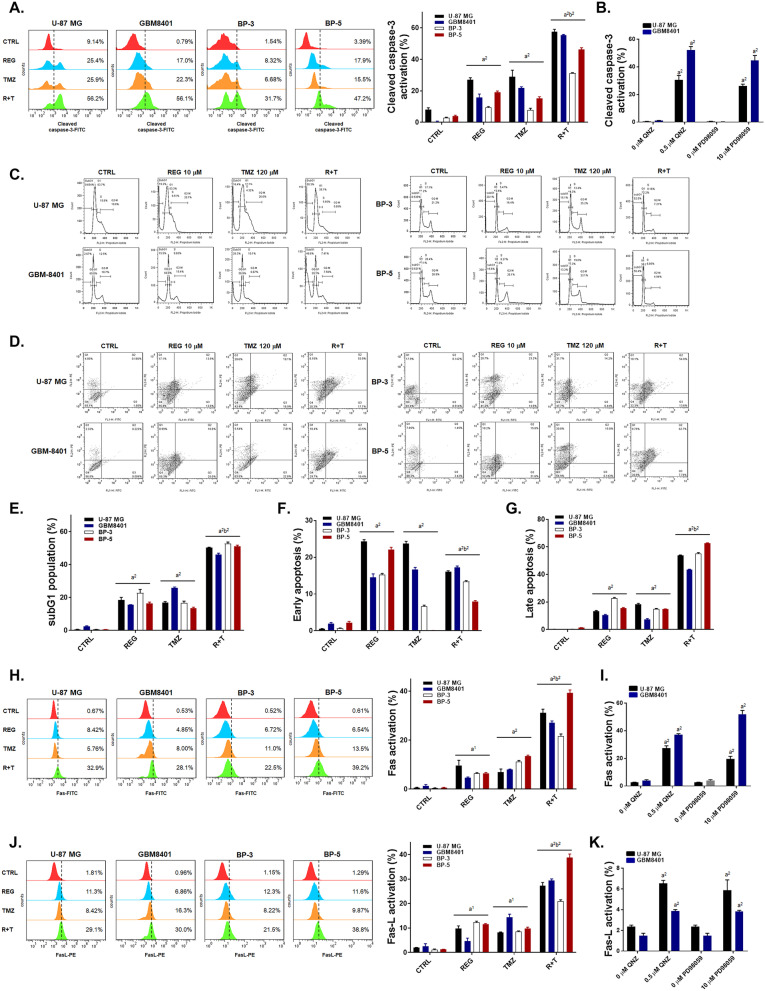
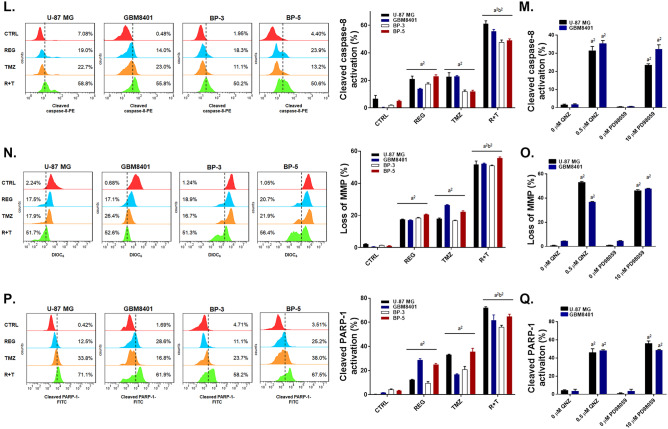
Regorafenib Significantly Enhances the Extrinsic/Intrinsic Apoptosis Pathway Induced by TMZ in GBM Cells
In Fig. 4A, the activation of cleaved caspase-3 was enhanced by the combination treatment of TMZ with regorafenib in the four GBM cells. Inhibition of either ERK or NF-κB signaling pathways by PD98059 and QNZ, respectively, also activated the cleaved caspase-3 (Fig. 4B). The highest subG1 accumulation was found in the combination treatment group (Fig. 4C, E). Moreover, in the annexin V/PI double stain experiments, early and late apoptosis populations were markedly increased by the combination therapy as compared to monotherapy (Fig. 4D, F–G). Next, we evaluated the Fas/Fas-L- and caspase-8-mediated extrinsic apoptosis pathways by flow cytometry. In Fig. 4H, J, activation of both Fas and Fas-L by the combination treatment was more significant than by the single treatment with any of the drugs. The inhibitors PD98059 (Fig. 4I, K) and QNZ (Fig. 4I, K) effectively activated the expression of Fas and Fas-L in U-87 MG and GBM8401 cells. Expression of caspase-8 which is downstream of Fas/Fas-L was effectively enhanced by 50–60% by combination treatment as showed in Fig. 4L. Through inhibition of ERK and NF-κB, the cleavage of caspase-8 was enhanced by 30–40% in U-87 MG and GBM8401 cells (Fig. 4M). In addition, regorafenib may enhance the TMZ-induced mitochondria-associated intrinsic apoptosis signaling pathway through the reduction of mitochondrial membrane potential (Fig. 4N). DNA fragment was also found to be increased in regorafenib or QNZ combined with TMZ (Supplementary Fig. 6). U-87 MG and GBM8401 cells treated with PD98059 and QNZ (Fig. 4O) showed marked mitochondrial membrane potential loss. Moreover, cleaved PARP-1 was markedly increased by the combination treatment of regorafenib with TMZ, and single treatment with PD98059 or QNZ as displayed in Fig. 4P–Q. Accordingly, we conclude that the extrinsic/intrinsic apoptosis effects which induced by the combination treatment of regorafenib with TMZ were majorly regulated by the inhibition of ERK/NF-κB signaling pathways.
Fig. 4.
Significant apoptosis effects were observed with the combination treatment of regorafenib with TMZ. Four GBM cells are treated with 10 μM regorafenib, 10 μM PD98059, 0.5 μM QNZ alone, or combined with 120 μM TMZ for 24 h. Cells are then detected A–B the activation of cleaved caspase-3, C–G the populations of subG1 phase and the activation of annexin V/PI, H–K the activation of Fas and Fas-L, L–M the activation of cleaved caspase-8, N–O the loss of the mitochondrial membrane potential (MMP) and P–Q the change of cleaved PARP-1 expression by flow cytometry. (a1 p < 0.05, a2 p < 0.01 vs. CTRL; b2 p < 0.01 vs. 120 μM TMZ)
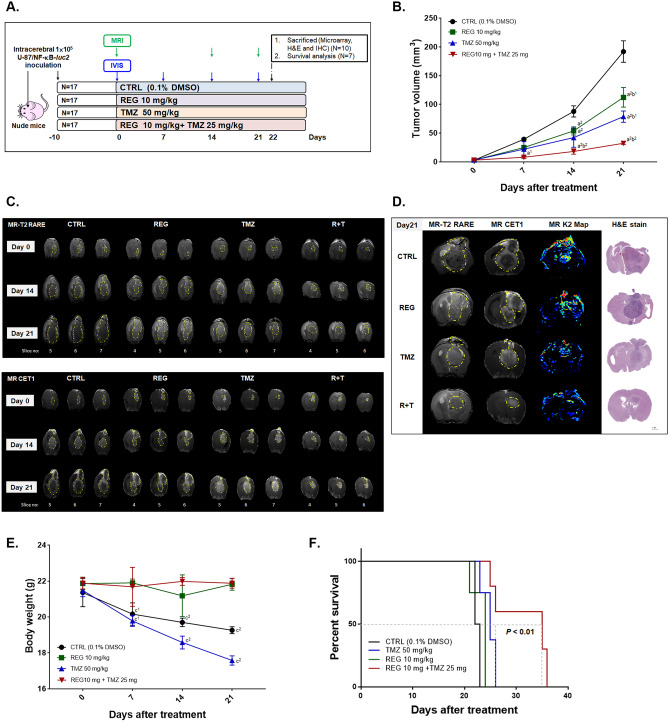
Regorafenib Combination with TMZ Effectively Suppressed the Tumor Growth and Prolonged the Overall Survival of Tumor-Bearing Mice
In orthotopic animal model experiments, U-87/NF-κB-luc2 bearing mice were divided into four different groups. Experiments were repeated twice with total 17 mice per experiment per group (Fig. 5A). Markedly tumor growth suppression was found in the combination treatment group as compared to single treatment groups (Fig. 5B). Importantly, the effective dosage of TMZ in the combination group was half of the one used in single treatment. Tumor growth was also monitored by MRI as well using CET1 and T2-RARE scan sequences (Fig. 5C). Representative whole slice images of mice brains from each group on days 0, 14, and 21 are shown in Supplementary Fig. 7–1 ~ 7–4. Smallest tumor size (from T2-RARE, CET1 images, and corresponding H&E stained tumor tissues), and lower angiogenesis (MRI K2 maps) were found in combination group on the end-point of experiment (day 21) (Fig. 5D). The mean tumor growth time (MTGT) was relatively long in combination group which represented as 54.52 days as compared to 9.56 days in CTRL group. The mean tumor growth delay time (MTGDT) is 44.76 days in combination groups (Supplementary table 5). The mean growth inhibition rate is represented as 1.74, 2.08, and 5.58 in REG, TMZ and R + T, respectively. The combination index in U-87 MG bearing animal of combination treatment was 0.47, which was considered as synergistic effect (Supplementary table 6). Although TMZ is the clinically approved drug for GBM patient, its toxicity is relatively high. As showed in Fig. 5E, the body weight of mice of the CTRL and TMZ groups decreased dramatically after 7 days of treatment, while that of the combination and regorafenib treatment groups were maintained similar. No obvious normal tissue damage was found in combination treatment group as compare to CTRL group (Supplementary Fig. 8). Additionally, the combination treatment markedly prolonged the survival of mice from 26 to 36 days as compared to TMZ single treatment (Fig. 5F). Taken together, regorafenib promote tumor inhibition capacity of TMZ on GBM in vivo model.
Fig. 5.
Tumor growth of our orthotopic animal model was inhibited by combination treatment of regorafenib with TMZ. A The experimental flow chart. B Tumor volumes were assessed by MRI once per week and calculated by customized MATLAB scripts from the T2-RARE images. C MRI T2-RARE and CET1 from each group on day 0, 14, and 24 are displayed. D MRI T2-RARE, CET1, and MR K2 map images and the corresponding H&E stained sections of the mice of each group on day 21 are displayed. E Mice body weight was measured by digital weight once per week. F Kaplan–Meier survival curve of each group. (a1 p < 0.05, a2 p < 0.01 vs. CTRL; b1 p < 0.05, b2 p < 0.01 vs. TMZ; c1 p < 0.05, c2 p < 0.01 vs. REG + TMZ)
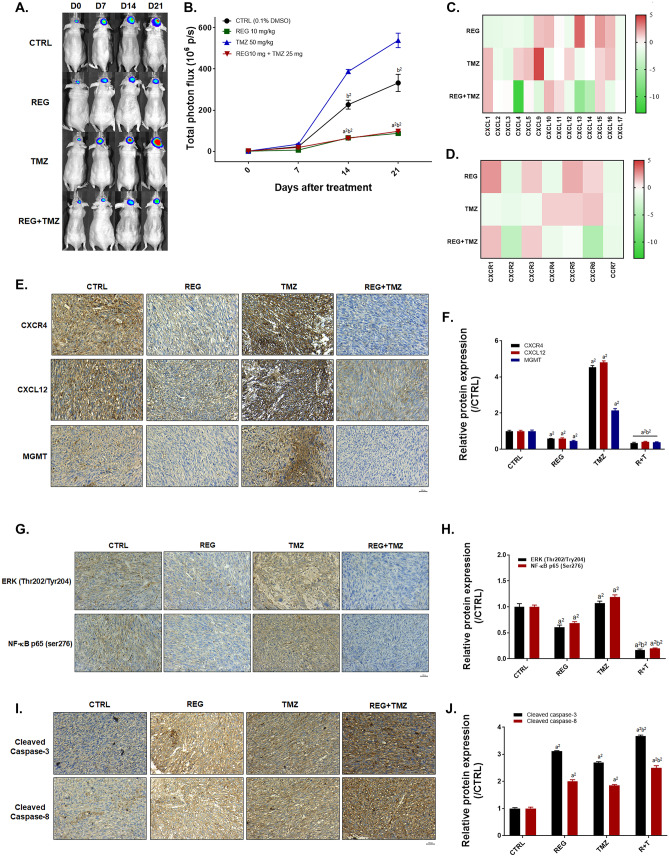
Regorafenib Markedly Diminished the TMZ-Induced CXCL-12/CXCR4/ERK/NF-κB-Mediated Signal Transduction
TMZ-induced NF-κB activation was diminished by the combination with regorafenib in vivo (Fig. 6A–B). The microarray assays of the mice tumor tissues after various treatment conditions showed that several CXC chemokines and CXC chemokine receptors were induced by TMZ, such as CXCL-1, -4, -5, -9, -12, -15, -16, CXCR-4, -5, and -6 (Fig. 6C–D). Importantly, the induction of CXC chemokines and CXC chemokine receptors by TMZ was diminished after combining with regorafenib. In Fig. 6E–F IHC results, combination group displayed the lowest CXCR4 and CXCL-12 expression levels; whereas, the highest expression level was found in TMZ single treatment group. Moreover, the TMZ-induced MGMT expression was significantly decreased by combined treatment of TMZ with regorafenib. Similarly, the protein expression of phosphorylated ERK and p65 subunit of NF-κB were decreased by the combination therapy (Fig. 6G, H). The caspase-mediated apoptosis was effectively enhanced by the combined treatment of TMZ with regorafenib as compared to single treatment which validated by both IHC staining (Fig. 6I–J) and ex vivo Western blotting (Supplementary Fig. 9). In summary, the combination treatment of regorafenib with TMZ may effectively reduce the TMZ-induced CXCR4/CXCL-12/ERK/NF-κB signaling transduction.
Fig. 6.
The TMZ-induced CXCR4/CXCL-12 signaling was suppressed by the combined treatment of regorafenib with TMZ. A Representative IVIS images and B quantification of each group are displayed. C–D CXC chemokines messenger RNAs and CXC chemokine receptors messenger RNAs assayed by microarray as compared to CTRL (0.1% DMSO) are displayed. E CXCR4, CXCL-12, and MGMT stained images and F quantification results from tumor tissue are displayed. G ERK(Thr202/Tyr204) and NF-κB p65 (Ser276) IHC stained images and H quantification results from tumor tissue are displayed. I IHC stained images of the cleaved caspase-3 and -8 of tumor tissues of each group and J their quantification values are displayed. (a1 p < 0.05, a2 p < 0.01 vs. CTRL; b1 p < 0.05, b2 p < 0.01 vs. TMZ 50 mg/kg)
Discussion
Chemokine-signaling network is required for glioblastoma progression. CXC chemokine ligands CXCL1, CXCL2, CXCL4, CXCL9, CXCL12, CXCL16, as well as CX3CL have been reported to participate in proliferation, survival, angiogenesis, invasion, and metastasis in glioblastoma [41]. Binding of the chemokine CXCL-12 with its receptor CXCR4 mediates progression of GBM through initiation of oncogenic signal transduction pathways [42]. Tabouret et al. found that the CXCL12/CXCR4 pathway was associated with recurrence of GBM after the combined treatment of radiotherapy with TMZ [10]. The information we retrieved from the TCGA databank indicates that CXCR4 expression is higher in tumor tissues of GBM patients than in normal brains (Fig. 1B). Furthermore, GBM patients with high CXCR4 expression levels suffer from poor survival compared to those with low expression levels (Fig. 1C), which makes CXCR4 a potential biomarker for GBM. The patients derived GBM BP5 cells obtained from human tumor tissues expressed higher endogenous CXCR4 levels and exhibit lower sensitivity to TMZ than other GBM cells (Fig. 1D–E). Then, chemokine array was utilized to detect effect of TMZ, regorafenib, or combination treatment on chemokine levels in U-87 MG cells. We found that TMZ significantly triggered expression of CXCL1, CXCL12, and CXCL16 compared to that of CTRL group (Fig. 1A). The increased expression of CXCL1 associates with unfavorable overall survival and mesenchymal transition–mediated radioresistance in glioblastoma [43]. In addition, CXCL16 attenuates expression of thrombospondin type 1 domain containing 4 (THSD4) and vascular endothelial growth factor-c (VEGFC) to facilitates TMZ-promoted cellular dormancy, which is one strategy for escaping from chemotoxicity [44]. Our results showed that TMZ-induced expression of CXCL1, CXCL12 CXCL16 was effectively reduced by regorafenib (Fig. 2A). Most important, the survival of mice observed in our study indicates that regorafenib and TMZ combination treatment resulted in the longest survival time periods compared to other treatments (Fig. 5F). The TMZ-induced proteins expression of CXCL12 and CXCR4 was significantly suppressed by regorafenib treatment in GBM cells and tumors (Figs. 2H and 6E–F).
Mitogen-activated protein kinases (MAPKs) and protein kinase B (AKT) signaling are activated by the CXCL12/CXCR4 interaction. MAPKs (ERK, P38, and JNK) and AKT upregulated NF-κB activation may lead to cell survival, invasion, and angiogenesis [45–47]. Haas et al. demonstrated that inhibition of phosphatidylinositol-3-kinase (PI3K) signaling enhanced the TMZ-inhibited cell viability [48]. We found both siCXCR4, AMD3100 (CXCR4 inhibitor), LIT-927 (CXCL12 neutraligand), PD98059 (MEK/ERK signaling inhibitor), and QNZ (NF-κB inhibitor) increased TMZ-induced toxicity (Figs. 1F–G, H–I, L–M and 3G–H), but decreased the endogenous NF-κB activation and suppressed the TMZ-induced NF-κB activation (Fig. 3D–F). Inhibition of ERK, p38, and AKT by regorafenib is associated with down-regulation of NF-κB activity in HCC, bladder, and non-small cell lung cancer (NSCLC) [19, 49, 50]. We found that regorafenib significantly reduced the TMZ-increased ERK phosphorylation (Fig. 3J). Fianco et al. demonstrated that high expression of caspase-8 modulated the angiogenesis and TMZ resistance through the activation of NF-κB in GBM [51]. We illustrated that regorafenib augmented the TMZ-induced caspase-8 activation; however; the TMZ-induced NF-κB activation and VEGF expression were blocked by regorafenib treatment (Fig. 3J–L). Regorafenib significantly increased the action of TMZ to inhibit GBM cell growth in vitro (Fig. 2C–F, Supplementary Fig. 3) and in vivo (Fig. 5C–F).
NF-κB p50/p65 heterodimers are the oncogenic transcription factors which regulates numerous oncogenes expression. Activation of NF-κB contributes significantly to tumor progression through upregulation of proliferative, anti-apoptotic, angiogenic, and metastasis-associated proteins encoded by NF-κB-regulated oncogenes [52, 53]. Activation of NF-κB, which is frequently observed in GBM, is related to the grade of glioma [54]. Previous studies showed that the inhibition of NF-κB by specific inhibitors suppressed the expression of anti-apoptotic, proliferative, and metastatic proteins and reduced the invasion capability of GBM [27, 55]. Yu et al. also demonstrated that inhibition of NF-κB by small interfering ribonucleic acid (siRNA) specific to p65 abrogates the protein levels of MGMT in GBM [55]. In this study, we indicated that regorafenib acts as an inhibitor to NF-κB signaling, which significantly diminishes the TMZ-induced NF-κB activation and expression of related proteins such as MMP-9, VEGF, Cyclin-D1, XIAP, C-FLIP, MCL-1, and MGMT (Fig. 3K–L). Regorafenib also promotes the effects of TMZ to inhibit the cell invasion and migration of GBM (Fig. 3M–P).
Apoptosis prevents the growth of tumor cells by induction of cell death through extrinsic/intrinsic (mitochondrial) apoptotic pathways. Apoptosis aids the chemotherapeutic agents to inhibit tumor progression, so inhibition of apoptosis by anti-apoptotic proteins limits the anticancer efficacy of chemotherapeutic agents [56]. The pro-apoptotic protein Bak mediates the loss of mitochondrial membrane potential (MMP), resulting in cytochrome-c and apoptotic protease activating factor 1 (APAF-1) release from mitochondria. The anti-apoptotic protein myeloid cell leukemia-1 (MCL-1) blocks the intrinsic apoptotic pathway by inhibiting the Bak-induced loss of MMP [57, 58]. Inhibition of MCL-1 expression augments the TMZ-induced apoptosis in GBM cells [59, 60]. Our results showed that regorafenib significantly enhances the loss of MMP and apoptosis (Fig. 4), DNA damage (Supplementary Fig. 6) and inhibit the TMZ-triggered MCL-1 expression. In this study, we provided data that regorafenib effectively enhances TMZ-induced apoptosis through extrinsic and intrinsic pathways in glioblastoma cells. Interestingly, Jiang et al. found that autophagy but not apoptosis is essential for GBM growth inhibition induced by regorafenib [61]. They proposed that regorafenib may elicits lethal autophagy arrest in TMZ-resistant GBM cells through stabilizing phosphoserine aminotransferase 1 (PSAT1).
Down-regulation of the pro-apoptotic proteins contribute to evasion of apoptosis and tumor progression [62]. Caspase family of proteins acts as a critical modulator in the apoptotic pathways. High expression of active-caspase-8 is necessary for the interaction of death receptors with their ligands which initiates the extrinsic apoptotic pathway. Active-caspase-3 modulates the formation of apoptotic deoxyribonucleic acid fragmentation and cleavage of PARP-1 [63]. The loss of caspase-3 and caspase-8 activation was linked to poor survival in patients with GBM [64, 65]. In this study, we show that regorafenib significantly increases the TMZ-induced Fas (Fig. 4H), Fas-L (Fig. 4J), caspase-8 (Fig. 4L), and caspase-3 (Fig. 4A) activation in GBM. The DNA repair proteins also abolished the chemotherapeutic agent–induced apoptosis by repairing DNA damage. Both of the DNA repair proteins PARP-1 and MGMT have been indicated to modulate the repair of TMZ-induced DNA damage [66, 67]. Spiegl-Kreinecker et al. revealed that the increased expression of MGMT protein is related to the poor outcome of TMZ treatment in GBM patients [68]. Decreased expression of MGMT protein and PARP inhibitors was found to sensitize GBM cells to TMZ [55, 69]. Here, we demonstrated that regorafenib significantly suppresses the TMZ-induced MGMT expression (Figs. 3K–L and 6E–F) and promotes the TMZ-induced PARP-1 cleavage (Fig. 4P).
In conclusion, the enhancing effects of regorafenib towards the TMZ anti-GBM capabilities may be due to the suppression of CXCL-12/CXCR4-mediated ERK/NF-κB signaling and induction of apoptosis through the extrinsic/intrinsic pathways. Our results suggest that regorafenib is a potential complementary agent that could effectively increase the therapeutic efficacy of TMZ in GBM.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
Acknowledgements
Experiments and data analysis were performed in part through the use of the Medical Research Core Facilities Center, Office of Research & Development at China Medical University, Taichung, Taiwan, R.O.C and Taipei Medical University (TMU Core Facility), Taipei, Taiwan, R.O.C. We would like to thank Mahmoud Ashraf Fikry Fathy Askar from the National Tsing-Hua University for helping with data calculations and statistical analysis.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author Contribution
Conception, study supervision, writing, and revision of the manuscript: F.T. Hsu, A.A.A. Ali, T.I. Hsu, Y.C. Liu, and I.T. Chiang. Methodology development: F.T. Hsu, H.S. Liu, and S.Y. Chou. Data acquisition, analysis and interpretation: F.T. Hsu, H.S. Liu, A.A.A. Ali, S.Y. Chou, T.I. Hsu, Y.C. Liu, and I.T. Chiang.
Funding
This study was financially supported by a grant from the Ministry of Science and Technology, Taipei, and Show Chwan Memorial Hospital, Changhua, Taiwan, respectively (grant number: MOST 108–2314-B-039–007-MY3, MOST 109-2314-B-039 -021-MY3 and SRD-109015). This work was also financially supported by the “Drug Development Center, China Medical University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
I.-Tsang Chiang and Yu-Chang Liu contributed equally to this work.
References
- 1.Merkel A, Soeldner D, Wendl C, et al. Early postoperative tumor progression predicts clinical outcome in glioblastoma-implication for clinical trials. J Neurooncol. 2017;132:249–254. doi: 10.1007/s11060-016-2362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry A, Wesseling P. Histologic classification of gliomas. Handb Clin Neurol. 2016;134:71–95. doi: 10.1016/B978-0-12-802997-8.00005-0. [DOI] [PubMed] [Google Scholar]
- 3.Oshiro S, Tsugu H, Komatsu F, et al. Efficacy of temozolomide treatment in patients with high-grade glioma. Anticancer Res. 2009;29:911–917. [PubMed] [Google Scholar]
- 4.Lee CY. Strategies of temozolomide in future glioblastoma treatment. Onco Targets Ther. 2017;10:265–270. doi: 10.2147/OTT.S120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm SM, Carter C, Tang L, et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 7.Roos WP, Batista LF, Naumann SC, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 8.Bruyere C, Mijatovic T, Lonez C, et al. Temozolomide-induced modification of the CXC chemokine network in experimental gliomas. Int J Oncol. 2011;38:1453–1464. doi: 10.3892/ijo.2011.964. [DOI] [PubMed] [Google Scholar]
- 9.Ehtesham M, Mapara KY, Stevenson CB, et al. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274:305–312. doi: 10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabouret E, Tchoghandjian A, Denicolai E, et al. Recurrence of glioblastoma after radio-chemotherapy is associated with an angiogenic switch to the CXCL12-CXCR4 pathway. Oncotarget. 2015;6:11664–11675. doi: 10.18632/oncotarget.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes & diseases. 2016;3:198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiapaer S, Furuta T, Tanaka S, et al. Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurol Med Chir (Tokyo) 2018;58:405–421. doi: 10.2176/nmc.ra.2018-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Cao Y, Chen C, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 14.Gong L, Giacomini MM, Giacomini C, et al. PharmGKB summary: sorafenib pathways. Pharmacogenet Genomics. 2017;27:240–246. doi: 10.1097/FPC.0000000000000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zustovich F, Landi L, Lombardi G, et al. Sorafenib plus daily low-dose temozolomide for relapsed glioblastoma: a phase II study. Anticancer Res. 2013;33:3487–3494. [PubMed] [Google Scholar]
- 16.Reardon DA, Vredenburgh JJ, Desjardins A, et al. Effect of CYP3A-inducing anti-epileptics on sorafenib exposure: results of a phase II study of sorafenib plus daily temozolomide in adults with recurrent glioblastoma. J Neurooncol. 2011;101:57–66. doi: 10.1007/s11060-010-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravi S, Singal AK. Regorafenib: an evidence-based review of its potential in patients with advanced liver cancer. Core Evid. 2014;9:81–87. doi: 10.2147/CE.S48626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YC, Wu RH, Wang WS. Regorafenib diminishes the expression and secretion of angiogenesis and metastasis associated proteins and inhibits cell invasion via NF-kappaB inactivation in SK-Hep1 cells. Oncology Lett. 2017;14:461–467. doi: 10.3892/ol.2017.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai JJ, Pan PJ, Hsu FT. Regorafenib induces extrinsic and intrinsic apoptosis through inhibition of ERK/NF-kappaB activation in hepatocellular carcinoma cells. Oncology Rep. 2017;37:1036–1044. doi: 10.3892/or.2016.5328. [DOI] [PubMed] [Google Scholar]
- 20.Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (London, England) 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 21.Lombardi G, De Salvo GL, Brandes AA, et al. Regorafenib compared with lomustine in patients with relapsed glioblastoma (REGOMA): a multicentre, open-label, randomised, controlled, phase 2 trial. Lancet Oncol. 2019;20:110–119. doi: 10.1016/S1470-2045(18)30675-2. [DOI] [PubMed] [Google Scholar]
- 22.Herrlinger U, Tzaridis T, Mack F, et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet (London, England) 2019;393:678–688. doi: 10.1016/S0140-6736(18)31791-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin XL, Xu Q, Tang L, et al. Regorafenib inhibited gastric cancer cells growth and invasion via CXCR4 activated Wnt pathway. PLoS One. 2017;12:e0177335-e. [DOI] [PMC free article] [PubMed]
- 24.Lin CY, Yang ST, Shen SC, et al. Serum amyloid A1 in combination with integrin alphaVbeta3 increases glioblastoma cells mobility and progression. Mol Oncol. 2018;12:756–771. doi: 10.1002/1878-0261.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TC. The mass-action law based algorithm for cost-effective approach for cancer drug discovery and development. Am J Cancer Res. 2011;1:925–954. [PMC free article] [PubMed] [Google Scholar]
- 26.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 27.Yen TH, Hsieh CL, Liu TT, et al. Amentoflavone induces apoptosis and inhibits NF-kB-modulated anti-apoptotic signaling in glioblastoma cells. In vivo (Athens, Greece) 2018;32:279–285. doi: 10.21873/invivo.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang WW, Zhang Y, Liu W, et al. CXCR4 induces cell autophagy and maintains EBV latent infection in EBVaGC. Theranostics. 2020;10:11549–11561. doi: 10.7150/thno.44251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen WT, Chen CH, Su HT, et al. Amentoflavone induces cell-cycle arrest, apoptosis, and invasion inhibition in non-small cell lung cancer cells. Anticancer Res. 2021;41:1357–1364. doi: 10.21873/anticanres.14893. [DOI] [PubMed] [Google Scholar]
- 30.Ting CY, Wang HE, Yu CC, et al. Curcumin triggers DNA damage and inhibits expression of DNA repair proteins in human lung cancer cells. Anticancer Res. 2015;35:3867–3873. [PubMed] [Google Scholar]
- 31.Chiang IT, Chen WT, Tseng CW, et al. Hyperforin inhibits cell growth by inducing intrinsic and extrinsic apoptotic pathways in hepatocellular carcinoma cells. Anticancer Res. 2017;37:161–167. doi: 10.21873/anticanres.11301. [DOI] [PubMed] [Google Scholar]
- 32.Singh NP, McCoy MT, Tice RR, et al. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 33.Chen CH, Hsu FT, Chen WL, et al. Induction of apoptosis, inhibition of MCL-1, and VEGF-A expression are associated with the anti-cancer efficacy of magnolol combined with regorafenib in hepatocellular carcinoma. Cancers (Basel). 2021;13. [DOI] [PMC free article] [PubMed]
- 34.Wu ZY, Lien JC, Huang YP, et al. Casticin inhibits A375.S2 human melanoma cell migration/invasion through downregulating NF-kappaB and matrix metalloproteinase-2 and -1. molecules (Basel, Switzerland). 2016;21:384. [DOI] [PMC free article] [PubMed]
- 35.Zhou J, Tryggestad E, Wen Z, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 2011;17:130–134. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics (Oxford, England) 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 37.Talloen W, Hochreiter S, Bijnens L, et al. Filtering data from high-throughput experiments based on measurement reliability. Proc Natl Acad Sci USA. 2010;107:E173–E174. doi: 10.1073/pnas.1010604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27:859–867. [PMC free article] [PubMed] [Google Scholar]
- 39.Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol (Clifton, NJ) 2014;1180:31–43. doi: 10.1007/978-1-4939-1050-2_3. [DOI] [PubMed] [Google Scholar]
- 40.Hofman F. Immunohistochemistry. Current protocols in immunology. 2002;Chapter 21:Unit 21.4. [DOI] [PubMed]
- 41.Urbantat RM, Vajkoczy P, Brandenburg S. Advances in chemokine signaling pathways as therapeutic targets in glioblastoma. Cancers (Basel) 2021;13:2983. doi: 10.3390/cancers13122983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurth R, Bajetto A, Harrison JK, et al. CXCL12 modulation of CXCR4 and CXCR7 activity in human glioblastoma stem-like cells and regulation of the tumor microenvironment. Front Cell Neurosci. 2014;8:144. doi: 10.3389/fncel.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alafate W, Li X, Zuo J, et al. Elevation of CXCL1 indicates poor prognosis and radioresistance by inducing mesenchymal transition in glioblastoma. CNS Neurosci Ther. 2020;26:475–485. doi: 10.1111/cns.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adamski V, Hattermann K, Kubelt C, et al. Entry and exit of chemotherapeutically-promoted cellular dormancy in glioblastoma cells is differentially affected by the chemokines CXCL12, CXCL16, and CX3CL1. Oncogene. 2020;39:4421–4435. doi: 10.1038/s41388-020-1302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao YX, Zhou CH, Zeng H, et al. The role of the CXCL12-CXCR4/CXCR7 axis in the progression and metastasis of bone sarcomas (Review) Int J Mol Med. 2013;32:1239–1246. doi: 10.3892/ijmm.2013.1521. [DOI] [PubMed] [Google Scholar]
- 46.Huang YC, Hsiao YC, Chen YJ, et al. Stromal cell-derived factor-1 enhances motility and integrin up-regulation through CXCR4, ERK and NF-kappaB-dependent pathway in human lung cancer cells. Biomed Pharmacother. 2007;74:1702–1712. doi: 10.1016/j.bcp.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Haas B, Klinger V, Keksel C, et al. Inhibition of the PI3K but not the MEK/ERK pathway sensitizes human glioma cells to alkylating drugs. Cancer Cell Int. 2018;18:69. doi: 10.1186/s12935-018-0565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang CH, Chung JG, Hsu FT. Regorefenib induces extrinsic/intrinsic apoptosis and inhibits MAPK/NF-kappaB-modulated tumor progression in bladder cancer in vitro and in vivo. Environ Toxicol. 2019;34:679–688. doi: 10.1002/tox.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng MC, Li MH, Chung JG, et al. Apoptosis induction and AKT/NF-kappaB inactivation are associated with regroafenib-inhibited tumor progression in non-small cell lung cancer in vitro and in vivo. Biomed Pharmacother. 2019;116:109032. [DOI] [PubMed]
- 50.Fianco G, Mongiardi MP, Levi A, et al. Caspase-8 contributes to angiogenesis and chemotherapy resistance in glioblastoma. Elife. 2017;6. [DOI] [PMC free article] [PubMed]
- 51.Cahill KE, Morshed RA, Yamini B. Nuclear factor-kappaB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol. 2016;18:329–339. doi: 10.1093/neuonc/nov265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puliyappadamba VT, Hatanpaa KJ, Chakraborty S, et al. The role of NF-κB in the pathogenesis of glioma. Mol Cell Oncol. 2014;1:e963478-e. [DOI] [PMC free article] [PubMed]
- 54.Hsu FT, Chiang IT, Kuo YC, et al. Amentoflavone effectively blocked the tumor progression of glioblastoma via suppression of ERK/NF-kappa B signaling pathway. Am J Chin Med. 2019;47:913–931. doi: 10.1142/S0192415X19500484. [DOI] [PubMed] [Google Scholar]
- 55.Yu Z, Chen Y, Wang S, et al. Inhibition of NF-kappaB results in anti-glioma activity and reduces temozolomide-induced chemoresistance by down-regulating MGMT gene expression. Cancer Lett. 2018;428:77–89. doi: 10.1016/j.canlet.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 56.Ricci MS, Zong WX. Chemotherapeutic approaches for targeting cell death pathways. Oncologist. 2006;11:342–357. doi: 10.1634/theoncologist.11-4-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis SN, Chen L, Dewson G, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gratas C, Sery Q, Rabe M, et al. Bak and Mcl-1 are essential for temozolomide induced cell death in human glioma. Oncotarget. 2014;5:2428–2435. doi: 10.18632/oncotarget.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li RY, Chen LC, Zhang HY, et al. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther. 2013;19:477–483. doi: 10.1111/cns.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang JW, Zhang L, Chen H, et al. Regorafenib induces lethal autophagy arrest by stabilizing PSAT1 in glioblastoma. Autophagy. 2020;16:106–122. doi: 10.1080/15548627.2019.1598752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pistritto G, Trisciuoglio D, Ceci C, et al. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging. 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. [DOI] [PMC free article] [PubMed]
- 64.Kobayashi T, Masumoto J, Tada T, et al. Prognostic significance of the immunohistochemical staining of cleaved caspase-3, an activated form of caspase-3, in gliomas. Clin Cancer Res. 2007;13:3868–3874. doi: 10.1158/1078-0432.CCR-06-2730. [DOI] [PubMed] [Google Scholar]
- 65.Saggioro FP, Neder L, Stávale JN, et al. Fas, Fas-L, and cleaved caspases 8 and 3 in glioblastomas: a tissue microarray-based study. Pathol Res Pract. 2014;210:267–273. doi: 10.1016/j.prp.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Murnyak B, Kouhsari MC, Hershkovitch R, et al. PARP1 expression and its correlation with survival is tumour molecular subtype dependent in glioblastoma. Oncotarget. 2017;8:46348–46362. doi: 10.18632/oncotarget.18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Zhang M, Gan H, et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun. 2018;9:2949. doi: 10.1038/s41467-018-05373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiegl-Kreinecker S, Pirker C, Filipits M, et al. O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol. 2010;12:28–36. doi: 10.1093/neuonc/nop003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta SK, Smith EJ, Mladek AC, Tian S, Decker PA, Kizilbash SH, et al. PARP inhibitors for sensitization of alkylation chemotherapy in glioblastoma: impact of blood-brain barrier and molecular heterogeneity. Front Oncol. 2018;8:670. doi: 10.3389/fonc.2018.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.