Abstract
During the onset of the cooperative association between the Hawaiian sepiolid squid Euprymna scolopes and the marine luminous bacterium Vibrio fischeri, the anatomy and morphology of the host's symbiotic organ undergo dramatic changes that require interaction with the bacteria. This morphogenetic process involves an array of tissues, including those in direct contact with, as well as those remote from, the symbiotic bacteria. The bacteria induce the developmental program soon after colonization of the organ, although complete morphogenesis requires 96 h. In this study, to determine critical time points, we examined the biochemistry underlying bacterium-induced host development using two-dimensional polyacrylamide gel electrophoresis. Specifically, V. fischeri-induced changes in the soluble proteome of the symbiotic organ during the first 96 h of symbiosis were identified by comparing the protein profiles of symbiont-colonized and uncolonized organs. Both symbiosis-related changes and age-related changes were analyzed to determine what proportion of the differences in the proteomes was the result of specific responses to interaction with bacteria. Although no differences were detected over the first 24 h, numerous symbiosis-related changes became apparent at 48 and 96 h and were more abundant than age-related changes. In addition, many age-related protein changes occurred 48 h sooner in symbiotic animals, suggesting that the interaction of squid tissue with V. fischeri cells accelerates certain developmental processes of the symbiotic organ. These data suggest that V. fischeri-induced modifications in host tissues that occur in the first 24 h of the symbiosis are independent of marked alterations in the patterns of abundant proteins but that the full 4-day morphogenetic program requires significant alteration of the host soluble proteome.
Substantial changes in the protein expression patterns of both host tissues and their associated symbiont partners have been demonstrated by two-dimensional (2-D) polyacrylamide gel electrophoresis (PAGE) in both pathogenic and cooperative symbiotic associations. Notably, Abshire and Neidhardt (1) described the proteins synthesized by Salmonella enterica serovar Typhimurium within human macrophage cells and found that the levels of approximately 140 proteins are changed, either induced or repressed, during growth within the host cell. Likewise, Abu Kwaik et al. (2) demonstrated the induction of 33 amoeba proteins, and the repression of 11 others, upon contact with a specific pathogenic bacterium. In legume symbioses, researchers have found dozens of symbiosis-induced plant nodulin proteins in response to symbiotic bacteria, as well as rhizobial proteins that are induced or repressed by interaction with the plant (7, 13, 16, 18). In these plant-bacterial symbioses, protein changes can be correlated to specific anatomical and morphological changes in the host and symbiont.
The symbiotic relationship between the Hawaiian sepiolid squid Euprymna scolopes and the marine luminous bacterium Vibrio fischeri is characterized by a complex multilevel communication between the two partners (19, 24), similar to that of rhizobium-legume symbiosis. In the squid-vibrio association, the bacterial symbionts produce luminescence that the nocturnal host animal uses in its behavior, although specific bioluminescent behaviors have not been well documented (19). During the initiation and early stages of the symbiosis following the onset of infection, dramatic changes occur in the anatomy and morphology of both the host and the symbiont that require the interaction of the partners (5, 20, 23). These events involve an array of host tissues that are both in direct contact with the bacterial symbionts and remote from this growing culture.
In the absence of V. fischeri, the light organ remains uncolonized, or aposymbiotic, but in the host's natural environment of Hawaiian nearshore waters, the light organ of the juvenile squid becomes infected with V. fischeri within hours. The bacteria enter via three pores on either side of the light organ and travel through ducts that lead to independent, internal epithelium-lined crypts (22). Within 12 h, the bacterial culture grows to fill the crypt spaces, and during these early hours of the symbiosis conspicuous changes in the host light organ are observable (Fig. 1). The bacterium-induced modifications in the polarized epithelial cells of the light organ, with which V. fischeri directly associates, include an increase in the density of the microvilli (17) and a swelling of these cells (23) (Fig. 1B). These changes are evident by microscopy as early as 12 to 18 h following infection (17; J. D. Lemus and J. F. Foster, unpublished data). In addition to these direct effects, the most obvious modification of the juvenile light organ is the loss of fields of ciliated, microvillous cells on the lateral surfaces of the light organ, remote from the bacterial population (Fig. 1A). These surface fields of cells surround the pores on either side of the organ, and each lateral field bears two appendages, one anterior and one posterior. The ciliary beat of these fields appears to facilitate the infection of the organ (21). Prior studies have shown that, although the complete loss of this superficial epithelium requires 96 h (23), the bacteria deliver an irreversible signal between 8 and 12 h following the first exposure to V. fischeri that triggers this protracted process (5).
FIG. 1.
Morphology and anatomy of the early developmental changes in the E. scolopes light organ. (A) Scanning electron micrographs of a hatchling (left half) and 96-h old symbiotic (right half) juvenile light organ. Bar = 100 μm. (B) Diagram depicting the juvenile light organ showing the interior crypts of a hatchling light organ (left half) and the changes that occur after colonization with symbiotic bacteria (right half), specifically, the increase in the size of the crypt epithelial cells and the density of microvilli.
The bacterial symbionts also display significant morphological and physiological changes over the first 96 h of the symbiosis (21, 26), which suggest responses to modified conditions in the light organ environment. Bacterial strains deficient in establishing a normal symbiosis have been identified and have been categorized into three types: initiation, colonization, and persistence mutants (26). Initiation mutants are unable to infect the squid light organ (10), colonization mutants are able to infect the light organ but do so at a significantly diminished extent compared to that of wild-type strains (9), and persistence mutants are initially able to reach wild-type colonization densities, but the colonization begins to decline significantly between 48 and 96 h postinoculation (8, 34). The existence of these phenotypes also suggests that concomitant changes in the environment of the light organ occur between 48 and 96 h postinoculation.
In this study, we focused specifically on the changes in host cell proteins associated with the bacterium-induced developmental modifications of squid light organ tissues. Given the spatial and temporal complexity of the biochemical interactions that occur between the partners, one of the goals was to identify critical time points during the early stages of the symbiosis. Using 2-D PAGE (25), we have investigated changes in the steady-state profiles of aqueous soluble (i.e., soluble) proteins within the light organ of E. scolopes in response to the presence of V. fischeri during the first 96 h of symbiosis. Specifically, we compared and contrasted both symbiosis-related changes and age-related changes. The results of this study revealed that, while the bacteria induce dramatic alterations in morphology in the first hours following symbiosis, marked changes in the soluble proteome do not occur until days after initiation of the symbiotic association.
MATERIALS AND METHODS
Maintenance of animals.
Juvenile squid were obtained from egg clutches that were laid by females maintained at 23°C in either a recirculating aquarium or open-flow seawater tables as previously described (5, 17). Newly hatched squid were rendered symbiotic by incubating them in seawater containing ∼103 V. fischeri cells/ml for approximately 12 h, followed by maintenance in ambient Hawaiian seawater. Aposymbiotic squid were maintained in V. fischeri-free seawater from the time of hatching. All animals were kept individually in glass scintillation vials, and the seawater was changed daily. Symbiotic infection was monitored daily throughout the experimental period by measuring the luminescence of squid with a photometer (Biospherical Instruments, San Diego, Calif., or Turner Designs, Sunnyvale, Calif.) (27).
Preparation of protein samples.
Whole squid from a given experiment were quick-frozen in liquid nitrogen and placed in aluminum foil packets at −80°C until their use for 2-D PAGE. Preliminary experiments revealed that freezing squid in this manner did not result in detectable degradation of the host proteins. The light organs from 50 frozen squid (on average, depending on their size and protein content) were dissected out of the mantle cavity and maintained on ice in a 50 mM Tris buffer (pH 7.2). Light organs were then homogenized in a glass micromortar and pestle, and the homogenate was centrifuged at 14,000 × g for 15 min at 4°C to remove bacterial cells, animal cell membranes, and ink. The supernatant fluid, which contained the aqueous soluble fraction of squid tissues and exported bacterial proteins, was removed, and its protein concentration was determined spectrophotometrically (37). Twenty micrograms of soluble protein sample was prepared for isoelectric focusing (IEF) by adding four parts lysis solution (9 M urea, 1% dithiothreitol [DTT], 2% Pharmalyte 3-10, 0.5% Triton X-100, 0.14% phenylmethylsulfonyl fluoride) to one part protein sample (volume per volume) by following the recommendations of the manufacturer of the 2-D gel system used in this study (Amersham Pharmacia Biotech, Piscataway, N.J.).
2-D PAGE methods.
To achieve high resolution and reproducibility, we used the Pharmacia LKB Multiphor II 2-D electrophoresis system with precast Immobiline Dry Strip IEF gels (linear pH gradient of 4 to 7) and ExcelGel (12 to 14% gradient) sodium dodecyl sulfate (SDS)-polyacrylamide gels. In this method, the proteins are initially separated in the first dimension based on their isoelectric points; the focused proteins of the first dimension are then separated in a second dimension based on their subunit molecular masses. A pH range of 4 to 7 was chosen for IEF because preliminary gels run in a gradient of pH 3 to 10 indicated that the highest resolution was required within the pH 4 to 7 range. Immobiline Dry Strips were first soaked overnight in rehydration solution (8 M urea, 0.5% Triton-X 100, 0.5% Pharmalyte 3-10, 0.2% DTT, and a few grains of Orange G dye). The first dimension was run at 20°C for a total of 46 to 56 kV · h (2 h at 500 V, a 5-h gradient to 3,500 V, followed by 10 to 13 h at 3,500 V). IEF gels not immediately used for the second dimension were frozen at −80°C without equilibration; preliminary experiments showed that freezing of the first dimension did not affect the quality of the second dimension. Immediately prior to the second dimension run, IEF gels were incubated successively in two equilibration solutions for 10 min each; the solutions consisted of (i) a 50 mM Tris-HCl buffer (pH 6.8) containing 6 M urea, 30% glycerol, 1% SDS, and 0.8% DTT and (ii) a 50 mM Tris-HCl buffer containing 6 M urea, 30% glycerol, 1% SDS, and 4.5% iodoacetamide. The excess liquid was then drained from the gel. The SDS-PAGE second dimension was run at 15°C for a total of 3,500 V · h (3.5 h at 1,000 V). Protein standards for 2-D gels (Bio-Rad Corp., Hercules, Calif.) were run on separate gels to provide landmarks for comparison of apparent molecular weights (Mr) and isoelectric points (pI). The IEF Dry Strips have a fixed pH gradient, which ensures the reproducibility of pI values between gels. However, because Mr might be affected by minor variations in the duration of the run, low-molecular-weight SDS-PAGE standards (Bio-Rad) were run alongside each sample on the second-dimension gels. Second-dimension gels were silver stained using a modified method of Heukeshoven and Dernick (11). Stained gels were covered with cellophane and air dried overnight at room temperature.
Analysis of resolved proteins on gels.
Gels were analyzed pairwise by eye for differences in their protein patterns by overlaying the gels on a light table. Because proteins were stained with silver, which does not exhibit a linear relationship between stained-spot density and protein concentration, a computer-generated densitometric analysis of protein spots was not performed. Unless the levels of abundance of a particular protein were dramatically different between two gels, only the presence and absence of proteins was recorded. Multiple (at least three) gel replicates of each light organ treatment were compared, and a composite gel for each treatment was generated. Specifically, each replicate gel was compared to the other replicates for the presence and absence of proteins. The presence or absence of any particular protein was considered valid only when at least two out of the three replicates displayed the same pattern for that protein (1). Gels were scanned into a computer graphics program (Adobe Photoshop 4.0), and one replicate was used as a reference gel. Protein spots were then added or subtracted manually to this reference gel according to their expression in two out of the three replicates. The resulting gel was defined as the composite gel for that treatment. Composite gels were then analyzed pairwise between treatments.
RESULTS
Patterns of host protein expression.
2-D gels of steady-state soluble proteins in the light organs of juvenile animals were highly reproducible and yet complex, averaging approximately 1,000 spots per gel. In initial experiments few, if any, reproducible differences were detected in steady-state proteins between aposymbiotic and symbiotic light organs at 12 or at 24 h, although previous studies had shown that this period is critical in signaling early events of the symbiosis (5) and is when conspicuous tissue remodeling has already begun (17, 23). We therefore focused subsequent analyses of proteins at 48 and 96 h postinoculation. For these comparisons, steady-state protein profiles were generated for aposymbiotic and symbiotic animals of three different ages: hatchling, 48 h, and 96 h. Analyses of these gels revealed numerous changes in the protein patterns of the light organs during these early stages of development and symbiosis. To distinguish changes in host proteins associated with the presence or absence of symbiotic bacteria within the light organ from those associated only with age or development, we categorized protein differences between gels into two major types: age related and symbiosis related (Fig. 2).
FIG. 2.
Number of changes in steady-state protein profiles in light organs as a function of both age and symbiotic state. Age-related protein changes at either 48 or 96 h are categorized as follows: induced (present in both aposymbiotic [APO] and symbiotic [SYM] light organs but absent from hatchling light organs) and repressed (present in hatchling light organs but absent from both aposymbiotic and symbiotic light organs). Symbiosis-related protein changes at 48 and 96 h are categorized as follows: induced in symbiotic (present in symbiotic but absent from aposymbiotic and hatchling), repressed in symbiotic (absent from symbiotic but present in aposymbiotic and hatchling), induced in aposymbiotic (present in aposymbiotic but absent from symbiotic and hatchling), and repressed in aposymbiotic (absent from aposymbiotic but present in symbiotic and hatchling) light organs.
Age-related changes in protein profiles.
An age-related protein change was defined as a difference in the levels of expression of a particular protein between hatchling and juvenile light organs, regardless of symbiotic state. The age-related changes could be divided into those present in both aposymbiotic and symbiotic squid at a given age but absent from hatchlings, i.e., induced proteins, and those present in hatchlings but absent from aposymbiotic and symbiotic squid, i.e., repressed proteins. In these comparisons between hatchling and 48- or 96-h-old animals, the total number of age-related changes that were detected increased more than fivefold between 48 h (n = 7) and 96 h (n = 40). Of these proteins with age-related changes, repressed proteins were more abundant at both 48 h (n = 4) and 96 h (n = 29) than induced proteins (n = 3 and 11, respectively) (Fig. 2; Table 1).
TABLE 1.
Molecular weight and isoelectric point estimations of age-related protein differences in juvenile light organs at 48 and 96 h after hatching
| Value for induced proteins at:
|
Value for repressed proteins at:
|
||||||
|---|---|---|---|---|---|---|---|
| 48 h
|
96 h
|
48 h
|
96 h
|
||||
| Mr | pI | Mr | pI | Mr | pI | Mr | pI |
| 85 | 6.25 | 89 | 6.55 | 51 | 5.35 | 90 | 6.50 |
| 84 | 6.20 | 55 | 6.10 | 43 | 6.15 | 66 | 6.45 |
| 34 | 5.45 | 41 | 6.60 | 26 | 5.05 | 57 | 6.50 |
| 40 | 6.50 | 25 | 5.00 | 54 | 6.45 | ||
| 40 | 6.55 | 53 | 5.30 | ||||
| 36 | 6.35 | 52 | 5.65 | ||||
| 30 | 6.00 | 50 | 4.45 | ||||
| 30 | 6.15 | 50 | 5.55 | ||||
| 26 | 6.75 | 48 | 4.40 | ||||
| 16 | 6.75 | 48 | 5.60 | ||||
| 13 | 5.20 | 46 | 5.60 | ||||
| 31 | 5.70 | ||||||
| 30 | 4.60 | ||||||
| 30 | 4.65 | ||||||
| 30 | 6.30 | ||||||
| 30 | 6.70 | ||||||
| 29 | 6.35 | ||||||
| 25 | 4.40 | ||||||
| 24 | 4.95 | ||||||
| 24 | 5.00 | ||||||
| 24 | 6.65 | ||||||
| 23 | 5.00 | ||||||
| 22 | 4.90 | ||||||
| 21 | 5.60 | ||||||
| 20 | 5.60 | ||||||
| 20 | 6.65 | ||||||
| 5 | 5.25 | ||||||
| 4 | 5.05 | ||||||
| 3 | 5.20 | ||||||
Symbiosis-related changes in protein profiles.
A symbiosis-related change was defined as a difference in the levels of expression of a particular protein between aposymbiotic and symbiotic light organs of the same age. Overall, symbiosis-related changes were more abundant than age-related changes, especially at 48 h (Fig. 2). Eighty-four symbiosis-related changes were detected at 48 h, and 54 symbiosis-related changes were detected at 96 h (Fig. 2 and 3). The most common type of symbiosis-related change at both 48 and 96 h was the presence of proteins in symbiotic animals that were absent from hatchling or aposymbiotic animals, i.e., induced in symbiotic animals (Fig. 3; Tables 2 and 3). Nine of the 49 proteins induced in symbiotic animals at 48 h were also induced in symbiotic animals at 96 h (Fig. 3), indicating that these proteins were synthesized early in the symbiosis and were either stable or continually synthesized through 96 h. Excluding the nine symbiosis-induced proteins that were identical in 48- and 96-h animals, 40 proteins were unique to 48-h symbiotic animals and 28 proteins were unique to 96-h symbiotic animals (Fig. 3).
FIG. 3.
Composite 2-D gels of steady-state proteins from 48-h aposymbiotic (APO) (A), 48-h symbiotic (SYM) (B), 96-h aposymbiotic (C), and 96-h symbiotic (D) light organs, identifying symbiosis-related changes in the proteome. Composite gels represent the reproducible proteins present in at least two out of three original replicate gels. Protein differences between aposymbiotic and symbiotic light organs at a given age (i.e., the data in panel A versus those in panel B or the data in panel C versus those in panel D) are depicted. All ovals indicate proteins induced in symbiotic animals, all rectangles indicate proteins repressed in aposymbiotic animals, diamonds indicate proteins induced in aposymbiotic animals, and triangles indicate proteins repressed in symbiotic animals. Blue ovals and rectangles represent symbiosis-related protein changes that were shared in 48- and 96-h symbiotic light organs. The horizontal dimension represents pH, and the vertical dimension represents Mr.
TABLE 2.
Molecular weight and isoelectric point estimations of symbiosis-related protein differences in juvenile light organs at 48 h postinoculation
| Induced in symbiotic light organ
|
Induced in aposymbiotic light organ
|
Repressed in symbiotic light organ
|
Repressed in aposymbiotic light organ
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mr | pI | Mr | pI | Mr | pI | Mr | pI | Mr | pI |
| 106 | 4.90 | 49 | 4.60 | 56 | 4.40 | 53 | 5.90 | 66 | 5.00 |
| 97 | 5.80 | 47 | 5.30 | 64 | 5.20 | ||||
| 97 | 5.85 | 46 | 4.70 | 62 | 5.80 | ||||
| 83 | 4.90 | 45 | 6.85 | 61 | 5.05 | ||||
| 80 | 6.45 | 44 | 4.60 | 60 | 5.70 | ||||
| 79 | 6.65 | 44 | 6.90 | 60 | 5.85 | ||||
| 79 | 6.70 | 43 | 4.60 | 59 | 5.50 | ||||
| 71 | 4.90 | 43 | 6.60 | 57 | 4.50 | ||||
| 71 | 5.20 | 43 | 6.70 | 55 | 5.70 | ||||
| 64 | 5.30 | 42 | 6.65 | 52 | 5.70 | ||||
| 64 | 6.05 | 40 | 5.80 | 51 | 6.30 | ||||
| 64 | 6.60 | 40 | 5.85 | 50 | 5.70 | ||||
| 64 | 6.65 | 37 | 6.70 | 45 | 5.60 | ||||
| 63 | 5.90 | 35 | 6.60 | 44 | 4.65 | ||||
| 62 | 6.60 | 32 | 6.40 | 44 | 5.70 | ||||
| 61 | 6.80 | 32 | 6.45 | 43 | 5.70 | ||||
| 61 | 6.85 | 31 | 4.35 | 35 | 5.80 | ||||
| 58 | 5.75 | 28 | 5.85 | 34 | 4.50 | ||||
| 57 | 6.80 | 24 | 6.80 | 31 | 6.85 | ||||
| 55 | 6.60 | 23 | 4.70 | 29 | 5.60 | ||||
| 52 | 5.45 | 20 | 4.20 | 28 | 4.60 | ||||
| 51 | 5.30 | 20 | 5.20 | 27 | 6.40 | ||||
| 50 | 5.45 | 12 | 6.30 | 23 | 4.95 | ||||
| 50 | 6.60 | 10 | 6.70 | 22 | 5.50 | ||||
| 21 | 5.50 | ||||||||
| 18 | 5.70 | ||||||||
| 15 | 4.85 | ||||||||
| 14 | 5.50 | ||||||||
TABLE 3.
Molecular weight and isoelectric point estimations of symbiosis-related protein differences in juvenile light organs at 96 h postinoculation
| Induced in symbiotic light organs
|
Induced in aposymbiotic light organs
|
Repressed in symbiotic light organs
|
Repressed in aposymbiotic light organs
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mr | pI | Mr | pI | Mr | pI | Mr | pI | Mr | pI |
| 90 | 5.4–5.7 | 51 | 6.60 | 32 | 6.10 | 37 | 5.65 | 91 | 6.55 |
| 68 | 5.80 | 50 | 4.75 | 27 | 5.85 | 18 | 6.65 | 91 | 6.60 |
| 67 | 5.90 | 50 | 5.40 | 18 | 4.30 | 12 | 5.30 | 90 | 6.45 |
| 65 | 6.00 | 50 | 5.60 | 16 | 6.40 | 90 | 6.50 | ||
| 64 | 5.90 | 49 | 5.65 | 54 | 5.50 | ||||
| 59 | 4.75 | 46 | 6.25 | 51 | 5.50 | ||||
| 59 | 4.80 | 45 | 4.45 | 51 | 5.55 | ||||
| 57 | 4.40 | 45 | 4.50 | 48 | 6.30 | ||||
| 56 | 4.65 | 45 | 6.50 | 25 | 4.35 | ||||
| 55 | 6.80 | 42 | 6.30 | 19 | 4.20 | ||||
| 54 | 5.50 | 41 | 5.90 | ||||||
| 54 | 5.60 | 36 | 6.50 | ||||||
| 54 | 5.80 | 32 | 4.60 | ||||||
| 54 | 6.90 | 32 | 6.05 | ||||||
| 51 | 5.30 | 29 | 4.50 | ||||||
| 51 | 5.35 | 28 | 6.70 | ||||||
| 51 | 5.45 | 22 | 4.20 | ||||||
| 51 | 5.50 | 16 | 5.20 | ||||||
| 51 | 5.60 | ||||||||
The secondmost common type of symbiosis-related change included proteins that were repressed in aposymbiotic animals but present both in symbiotic animals of the same age and in hatchlings (Fig. 2 and Fig. 3). Thirty-three proteins were repressed in aposymbiotic light organs at 48 h (Table 2), and 10 proteins were repressed in aposymbiotic light organs at 96 h (Table 3), with 1 of these proteins being identical between the two different ages (Fig. 3). In addition, we detected one protein at 48 h (Table 2) and four proteins at 96 h (Table 3) that were induced in aposymbiotic animals and not present in either hatchling or symbiotic animals (Fig. 3). Finally, one protein at 48 h (Table 2) and three proteins at 96 h (Table 3) were repressed in symbiotic animals (Fig. 3).
Early induction of age-related proteins in symbiotic light organs.
Of the 49 symbiosis-induced proteins at 48 h, 20 were also present in aposymbiotic light organs at 96 h (Fig. 4). All of these 20 proteins were also present in 96-h symbiotic light organs, indicating that they were not symbiosis-specific proteins at 96 h but rather that they were age related. These data suggest that symbiosis induces the early expression at 48 h of proteins that are later needed for age-related development at 96 h. Conversely, of the 37 symbiosis-induced proteins in light organs at 96 h, none was present in aposymbiotic light organs at 48 h.
FIG. 4.
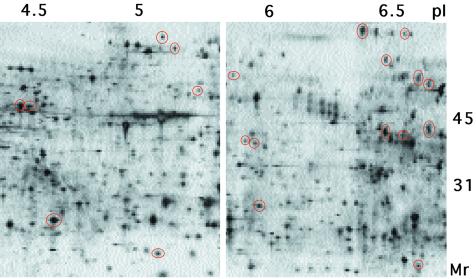
Precocious expression of proteins induced by symbiosis. Magnification of two areas from a composite 2-D gel of 48-h symbiotic light organs showing proteins (red ovals) that were specific to symbiosis at 48 h but were present at 96 h in both aposymbiotic and symbiotic light organs.
DISCUSSION
The results of this study provide evidence that (i) the dramatic light organ morphogenesis in E. scolopes is accompanied by complex changes in the soluble proteomes of associated tissues; (ii) the bacteria play a critical role in the induction of these changes; (iii) although the bacterial induction of morphogenetic changes occurs within hours of exposure to the symbiont, the very early stages of light organ development are not associated with substantial changes in the soluble protein patterns; (iv) interaction with the bacterial symbionts is required for the continued production of some subset of the light organ proteins; and (v) bacteria accelerate the developmental program of the host light organ tissue.
These data estimate the magnitude of overall change in the soluble proteome of the host squid during these early developmental stages of symbiosis. In most cases, gain or loss was always reproducible, but for the few proteins (less than 10 in all cases) that were present in only two of the three gels of a given treatment, a closer examination was necessary. While some variation in 2-D gel results may be due to differences in the reagents between runs, we believe that most of the variation that we observed between gels is likely to reflect actual differences in the samples; the squid vary at hatching in maturity and activity, as well as in how readily they become infected and the extent to which they become symbiotic. Such variation in 2-D gel patterns has been reported in other studies using these methods (1, 28, 36).
A theoretical source of contamination of host proteins in the samples of symbiotic juvenile light organs was exported bacterial proteins that could not be separated from the aqueous soluble proteins of the animal cells. However, the light organs of 50 symbiotic juveniles harbor only about 5 million bacteria, which contain approximately 0.5 μg of total protein in all protein pools (E. G. Ruby, personal communication). The exported proteins most likely represent only a small proportion of this total pool and thus should be present in quantities well below the detection limit of the 2-D gel method used.
Symbiosis-related changes in the light organ proteome outnumbered age-related changes at both 48 and 96 h, suggesting that a complex sequential dialogue occurs between the partners over the first few days of the symbiosis, similar to what has been reported in legume-rhizobium symbioses (7, 12, 29). Within the initial 96-h period of light organ development, both early (48 h) and late (96 h) proteins could be distinguished. Proteins that are induced earlier would be more likely to play a role in establishing a stable symbiosis and contribute to morphogenetic processes, whereas proteins induced later would be more likely to be associated with the maintenance and functioning of stable symbiosis. Similarly, the production of what have been designated early and late host proteins has also been noted for legume-rhizobium symbioses; proteins produced early are associated with invasion of the root hair and early cortical cell division, whereas those produced later are involved in the nitrogen-fixing function of the mature nodule (3, 6, 16). In light of the mutant phenotypes that have been generated in V. fischeri, it is not surprising to see differences between the samples at 48 and 96 h; the persistence mutants, by definition, colonize normally until around 48 h and then are eliminated from the light organ crypts (8, 34), suggesting that critical interactions occur in this time frame. Comparative analyses of the protein profiles of animals infected with various persistence mutants and wild-type parent strains should reveal protein species important during this period of the development of the symbiosis.
Because the observable morphological changes in the light organ are so dramatic in the first 24 h of symbiosis even though we detected no changes in the soluble proteome, the bacteria must effect these alterations by processes undetected by our methods. In this study, we noted only the presence or absence of a protein or very significant increases and decreases in the abundance of a particular protein. Minor changes in the levels of potentially critical proteins, or the appearance and disappearance of low-abundance species (e.g., transcription factors), were not noted, an issue that may be resolved in future studies by using radiolabels to identify bacterium-induced changes in the patterns of protein synthesis (1, 36). Alternatively, some or all of these early developmental events may be effected through posttranslational modifications, such as phosphorylations and glycosylations, of proteins that already exist in the hatchling protein pool. The scope of this study also did not include an analysis of host membrane proteins, which may exhibit significant changes during the first interactions of the host and symbionts. In a study of legume-rhizobium symbioses, 2-D gel analyses of root and root hair membrane proteins has shown several conspicuous changes in this fraction of the root hair proteome in the early hours of the symbiosis (16).
The identification of shared proteins between hatchling and symbiotic animals (33 proteins at 48 h and 10 at 96 h) provided evidence for the prehatch production of symbiosis-specific proteins. The loss or repression of production of these proteins in 48- and 96-h aposymbiotic animals suggested that interaction with the bacteria is essential for continued production of these proteins in the days following hatching. Their presence in hatchling light organs and their continued production in symbiotic organs suggest that the stages of establishment and maintenance of the symbiosis share certain biochemical mechanisms. Since aposymbiotic squid have never been observed in the field, the function of these downregulated proteins in a natural setting remains uncertain. However, the requirement for symbiotic bacteria for sustained protein production is not unprecedented; production of fucosylated glycoconjugates, which are apparent in both conventional and gnotobiotic mice between postnatal days 17 and 21, is repressed in gnotobiotic mice thereafter but increases dramatically in conventional mice (4). These data suggest that the presence of bacteria is essential for the persistence of these biomolecules.
We also observed a few proteins (one at 48 h and three at 96 h) that were present in hatchling and aposymbiotic squid but repressed in symbiotic squid; i.e., these proteins appear to be synthesized prior to hatching and continue to be expressed in the absence of interaction with V. fischeri, but their production is repressed, or they are degraded, once a symbiosis has been established. The pattern of expression in these proteins suggests that they are involved either actively or passively in initiating the symbiosis but apparently not in its long-term maintenance. For example, the squid may produce specific compounds that attract V. fischeri and/or promote colonization, as occurs in rhizobium-legume symbiosis (32). Alternatively, proteins may be involved with maintaining symbiotic specificity, selectively excluding nonsymbiotic bacteria. The repression of such “defense-related” proteins may provide a mechanism by which V. fischeri is able to colonize the light organ. One such protein may be squid halide peroxidase, the expression of which is repressed in the light organs of symbiotic juveniles (31). A dual role for this enzyme in both the establishment of a symbiosis and defense against potential pathogens has been suggested (30, 31, 35). A defense-related mechanism has also been suggested with plant-rhizobium symbiosis to explain the symbiotic repression of certain root hair proteins (16). A final possibility is that these proteins become associated with the membrane fraction of the cell upon initiation of a symbiosis, in which case they “disappear” from the soluble fraction. (Likewise, a protein that is induced in the soluble fraction of aposymbiotic or symbiotic animals may have become disassociated from the membrane.) A study of the animal membrane proteins (separated from the bacteria) during the initial stages of a symbiosis should reveal the validity of this hypothesis.
Two lines of evidence suggest that V. fischeri accelerates the developmental program of the host. In the present study, many proteins that were induced in 48-h symbiotic light organs were later present in both aposymbiotic and symbiotic animals at 96 h. We are unaware of any reports of such an expression pattern in any other symbiotic system. However, within an evolutionary context, it has been proposed that precocious maturation may confer a selective advantage early in life (15, 33). Although at present this idea is speculative, if symbiosis induces the early synthesis of certain developmental proteins that accelerate maturation, then it may be advantageous to the squid to develop a stable symbiosis as early as possible. A similar phenomenon is observed in the morphology of light organ tissues; in natural ambient seawater that contains nonsymbiotically competent bacteria, very subtle cellular regression of the superficial ciliated epithelial fields of juvenile light organs occurs, but animals reared in axenic seawater show no such regression (M. K. Montgomery and M. J. McFall-Ngai, unpublished data). The presence of symbiotic bacteria inside the light organ appears to accelerate this low-level regression process such that the ciliated fields have completely disappeared by 96 h postinoculation.
In conclusion, we have demonstrated the magnitude and complexity of the changes in soluble proteins that occur during the early stages of bacterium-induced light organ morphogenesis in E. scolopes. The fact that both partners of the symbiosis can be cultured independently of one another has made it possible to investigate both age- and symbiosis-induced changes in light organ proteins over time. Although other symbiosis-induced changes occur within hours, biochemical changes in soluble proteins do not become apparent in the light organ until 48 h postinoculation. Because there are a great many protein differences between aposymbiotic and symbiotic squid at 48 h, this period may be a critical time in the biochemical response of the squid to symbiotic bacteria that will allow a stable association to develop. In support of this hypothesis, 48 h postinoculation is also the time that the colonization levels of persistent V. fischeri mutants in the light organ begin to decline. As 2-D PAGE is now the method of choice for mapping the proteome complement of the cell (14), this study should provide a valuable database for researchers interested in symbiont-induced protein expression in eukaryotic hosts. The data presented here also lay the groundwork for future studies of the proteomes of animals infected with colonization and persistence mutants of V. fischeri to help identify specific light organ proteins that may be involved in the biochemical dialogue between the two partners.
ACKNOWLEDGMENTS
We thank the members of the McFall-Ngai and Ruby laboratories for helpful comments on the manuscript. We thank V. Weis, J. Vavra, and L. Gilbane for technical assistance.
This work was funded by NSF grant IBN 9904601 to M.J.M.-N. and E. G. Ruby and by NIH grant RO1-RR12294 to E. G. Ruby and M.J.M.-N.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y A, Fields B S, Engleberg N C. Protein expression by the protozoan Hartmannella vermiformis upon contact with its bacterial parasite Legionella pneumophila. Infect Immun. 1994;62:1860–1866. doi: 10.1128/iai.62.5.1860-1866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisseling T, Been C, Van Kammen A, Nadler K. Nodule-specific host proteins in effective and ineffective root nodules of Pisum sativum. EMBO J. 1983;2:961–966. doi: 10.1002/j.1460-2075.1983.tb01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bry L, Falk P G, Midtvedt T, Gordon J I. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 5.Doino J A, McFall-Ngai M J. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- 6.Gloudemans T, Bisseling T. Plant gene expression in early stages of Rhizobium-legume symbiosis. Plant Sci. 1989;65:1–14. [Google Scholar]
- 7.Govers F, Gloudemans T, Moerman M, Van Kammen A, Bisseling T. Expression of plant genes during the development of pea root nodules. EMBO J. 1985;4:861–867. doi: 10.1002/j.1460-2075.1985.tb03711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graf J, Ruby E G. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 9.Graf J, Ruby E G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heukeshoven J, Dernick R. Neue Ergebnisse zum Mechanismus der Silberfarbung. Electrophor Forum. 1986;86:22–27. [Google Scholar]
- 12.Hirsch A M. Tansley review no. 40. Developmental biology of legume modulation. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch A M, La Rue T A. Is the legume nodule a modified root or stem or an organ sui generis? Crit Rev Plant Sci. 1997;16:361–392. [Google Scholar]
- 14.James P. Of genomes and proteomes. Biochem Biophys Res Commun. 1997;231:1–6. doi: 10.1006/bbrc.1996.6045. [DOI] [PubMed] [Google Scholar]
- 15.Kirkwood T B L, Rose M R. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond B. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- 16.Krause A, Broughton W J. Proteins associated with root-hair deformation and nodule initiation in Vigna unguiculata. Mol Plant-Microbe Interact. 1992;5:96–103. [Google Scholar]
- 17.Lamarcq L H, McFall-Ngai M J. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legocki R P, Verma D P S. Identification of “nodule-specific” host proteins (nodulins) involved in the development of Rhizobium-legume symbiosis. Cell. 1980;20:153–163. doi: 10.1016/0092-8674(80)90243-3. [DOI] [PubMed] [Google Scholar]
- 19.McFall-Ngai M J. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu Rev Ecol Syst. 1999;30:235–256. [Google Scholar]
- 20.McFall-Ngai M J, Ruby E G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 21.McFall-Ngai M J, Ruby E G. Sepiolids and vibrios: when first they meet. BioScience. 1998;48:257–265. [Google Scholar]
- 22.Montgomery M K, McFall-Ngai M J. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery M K, McFall-Ngai M J. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery M K, McFall-Ngai M J. Late postembryonic development of the symbiotic light organ of Euprymna scolopes (Cephalopoda:Sepiolidae) Biol Bull. 1998;195:326–336. doi: 10.2307/1543144. [DOI] [PubMed] [Google Scholar]
- 25.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby E G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 27.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 28.Santaren J F, Garcia-Bellido A. 2D gene expression parameters of wing imaginal disc of Drosophila for developmental analysis. Dev Genes Evol. 1996;206:349–354. doi: 10.1007/s004270050063. [DOI] [PubMed] [Google Scholar]
- 29.Scheres B, Van Engelen F, Van Der Knaap E, Van De Wiel C, Van Kammen A, Bisseling T. Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell. 1990;2:687–700. doi: 10.1105/tpc.2.8.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Small A L, McFall-Ngai M J. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- 31.Small A L, McFall-Ngai M J. Halide peroxidase gene expression in cooperative and potentially pathogenic associations of Euprymna scolopes with Vibrio fischeri. Am Zool. 1998;38:93A. [Google Scholar]
- 32.van Brussel A A N. Symbiotic signals in early stages of the morphogenesis of rhizobium-induced root nodules. Symbiosis. 1990;9:135–146. [Google Scholar]
- 33.Vanfleteren J R, De Vreese A. Analysis of the proteins of aging Caenorhabditis elegans by high resolution two-dimensional gel electrophoresis. Electrophoresis. 1994;15:289–296. doi: 10.1002/elps.1150150149. [DOI] [PubMed] [Google Scholar]
- 34.Visick K L, McFall-Ngai M J. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weis V M, Small A L, McFall-Ngai M J. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis V M, Levine R P. Differential protein profiles reflect the different lifestyles of symbiotic and aposymbiotic Anthopleura elegantissima, a sea anemone from temperate waters. J Exp Biol. 1996;199:883–892. doi: 10.1242/jeb.199.4.883. [DOI] [PubMed] [Google Scholar]
- 37.Whittaker J, Granum P. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]