Abstract
The bioremediation of polluted groundwater and toxic waste sites requires that bacteria come into close physical contact with pollutants. This can be accomplished by chemotaxis. Five motile strains of bacteria that use five different pathways to degrade toluene were tested for their ability to detect and swim towards this pollutant. Three of the five strains (Pseudomonas putida F1, Ralstonia pickettii PKO1, and Burkholderia cepacia G4) were attracted to toluene. In each case, the response was dependent on induction by growth with toluene. Pseudomonas mendocina KR1 and P. putida PaW15 did not show a convincing response. The chemotactic responses of P. putida F1 to a variety of toxic aromatic hydrocarbons and chlorinated aliphatic compounds were examined. Compounds that are growth substrates for P. putida F1, including benzene and ethylbenzene, were chemoattractants. P. putida F1 was also attracted to trichloroethylene (TCE), which is not a growth substrate but is dechlorinated and detoxified by P. putida F1. Mutant strains of P. putida F1 that do not oxidize toluene were attracted to toluene, indicating that toluene itself and not a metabolite was the compound detected. The two-component response regulator pair TodS and TodT, which control expression of the toluene degradation genes in P. putida F1, were required for the response. This demonstration that soil bacteria can sense and swim towards the toxic compounds toluene, benzene, TCE, and related chemicals suggests that the introduction of chemotactic bacteria into selected polluted sites may accelerate bioremediation processes.
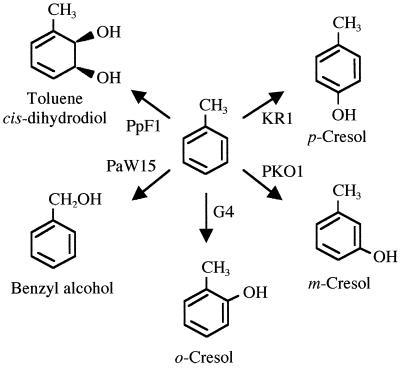
Bacterial chemotaxis has been studied in detail for Escherichia coli and Salmonella enterica serovar Typhimurium (35). Simple sugars, amino acids, and organic acids are chemoattractants for these enteric bacteria. Aromatic acids such as benzoate, 4-hydroxybenzoate, and salicylate are attractants for Pseudomonas putida PRS2000 (15). Recently, the soil bacterium P. putida G7 was reported to be attracted to the pollutant naphthalene (12, 24, 31). This expanded the range of organic compounds that are known to serve as bacterial chemoattractants to include aromatic hydrocarbons. However, nothing is known about chemotaxis towards other common aromatic hydrocarbons such as toluene and benzene. Five distinct pathways have been described for the aerobic degradation of toluene. All pathways are initiated with the oxidation of toluene, but five different oxidation products are formed (Fig. 1). P. putida F1 contains toluene 2,3-dioxygenase, an enzyme that oxidizes the aromatic ring of toluene, incorporating both atoms of molecular oxygen. After a dehydrogenation step, 3-methylcatechol is formed. This compound is further degraded via meta ring fission (8, 10, 11). P. putida PaW15 (a leucine auxotroph of strain mt-2) initiates degradation at the methyl group of toluene, eventually forming benzoate. Benzoate is converted to catechol, which is also degraded by a meta cleavage route (41). Strains that monooxygenate the aromatic ring of toluene have also been described. Burkholderia cepacia G4 utilizes a toluene 2-monooxygenase that sequentially oxidizes the aromatic ring at the 2- and 3-positions to form o-cresol and then 3-methylcatechol (32). Ralstonia pickettii PKO1 (formerly Burkholderia pickettii PKO1 [42]), has a toluene 3-monooxygenase that attacks initially at the 3-position of toluene, forming m-cresol, and a second monooxygenase attacks at the 2-position of m-cresol, to form 3-methylcatechol (30). 3-Methylcatechol is degraded by a meta cleavage pathway in strains G4 and PKO1. Finally, Pseudomonas mendocina KR1 has a toluene 4-monooxygenase that oxidizes toluene to form p-cresol. The methyl group of p-cresol is then sequentially oxidized to form 4-hydroxybenzoate, which is further degraded by the β-ketoadipate (ortho) pathway (39, 40). In each of these five strains, toluene degradation genes are expressed during growth with toluene (7, 18, 26, 28, 40). Here, five strains of bacteria that degrade the aromatic hydrocarbon toluene by these five different oxidative pathways were screened for their ability to sense and respond behaviorally to a concentration gradient of toluene. Chemotaxis of one of these strains, P. putida F1, towards 12 organic pollutants was tested.
FIG. 1.
Initial reactions in the five bacterial pathways for aerobic degradation of toluene in strains P. putida F1, P. putida PaW15 (a leucine auxotroph of strain mt-2 [41]), B. cepacia G4, R. pickettii PKO1, and P. mendocina KR1. P. putida F1 utilizes a dioxygenase-initiated pathway for toluene degradation. G4, PKO1, and KR1 initiate toluene degradation with toluene 2-, 3-, and 4-monooxygenases, respectively. PaW15 carries the TOL plasmid and oxidizes the methyl group of toluene.
MATERIALS AND METHODS
Growth of bacterial strains.
For chemotaxis assays, P. putida F1 (8, 10) was grown in minimal medium (MSB) (34) with 40 mM pyruvate and toluene provided as a vapor (induced) or with 40 mM pyruvate only (uninduced). P. putida F1 and mutant derivatives were kindly provided by D. T. Gibson. P. putida F1 mutant strains were grown under identical conditions. B. cepacia G4 (32), R. pickettii PKO1 (30), P. mendocina KR1 (39), and P. putida PaW15 (41) were grown in MSB with 10 mM succinate with (induced) or without (uninduced) toluene present. PaW15 cultures were supplemented with 50 μg of leucine per ml. Growth with alternative carbon sources was determined in MSB liquid medium or on MSB agar plates. Volatile compounds were supplied in the vapor phase. Growth was reported as positive if the culture could be sequentially transferred on the substrate when it was provided as the sole carbon source.
Chemotaxis assays.
Agarose plug assays were carried out as previously described (44) with slight modifications. Plugs contained 2% low-melting-temperature agarose (NuSieve GTG Agarose; FMC Bioproducts, Rockland, Maine) in chemotaxis buffer (40 mM potassium phosphate [pH 7.0], 0.05% glycerol, 10 mM EDTA), 10% (vol/vol) toluene, and a few crystals of Coomassie blue to provide contrast. A drop (10 μl) of the melted agarose mixture was placed on a microscope slide, and a coverslip supported by two plastic strips was then placed on top to form a chamber. Cells were harvested in log phase (optical density at 600 nm [OD600] of between 0.3 and 0.7), resuspended in chemotaxis buffer to an OD660 of approximately 0.7, and flooded into the chamber to surround the agarose plug. Compounds other than toluene to be tested as chemoattractants were also provided at 10% (wt/vol or vol/vol) in plug assays, except toluene cis-dihydrodiol and α,α,α-trifluorotoluene (TFT) cis-dihydrodiol (1 mg/ml), succinate (1 mM), and Casamino Acids (2% [wt/vol]). Control plugs contained no attractant, and no response was seen. Modified capillary assays were carried out essentially as previously described (12). Capillaries (1 μl) contained attractant in 1% low-melting-temperature agarose dissolved in chemotaxis buffer. Attractants were supplied at between 1 and 3 mM (toluene, benzene, TFT, and trichloroethylene [TCE]), 2% Casamino Acids, or 1 mg of TFT cis-dihydrodiol per ml. Freshly grown cells were suspended in chemotaxis buffer to an OD660 of approximately 0.05 and placed in a chamber formed by a microscope slide, a glass U-tube, and a coverslip, and the capillary containing the attractant was inserted into the pool of cells. Cell behavior was observed at an ×40 magnification.
RESULTS
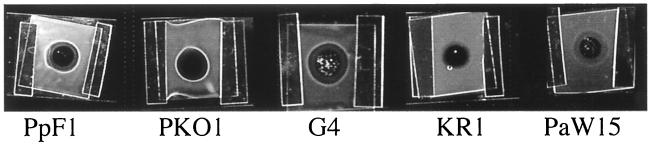
Using an agarose plug assay (44), a chemotactic response was observed in the form of a band of cells that accumulated in a ring surrounding, but not touching, the toluene-containing agarose plug (Fig. 2). Diffusion of toluene from the agarose plug into the surrounding pool of cells results in the development of a concentration gradient of dissolved toluene. This assay is particularly useful for testing responses to volatile compounds, since the incorporation of the chemoeffector in the agarose in this partially closed system reduced the loss of the aromatic hydrocarbon by volatilization. Toluene is soluble in aqueous media to a concentration of about 6 mM (33). Toluene-grown cells of three of the five strains, P. putida F1, R. pickettii PKO1, and B. cepacia G4, accumulated near agarose plugs containing toluene (Fig. 2), but strains grown with pyruvate or succinate did not respond (data not shown). A weak response was occasionally seen with toluene-grown P. putida PaW15, which carries a catabolic plasmid (the TOL plasmid) that encodes toluene degradation genes. No response to toluene was observed with P. mendocina KR1 (Fig. 2). When grown with either toluene or an organic acid, cells of each strain had a strong chemotactic response to Casamino Acids. This indicates that all of the strains have a general chemotaxis system for the detection of amino acids and that all strains were sufficiently motile to mount a chemotactic response. These results show that some, but not all, toluene-degrading strains are chemotactic towards toluene and that the presumed receptor(s) responsible for detecting toluene is expressed only in response to growth in the presence of toluene.
FIG. 2.
Chemotactic responses of the five bacterial strains (see the legend to Fig. 1) to toluene in agarose plug assays 5 min after addition of cells. Cells were grown in the presence of toluene as described in Materials and Methods.
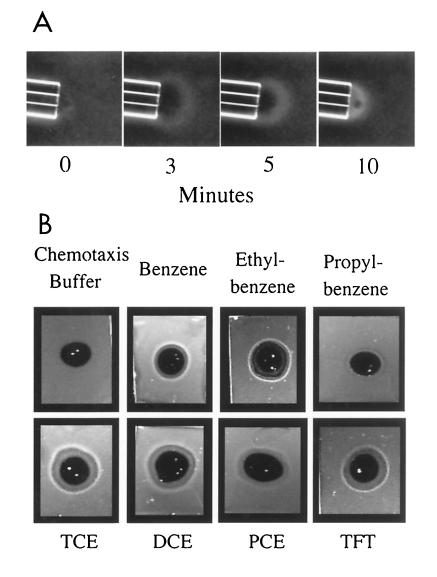
A time course of the chemotactic response of P. putida F1 to toluene in a modified capillary assay (12) is shown in Fig. 3A. The response was apparent at 1 min and continued to develop over 10 to 15 min. Pyruvate-grown P. putida F1 did not respond (data not shown). In the modified capillary assays, there was an initial clearing at the mouth of the capillary and cells accumulated at a short distance from the capillary (Fig. 3A). Cells present in the band of accumulation were observed to dart rapidly back and forth within the band. In contrast, when Casamino Acids were present in the agarose plug or the capillary, cells accumulated as close to the source as possible. In chemotaxis systems studied to date, the addition of a chemoattractant to cells results in a short-lived smooth-swimming response, where changes of swimming direction are less frequent (35). Although the results of two different assays indicate that toluene elicited a positive chemotactic response, we have been unable to consistently observe smooth-swimming cells upon addition of toluene. In fact, in temporal assays (14) with high concentrations of toluene (>500 μM), a repellent response (increased frequency of changing direction) was observed with both induced and uninduced cells. Due to the high volatility of toluene, it is difficult to reproducibly deliver precise amounts. In plug assays, toluene diffuses into the pool of cells and bacteria accumulate at a short distance from the toluene-containing plug, suggesting that the cells are seeking an optimal concentration of toluene. These observations suggest that P. putida F1 is attracted to a very narrow range of toluene concentrations.
FIG. 3.
(A) Time course of the chemotactic response of P. putida F1 to toluene in modified capillary assays at an ×40 magnification. P. putida F1 was grown in the presence of toluene as described in Materials and Methods. Capillaries contained 1.4 mM toluene. No response was seen when capillaries contained only agarose and chemotaxis buffer. (B) Chemotactic responses of P. putida F1 to benzene, ethylbenzene, propylbenzene, TFT, TCE, DCE, and PCE after growth in the presence of toluene. Plug assays were carried out as described in Materials and Methods. Cells were incubated for 5 min in the presence of the agarose plugs. Images were cropped.
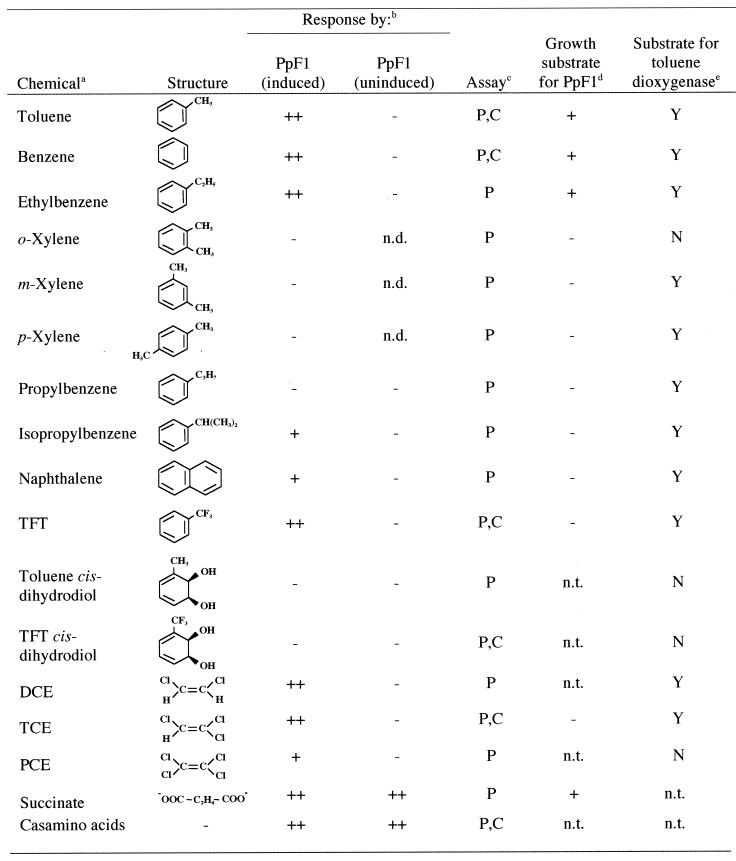
The chemotactic responses of P. putida F1 to benzene, ethylbenzene, and TFT were demonstrated in agarose plug assays (Fig. 3B). A wide range of substituted benzenes were shown to be chemoattractants in agarose plug assays and modified capillary assays (Table 1). However, the response was specific for a subset of related compounds. For example, the monosubstituted compounds toluene, ethylbenzene, and isopropylbenzene were good attractants, but propylbenzene and the disubstituted xylene isomers were not attractants (Fig. 3B; Table 1). The chlorinated aliphatic compounds TCE, cis-1,2-dichloroethylene (DCE), and perchloroethylene (PCE) elicited positive responses (Table 1; Fig. 3B). Responses to all aromatic hydrocarbons as well as to chlorinated aliphatic compounds were induced during growth of P. putida F1 with toluene. In contrast, the response to succinate and Casamino Acids was constitutive (Table 1).
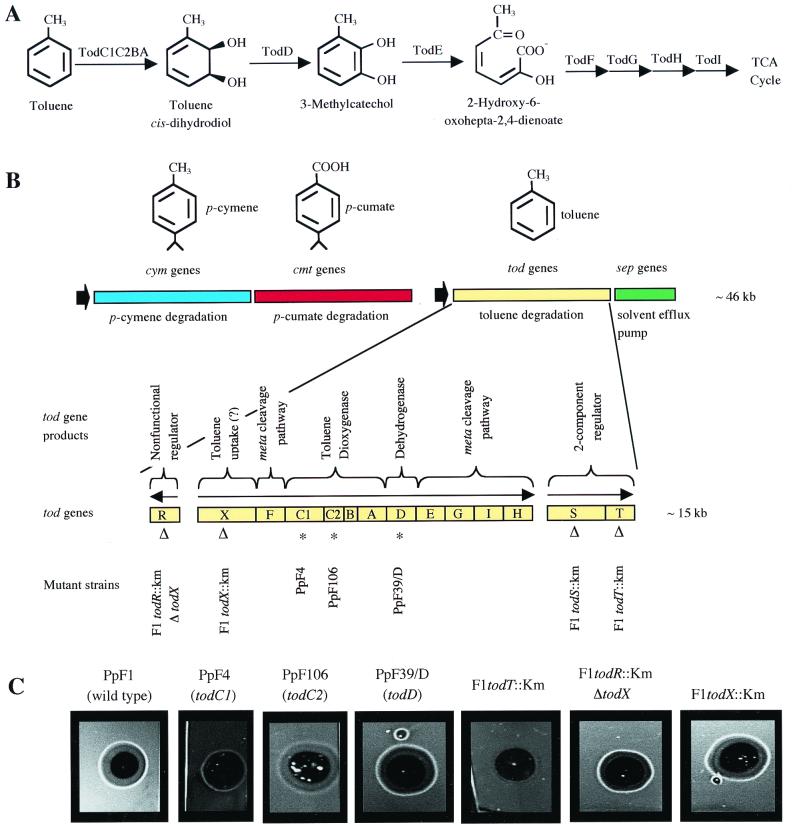
P. putida F1 degrades toluene through an initial dioxygenation reaction to form toluene cis-dihydrodiol, followed by dehydrogenation to form 3-methylcatechol, a compound that is converted to tricarboxylic acid cycle intermediates by a meta ring-cleavage pathway (Fig. 4A) (45). The genes for these reactions have been cloned and sequenced and are arranged in what is likely to be a single transcriptional unit, todXFC1C2BADEGIH, on the P. putida F1 chromosome (Fig. 4B) (20, 27, 38, 46). Strains with mutations in various toluene degradation genes (Fig. 4B) were tested for the ability to respond to toluene. P. putida F4 and P. putida F106 (8), which have mutations in todC1 and todC2, the genes encoding the toluene dioxygenase α and β subunits, responded to toluene, as did strain P. putida F39/D (10), a toluene cis-dihydrodiol dehydrogenase (TodD) mutant (Fig. 4C). None of these strains grows on toluene. The todR gene, which is located upstream of todX (Fig. 4B), appears to encode a nonfunctional truncated LysR-type regulator. TodX is a membrane protein that has been implicated in toluene transport (38). Strain F1todX::Km has a polar mutation in todX that blocks growth on toluene. Strain F1todR::KmΔtodX, which has a nonpolar mutation affecting todR and todX only (38), grows at wild-type rates with toluene. Both of these strains responded to toluene (Fig. 4C). These results show that the P. putida F1 chemotactic response to toluene results from direct detection of the aromatic hydrocarbon itself and not a metabolite, and they also show that the membrane protein TodX is not required for the chemotactic response. TFT is a structural analog of toluene that can be converted to a cis-dihydrodiol by toluene dioxygenase (2). TFT itself elicited a chemotactic response, but neither toluene cis-dihydrodiol nor TFT cis-dihydrodiol was an attractant for P. putida F1 (Table 1). This is further evidence that toluene itself is directly detected by strain P. putida F1 as the chemoattractant.
FIG. 4.
(A) Pathway for the degradation of toluene in P. putida F1. Gene products catalyzing each step are indicated in panel B. (B) Genetic organization of the toluene degradation gene cluster and surrounding catabolic genes in P. putida F1. Solid arrows indicate repeated sequences (4). Also shown are the functions of the tod gene products and the locations of mutations of interest. Triangles indicate the insertion of a kanamycin cassette, and asterisks indicate spontaneous mutations. (C) Chemotactic responses of P. putida F1 and mutant strains after 5 min in the presence of toluene-containing agarose plugs. Strains were grown with pyruvate and toluene. Plug assays were performed as described in Materials and Methods; images were cropped. Cells grown with pyruvate alone did not respond (data not shown). No response was observed when toluene was omitted from the plug (data not shown). The response of each strain was tested in the modified capillary assay (data not shown), and results were consistent with the plug assay results.
In the P. putida F1 chromosome, the todRXFC1C2BADEGIH gene cluster is followed by two genes, todST (Fig. 4B), which encode a two-component sensory transduction system that is required for induction of the tod structural genes in the presence of toluene (21). Inactivation of todS blocks growth on toluene, and a mutation in todT results in very slow growth with toluene. Neither strain F1todS::Km nor strain F1todT::Km (21) responded to toluene in agarose plug or modified capillary assays (Fig. 4C), suggesting that genes required for the chemotactic response to toluene are coordinately controlled with those for toluene degradation by TodS and TodT in the presence of toluene.
DISCUSSION
We expect that one or more receptor proteins bind the aromatic hydrocarbons and chlorinated solvents that are chemoattractants for P. putida F1 and that this initiates a sensory signal transduction cascade that modulates swimming behavior. Methyl-accepting chemotaxis proteins (MCPs) are cell surface receptors for sugars and amino acids that have been extensively studied in E. coli and are found in other motile bacteria (1, 6). P. putida G7 detects naphthalene with an MCP that is induced by naphthalene and is cotranscribed with naphthalene degradation genes harbored on a large catabolic plasmid (13).
It may be that an MCP is also responsible for the toluene-induced response to toluene exhibited by strain P. putida F1. The genes for toluene degradation are located on what is probably a large catabolic transposon on the P. putida F1 chromosome. Nearby are genes for the degradation of p-cymene (isopropyltoluene) and p-cumate (isopropylbenzoate), and repeated sequences that may have been involved in an insertion event have been identified (Fig. 4B) (3, 4). However, there are no genes in the 46-kb sequenced region of P. putida F1 that would be predicted to encode an MCP. Genes encoding aromatic hydrocarbon-specific MCPs may exist but have not yet been found, or other proteins could play a direct role in the detection and chemotactic response to toluene. Proteins encoded by genes in or near the tod gene cluster that are possible candidates for the P. putida F1 toluene chemoreceptor are SepABC, a solvent efflux pump encoded by genes that are located downstream of todT (Fig. 4B) (P. Phoenix, H. Bergeron, A. Patel, and P. C. K. Lau, Abstr. Pseudomonas '99: Biotechnology and Pathogenesis, abstr. 137, 1999). It is also possible that the TodS sensor functions as a receptor to initiate chemosensory signal transduction. A wide range of aromatic compounds has been reported to induce the toluene dioxygenase genes (36), and TodS is probably the protein that initially senses these effectors. This makes TodS an attractive candidate for a chemoreceptor. However, only a subset of those compounds that have been shown to induce toluene dioxygenase expression (i.e., benzene, toluene, ethylbenzene, TFT, and m- and p-xylene) were also found to be chemoattractants.
Chemotaxis gives motile bacteria the advantage of being able to locate compounds such as toluene that can support their growth. Chemotactic cells would be especially efficient at sensing and swimming towards chemicals that are present at point sources, for example, absorbed to soil particles in groundwater or within slowly moving pollutant plumes. In this way, chemotactic bacteria can overcome mass-transfer limitations that impede bioremediation processes. Once cells are brought into close contact with pollutants, mechanisms like biofilm formation and surfactant production can come into play to increase the bioavailability and biodegradation of absorbed chemicals. Chemotaxis of P. putida F1 towards compounds that are not growth substrates is probably a fortuitous consequence of a broad-substrate-specificity chemoreceptor that detects a variety of chlorinated aliphatic compounds and aromatic hydrocarbons, in addition to toluene.
Although it is clear from our analysis of toluene degradation mutants that the toluene dioxygenase is not required for chemotaxis towards toluene, it is noteworthy that most of the compounds that are detected by P. putida F1 as part of its toluene-inducible repertoire of chemotactic responses are also substrates for its toluene dioxygenase (Table 1) (16, 19). Toluene dioxygenase and other toluene-oxidizing enzymes detoxify the suspected carcinogen and U.S. Environmental Protection Agency priority pollutant TCE (22, 23, 28, 29, 37, 47). TCE is the most commonly reported groundwater contaminant, and aromatic hydrocarbon-degrading bacteria are believed to be responsible for the endogenous remediation of TCE that is seen at contaminated sites. A persistent challenge to bioremediation as a waste treatment technology is to find ways to accelerate this process, either by stimulating the activities of indigenous microorganisms or by directly introducing strains that have enhanced biodegradation capabilities. To date, strains G4, KR1, and P. putida F1 have been used in pilot and field-scale bioremediation applications for removal of TCE and benzene–toluene–p-xylene mixtures (5, 9, 17, 25, 43). The results reported here raise the possibility that bacteria with strong chemotactic responses to environmental pollutants may speed biodegradation processes. In selecting bacterial strains to be used to treat contaminated sites, it may be worthwhile to consider their chemotactic, as well as their biodegradative, capabilities.
TABLE 1.
Chemotactic response of P. putida F1 to aromatic hydrocarbons, substituted aromatic compounds, and chlorinated aliphatic compounds
All compounds were provided at the concentrations indicated in Materials and Methods.
P. putida F1 (PpF1) was grown as described in Materials and Methods. ++, strong response; +, weak response; −, no response; n.d., not determined.
Assay used: P, agarose plug assay; C, modified capillary assay.
Growth was determined as described in Materials and Methods. +, growth; −, no growth; n.t., not tested. Growth with benzene, toluene, and ethylbenzene was previously reported (11). Spontaneous mutants that grew with propylbenzene and isopropylbenzene were observed.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM56665 from the U.S. National Institute of General Medical Sciences.
We thank J. Parales for generating figures and D. T. Gibson, L. McCarter, and E. P. Greenberg for helpful discussions. Toluene cis-dihydrodiol and TFT cis-dihydrodiol were kindly provided by S. M. Resnick and D. T. Gibson.
REFERENCES
- 1.Aizawa S-I, Harwood C S, Kadner R J. Signalling components in bacterial locomotion and sensory reception. J Bacteriol. 2000;182:1459–1471. doi: 10.1128/jb.182.6.1459-1471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd D R, Sharma N D, Byrne B, Hand M V, Malone J F, Sheldrake G N, Blacker J, Dalton H. Enzymatic and chemoenzymatic synthesis and stereochemical assignment of cis-dihydrodiol derivatives of monosubstituted benzenes. J Chem Soc Perkin Trans 1. 1998;1998:1935–1943. [Google Scholar]
- 3.Eaton R W. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton R W. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ensley B D, Kurisko P R. A gas lift bioreactor for removal of contaminants from the vapor phase. Appl Environ Microbiol. 1994;60:285–290. doi: 10.1128/aem.60.1.285-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finette B A, Gibson D T. Initial studies on the regulation of toluene degradation by Pseudomonas putida F1. Biocatalysis. 1988;2:29–37. [Google Scholar]
- 8.Finette B A, Subramanian V, Gibson D T. Isolation and characterization of Pseudomonas putida PpF1 mutants defective in the toluene dioxygenase enzyme system. J Bacteriol. 1984;160:1003–1009. doi: 10.1128/jb.160.3.1003-1009.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folsom B R, Chapman P J. Performance characterization of a model bioreactor for the biodegradation of trichloroethylene by Pseudomonas cepacia G4. Appl Environ Microbiol. 1991;57:1602–1608. doi: 10.1128/aem.57.6.1602-1608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson D T, Hensley M, Yoshioka H, Mabry T J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970;9:1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 11.Gibson D T, Koch J R, Kallio R E. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry. 1968;7:2653–2661. doi: 10.1021/bi00847a031. [DOI] [PubMed] [Google Scholar]
- 12.Grimm A C, Harwood C S. Chemotaxis of Pseudomonas putida to the polyaromatic hydrocarbon naphthalene. Appl Environ Microbiol. 1997;63:4111–4115. doi: 10.1128/aem.63.10.4111-4115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm A C, Harwood C S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol. 1999;181:3310–3316. doi: 10.1128/jb.181.10.3310-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood C S, Fosnaugh K, Dispensa M. Flagellation of Pseudomonas putida and analysis of its motile behavior. J Bacteriol. 1989;171:4063–4066. doi: 10.1128/jb.171.7.4063-4066.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harwood C S, Rivelli M, Ornston L N. Aromatic acids are chemoattractants for Pseudomonas putida. J Bacteriol. 1984;160:622–628. doi: 10.1128/jb.160.2.622-628.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudlicky T, Gonzalez D, Gibson D T. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichim Acta. 1999;32:35–62. [Google Scholar]
- 17.Krumme M L, Timmis K N, Dwyer D F. Degradation of trichloroethylene by Pseudomonas cepacia G4 and the constitutive mutant strain G4 5223 PR1 in aquifer microcosms. Appl Environ Microbiol. 1993;59:2746–2749. doi: 10.1128/aem.59.8.2746-2749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukor J J. Diversity of toluene degradation following long term exposure to BTEX in situ. In: Kamely D, Chakrabarty A, Omenn G S, editors; Kamely D, Chakrabarty A, Omenn G S, editors. Biotechnology and biodegradation. The Woodlands, Tex: Portfolio Publishing Co.; 1990. pp. 405–421. [Google Scholar]
- 19.Lange C C, Wackett L P. Oxidation of aliphatic olefins by toluene dioxygenase: enzyme rates and product identification. J Bacteriol. 1997;179:3858–3865. doi: 10.1128/jb.179.12.3858-3865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau P C K, Bergeron H, Labbé D, Wang Y, Brousseau R, Gibson D T. Sequence and expression of the todGIH genes involved in the last three steps of toluene degradation by Pseudomonas putida F1. Gene. 1994;146:7–13. doi: 10.1016/0378-1119(94)90827-3. [DOI] [PubMed] [Google Scholar]
- 21.Lau P C K, Wang Y, Patel A, Labbé D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leahy J G, Byrne A M, Olsen R H. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl Environ Microbiol. 1996;62:825–833. doi: 10.1128/aem.62.3.825-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Wackett L P. Trichloroethylene oxidation by toluene dioxygenase. Biochem Biophys Res Commun. 1992;185:443–451. doi: 10.1016/s0006-291x(05)81005-8. [DOI] [PubMed] [Google Scholar]
- 24.Marx R B, Aitken M D. Quantification of chemotaxis to naphthalene by Pseudomonas putida G7. Appl Environ Microbiol. 1999;65:2847–2852. doi: 10.1128/aem.65.7.2847-2852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massol-Deyá A, Weller R, Ríos-Hernández L, Zhou J-Z, Hickey R F, Tiedje J M. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol. 1997;63:270–276. doi: 10.1128/aem.63.1.270-276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClay K, Streger S H, Steffan R J. Induction of toluene oxidation activity in Pseudomonas mendocina KR1 and Pseudomonas sp. strain ENVPC5 by chlorinated solvents and alkanes. Appl Environ Microbiol. 1995;61:3479–3481. doi: 10.1128/aem.61.9.3479-3481.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menn F-M, Zylstra G J, Gibson D T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase in Pseudomonas putida F1. Gene. 1991;104:91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 28.Nelson M J K, Montgomery S O, Mahaffey W R, Pritchard P H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol. 1987;53:949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman L M, Wackett L P. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J Bacteriol. 1997;179:90–96. doi: 10.1128/jb.179.1.90-96.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen R H, Kukor J J, Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994;176:3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samanta S K, Jain R K. Evidence for plasmid-mediated chemotaxis of Pseudomonas putida towards naphthalene and salicylate. Can J Microbiol. 2000;46:1–6. doi: 10.1139/cjm-46-1-1. [DOI] [PubMed] [Google Scholar]
- 32.Shields M S, Montgomery S O, Chapman P J, Cuskey S M, Pritchard P H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl Environ Microbiol. 1989;55:1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikkema J, de Bont J A M, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads; a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 35.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors; Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 36.Wackett L P. Ph.D. dissertation. Austin: The University of Texas; 1984. [Google Scholar]
- 37.Wackett L P, Gibson D T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Rawlings M, Gibson D T, Labbé D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 39.Whited G M, Gibson D T. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3017–3020. doi: 10.1128/jb.173.9.3017-3020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whited G M, Gibson D T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams P, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 43.Yee D C, Maynard J A, Wood T K. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl Environ Microbiol. 1998;64:112–118. doi: 10.1128/aem.64.1.112-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H S, Alam M. An agarose-in-plug bridge method to study chemotaxis in the archaeon Halobacterium salinarum. FEMS Microbiol Lett. 1997;156:265–269. doi: 10.1111/j.1574-6968.1997.tb12738.x. [DOI] [PubMed] [Google Scholar]
- 45.Zylstra G J, Gibson D T. Aromatic hydrocarbon degradation: a molecular approach. Genet Eng. 1991;13:183–203. doi: 10.1007/978-1-4615-3760-1_8. [DOI] [PubMed] [Google Scholar]
- 46.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todC1C2BADE genes and their expression in E. coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 47.Zylstra G J, Wackett L P, Gibson D T. Trichloroethylene degradation by Escherichia coli containing the cloned Pseudomonas putida F1 toluene dioxygenase genes. Appl Environ Microbiol. 1989;55:3162–3166. doi: 10.1128/aem.55.12.3162-3166.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]