Abstract
Antisense peptide nucleic acids (PNAs) that target mRNAs of essential bacterial genes exhibit specific bactericidal effects in several microbial species, but our mechanistic understanding of PNA activity and their target gene spectrum is limited. Here, we present a systematic analysis of PNAs targeting 11 essential genes with varying expression levels in uropathogenic Escherichia coli (UPEC). We demonstrate that UPEC is susceptible to killing by peptide-conjugated PNAs, especially when targeting the widely-used essential gene acpP. Our evaluation yields three additional promising target mRNAs for effective growth inhibition, i.e.dnaB, ftsZ and rpsH. The analysis also shows that transcript abundance does not predict target vulnerability and that PNA-mediated growth inhibition is not universally associated with target mRNA depletion. Global transcriptomic analyses further reveal PNA sequence-dependent but also -independent responses, including the induction of envelope stress response pathways. Importantly, we show that 9mer PNAs are generally as effective in inhibiting bacterial growth as their 10mer counterparts. Overall, our systematic comparison of a range of PNAs targeting mRNAs of different essential genes in UPEC suggests important features for PNA design, reveals a general bacterial response to PNA conjugates and establishes the feasibility of using PNA antibacterials to combat UPEC.
INTRODUCTION
Short antisense oligomers (ASOs) designed to target mRNAs of essential bacterial genes have emerged as attractive species-specific programmable RNA antibiotics and have been successfully applied against several bacterial species (1–5). Popular ASOs include peptide nucleic acids (PNAs), which harbor a pseudo-peptide backbone that links the four natural nucleobases. This chemical structure confers resistance towards nucleases and proteases. Since the neutral backbone also prevents charge repulsion, PNAs have strong binding affinities to complementary target sequences (4,5). These properties allow the use of short oligomers—usually in the range of 9–12mers, to block a target (6–8). The capacity of PNAs to base-pair with complementary sequences makes them bona fide antisense drugs, with the ability to inhibit gene expression both on the transcriptional (9–12) and posttranscriptional level (6,8,13,14). PNAs are generally designed to be complementary to the translation start site of a target mRNA and sterically block ribosome binding and initiation of translation (1,6,13). In order to facilitate intrabacterial delivery, PNAs are commonly conjugated to short, usually cationic or amphiphilic cell-penetrating peptides (CPPs) that facilitate crossing of the bacterial envelope (5,15–17). CPPs possess different penetration efficiencies depending on the targeted bacterial species (15–17). In gram-negative bacteria, it is generally assumed that CPPs cross the outer membrane via an autonomous mechanism. Translocation across the inner membrane can either occur autonomously or involves specific transporters, but this has been characterized only for a few CPPs (18–20). Specifically, uptake of (KFF)3K-conjugated PNAs occurs via the inner membrane transporter SbmA (21,22), whereas arginine-rich peptides, such as (RXR)4XB, follow a self-promoted and membrane potential-dependent mechanism (23).
Guidelines for the design of antisense PNAs take into consideration mostly features of the PNA molecule itself, such as its optimal length, its target region, infrequent off-targeting, and avoiding self-complementarity or long guanosine (G)-stretches (1,6–8,13,24). However, these guidelines are based on the analysis of a small number of PNA target genes, and generally do not take into account the characteristics of the PNA target genes themselves. This is also true for a recent side-by-side analysis of various PNAs designed with a novel semi-automated pipeline, which focused mainly on growth inhibition as a hallmark of PNA activity (25). This prompted us to systematically investigate PNA activity with respect to the choice of the PNA target gene, the characteristics of the applied PNA, and the physiological consequences of PNA treatment. We chose uropathogenic Escherichia coli (UPEC) as a model organism, due to its clinical relevance. UPEC is the primary cause of urinary tract infections, with ∼150 million cases each year, and the emergence of antibiotic resistant UPEC strains creates the need for the development of alternative treatment options (26–30).
Firstly, we aimed to define the characteristics of an effective PNA target. One widely-used PNA target is the essential gene acpP (acyl carrier protein), which is involved in fatty acid biosynthesis (31). PNA-mediated silencing of acpP mRNA, using concentrations similar to conventional antibiotics, causes strong bactericidal effects in several bacterial species, including E. coli and Salmonella enterica (6–8,21,32). Although additional PNA targets that mediate bacterial growth inhibition have been identified, as reviewed elsewhere (4,5), acpP is the most promising one due to its exceptional antibacterial activity. What makes acpP such an efficient target is still unclear; in fact, the high cellular abundance of the acpP mRNAs (33) makes it an unlikely target, given the unfavorable PNA/mRNA stoichiometry. However, the effect of target mRNA abundance on PNA activity has not been extensively investigated yet, and we therefore explored this hypothesis by designing PNAs targeting eleven essential genes with varying expression levels, including acpP.
Secondly, growth inhibition is the primary measure of efficacy of essential gene silencing by PNAs, generally measured using the endpoint-driven minimal inhibitory concentration (MIC) assay. However, growth inhibition does not provide information on wider PNA-triggered cellular responses. We therefore recently employed RNA-sequencing (RNA-seq) as a readout for how PNAs affect target mRNA level, and how PNA treatment globally reshapes the transcriptome in Salmonella (32). Although PNAs are designed to block translation of their target, there is growing evidence that PNAs also cause target mRNA depletion (21,24,32,34–36). However, PNA-triggered target mRNA decay and its relation to the efficacy of growth inhibition have not been systematically analyzed yet. Additionally, an analysis of the global transcriptomic landscape triggered by PNAs targeting different mRNAs to identify common responses is currently missing.
Finally, we focused on one important aspect which strongly influences the in cellulo antisense efficacy of a PNA, namely its length. Specifically, while short PNA molecules are taken up by the cell more easily, long PNA molecules are thought to have better binding affinities (6–8,37–39). In E. coli, optimal PNA length has been proposed to be 10 nucleobases, but this is mostly based on the analyses of two genes, acpP (7) and lacZ (β-galactosidase) (8). Currently, it has not been comprehensively tested whether this PNA length optimum is target gene-dependent or universally valid.
In the present study, our investigation of eleven essential genes identified several promising new targets, while also reinforcing the unique antibacterial activity of acpP PNA. Further, we demonstrate that target gene abundance is not predictive of PNA target susceptibility. Our global RNA-seq analysis reveals that PNA-mediated target mRNA depletion is not a universal trait and is not essential for the antibacterial activity of a PNA. This analysis also identified common transcriptome-wide responses upon PNA treatment, such as induction of envelope stress response pathways. Finally, we show that 9mer PNAs, but not 8mers, are at least as efficient in growth inhibition as their commonly used 10mer counterparts. We consider this an important finding in the quest to reduce the off-target potential of antisense PNAs. In summary, our results provide insights into the druggable target gene spectrum, important aspects of PNA activity and global transcriptomic remodeling triggered by PNAs in UPEC.
MATERIALS AND METHODS
Bacterial strains, oligonucleotides and PNAs
The UPEC strain E. coli 536 was used throughout this study (internal strain number JVS-12054; NCBI GenBank: CP000247.1). The strain was streaked on Luria-Bertani plates and cultured in non-cation adjusted Mueller-Hinton Broth (MHB, BD Difco™, Thermo Fisher Scientific), with aeration at 37°C and 220 rpm shaking.
Oligonucleotides were purchased from Eurofins (Eurofins Genomics) and dissolved in nuclease-free water. Sequences of all oligonucleotides used for amplifying the target region of the respective essential gene for generation of green fluorescence protein (gfp) fusion constructs (see ‘Overlap polymerase chain reaction (PCR) for generation of target gene-gfp fusion constructs’) are listed in Supplementary Table S1.
Peptide-conjugated PNAs (PPNA), i.e. conjugated to (KFF)3K, (RXR)4XB, Tat or 2,3-diaminopropionic acid nonamer (Dap9), were obtained from Peps4LS GmbH. Sufficient quality and purity of these constructs was confirmed by mass spectrometry and HPLC (purity > 98%). PPNAs (Supplementary Table S2) were dissolved in ultrapure water and heated at 55°C for 5 min. Concentrations were determined by using a NanoDrop spectrophotometer (A260 nm; ThermoFisher) and calculated based on the extinction coefficient. PPNAs were stored at –20°C and incubated at 55°C for 5 min before preparing the respective working dilutions. Low retention pipette tips and low binding Eppendorf tubes (Sarstedt) were used throughout.
Minimal inhibitory concentration (MIC) assay
The broth microdilution method was applied for determination of MIC values according to the Clinical and Laboratory Standards Institute and a recently published protocol with a few modifications (40). Briefly, an overnight bacterial culture was diluted 1:100 in fresh MHB and grown to OD600 0.5 (∼2–2.5 h). The obtained culture was diluted 1:2000 in fresh MHB to adjust a final cell concentration of ∼105 cfu/ml. Subsequently, 190 μl of the diluted bacterial solution was dispensed into a 96-well plate (Thermo Fisher Scientific). After adding 10 μl of 20x PPNA working solutions (ranging from 200 to 6.25 μM), or an equivalent volume of water as negative control, growth was monitored in a Synergy H1 plate reader (Biotek) by measuring the OD at 600 nm every 20 min with continuous double-orbital shaking (237 cpm) at 37°C for 24 h. The MIC was determined as the lowest concentration of PPNA, which inhibited visible growth in the wells (OD600 < 0.1).
Killing time kinetics (bactericidal effects)
An overnight bacterial culture was diluted 1:100 in fresh MHB and grown to OD600 0.5. The obtained culture was diluted 1:2000 in fresh MHB to adjust a final cell concentration of approximately 105 cfu/ml. After pre-incubating the sample at 37°C for 5 min, 10 μl of this diluted bacterial culture as well as serial dilutions (10–1, 10–2, 10–3) were directly spotted onto LB agar plates. Simultaneously, an adequate dilution was prepared and streaked on LB plates, serving as input condition for cfu determination. Further, 190 μl of the bacterial solution was transferred into 2 ml eppendorf tubes. Immediately, 10 μl of 20× PPNA solutions were added, adjusting the respective MIC. An equivalent volume of water was used as negative control, whereas a scrambled PPNA served as sequence-unrelated control. At the indicated time points post treatment, aliquots were used for cfu determination and spot assay.
Overlap polymerase chain reaction (PCR) for generation of gfp fusion constructs
For the generation of target gene-gfp fusion constructs, oligonucleotides were designed to amplify the genomic region spanning nucleotides –40 to + 51 relative to the translational start codon of each PNA’s target gene. This PCR step involved the attachment of the T7 promoter sequence to the 5′ end, to allow following transcription of the respective gfp fusion products, and the attachment of a 30 nucleotide gfp-overlap at the 3′ end, for PCR-aided fusion to gfp. A colony of UPEC 536 was resuspended in water and used as DNA template. In a separate PCR, gfp was amplified from pXG-10 plasmid (41) DNA and subsequently used for overlap PCR with each individual target gene region. All oligonucleotide sequences are listed in Supplementary Table S1.
The PCR cycles for the primary amplification of target gene fragments and gfp were selected as follows: 98°C for 1 min, 30 × 98°C for 10 s – 60°C for 20 s – 72°C for 20 s, 72°C for 5 min, 10°C ∞. Subsequently, the secondary overlap fusion PCRs were performed by using the oligonucleotides annealing at the 5′ end of the target gene and the 3′ end of gfp, with each 10–50 ng per PCR product as template. Conditions for this PCR program were set as follows: 98°C for 1 min, 40 × 98°C for 10 s – 60°C for 20 s – 72°C for 40 s, 72°C for 10 min, 10°C ∞. Afterwards, overlap PCR products were purified with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel) according to the manufacturer's instructions and DNA concentration was quantified by using NanoDrop spectrophotometer (A260 nm).
In vitro transcription
Templates for T7 RNA polymerase-driven transcription were produced via PCR as described above. After column-based purification, ∼800 ng of DNA products were subjected to 20 μl in vitro transcription reactions according to the manufacturer's instructions (MEGAscript T7 kit, Ambion/Thermo Scientific). After incubating the samples at 37°C for 3 h and 45 min, 2 units (U) of Turbo DNase were added per reaction for additional 15 min. Subsequently, 115 μl water, 15 μl Ammonium Acetate Stop solution and 3 volumes of ethanol were added for RNA precipitation at - 80°C overnight. After centrifugation, pellets were washed using 70% ethanol and finally resuspended in ultrapure water. RNA concentration was measured with Qubit (Fisher Scientific). For verification of the expected product size and RNA integrity, RNA gels (6% PAA, 7 M urea) were prepared with subsequent staining using StainsAll (Sigma-Aldrich).
In vitro translation, titration of PPNA-inherent inhibitory effects and western blotting
For in vitro translation of target gene-gfp fusion constructs, the PURExpress® In Vitro Protein Synthesis Kit (New England Biolabs, E6800L) was used as described in the instructions with minor modifications. To ensure a precisely adjusted final RNA concentration of 100 nM in all samples, in vitro transcribed RNA instead of T7 promoter-containing template DNA was used in all translation reactions. Briefly, the final volume of each in vitro translation reaction was set to 10 μl, each containing 4 μl Solution A, 3 μl Solution B, and 1 pmol of in vitro transcribed heat-denatured RNA to adjust a final concentration of 100 nM. After 2 h at 37°C, the tubes were immediately placed on ice. For denaturation, protein samples were diluted in 1x reducing protein loading buffer (62.6 mM Tris–HCl pH 6.8, 2% SDS, 0.1 mg/ml bromophenol blue, 15.4 mg/ml DTT, 10 % glycerol) and the tubes were incubated at 95°C for 5 min.
To analyze whether sequence-specific or sequence-unrelated PPNAs are capable of inhibiting in vitro translation of target gene-GFP constructs, RNA samples were pre-incubated with defined PPNA solutions at 37°C for 5 min. In particular, PPNA-inherent effects were titrated using the following conditions: 1 pmol of the respective RNA was in vitro translated in reactions containing 1 μM, 500 nM, 200 nM and 100 nM PPNA, corresponding to 10:1, 5:1, 2:1 and 1:1 ratios of PPNA:RNA. After this pre-annealing step, PURExpress®in vitro translation mix was added and samples were handled as described above.
For visualization of in vitro translated protein amounts, samples were separated on SDS-PAA (12% PAA) gels, with subsequent semi-dry western blot transfer on polyvinylidene fluoride (PVDF) membranes. To verify equal loading, membranes were first stained with Ponceau S (Sigma-Aldrich) and then probed with 5% skim milk (in 1× TBS-T) containing anti-GFP antibody (1:1000; Sigma-Aldrich) overnight. After incubation with an HRP-conjugated secondary antibody (1:10 000; ThermoScientific) in 1× TBS-T, the membrane was incubated with a self-made developing solution and protein levels were detected using an ImageQuant LAS 500 (GE Healthcare Life Sciences). Images were processed and band intensities quantified using ImageJ (42).
PPNA treatment of UPEC 536 and isolation of total RNA
Bacterial overnight cultures were diluted 1:100 in fresh MHB and grown to an OD600 of 0.5. Subsequently, obtained cultures were again diluted 1:100 in fresh MHB to adjust a cell concentration of ∼106 cfu/ml. After transferring 1.9 ml of the bacterial solution into 5 ml low-binding tubes (LABsolute), 100 μl of 20× PPNA stock solutions were added to reach a final concentration of 5 μM for all (KFF)3K-conjugated PNAs. In parallel, an equal amount of cells was treated with the respective volume of sterile nuclease-free water, which was used as solvent for the test compounds, and served as negative control. After incubating the samples for 15 min at 37°C, RNAprotect Bacteria (Qiagen) was added according to the manufacturer's instructions. Following a 10-min incubation, cells were pelleted at 4°C and 21,100 × g for 20 min. The supernatant was discarded and pellets were either directly used or stored at –20°C (<1 day) for subsequent bacterial RNA isolation.
Total RNA was purified from bacterial pellets using the miRNeasy Mini kit (Qiagen) according to the protocol #3 described in Popella et al. (32). Briefly, cells were resuspended in 0.5 mg/ml lysozyme (Roth) in TE buffer (pH 8.0) and incubated for 5 min. Afterwards, RLT buffer supplemented with β-mercaptoethanol, and ethanol were added according to the manufacturer's instructions. After sample loading, column wash-steps were performed according to the manual. RNA concentration was measured with a NanoDrop spectrophotometer.
RNA-seq
For transcriptomic analyses, RNA samples were processed and subjected to RNA-seq at Core Unit SysMed (University of Würzburg, Germany). Briefly, RNA quality was checked using a 2100 Bioanalyzer with the RNA 6000 Pico/ Nano kit (Agilent Technologies). RNA samples were DNase-treated using DNAse I kit (Thermo Fisher), followed by ribosomal RNA depletion using Lexogen's RiboCop META rRNA Depletion Kit protocol according to manufacturer's recommendation. Subsequently, cDNA libraries suitable for sequencing were prepared using CORALL Total RNA-Seq Library Prep protocol (Lexogen) according to manufacturer's recommendation with 14–25 PCR cycles. Library quality was checked using a 2100 Bioanalyzer with the DNA High Sensitivity kit (Agilent Technologies). Sequencing of pooled libraries, spiked with 5% PhiX control library, was performed at 10 million reads/sample in single-end mode with 75 nt read length on the NextSeq 500 platform (Illumina) using High output sequencing kits. Demultiplexed FASTQ files were generated with bcl2fastq2 v2.20.0.422 (Illumina).
Quantification of RNA-seq data
RNA-Seq data analysis was performed similarly to our previous analysis of PNA activity in S. enterica serovar Typhimurium (32). Small RNAs (sRNAs) were added to the UPEC annotation file. Specifically, sRNA sequences of E. coli str. K-12 substr. MG1655 were aligned to the UPEC genome using BLASTN (v2.9.0), and the alignment match with the lowest E-value was added as an sRNA to the UPEC annotation. All added sRNAs had more than 89% nucleotide identity and the largest number of sequence mismatches was 4. Raw RNA-Seq reads were trimmed, filtered and mapped to the UPEC genome (E. coli 536, CP000247.1) (43). BBduk was used to remove adapter sequences and trim nucleobases with Phred quality scores <10. Reads were then mapped against the UPEC genome using BBMap (v38.84) and the resulting alignments were quantified against both coding sequences and sRNAs using the featureCounts methods of the Subread (2.0.1) package (44).
Normalization and differential expression analysis
R/Bioconductor packages were used for the downstream RNA-Seq analysis. Raw read counts from all conditions were used with edgeR (v3.34.1) for differential expression analysis (45). Prior to analysis, reads which did not meet the threshold of 1.48 counts per million (CPM) in at least 10 libraries were filtered out. The cutoff was set as 10/L, where L is the minimum library size across all samples in millions, as proposed in (46).
The remaining libraries were normalized by the trimmed mean of M values (TMM) normalization (47). Then, libraries were assessed for batch effects using principal component analysis (PCA) plots. A consistent batch effect was observed between different sequencing runs. As described in the edgeR manual (https://bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf), batch effects were corrected by adding a batch variable to the design matrix before differential expression analysis. Quasi-likelihood dispersions were estimated using the glmQLFit function. Then contrasts were created and used to test for differential expression with the glmQLFTest function.
Features with absolute fold changes >2 and false discovery rate (FDR) adjusted P-values (48) <0.001 were considered differentially expressed. The results were plotted as heatmaps using the ComplexHeatmap (v2.8.0) package.
KEGG pathway analysis
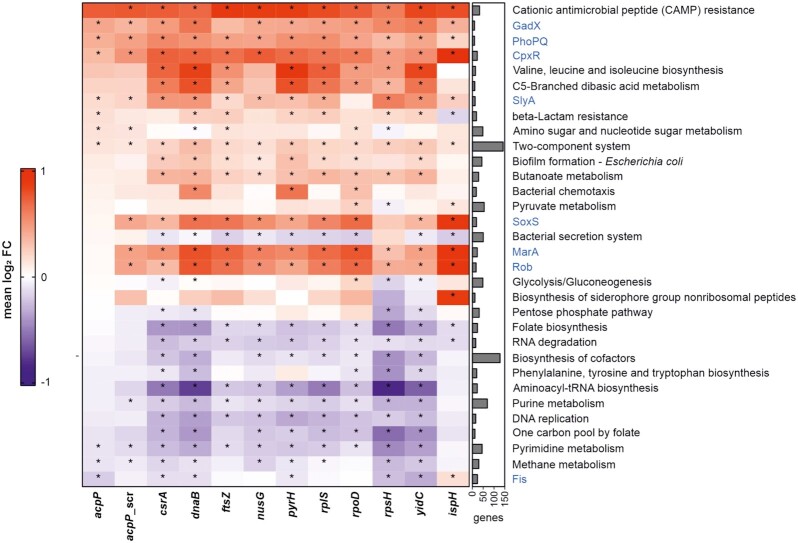
For assigning genes to KEGG pathways (49), the R package KEGGREST (1.32.0) was used. Additionally, gene sets of regulons were added. To assign genes to regulons, the E. coli K12 RegulonDB (v10.9) was parsed and genes appearing in both UPEC and K12 were assigned to regulons (50). Rotation gene set testing (FRY version of ROAST gene set testing, (51)) was applied to assess enrichments of pathways and regulons in differentially expressed genes. Gene sets with >10 genes and an FDR-adjusted P-value <0.001 (marked with an asterisk, *) were visualized in Figure 7, the color indicating the median log2 FC of the respective gene set. Figure 7 shows all gene sets which rank among the top 10 significant gene sets with the lowest P-value for at least one sample.
Figure 7.
Gene set enrichment analysis. Global enrichment analysis of differentially expressed genes classified according to annotated KEGG pathways and manually added regulons (marked in blue, retrieved from RegulonDB annotations for E. coli K-12). Enrichment analysis shows a common induction of cationic antimicrobial peptide (CAMP) resistance, GadX, PhoPQ, CpxR and two-component system regulons. Asterisks (*) indicate statistically significant (P-value adj < 0.05) gene sets. Right bar chart shows the number of genes included within the single KEGG pathways/regulons. Up- (red) and downregulated (blue) pathways are indicated as median log2 fold change (FC).
Prediction of PNA/RNA melting temperature
Since there is no reliable bioinformatic algorithm for prediction of the melting temperature (Tm) of PNA/RNA duplexes currently available, we used the MELTING package to predict Tms for the corresponding RNA/RNA duplexes (52). We used our predicted values as estimation of the actual PNA/RNA Tm, which are known to be higher compared to the respective RNA/RNA duplexes (53). As proof of concept, we verified our Tm prediction based on previously published experimental Tms of PNA/RNA duplexes (7).
RESULTS
acpP PNA conjugated to (KFF)3K, (RXR)4XB or Tat peptides inhibits growth of UPEC 536
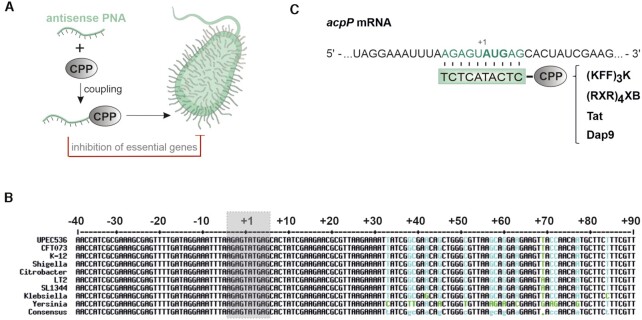
To explore the application of PNAs to combat UPEC (Figure 1A), we took advantage of a commonly used acpP PNA with proven efficacy in numerous bacterial species (7,21,23,24,32,34,35,54). The PNA has full complementarity to 10 nucleotides located around the start codon of acpP, a site that is highly conserved in several γ-proteobacteria including the pyelonephritis isolate UPEC 536, widely used as model for extraintestinal E. coli (43) (Figure 1B, C).
Figure 1.
Schematic illustration of the applied PNA technology and the PNA target region of the conserved essential gene, acpP. (A) An antisense PNA is coupled to a CPP to facilitate intrabacterial delivery. When targeting essential bacterial genes, CPP-PNA conjugates can serve as potent antimicrobial molecules. Parts of this image have been created with Smart (Servier medical art). (B) Multiple sequence alignment (created with http://multalin.toulouse.inra.fr/multalin/; (118)) of the essential gene acpP including the following γ-proteobacteria: UPEC 536 (CP000247.1), E. coli CFT073 (CP051263.1), E. coli K-12 (CP032667.1), Shigella dysenteriae (CP000034.1), Citrobacter rodentium (CP038008.1), Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (AE006468.2), Salmonella enterica subsp. enterica serovar Typhimurium str. SL1344 (FQ312003.1), Klebsiella pneumoniae (CP003200.1) and Yersinia pestis (NC_003143.1). A defined section (–40 to +90 nt) including the region around the PNA binding site (grey box) is shown. The following color-code has been applied: perfect consensus black, varying alignment columns cyan (nucleotide substitution green). (C) Relevant region of acpP mRNA in UPEC 536, with the PNA target sequence shaded in green. The start codon (AUG) is shown in bold type. Below, the PNA sequence is shown (green box) with the different conjugated CPPs for delivery into UPEC ((KFF)3K, (RXR)4XB, Tat, Dap9).
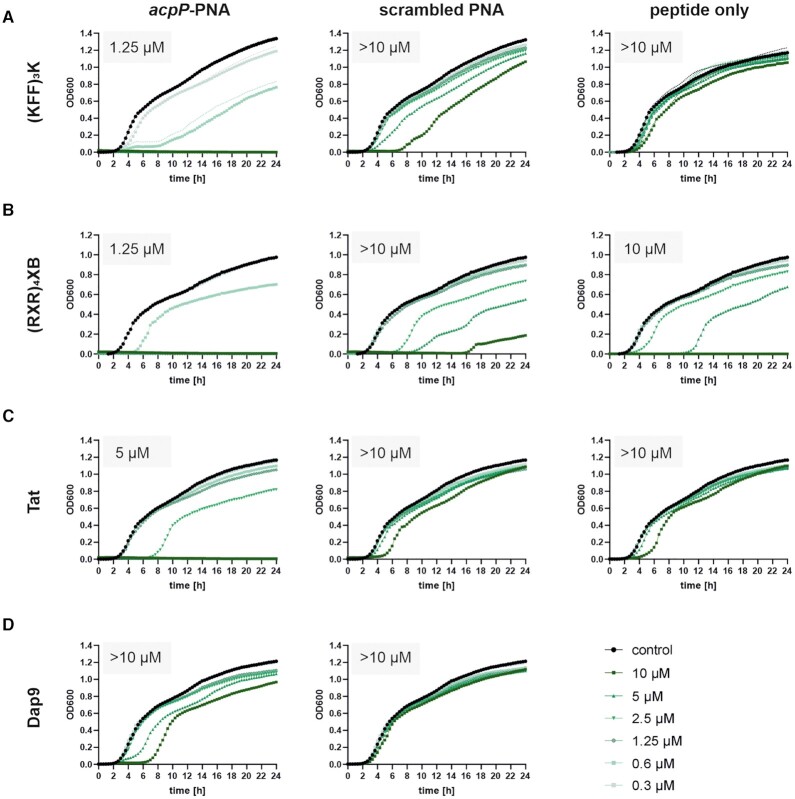
Using UPEC 536, we determined the MIC of acpP PNA coupled to the CPPs (KFF)3K, (RXR)4XB, Tat, or 2,3-diaminopropionic acid nonamer (Dap9), which have previously been shown to mediate efficient bacterial permeation (16,32) (Figure 2). We included scrambled PNA and peptide only controls in our analyses to differentiate between sequence-independent effects triggered by either the PNA module or by the carrier peptide itself. (KFF)3K-, (RXR)4XB- and Tat-coupled acpP PNAs all lead to efficient in vitro growth inhibition, with (KFF)3K- and (RXR)4XB-conjugated acpP PNA being more potent (MIC of 1.25 μM) than Tat-coupled acpP PNA (MIC 5 μM) (Figure 2, left panels). However, while (KFF)3K- or Tat-conjugated scrambled PNAs or the respective ′peptide only′ controls did not cause substantial growth inhibition, the (RXR)4XB compounds all impaired UPEC 536 growth. Thus, the (RXR)4XB peptide itself seems to possess antibacterial activity against UPEC 536 (MIC of 10 μM). On the other hand, the Dap9-conjugated acpP 10mer PNA did not mediate substantial growth inhibition of UPEC 536 (MIC > 10 μM; Figure 2D). Based on these data, we conclude that the (KFF)3K peptide is a potent carrier for PNA delivery into UPEC 536 and we therefore used it in all subsequent analyses.
Figure 2.
An antisense PPNA targeting acpP shows potent growth inhibitory capacity against UPEC. Determination of growth kinetics and MICs of UPEC 536 upon exposure to acpP-specific (left panels) and scrambled control (middle panels) PNAs, conjugated to (A) (KFF)3K, (B) (RXR)4XB, (C) Tat or (D) Dap9 peptide. (A–D) UPEC 536 (105 cfu/ml) were exposed to gradually decreasing concentrations (starting from 10 μM) of each PPNA or peptide. Treatment with an equal volume of water served as control (black curves). (A–C) CPPs (right panels) were used as PNA-independent controls. Growth is depicted as OD600 (y-axis) and was monitored over 24 h (x-axis). Each experiment was performed two to four times. Curves represent the mean of all biological replicates and dashed lines indicate standard error of the mean.
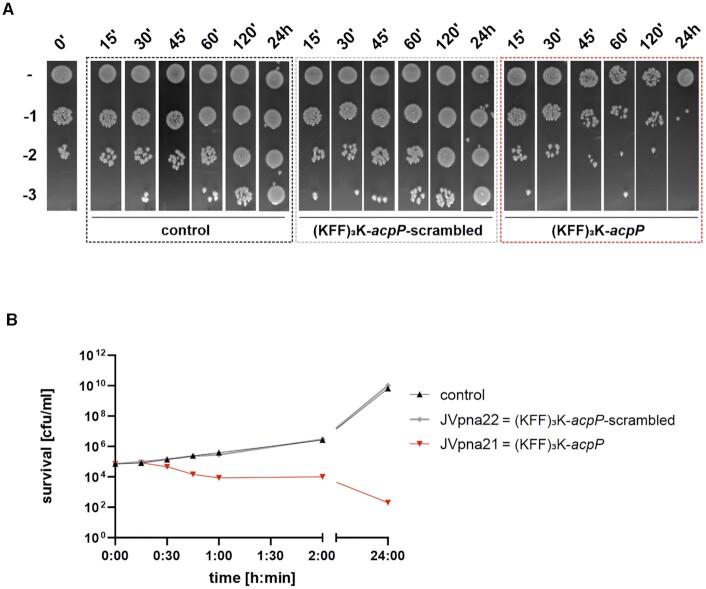
(KFF)3K-acpP mediates bactericidal effects in UPEC 536
To clarify if the observed growth inhibitory capacity of (KFF)3K-acpP was an indication of bacteriostatic or bactericidal effects, UPEC 536 cells were challenged with the MIC of 1.25 μM for different time intervals, with water or (KFF)3K-acpP-scrambled serving as controls (Figure 3). For all conditions, spot assays with serial dilutions (Figure 3A) as well as colony forming units (cfu) per ml (Figure 3B) were used to measure bacterial survival. As expected, scrambled (KFF)3K-acpP did not affect the abundance of living bacterial cells compared to the water control within 24 h. In sharp contrast, treatment of UPEC 536 with (KFF)3K-acpP strongly decreased the number of cfu, evident from 30 min post treatment (mpt) onwards. A plateau of the bactericidal effect was observed from 60 to 120 mpt, revealing a 1 log reduction in living cells. This effect was even more pronounced after 24 h of treatment, showing a 2–3 log decrease in cfu/ml. These data demonstrate that (KFF)3K-acpP acts as a potent antibacterial agent with sequence-specific activity not only against E. coli K-12 (21), but also against UPEC 536.
Figure 3.
An antisense PPNA targeting acpP shows antibacterial activity against UPEC 536 at 1× MIC. Bactericidal effects of (KFF)3K-acpP (red) were determined at 1× MIC (1.25 μM) against UPEC 536 (105 cfu/ml) during a 24 h time course. (KFF)3K-acpP-scrambled (grey) was used as control PPNA, whereas exposure to an equal volume of water served as additional control (black). After the indicated time points, aliquots of each condition were subjected to (A) spot assays or (B) cfu determination on LB plates to investigate the amount of viable cells. (A) Serial dilutions (10–1 to 10–3) were prepared, as indicated on the left. (B) Curves represent the mean of the biological replicates. The experiments were performed twice.
(KFF)3K-acpP inhibits in vitro synthesis of AcpP1-17-GFP
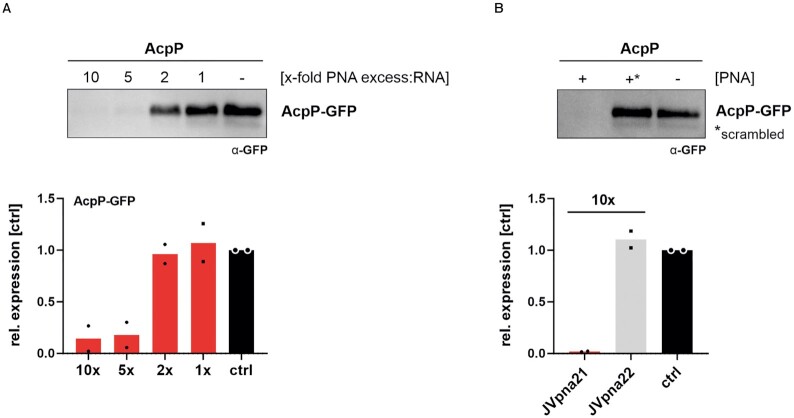
Next, we investigated whether (KFF)3K-acpP inhibits acpP translation, as would be expected from the sequence-complementarity of the PNA to the translational start site of its target mRNA and the observed in cellulo PNA-mediated reduction of AcpP protein levels in E. coli (21). To this aim, we applied a cell-free in vitro translation system, which constitutes a direct readout of translational control independent of confounding factors, e.g. mRNA degradation or loss of cell viability. Specifically, the genomic region spanning −40 to + 51 of the acpP mRNA relative to the start site, including the PNA target region, was fused to gfp (acpP91::gfp). After PCR amplification and in vitro transcription, the acpP91::gfp construct was translated in vitro and AcpP1-17-GFP protein abundance was determined via western blotting (Figure 4). Titration of the (KFF)3K-acpP construct from 10:1, 5:1, 2:1 and 1:1 molar ratios to acpP91::gfp RNA showed that a 5-fold molar excess of (KFF)3K-acpP over acpP91::gfp RNA was already sufficient to trigger an 82% reduction in translation efficiency on average, compared to the water-treated control (Figure 4A). We also included (KFF)3K-acpP-scrambled control at 10-molar excess over acpP91::gfp RNA, to confirm sequence-specific inhibition of translation in vitro. While (KFF)3K-acpP substantially inhibited in vitro synthesis of AcpP1-17-GFP, the scrambled peptide-conjugated PNA (PPNA) construct did not affect translation efficacy (Figure 4B). These results demonstrate that (KFF)3K-acpP is capable of inhibiting cell-free translation of a truncated acpP-target fused to GFP.
Figure 4.
An antisense PPNA targeting acpP inhibits translation of its target in vitro. In vitro transcribed acpP::gfp RNA was subjected to in vitro translation assays. (A) Titration of the molar excess of (KFF)3K-acpP over template mRNA was performed to determine the inhibitory efficacy. As control, an equal volume of water was added (−, ctrl). Western blotting was performed for detection of the AcpP1-17-GFP fusion (∼25 kDa) by using an anti-GFP antibody. Signal intensities were quantified using ImageJ and protein expression is shown relative to the ‘untreated’ control. Black dots indicate individual protein expression levels of the experimental duplicates and bars show the mean. (B) In vitro translation was performed in the presence or absence of (KFF)3K-acpP (+; JVpna21) or its scrambled control (+*; JVpna22). A 10-molar excess of both PPNA constructs over template RNA was applied. The experiments were performed two times.
Expanding the essential gene spectrum for PPNA targeting of UPEC 536
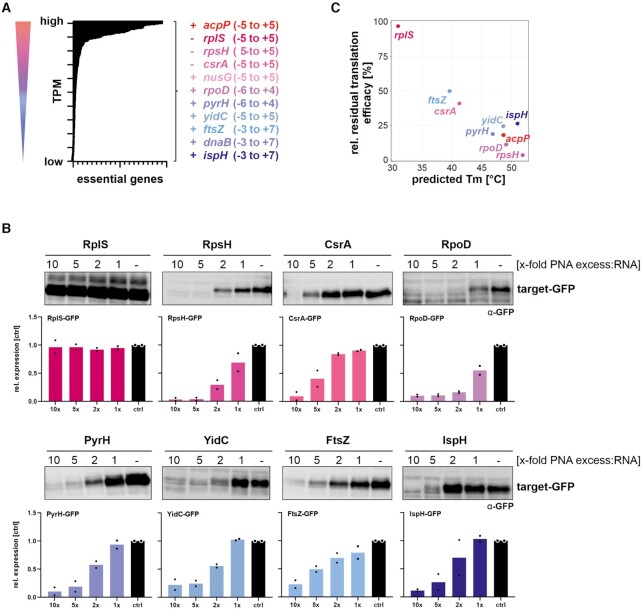
In order to investigate if PNA-mediated antibacterial activity can be harnessed more widely and to clarify whether transcript abundance influences the efficacy by which PNAs deplete their target, we selected additional essential genes with differing expression levels for PNA-based targeting. We first ranked genes previously shown to be essential in a variety of E. coli strains (55–57) by expression level, and chose ten target genes covering the range of observed expression (Figure 5A). We designed 10mer antisense PNA sequences to target the translational start sites of these genes in a window spanning nucleotides −6 to + 7. With the exception of the PNAs for ispH (4-hydroxy-3-methylbut-2-enyl diphosphate reductase), pyrH (uridine monophosphate kinase) or rplS (50S ribosomal subunit protein L19), the PNAs we designed do not have predicted off-target sites within the translation initiation regions of other genes. We then performed in vitro translation assays in the presence or absence of the respective sequence-specific PPNA for each selected target gene as described above for acpP (Figure 4). In vitro translation of dnaB and nusG was unsuccessful in our assay conditions, and we therefore excluded both genes from this analysis. With the exception of rplS, all PPNAs strongly decreased the protein abundance of the respective target fusion (Figure 5B). Since the PPNA/RNA duplex stability affects the antisense efficacy of a PNA (7,8,37–39), we analyzed whether the stability also affects the inhibition of translation in vitro. Our analysis confirms a direct association of the predicted melting temperature,i.e. PPNA/RNA duplex stability, with the capacity of a PPNA to inhibit translation of its target in vitro (Figure 5C).
Figure 5.
Expanding the target spectrum of essential gene-specific antisense PNAs in UPEC. (A) All essential genes, annotated in E. coli (55–57), were ranked according to their expression levels in our UPEC 536 RNA-seq data set (untreated control samples, averaged from three independent biological replicates). High to low expression levels are indicated by red to blue coloring. Essential gene candidates for PNA targeting were selected to cover a broad range of mRNA abundances (indicated on the right side). The coding strand of each gene is indicated with (+) or (−). The target window is indicated as nucleotide positions relative to the start codon. (B) In vitro transcribed gfp-fusion RNAs for each essential gene were subjected to in vitro translation assays, in the presence or absence of the cognate PPNA. The capacity of the tested PPNAs to inhibit in vitro translation was analyzed in 10:1, 5:1, 2:1 to 1:1 molar ratios of PPNA:RNA. An equal volume of water was added to serve as control condition (−, ctrl). Samples were subjected to Western blotting and membranes were probed with an anti-GFP antibody to determine the expression levels of the GFP fusion proteins (∼25 kDa). Signal intensities were quantified using ImageJ. Protein expression levels are shown relative to the respective control sample. Black dots indicate individual protein expression levels of the experimental duplicates and bars show the mean. Each experiment was performed twice. (C) Averaged residual translation efficacy, relative to the respective control condition, at 5-molar excess of PPNA over RNA, plotted against the predicted melting temperature (Tm,°C) of each PNA/RNA duplex.
Target mRNA abundance does not dictate the growth inhibitory capacity of a PPNA
Next, we determined the MIC of each PPNA and observed that 7 out of 10 tested (KFF)3K-conjugated 10mer PNAs are capable of inhibiting visible growth of UPEC 536 at concentrations ≤10 μM (Table 1). PPNAs targeting yidC (membrane protein insertase), csrA (carbon storage regulator) and nusG (transcription termination/antitermination factor) did not trigger complete growth inhibition, although the two former did cause growth reduction at 10 μM (Supplementary Figure S1). These effects were not correlated with target gene abundance (Figure 6A).
Table 1.
Analyzing the growth inhibitory capacity of 8–11mer PPNAs targeting selected essential genes with varying expression levels. UPEC 536 cells (105 cfu/ml) were exposed to serial dilutions of (KFF)3K-coupled 8mer, 9mer, 10mer or 11mer PPNAs and growth was monitored after 24 h. Minimum inhibitory concentrations (MICs) were determined. The PPNAs are ordered according to the target transcript abundance from low to high expression levels (transcript per million, TPM). * Averaged value from RNA-seq analyses of three untreated control samples. # Sequences are shown from N- to C-termini orientation
| 8mer PPNAs | 9mer PPNAs | 10mer PPNAs | 11mer PPNAs | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Locus tag | TPM* | Peptide sequence# | PNA sequence# | Target | MIC [μM] | PNA sequence# | Target | MIC [μM] | PNA sequence# | Target | MIC [μM] | PNA sequence# | Target | MIC [μM] |
| ispH | ECP_0027 | 90.3 | KFFKFFKFFK | - | - | - | TCTGCATGT | -2 to + 7 | 10 | TCTGCATGTT | -3 to + 7 | 10 | ATCTGCATGTT | -3 to + 8 | >10 |
| dnaB | ECP_4268 | 104.4 | KFFKFFKFFK | CTGCCATA | -1 to + 7 | 5 | CTGCCATAG | -2 to + 7 | 5 | CTGCCATAGT | -3 to + 7 | 5–10 | CTGCCATAGTG | -4 to + 7 | 10 |
| ftsZ | ECP_0097 | 323.5 | KFFKFFKFFK | CAAACATA | -1 to + 7 | >10 | CAAACATAG | -2 to + 7 | 2.5 | CAAACATAGT | -3 to + 7 | 2.5–5 | CAAACATAGTT | -4 to + 7 | 10 |
| yidC | ECP_3906 | 478.8 | KFFKFFKFFK | - | - | - | TCCATCGTT | -4 to + 5 | 5–10 | TCCATCGTTA | -5 to + 5 | > 10 | TCCATCGTTAG | -6 to + 5 | > 10 |
| pyrH | ECP_0179 | 580 | KFFKFFKFFK | - | - | - | CCATGTTTC | -5 to + 4 | 10 | CCATGTTTCT | -6 to + 4 | 10 | CCATGTTTCTT | -7 to + 4 | 10 |
| rpoD | ECP_3157 | 870.5 | KFFKFFKFFK | - | - | - | CCATAAGAC | -5 to + 4 | 10 | CCATAAGACG | -6 to + 4 | 10 | CCATAAGACGG | -7 to + 4 | 10 |
| nusG | ECP_4195 | 909.7 | KFFKFFKFFK | - | - | - | GACATCTCA | -4 to + 5 | > 10 | GACATCTCAG | -5 to + 5 | > 10 | GACATCTCAGA | -6 to + 5 | > 10 |
| csrA | ECP_2656 | 1259.7 | KFFKFFKFFK | - | - | - | AGCATTCTT | -4 to + 5 | > 10 | AGCATTCTTT | -5 to + 5 | > 10 | AGCATTCTTTG | -6 to + 5 | 10 |
| rpsH | ECP_3394 | 5288.6 | KFFKFFKFFK | CTCATCTG | -3 to + 5 | 5 | CTCATCTGT | -4 to + 5 | 2.5 | CTCATCTGTC | -5 to + 5 | 2.5–5 | CTCATCTGTCT | -6 to + 5 | 2.5–5 |
| rplS | ECP_2607 | 7495.7 | KFFKFFKFFK | - | - | - | CTCATAATT | -4 to + 5 | > 10 | CTCATAATTT | -5 to + 5 | ≥ 10 | CTCATAATTTA | -6 to + 5 | 10 |
| acpP | ECP_1086 | 12828.1 | KFFKFFKFFK | CTCATACT | -3 to + 5 | 5 | CTCATACTC | -4 to + 5 | 1.25 | CTCATACTCT | -5 to + 5 | 1.25 | CTCATACTCTT | -6 to + 5 | 1.25 |
| scrambled | - | - | KFFKFFKFFK | CACTATCT | - | > 10 | CACTATCTC | - | > 10 | TCACTATCTC | - | > 10 | TTCACTATCTC | - | > 10 |
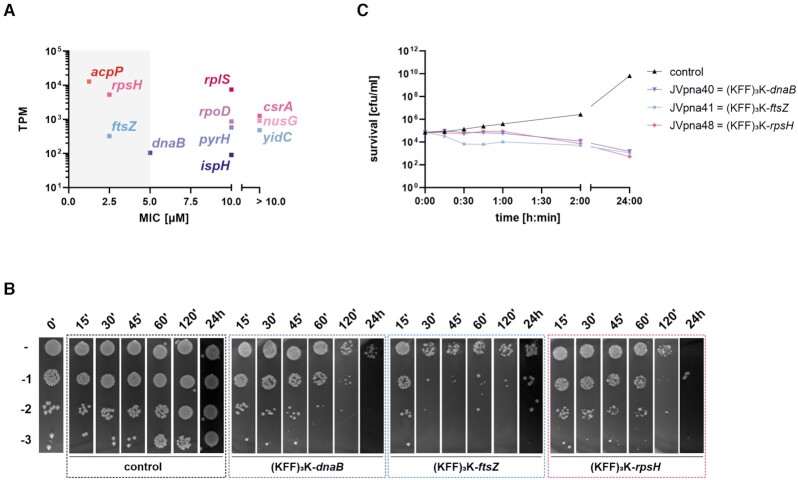
Figure 6.
Antisense PPNAs targeting dnaB, ftsZ, and rpsH confer antibacterial activity against UPEC 536 at 1× MIC. (A) MICs for each 10mer PPNA, listed in Table 1, were plotted against the abundance (transcripts per million, TPM) of the respective PNA’s target. Grey shaded area highlights the four PPNAs with the strongest antibacterial efficacy against UPEC 536. (B, C) Bactericidal effects were determined for (KFF)3K-dnaB (5 μM), (KFF)3K-ftsZ (2.5 μM) and (KFF)3K-rpsH (2.5 μM) at 1× MIC against UPEC 536 (105 cfu/ml). Exposure to an equal volume of water served as control (black). Samples were taken at the indicated time points post treatment. (B) Spot assays with serial dilutions (10–1 to 10–3) and (C) cfu evaluation on LB plates was performed to determine the number of viable cells. Curves represent the mean of the biological replicates. Each experiment was performed twice.
For further characterization, we focused on PNAs targeting ftsZ (cell division protein), dnaB (replicative DNA helicase) and rpsH (30S ribosomal subunit protein S8), which showed MICs of less than 10 μM (Figure 6A). Notably, while PNAs targeting ftsZ have previously been shown to inhibit growth of E. coli, Enterococcus faecalis and Acinetobacter baumannii (21,34,54,58), dnaB and rpsH are novel PNA targets. We assayed bactericidal effects of these PPNAs against UPEC 536 at their MIC, via spot assays (Figure 6B) and cfu determination (Figure 6C). Similar to our observations for (KFF)3K-acpP (Figure 3), treatment with the three PPNAs targeting dnaB, ftsZ or rpsH caused reduction in bacterial survival, with (KFF)3K-ftsZ exhibiting an accelerated effect compared to both other constructs. All three tested PPNAs led to a 1 to 2 log reduction in cfu/ml evident from 60–120 min onward.
Overall, these results exclude target mRNA abundance as a key determinant for the growth inhibitory capacity of a PPNA in UPEC, consistent with a recent study in multidrug resistant Enterobacteriaceae (25). Surprisingly, the efficacy of in vitro inhibition of translation (Figures 4 and 5B) also does not correlate with PPNA activity against UPEC.
RNA-seq analysis reveals specific and common (KFF)3K-PNA-mediated transcriptional changes
In order to investigate the global transcriptomic response of UPEC when challenged with different PPNAs and to test if the previously observed target transcript depletion (32) is a universal effect of PPNA activity, we performed RNA-seq analysis. Specifically, UPEC 536 was exposed to equimolar concentrations of the 10mer (KFF)3K-coupled PNAs listed in Table 1. After 15 min, cells were harvested and RNA was isolated for subsequent RNA-seq analyses. As a quality control, we evaluated the RNA-seq dataset by principal component analysis (PCA) and hierarchical cluster analysis (HCA) (Supplementary Figure S2). After removing batch effects within our triplicates, we found that all biological replicates of one condition clustered together.
We initially performed a gene set enrichment analysis to identify globally regulated pathways (Figure 7). We included annotated KEGG pathways (49) and manually added the virulence and stress response-associated regulons GadX, PhoPQ, CpxR, SlyA, SoxS, MarA, Rob and Fis (marked in blue) retrieved from RegulonDB (50). Across the analyzed PPNAs, we observed a significant induction (FDR-adjusted P-value < 0.001) of the cationic antimicrobial peptide resistance (CAMP), GadX (note: for (KFF)3K-ftsZ below the significance threshold), PhoPQ and CpxR pathways in PPNA-treated UPEC 536. This indicates that PPNAs trigger several common stress response pathways in UPEC.
We then analyzed the regulation of the direct PPNA targets in each condition (Figure 8, ‘targets’). In addition to (KFF)3K-acpP (4.8-fold change), only 4 out of the 10 tested PPNAs triggered a substantial reduction of their target mRNA level, namely dnaB (2.5-fold change), pyrH (4.9-fold change), rpoD (8.2-fold change), and yidC (3.5-fold change). Notably, the latter three were not promising candidates based on MIC analysis (≥ 10 μM; Table 1). Most other PPNAs showed moderate but non-significant reduction of their respective target, except (KFF)3K-rpsH which did not appear to impact rpsH mRNA levels, despite blocking in vitro translation of its target (Figure 5B) and inhibiting growth of UPEC 536 (Table 1). Importantly, the scrambled control did not mediate mRNA depletion for any of the essential gene targets. Based on these findings we conclude that rapid PPNA-triggered reduction of target transcript levels is not a universal feature of PPNA antisense inhibition and is also not associated with MIC (Table 1).
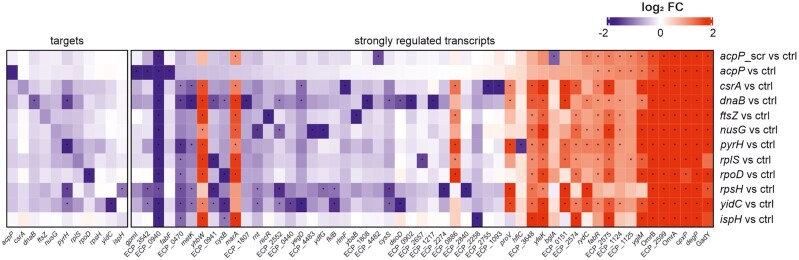
Figure 8.
Transcriptomic profiling shows that substantial PPNA-triggered target mRNA depletion is not a universal mechanism, but reveals the induction of a common PNA sequence-independent response. UPEC 536 cells (106 cfu/ml) were challenged with each of the PPNA conjugates indicated on the right (5 μM), or an equal volume of water as control. Cells were harvested 15 min post treatment for RNA isolation and subsequent RNA-seq analyses. Besides each PPNA’s target gene (left panel), top-regulated transcripts for each condition were included into the heatmap (right panel). Differential expression (log2 fold changes (FC), red and blue color code denotes up- and down-regulation, respectively) of each gene is shown relative to the untreated control. Asterisks (*) indicate significantly regulated genes (absolute FC > 2) with a false discovery rate (FDR) adjusted P-value <0.001. Comparisons for all conditions include triplicate RNA-seq samples.
Decoding direct and indirect PPNA-triggered transcriptomic responses
Finally, we extended the analysis to the five top up- and downregulated transcripts for each condition, shown in one heatmap (Figure 8, ‘strongly regulated transcripts’). Closer inspection of the gold standard (KFF)3K-acpP PNA, which has been recently employed to analyze transcriptomic changes of PPNA-treated Salmonella (32), revealed a limited transcriptomic response in UPEC (Figure 8; Dataset S1). While 43 genes were significantly induced, only 16 genes were significantly reduced upon acpP PNA treatment. Specifically, in addition to acpP itself, (KFF)3K-acpP triggered pronounced depletion of fabF (3-oxoacyl-[acyl carrier protein] synthase 2; 3.5-fold change), a gene co-transcribed with acpP. (KFF)3K-acpP also specifically depleted gpmI (2,3-bisphosphoglycerate-independent phosphoglycerate mutase; 5.5-fold change) and ECP_3542 (glycerophosphoryl diester phosphodiesterase; 4.7-fold change). This is likely due to the presence of a fully complementary 9 base PNA binding site within the translation initiation sites of gpmI. Similarly, two transcripts, ECP_4482 (UDP-N-acetylmuramate-l-alanyl-γ-d-glutamyl-meso-2,6-diaminoheptanedioate ligase) and bglA (6-phospho-beta-glucosidase), were exclusively depleted by the (KFF)3K-acpP-scrambled PNA and both have 8 base complementary binding sites in proximity to their respective annotated translational start sites (Figure 8). These observations suggest that an 8- to 9mer PNA can be sufficient to trigger depletion of a transcript.
PPNAs also elicited a common and thus sequence-independent regulation of certain transcripts. We identified ompF (outer membrane protein F, ECP_0940), one of the most abundant porins in the outer membrane (OM) of E. coli (59), among the mRNAs depleted in all conditions (∼3.6-fold change). This suggests that ompF is part of a global and likely indirect transcriptional response to PPNA exposure of UPEC. The strongest increase in expression among all conditions was found for the three small RNAs (sRNAs) GadY, OmrA and OmrB, as well as the mRNAs degP, cpxP and ECP_2599 (RaiA). Since the sRNAs OmrA and OmrB are known to regulate the synthesis of several abundant OMPs (60–62), we predict that their activity may alter the composition of the UPEC envelope following PPNA exposure. The transcript degP, also known as htrA, encodes a periplasmic serine endoprotease involved in protein quality control and stress response that has both chaperone and protease activities (63,64). CpxP—regulated by the CpxA/R inner membrane stress response—is a periplasmic adaptor protein and the negative feedback regulator of CpxA, which fine tunes the CpxA/R pathway (65–67). Further, CpxP acts as a scaffold protein for delivery of misfolded periplasmic proteins for DegP-mediated proteolysis (68,69). We also observed significant upregulation of ECP_2575, encoding the extracytoplasmic function sigma-E factor (σE), a master regulator of the OM stress response in Gram-negative bacteria (67,70). Taken together, our findings demonstrate that treatment of UPEC 536 with different PPNA constructs elicits a common envelope stress response, driven by σE and Cpx, induces the sRNAs OmrA/B and triggers depletion of ompF.
The growth inhibitory capacity of a PNA is affected by its length
It has been experimentally shown that 9–11mer PNAs have the highest efficacy in E. coli (6–8). The optimal length was proposed to be 10 nucleobases, since longer oligomers possess suboptimal uptake properties while shorter ones show reduced duplex stability (7). However, this conclusion is almost exclusively based on the analysis of PNAs that target lacZ or acpP (6–8). We therefore set out to comprehensively analyze PNA length effects and to elucidate whether the proposed optimal length of a 10mer PNA has general validity (7) (Table 1). To this aim, we systematically analyzed the growth inhibitory capacity of 8mer, 9mer, and 11mer PNAs, using the same assay conditions as described for their 10mer counterparts.
Surprisingly, MIC assays using 9mer PNAs targeting each of the essential genes listed in Table 1 revealed that almost all 9mer PPNAs caused a similar, in some cases even stronger, growth inhibition compared to their 10mer counterparts (Table 1; Supplementary Figures S and S3). Specifically, 9mer PPNAs targeting dnaB, ftsZ, or rpsH led to slightly improved growth inhibition compared to 10mers. To determine if the PNA length could be reduced even further, we tested 8mer PPNAs for the most efficient PNA targets, i.e.dnaB, ftsZ, rpsH and acpP. Three out of four constructs showed a 2–4 fold higher MIC than the respective 9mer PPNA (Table 1; Supplementary Figure S4), indicating that 9mer, but not 8mer, PPNAs can be more efficient in mediating growth inhibition compared to their 10mer counterparts. This might be attributed to either improved uptake or to increased target specificity, due to less frequent off-targeting (8).
To evaluate whether increased binding affinity improves growth inhibition (8), we also tested extended 11mer PPNAs in MIC assays (Table 1; Supplementary Figure S5). We observed similar or less efficient growth inhibition for the majority of tested 11mer PPNAs compared to 10mer molecules (Table 1). One notable exception is (KFF)3K-acpP, which had an identical MIC regardless of length. Overall, we conclude based on these data that 9mer PNA molecules can be at least as efficient in mediating growth inhibition as their 10mer counterparts (Table 1).
DISCUSSION
Our current mechanistic understanding of PNA function is mostly derived from studies involving a small number of PNA targets (1,6–8,13,24,32,34,35,71). Here, we present a systematic investigation of the antibacterial activity and mode of action of eleven PPNAs targeting different essential genes in UPEC. Our analysis revealed a broad distribution of growth inhibitory capacities. Below, we will discuss specific features of PPNA activity and PPNA design that might impact their antibacterial efficacy.
Aspects of PNA activity and implications for PNA design
Antisense PNAs targeting essential bacterial genes have been shown to possess antibacterial activity via selective inhibition of their targets (reviewed elsewhere (4,5)). The most commonly studied PNA target is acpP (5,8), which is strongly conserved and one of the most highly expressed genes in UPEC (Figures 1B and 5A). Importantly, acpP also emerged as the most vulnerable essential target gene in our systematic analysis, based on the level of growth inhibition upon acpP targeting. What makes then acpP PNA so potent? One obvious explanation could be that the PNA used here also depletes the mRNAs of three additional genes, i.e.fabF, gpmI and ECP_3542 (ugpQ), as part of its off-target activity. However, to the best of our knowledge, neither of the encoded proteins, i.e. 3-oxoacyl-[acyl carrier protein] synthase 2, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase and glycerophosphoryl diester phosphodiesterase, respectively, is essential for the growth of E. coli (57,72). Further, the lethal phenotype triggered by acpP PNA treatment in E. coli can be rescued by a plasmid-encoded acpP allele designed to have low complementarity to acpP PNA, confirming that depletion of acpP causes growth inhibition (8).
Another explanation might be the strong vulnerability of bacterial cells to acpP depletion, referred to as gene stringency, which exceeds that of other essential genes in E. coli (34). In particular, a 40% reduction in acpP mRNA levels leads to a 50% growth inhibition, whereas less stringency was demonstrated for ftsZ, murA and fabI (34). The vulnerability of acpP is further supported by a recent CRISPR interference (CRISPRi) study in Mycobacterium tuberculosis, which demonstrated that even incomplete inhibition of particular essential genes can severely affect bacterial fitness, even killing, while the bacteria are less sensitive to the depletion of other essential genes (73). The fatty acid biosynthesis pathway was highly enriched for vulnerable genes, including the acpP ortholog acpM, in Mycobacterium tuberculosis (https://pebble.rockefeller.edu/; (73)), suggesting that this trait could be deeply conserved across the bacterial phylogeny. Similarly, a recent CRISPRi study in E. coli also found differential vulnerability to transcript depletion among essential genes (74). Follow-up studies on the full essential gene targetome would be needed to classify particularly vulnerable pathways and genes for PNA treatment and to identify the most attractive targets, similar to the CRISPRi studies performed in Mycobacterium (73) or E. coli (74). Such analyses would also clarify whether the connectedness of a target protein's interaction network is a universal predictor of the growth inhibitory activity of a PNA, as recently suggested (25).
Designed to target the translational start site, the mechanism of action of PNAs is generally ascribed to the inhibition of target mRNA translation (6,8,13). We measured the effects of PNAs on mRNA translation using an in vitro translation system, which has been applied to monitor the translation rate of a target mRNA in the presence of its antisense sRNA (75–79). Previously, a similar approach was adopted to verify PNA-mediated translational inhibition of a Rickettsia-encoded gene (80). The in vitro translation system constitutes a straightforward method to predict the efficacy of PNA-mediated inhibition of translation in vitro. Additionally, it enables the direct analysis of PNA-mediated translational inhibition independent of in cellulo target mRNA stability. However, this system may not fully reconstitute intracellular target mRNA conformation or accessibility. In addition, the in vitro system is independent of the PNA uptake efficiency into the bacterial cell, which is a critical step that contributes to the activity of a PPNA. These considerations might explain why the ability to inhibit in vitro translation is not directly linked to PNA-mediated growth inhibition. In particular, the absence of an in vitro effect on rplS translation is puzzling, given the PNA-mediated growth reduction at high concentrations (Figure 5B, Table 1), though this may reflect a higher intracellular molar ratio than tested in our in vitro translation assay. Besides, it is also conceivable that the growth inhibition mediated by the 10mer rplS PNA at ≥ 10 μM (Table 1) is due to the off-target depletion of ECP_2657 (Figure 8), which encodes the essential protein alanyl-tRNA synthetase. While this possibility remains to be experimentally tested, the complementarity of the rplS PNA to 9 nucleotides within the translation initiation site of ECP_2657 makes this a likely scenario. An alternative strategy to monitor PNA-mediated effects on translation in vivo is ribosome profiling (Ribo-seq) (81), but due to the current costs of synthesizing PNA molecules, the establishment of this approach will require further optimization to minimize the starting material needed (82).
Apart from their predicted mode of action, i.e. sterically blocking ribosome binding and thus inhibiting translation, several studies in E. coli and other gram-negative bacteria have shown that PNAs concomitantly trigger target mRNA depletion, including PNAs targeting acpP, fabI, ftsZ, murA and rpoD (21,24,32,34–36). Since we recently demonstrated rapid PPNA-triggered depletion of acpP in Salmonella within 5 min of exposure (32), we reasoned that target mRNA depletion might be a universal rapid response caused by translational inhibition. However, based on our RNA-seq data a significant depletion of the PNA target is not universally observed after 15 min PPNA-exposure in UPEC (Figure 8) and is not associated with the PPNA’s growth inhibitory capacity (Table 1). Based on previous genome-wide studies of mRNA half-lives and translation rates in E. coli, we exclude a strong contribution of target mRNA stability (83) and translation efficiency (33) in PPNA-mediated depletion (Figure 8). It thus remains unclear what factors might determine the degree of target mRNA depletion upon PPNA treatment or whether some target mRNAs might have a delayed decay. Nevertheless, our data show that rapid target mRNA depletion is not a universal consequence of PPNA treatment.
Previous reports suggested that 10mer PNAs confer maximal activity, at least when targeting E. coli acpP (7) or lacZ (8). This length optimum reflects a balance of (i) PNA binding affinity, (ii) PPNA uptake efficiency, and (iii) the probability of off-target binding (7,8,37–39). Unexpectedly, our analysis revealed that all but one 9mer PNAs (i.e.rplS PNA) possess a similar or even superior ability to inhibit growth compared to 10mers (Table 1). Therefore, generalizations on an optimal PNA length based on one individual gene are not universally valid. Curiously, generation of a 9mer PNA by truncating the commonly used 10mer acpP PNA on the N-terminus dampens its antibacterial activity against E. coli K-12 (7), while truncation of the same 10mer acpP PNA on its C-terminus does not affect the growth inhibitory capacity of the resulting construct in UPEC (Table 1). Whether these two 9mer acpP PNAs would have different antibacterial activities if tested in the same organism is unclear. Nevertheless, our results demonstrate that using 9mer instead of 10mer PNAs can improve antibacterial activity, although this may require optimization of the truncated PNA sequence. A one-nucleobase difference might seem incremental but we expect 9mer PNAs to have fewer off-targets, which will be important for PNA applications seeking to rewire bacterial gene expression with high specificity.
The stability of a PNA/RNA duplex, measured by its melting temperature, has been proposed to affect PNA efficacy (7,8,37–39). Specifically, a positive correlation of the melting temperature and growth inhibitory capacity has been demonstrated for 8–10mer acpP PNAs (7). However, our systematic analysis of 8–10mer PNAs targeting 11 essential genes does not support a direct correlation between the predicted melting temperature and the growth inhibitory capacity (Supplementary Figure S6), although we find that the predicted melting temperature is associated with the capacity of a PNA to inhibit translation of its target in vitro. This suggests that other factors dictate the antisense efficacy and thus antibacterial activity of a PNA in cellulo. While target mRNA secondary structure has been assumed to play a key role, it has recently been excluded as a crucial determinant of a PNA’s growth inhibitory activity (25).
Apart from targeting essential genes, PNAs can also be applied to reduce the expression of antibiotic resistance genes and thereby to resensitize (multi)drug resistant bacteria to antibiotics (84–86). In addition, combined administration of PNAs targeting essential but also non-essential genes together with conventional antibiotics has been shown to increase the sensitivity of multidrug resistant bacteria to the respective compound (25). These types of combination treatment may be especially important to combat UPEC, due to the frequent emergence of drug resistant clinical isolates (29,30).
The CPP-moiety also influences the antibacterial efficacy of its conjugated PNA (Figure 2), because different CPPs use different modes of translocation into the cytoplasm of the target bacterium. (KFF)3K constitutes a promising vehicle for mechanistic studies, not only due to the efficient delivery of PNAs, but also because of the limited transcriptomic response triggered by this peptide, compared to arginine-rich CPPs, such as (RXR)4XB or Tat (32). Nevertheless, the emergence of sbmA-driven resistance, discussed below, is an important concern in (KFF)3K-mediated PNA translocation, likely rendering (KFF)3K inappropriate as suitable carrier peptide for clinical applications (21).
Defining the PPNA-triggered transcriptomic landscape in UPEC
PNAs conjugated to (KFF)3K, which were used throughout this study, are around 4 kDa in size and are thus substantially larger than conventional antibiotics (87,88). In gram-negative bacteria, uptake of several cationic antibiotics, such as kanamycin, beta-lactams or quinolones, is facilitated by passive diffusion through OM porins, such as OmpF (89–95). In contrast, (KFF)3K-coupled PNAs follow a different mechanism. Specifically, positively charged (KFF)3K-PNA molecules bind to and destabilize the negatively charged LPS layer of the OM of gram-negative bacteria to facilitate their periplasmic translocation (22,96). A yet unknown peptidase partially cleaves (KFF)3K in the extracellular and periplasmic space (22). The truncated PPNA molecules can then cross the inner membrane via the SbmA transporter in E. coli (21,22). Curiously, instead of a depletion of sbmA transcript levels to promote resistance, our RNA-seq data revealed increased mRNA levels of sbmA in all but one of the analyzed PPNA conditions. It is tempting to speculate that this is caused by a strong induction of σE and Cpx stress responses (97,98), which will be discussed in more detail below.
Following SbmA-mediated uptake, PNAs are released into the cytoplasm and can exert their function as antisense drugs (21,22). Due to their size and capacity for membrane permeabilization (99), it is not surprising that PPNAs induced a set of genes implicated in two well-known envelope stress pathways. Specifically, we found strong upregulation of members of the σE and Cpx responses (Figures 7 and 8), which, together with the Rcs (regulator of capsule synthesis), Bae (bacterial adaptive response), and Psp (phage shock protein) responses, form protective signal transduction cascades that ameliorate the detrimental effects of specific stressors (reviewed in (67)). While the σE response is triggered upon OM stress, usually via sensing of misfolded OMPs or aberrant forms of LPS, the Cpx response is related to inner membrane stress (67,100,101). Since excessive activation of envelope stress pathways can have toxic effects on the cell, their activation is balanced via negative feedback regulators (67). In particular, RseA (ECP_2574) and CpxP counterbalance activation of the σE and Cpx response, respectively, and are part of the respective stress response regulons (98,102–104). PPNA-induced transcript levels of rseA, cpxP and degP, which encodes a periplasmic protease that is transcriptionally activated by both σE and Cpx, reflect at least a partial activation of both pathways (Figure 8) (97,98,105–108). Thus, UPEC rapidly reacts to PPNA treatment via outer and inner membrane stress response pathways.
Recently, constitutive induction of the Cpx response has been linked to the development of tolerance to (RXR)4XB-coupled PNAs in E. coli. However, efficacy was unaffected under the tested conditions if the PNAs were coupled to (KFF)3K instead (23). This difference is very likely based on their distinct translocation mechanisms across the inner membrane (21–23). In addition, aberrant activation of σE in E. coli has been shown not to confer tolerance to antimicrobial peptides (AMPs), such as arginine-rich CPPs coupled to PNAs, but there is lack of information regarding the effect of σE on (KFF)3K-PNA activity (23,109). Therefore, it remains to be seen whether induction of the Cpx and σE stress response pathways can confer some degree of tolerance or resistance to PPNAs in UPEC.
We also observed universal PPNA-mediated depletion of the ompF mRNA (Figure 8), very likely due to its direct regulation by both the σE and Cpx stress responses (98,110–112). The Cpx response is also known to indirectly dampen the expression of ompF. In particular, Cpx induces the expression of marA, which in turn triggers the expression of the ompF-targeting sRNA MicF (61,113–117). Both marA and MicF were found to be highly induced in most of the PPNA conditions in our data sets (Figures 8 and S7). Although deletion of ompF did not confer resistance to (KFF)3K-acpP in E. coli K-12, in contrast to deletion of sbmA (21), it remains to be determined whether ompF mRNA depletion has physiological consequences or may influence PPNA uptake in UPEC.
Overall, our study reveals the efficacy of PPNAs against UPEC and presents a systematic analysis of eleven PNAs targeting different essential genes. We observe strong antibacterial effects for certain 10mer PPNAs independent of target gene abundance and provide evidence that 9mer PNAs, instead of their commonly used 10mer counterparts, can alternatively be used to inhibit bacterial growth. Global RNA-seq analysis indicate that rapid PPNA-mediated target mRNA depletion is not universal, but that PPNA treatment induces a common set of envelope stress responses and membrane remodeling factors. Further mechanistic studies will be needed to determine whether these responses may influence resistance to or tolerance of PPNAs, and whether the transcriptomic changes observed here indeed reflect a remodeling of the bacterial envelope. Nonetheless, our data will guide future PPNA design and help to better understand the physiological changes triggered by PPNA exposure.
DATA AVAILABILITY
Our RNA-seq data set has been deposited with GEO under accession number GSE191313 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE191313). The code used for data analysis is freely available at https://github.com/BarquistLab/UPEC_PNA_2021_11.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Barbara Plaschke for extensive technical assistance and Tobias Kerrinnes for laboratory organization. We especially thank Anke Sparmann for editing the manuscript. We further thank Tina Lence and Svetlana Ðurica-Mitić for constructive comments on the manuscript. We thank the Core Unit SysMed at the University of Würzburg for excellent technical support, library preparation and RNA-seq data generation (supported by the IZKF at the University of Würzburg, project Z-6).
Contributor Information
Linda Popella, Institute of Molecular Infection Biology (IMIB), University of Würzburg, D-97080, Würzburg, Germany.
Jakob Jung, Institute of Molecular Infection Biology (IMIB), University of Würzburg, D-97080, Würzburg, Germany.
Phuong Thao Do, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), D-97080, Würzburg, Germany.
Regan J Hayward, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), D-97080, Würzburg, Germany.
Lars Barquist, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), D-97080, Würzburg, Germany; Faculty of Medicine, University of Würzburg, D-97080, Würzburg, Germany.
Jörg Vogel, Institute of Molecular Infection Biology (IMIB), University of Würzburg, D-97080, Würzburg, Germany; Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), D-97080, Würzburg, Germany; Faculty of Medicine, University of Würzburg, D-97080, Würzburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bavarian Bayresq.net (to L.B., J.V.). The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Good L., Nielsen P.E.. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl Acad. Sci. U.S.A. 1998; 95:2073–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nielsen P.E., Egholm M.. An introduction to peptide nucleic acid. Curr. Issue Mol. Biol. 1999; 1:89–104. [PubMed] [Google Scholar]
- 3. Sully E.K., Geller B.L.. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016; 33:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogel J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 2020; 113:550–559. [DOI] [PubMed] [Google Scholar]
- 5. Pifer R., Greenberg D.E.. Antisense antibacterial compounds. Transl. Res. 2020; 223:89–106. [DOI] [PubMed] [Google Scholar]
- 6. Dryselius R., Aswasti S.K., Rajarao G.K., Nielsen P.E., Good L.. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides. 2003; 13:427–433. [DOI] [PubMed] [Google Scholar]
- 7. Goltermann L., Yavari N., Zhang M., Ghosal A., Nielsen P.E.. PNA length restriction of antibacterial activity of Peptide-PNA conjugates in Escherichia coli through effects of the inner membrane. Front. Microbiol. 2019; 10:1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Good L., Awasthi S.K., Dryselius R., Larsson O., Nielsen P.E.. Bactericidal antisense effects of peptide–PNA conjugates. Nat. Biotechnol. 2001; 19:360–364. [DOI] [PubMed] [Google Scholar]
- 9. Nielsen P.E., Egholm M., Berg R.H., Buchardt O.. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science (New York, N.Y.). 1991; 254:1497–1500. [DOI] [PubMed] [Google Scholar]
- 10. Betts L., Josey J.A., Veal J.M., Jordan S.R.. A nucleic acid triple helix formed by a peptide nucleic acid-DNA complex. Science (New York, N.Y.). 1995; 270:1838–1841. [DOI] [PubMed] [Google Scholar]
- 11. Nielsen P.E., Egholm M., Buchardt O.. Sequence-specific transcription arrest by peptide nucleic acid bound to the DNA template strand*. Gene. 1994; 149:139–145. [DOI] [PubMed] [Google Scholar]
- 12. Uhlmann E. Peptide nucleic acids (PNA) and PNA-DNA chimeras: from high binding affinity towards biological function. Biol. Chem. 1998; 379:1045–1052. [PubMed] [Google Scholar]
- 13. Good L., Nielsen P.E.. Antisense inhibition of gene expression in bacteria by PNA targeted to n1RNA. Nat. Biotechnol. 1998; 16:355–358. [DOI] [PubMed] [Google Scholar]
- 14. Moccia M., Adamo M.F.A., Saviano M.. Insights on chiral, backbone modified peptide nucleic acids: properties and biological activity. Artif. DNA, PNA & XNA. 2014; 5:e1107176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barkowsky G., Lemster A.-L., Pappesch R., Jacob A., Krüger S., Schröder A., Kreikemeyer B., Patenge N.. Influence of different cell-penetrating peptides on the antimicrobial efficiency of PNAs in Streptococcus pyogenes. Mol. Ther. Nucleic Acids. 2019; 18:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inoue G., Toyohara D., Mori T., Muraoka T.. Critical side chain effects of cell-penetrating peptides for transporting oligo peptide nucleic acids in bacteria. ACS Appl. Biol. Mater. 2021; 4:3462–3468. [DOI] [PubMed] [Google Scholar]
- 17. Oikawa K., Islam M.M., Horii Y., Yoshizumi T., Numata K.. Screening of a cell-penetrating peptide library in Escherichia coli: relationship between cell penetration efficiency and cytotoxicity. ACS Omega. 2018; 3:16489–16499. [Google Scholar]
- 18. Li J., Koh J.-J., Liu S., Lakshminarayanan R., Verma C.S., Beuerman R.W.. Membrane active antimicrobial peptides: translating mechanistic insights to design. Front. Neurosci. 2017; 11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hancock RobertE.W., Chapple DanielS.. Peptide antibiotics. Antimicrob. Agents Chemother. 1999; 43:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frimodt-Møller J., Campion C., Nielsen P.E., Løbner-Olesen A.. Translocation of non-lytic antimicrobial peptides and bacteria penetrating peptides across the inner membrane of the bacterial envelope. Curr. Genet. 2022; 68:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghosal A., Vitali A., Stach J.E.M., Nielsen P.E.. Role of SbmA in the uptake of peptide nucleic acid (PNA)-peptide conjugates in E. coli. ACS Chem. Biol. 2013; 8:360–367. [DOI] [PubMed] [Google Scholar]
- 22. Yavari N., Goltermann L., Nielsen P.E.. Uptake, stability, and activity of antisense Anti-acpP PNA-Peptide conjugates in Escherichia coli and the role of sbmA. ACS Chem. Biol. 2021; 16:471–479. [DOI] [PubMed] [Google Scholar]
- 23. Frimodt-Møller J., Koulouktsis A., Charbon G., Otterlei M., Nielsen P.E., Løbner-Olesen A.. Activating the cpx response induces tolerance to antisense PNA delivered by an arginine-rich peptide in Escherichia coli. Mol. Ther. Nucleic Acids. 2021; 25:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosal A., Nielsen P.E.. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther. 2012; 22:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eller K.A., Aunins T.R., Courtney C.M., Campos J.K., Otoupal P.B., Erickson K.E., Madinger N.E., Chatterjee A.. Facile accelerated specific therapeutic (FAST) platform develops antisense therapies to counter multidrug-resistant bacteria. Commun. Biol. 2021; 4:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J.. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015; 13:269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tabasi M., Karam M.R.A., Habibi M., Mostafavi E., Bouzari S.. Genotypic characterization of virulence factors in Escherichia coli isolated from patients with acute cystitis, pyelonephritis and asymptomatic bacteriuria. J. Clin. Diagn. Res. 2016; 10:DC01–DC07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiles T.J., Kulesus R.R., Mulvey M.A.. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008; 85:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blango M.G., Mulvey M.A.. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob. Agents Chemother. 2010; 54:1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adamus-Białek W., Baraniak A., Wawszczak M., Głuszek S., Gad B., Wróbel K., Bator P., Majchrzak M., Parniewski P.. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Mol. Biol. Rep. 2018; 45:1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rawlings M., Cronan J.E.. The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J. Biol. Chem. 1992; 267:5751–5754. [PubMed] [Google Scholar]
- 32. Popella L., Jung J., Popova K., Ðurica-Mitić S., Barquist L., Vogel J.. Global RNA profiles show target selectivity and physiological effects of peptide-delivered antisense antibiotics. Nucleic Acids Res. 2021; 49:4705–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li G.-W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goh S., Boberek J.M., Nakashima N., Stach J., Good L.. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One. 2009; 4:e6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nikravesh A., Dryselius R., Faridani O.R., Goh S., Sadeghizadeh M., Behmanesh M., Ganyu A., Klok E.J., Zain R., Good L.. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol. Ther. 2007; 15:1537–1542. [DOI] [PubMed] [Google Scholar]
- 36. Bai H., You Y., Yan H., Meng J., Xue X., Hou Z., Zhou Y., Ma X., Sang G., Luo X.. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012; 33:659–667. [DOI] [PubMed] [Google Scholar]
- 37. Ray A., Nordén B.. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 2000; 14:1041–1060. [DOI] [PubMed] [Google Scholar]
- 38. Giesen U., Kleider W., Berding C., Geiger A., Ørum H., Nielsen P.E.. A formula for thermal stability (Tm) prediction of PNA/DNA duplexes. Nucleic Acids Res. 1998; 26:5004–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jensen K.K., Ørum H., Nielsen P.E., Nordén B.. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997; 36:5072–5077. [DOI] [PubMed] [Google Scholar]
- 40. Goltermann L., Nielsen P.E.. PNA antisense targeting in bacteria: determination of antibacterial activity (MIC) of PNA-Peptide conjugates. Methods Mol. Biol. 2020; 2105:231–239. [DOI] [PubMed] [Google Scholar]
- 41. Urban J.H., Vogel J.. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007; 35:1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abramoff M.D., Magalhaes P.J., Ram S.J.. Image processing with imageJ. Biophoton. Int. 2004; 11:36–42. [Google Scholar]
- 43. Brzuszkiewicz E., Brüggemann H., Liesegang H., Emmerth M., Ölschläger T., Nagy G., Albermann K., Wagner C., Buchrieser C., Emody L.et al.. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad, Sci. U.S.A. 2006; 103:12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 45. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y., Lun A.T.L., Smyth G.K.. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016; 5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson M.D., Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. C. 1995; 57:289–300. [Google Scholar]
- 49. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santos-Zavaleta A., Salgado H., Gama-Castro S., Sánchez-Pérez M., Gómez-Romero L., Ledezma-Tejeida D., García-Sotelo J.S., Alquicira-Hernández K., Muñiz-Rascado L.J., Peña-Loredo P.et al.. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019; 47:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu D., Lim E., Vaillant F., Asselin-Labat M.-L., Visvader J.E., Smyth G.K.. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010; 26:2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dumousseau M., Rodriguez N., Juty N., Le Novère N.. MELTING, a flexible platform to predict the melting temperatures of nucleic acids. BMC Bioinf. 2012; 13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jasiński M., Miszkiewicz J., Feig M., Trylska J.. Thermal stability of peptide nucleic acid complexes. J. Biol. Chem. 2019; 123:8168–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nejad A.J., Shahrokhi N., Nielsen P.E.. Targeting of the essential acpP, ftsZ, and rne genes in carbapenem-resistant Acinetobacter baumannii by antisense PNA precision antibacterials. Biomedicines. 2021; 9:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phan M.-D., Peters K.M., Sarkar S., Lukowski S.W., Allsopp L.P., Gomes Moriel D., Achard M.E.S., Totsika M., Marshall V.M., Upton M.et al.. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 2013; 9:e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wetmore K.M., Price M.N., Waters R.J., Lamson J.S., He J., Hoover C.A., Blow M.J., Bristow J., Butland G., Arkin A.P.et al.. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. Mbio. 2015; 6:e00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou J., Rudd K.E.. EcoGene 3.0. Nucleic Acids Res. 2013; 41:D613–D624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Narenji H., Teymournejad O., Rezaee M.A., Taghizadeh S., Mehramuz B., Aghazadeh M., Asgharzadeh M., Madhi M., Gholizadeh P., Ganbarov K.et al.. Antisense peptide nucleic acids against ftsZ and efaA genes inhibit growth and biofilm formation of Enterococcus faecalis. Microb. Pathog. 2020; 139:103907. [DOI] [PubMed] [Google Scholar]
- 59. Inokuchi K., Mutoh N., Matsuyama S.-i., Mizushirna S.. Primary structure of the ompF gene that codes for a major outer membrane protein of Escherichia coli K-12. Nucleic Acids Res. 1982; 10:6957–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guillier M., Gottesman S.. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006; 59:231–247. [DOI] [PubMed] [Google Scholar]
- 61. Guillier M., Gottesman S., Storz G.. Modulating the outer membrane with small RNAs. Genes Dev. 2006; 20:2338–2348. [DOI] [PubMed] [Google Scholar]
- 62. Vogel J., Papenfort K.. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006; 9:605–611. [DOI] [PubMed] [Google Scholar]
- 63. Ge X., Wang R., Ma J., Liu Y., Ezemaduka A.N., Chen P.R., Fu X., Chang Z.. DegP primarily functions as a protease for the biogenesis of β-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J. 2014; 281:1226–1240. [DOI] [PubMed] [Google Scholar]
- 64. Krojer T., Sawa J., Schäfer E., Saibil H.R., Ehrmann M., Clausen T.. Structural basis for the regulated protease and chaperone function of degP. Nature. 2008; 453:885–890. [DOI] [PubMed] [Google Scholar]
- 65. Vogt S.L., Raivio T.L.. Just scratching the surface: an expanding view of the cpx envelope stress response. FEMS Microbiol. Lett. 2012; 326:2–11. [DOI] [PubMed] [Google Scholar]
- 66. Chao Y., Vogel J.. A 3′ UTR-Derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell. 2016; 61:352–363. [DOI] [PubMed] [Google Scholar]
- 67. Mitchell A.M., Silhavy T.J.. Envelope stress responses: balancing damage repair and toxicity. Nat. Rev. Microbiol. 2019; 17:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Debnath I., Norton J.P., Barber A.E., Ott E.M., Dhakal B.K., Kulesus R.R., Mulvey M.A.. The cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect. Immun. 2013; 81:1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Isaac D.D., Pinkner J.S., Hultgren S.J., Silhavy T.J.. The extracytoplasmic adaptor protein CpxP is degraded with substrate by degP. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17775–17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johansen J., Rasmussen A.A., Overgaard M., Valentin-Hansen P.. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006; 364:1–8. [DOI] [PubMed] [Google Scholar]
- 71. Campion C., Charbon G., Thomsen T.T., Nielsen P.E., Løbner-Olesen A.. Antisense inhibition of the Escherichia coli NrdAB aerobic ribonucleotide reductase is bactericidal due to induction of DNA strand breaks. J. Antimicrob. Chemother. 2021; 76:2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goodall E.C.A., Robinson A., Johnston I.G., Jabbari S., Turner K.A., Cunningham A.F., Lund P.A., Cole J.A., Henderson I.R.. The essential genome of Escherichia coli K-12. Mbio. 2018; 9:e02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bosch B., DeJesus M.A., Poulton N.C., Zhang W., Engelhart C.A., Zaveri A., Lavalette S., Ruecker N., Trujillo C., Wallach J.B.et al.. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell. 2021; 184:4579–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Silvis M.R., Rajendram M., Shi H., Osadnik H., Gray A.N., Cesar S., Peters J.M., Hearne C.C., Kumar P., Todor H.et al.. Morphological and transcriptional responses to CRISPRi knockdown of essential genes in Escherichia coli. Mbio. 2021; 12:e0256121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sharma C.M., Darfeuille F., Plantinga T.H., Vogel J.. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007; 21:2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Urban J.H., Vogel J.. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008; 6:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hoekzema M., Romilly C., Holmqvist E., Wagner E.G.H.. Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation. EMBO J. 2019; 38:e101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Holmqvist E., Unoson C., Reimegård J., Wagner E.G.H.. A mixed double negative feedback loop between the sRNA MicF and the global regulator lrp. Mol. Microbiol. 2012; 84:414–427. [DOI] [PubMed] [Google Scholar]
- 79. Maki K., Uno K., Morita T., Aiba H.. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc. Natl Acad. Sci. U.S.A. 2008; 105:10332–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pelc R.S., McClure J.C., Kaur S.J., Sears K.T., Rahman M.S., Ceraul S.M.. Disrupting protein expression with peptide nucleic acids reduces infection by obligate intracellular Rickettsia. PLoS One. 2015; 10:e0119283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-Wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Venturini E., Svensson S.L., Maaß S., Gelhausen R., Eggenhofer F., Li L., Cain A.K., Parkhill J., Becher D., Backofen R.et al.. A global data-driven census of Salmonella small proteins and their potential functions in bacterial virulence. Microlife. 2020; 1:uqaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Esquerré T., Laguerre S., Turlan C., Carpousis A.J., Girbal L., Cocaign-Bousquet M.. Dual role of transcription and transcript stability in the regulation of gene expression in Escherichia coli cells cultured on glucose at different growth rates. Nucleic Acids Res. 2014; 42:2460–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Aunins T.R., Erickson K.E., Chatterjee A.. Transcriptome-based design of antisense inhibitors potentiates carbapenem efficacy in CRE Escherichia coli. Proc. Nat. Acad. Sci. U.S.A. 2020; 117:30699–30709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Courtney C.M., Chatterjee A.. Sequence-Specific peptide nucleic acid-based antisense inhibitors of TEM-1 β-Lactamase and mechanism of adaptive resistance. ACS Infect. Dis. 2015; 1:253–263. [DOI] [PubMed] [Google Scholar]
- 86. Nezhadi J., Narenji H., Soroush Barhaghi M.H., Rezaee M.A., Ghotaslou R., Pirzadeh T., Tanomand A., Ganbarov K., Bastami M., Madhi M.et al.. Peptide nucleic acid-mediated re-sensitization of colistin resistance Escherichia coli KP81 harboring mcr-1 plasmid. Microb. Pathog. 2019; 135:103646. [DOI] [PubMed] [Google Scholar]
- 87. Winterhalter M. Antibiotic uptake through porins located in the outer membrane of Gram-negative bacteria. Expert Opin. Drug Deliv. 2021; 18:449–457. [DOI] [PubMed] [Google Scholar]
- 88. Jones S. Permeability rules for antibiotic design. Nat. Biotechnol. 2017; 35:639. [DOI] [PubMed] [Google Scholar]
- 89. Bafna J.A., Sans-Serramitjana E., Acosta-Gutiérrez S., Bodrenko I.V., Hörömpöli D., Berscheid A., Brötz-Oesterhelt H., Winterhalter M., Ceccarelli M.. Kanamycin uptake into Escherichia coli is facilitated by OmpF and OmpC porin channels located in the outer membrane. ACS Infect. Dis. 2020; 6:1855–1865. [DOI] [PubMed] [Google Scholar]
- 90. Hancock R.E.W., Bell A.. Antibiotic uptake into gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 1988; 7:713–720. [DOI] [PubMed] [Google Scholar]
- 91. Nestorovich E.M., Danelon C., Winterhalter M., Bezrukov S.M.. Designed to penetrate: Time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl Acad. Sci. U.S.A. 2002; 99:9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ghai I., Bajaj H., Arun Bafna J., El Damrany Hussein H.A., Winterhalter M., Wagner R. Ampicillin permeation across OmpF, the major outer-membrane channel in Escherichia coli. J. Biol. Chem. 2018; 293:7030–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sawai T., Yamaguchi A., Saiki A., Hoshino K.. OmpF channel permeability of quinolones and their comparison with β-lactams. FEMS Microbiol. Lett. 1992; 95:105–108. [DOI] [PubMed] [Google Scholar]
- 94. Cama J., Bajaj H., Pagliara S., Maier T., Braun Y., Winterhalter M., Keyser U.F.. Quantification of fluoroquinolone uptake through the outer membrane channel OmpF of Escherichia coli. J. Am. Chem. Soc. 2015; 137:13836–13843. [DOI] [PubMed] [Google Scholar]
- 95. Vergalli J., Bodrenko I.V., Masi M., Moynié L., Acosta-Gutiérrez S., Naismith J.H., Davin-Regli A., Ceccarelli M., van den Berg B., Winterhalter M.et al.. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020; 18:164–176. [DOI] [PubMed] [Google Scholar]
- 96. Vaara M., Porro M.. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996; 40:1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Raivio T.L., Leblanc S.K.D., Price N.L.. The Escherichia coli cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 2013; 195:2755–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rhodius V.A., Suh W.C., Nonaka G., West J., Gross C.A.. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006; 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Eriksson M., Nielsen P.E., Good L.. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 2002; 277:7144–7147. [DOI] [PubMed] [Google Scholar]
- 100. Ades S.E. Regulation by destruction: design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 2008; 11:535–540. [DOI] [PubMed] [Google Scholar]
- 101. Raivio T.L. Everything old is new again: an update on current research on the cpx envelope stress response. Biochim. Biophys. Acta. 2014; 1843:1529–1541. [DOI] [PubMed] [Google Scholar]
- 102. Hayden J.D., Ades S.E.. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One. 2008; 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Buelow D.R., Raivio T.L.. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease degP. J. Bacteriol. 2005; 187:6622–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ruiz N., Silhavy T.J.. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 2005; 8:122–126. [DOI] [PubMed] [Google Scholar]
- 105. Danese P.N., Silhavy T.J.. The sigma-E and the cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997; 11:1183–1193. [DOI] [PubMed] [Google Scholar]
- 106. Danese P.N., Snyder W.B., Cosma C.L., Davis L.J.B., Silhavy T.J.. The cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, degP. Genes Dev. 1995; 9:387–398. [DOI] [PubMed] [Google Scholar]
- 107. Erickson J.W., Gross C.A.. Identification of the sigma-E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989; 3:1462–1471. [DOI] [PubMed] [Google Scholar]
- 108. Danese P.N., Silhavy T.J.. CpxP, a stress-combative member of the cpx regulon. J. Bacteriol. 1998; 180:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Audrain B., Ferrières L., Zairi A., Soubigou G., Dobson C., Coppée J.-Y., Beloin C., Ghigo J.-M.. Induction of the cpx envelope stress pathway contributes to Escherichia coli tolerance to antimicrobial peptides. Appl. Environ. Microbiol. 2013; 79:7770–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Alba B.M., Gross C.A.. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 2004; 52:613–619. [DOI] [PubMed] [Google Scholar]
- 111. Batchelor E., Walthers D., Kenney L.J., Goulian M.. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J. Bacteriol. 2005; 187:5723–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Matera G., Altuvia Y., Gerovac M., El Mouali Y., Margalit H., Vogel J.. Global RNA interactome of salmonella discovers a 5′ UTR sponge for the MicF small RNA that connects membrane permeability to transport capacity. Mol. Cell. 2022; 82:629–644. [DOI] [PubMed] [Google Scholar]
- 113. Andersen J., Forst S.A., Zhao K., Inouye M., Delihas N.. The function of micF RNA. J. Biol. Chem. 1989; 264:17961–17970. [PubMed] [Google Scholar]
- 114. Cohen S.P., McMurry L.M., Levy S.B.. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 1988; 170:5416–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cohen S.P., McMurry L.M., Hooper D.C., Wolfson J.S., Levy S.B.. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 1989; 33:1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Delihas N., Forst S.. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 2001; 313:1–12. [DOI] [PubMed] [Google Scholar]
- 117. Weatherspoon-Griffin N., Yang D., Kong W., Hua Z., Shi Y.. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 2014; 289:32571–32582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988; 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our RNA-seq data set has been deposited with GEO under accession number GSE191313 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE191313). The code used for data analysis is freely available at https://github.com/BarquistLab/UPEC_PNA_2021_11.