Abstract
ARMC5 is implicated in several pathological conditions, but its function remains unknown. We have previously identified CUL3 and RPB1 (the largest subunit of RNA polymerase II (Pol II) as potential ARMC5-interacting proteins. Here, we show that ARMC5, CUL3 and RBX1 form an active E3 ligase complex specific for RPB1. ARMC5, CUL3, and RBX1 formed an active E3 specific for RPB1. Armc5 deletion caused a significant reduction in RPB1 ubiquitination and an increase in an accumulation of RPB1, and hence an enlarged Pol II pool in normal tissues and organs. The compromised RPB1 degradation did not cause generalized Pol II stalling nor depressed transcription in the adrenal glands but did result in dysregulation of a subset of genes, with most upregulated. We found RPB1 to be highly expressed in the adrenal nodules from patients with primary bilateral macronodular adrenal hyperplasia (PBMAH) harboring germline ARMC5 mutations. Mutant ARMC5 had altered binding with RPB1. In summary, we discovered that wildtype ARMC5 was part of a novel RPB1-specific E3. ARMC5 mutations resulted in an enlarged Pol II pool, which dysregulated a subset of effector genes. Such an enlarged Pol II pool and gene dysregulation was correlated to adrenal hyperplasia in humans and KO mice.
INTRODUCTION
DNA-dependent RNA polymerase II (Pol II) is responsible for the synthesis of all mRNA and some small RNAs (1). It is comprised of 12 subunits, with RPB1 being the largest one (2). Protein biosynthesis and degradation determine intracellular protein homeostasis. Thus, it is logical to assume that RPB1 degradation is involved in determining the homeostasis of the Pol II pool size. The effect of an abnormal Pol II pool size is poorly understood. It is assumed that since Pol II is implicated in the transcription of all genes, its pool size will affect the transcription of all genes which are expressed. This assumption has not been confirmed.
During transcription, if template DNA is damaged or the cells are under some other stress, Pol II will stall until the damage is repaired or the stress is relieved (3–6). In the case of prolonged stalling, experimental data suggest that Pol II will be channeled to proteasomes for degradation so that transcription may resume (5,7–13).
Proteins need to be ubiquitinated before being degraded in proteasomes. Such ubiquitination depends on a cascade of three enzymes, that is, E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme), and E3 (Ub ligase) (14). There are two E1s (UBA1 and UBA6) in humans, but they lack substrate specificity (15). There are a total of 40 known E2s with limited specificity (16). E3 determines the substrate specificity. Each protein has a specific, or sometimes several E3s, and each E3 can have several substrates (14). There are three families of E3s: Really Interesting New Gene (RING)-type E3s (single or multiple subunits), Homologous to the E6-AP Carboxyl Terminus (HECT)-type E3s, and RING-between-RINGS (RBR)-type E3s (17). The RING-type E3s are the largest family. A multiple subunit RING E3 contains a RING-finger protein (e.g. ROC1/RBX1), a cullin (CUL) protein (CUL1, 2, 3, 4A, 4B, 5 and 7), and a substrate recognition unit (18). CUL3 interacts with a RING-finger protein RBX1. CUL3 has a BTB-interacting domain, and it recruits a BTB domain-containing protein as its substrate recognition unit to form an active E3 (19).
Given the critical roles of RPB1 degradation, there is much interest in identifying RPB1-specific E3s (11,13). Several such E3s have been reported in yeast and mammalian cells (11,20–26). However, most of these E3s only have proven functions in irradiated cells or those treated with DNA-damaging agents. E3 activity of a few of them in unperturbed cell lines is suggested by some knockdown studies, but these have not been studied at the tissue or whole-organ level (23,26).
ARMC5 is a protein containing an armadillo (ARM) domain, which comprises multiple ARM repeats in its N-terminus and a BTB domain towards its C-terminus. Human and mouse ARMC5 proteins share 90% amino acid (aa) sequence homology and also have similar tertiary structures. Mouse ARMC5 is 926 aa in length (NP_666317.2). Human ARMC5 has several isoforms due to alternative splicing or the use of additional exons (27). The longest human ARMC5 isoform has 1030 aa (NP_001275696.1), and the most highly and abundantly expressed isoform is 935 aa long (NP_001098717.1) (27).
Primary bilateral macronodular adrenal hyperplasia (PBMAH) is a rare disease accounting for less than 1% of all Cushing's syndrome, itself a disease with an incidence of between 1 and 3 per million per year (28). Cushing's syndrome due to PBMAH has an insidious onset and is usually diagnosed in the fifth and sixth decades of life (29). Whilst we were investigating the function of ARMC5 in knockout (KO) mice, several groups identified ARMC5 mutations in about 21–26% of PBMAH patients (28–33). The biosynthesis of cortisol per adrenocortical cell is relatively inefficient due to partial deficiency of several steroidogenic enzymes (30,34–36). Hypercortisolism ultimately develops because of the massively enlarged nodular adrenals (28,30). Recently we reported that Armc5 bi-allelic KO mice were small and had compromised T-cell proliferative capacity and impaired T-cell immune responses (37). Aged KO mice had adrenal hypertrophy accompanied by moderately augmented blood cortisol levels (37), similar to that observed in PBMAH patients, suggesting that Armc5 deletion alone is sufficient to cause a PBMAH-like condition. Consistent with our data, blood cortisol levels were also elevated in 30% of aged mice with monoallelic Armc5 deletion in another study (38).
The mechanisms of action of ARMC5 are unknown. ARMC5 does not contain any conserved enzymatic motifs or domains and is thus unlikely an enzyme per se. Its function is likely to depend on its interaction with other molecules. To identify binding partners of ARMC5, we conducted a yeast 2-hybrid assay (Y2H), using human ARMC5 as bait. RPB1, CUL3 and ARMC5 itself were among the binding partners of the highest scores (37).
In the present study, we demonstrated that ARMC5 physically interacted with CUL3 and RPB1 and was the substrate recognition subunit of a novel multiple-unit RING-finger E3. This E3 is largely responsible for RPB1 ubiquitination in normal cells and tissues. Armc5 deletion led to compromised RPB1 degradation. It is widely believed that RPB1 degradation is required to resolve stalled Pol II. Interfering with RPB1 degradation might cause increased stalling and hence reduced transcription. However, we did not observe augmented Pol II stalling nor a generally reduced transcription in Armc5 KO cells in spite of the failed RPB1 degradation. This implies either RPB1 degradation is not the only way or other E3s are compensating to resolve the stalling. Among 1486 differentially expressed genes in KO adrenal glands, most of them were up-regulated, presumably due to an enlarged Pol II pool size. We further showed that the adrenal gland nodules from PBMAH patients carrying ARMC5 mutations presented highly elevated RPB1 protein levels, demonstrating the relevance of our findings to human PBMAH.
MATERIALS AND METHODS
Armc5 KO mice
Armc5 KO mice and their littermates used in this report were of 129/sv × CD1 background. Armc5 KO mice were generated as described previously (37). All mice were housed and handled in accordance with a protocol approved by the Institutional Animal Protection Committees of the CRCHUM and INRS-IAF. All the murine studies were approved by the Animal Protection Committee (Comité institutionnel d’intégration de la protection des animaux) of the CRCHUM.
Cell culture and transfection
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM). Mouse embryonic fibroblasts (MEFs) were derived from E12.5 KO or WT fetuses and cultured in DMEM. SW-13 adrenal gland carcinoma cells were cultured in L-15 medium at 37°C. All culture media were supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 U/ml). Transfection of HEK293 cells and SW-13 cells was performed using Lipofectamine 2000 and Lipofectin (both from Invitrogen), respectively. Proteasome inhibitor MG132 (10 μM; Cayman Chemical) or nuclear export inhibitor leptomycin B (20 nM; Cell Signaling Technology) was added to cell culture as required. Cells were analyzed 48 h after the transfection.
Plasmids
Plasmid ARMC5-HA (EX-H0661-M07) expressing full-length ARMC5 (aa1-935) with HA at its C-terminus and plasmid ARCM5-FLAG (EX-H0661-M14) expressing full-length ARMC5 (aa1-935) with FLAG at its C-terminus were obtained from GeneCopeia. Plasmid CUL3-Myc (RC208066) expressing full-length human CUL3 with Myc at its C-terminus was obtained from OriGene. Plasmid CUL3-HA expressing full-length human CUL3 with HA tag at its C-terminus was cloned by restriction enzyme-based method using the insert from plasmid CUL3-Myc and the vector from plasmid ARMC5-HA. Plasmid FLAG-RPB1 (Plasmid #35175) expressing full-length human RPB1 with FLAG at its N-terminus, plasmid FLAG-RPB1-ΔCTD (C-terminal domain; Plasmid #35176) expressing CTD-deleted human RPB1 with FLAG at its N-terminus, and plasmid HA-Ubiquitin (Plasmid #18712) were obtained from Addgene. Following plasmids expressing human ARMC5 and CUL3 deletion mutants were generated using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs): ARMC5(Δaa2–142)-HA with aa 2-142 deleted, ARMC5(Δaa143–444)-HA with aa 143–444 deleted, ARMC5(Δaa445–747)-HA with aa 445–747 deleted, ARMC5(Δaa748–816)-HA with aa 748–816 deleted, ARMC5(Δaa817–934)-HA with aa 817–934 deleted, CUL3(Δaa31–385)-HA with aa 31–385 deleted, CUL3(Δaa377–675)-Myc with aa 377–765 deleted, and CUL3(Δaa695–762)-Myc with the neddylation site (aa 695–762) deleted. Plasmids ARMC5-BTB-HA expressing the ARMC5 BTB domain (aa 748–935) with HA at its C-terminus, and CUL3(aa1–376)-Myc expressing CUL3 N-terminal culling repeats (aa 1–376) with Myc at its C-terminus were generated by retrieving the needed fragments with PCR using Q5 Hot Start High-Fidelity DNA polymerase (New England Biolabs) from full-length ARMC5 or CUL3 cDNA and re-cloned them to the original vectors.
Antibodies (Abs), bacteria, chemicals, peptides, recombinant proteins, commercial assay kits, plasmids , and software
These reagents and software and their sources are listed in Supplementary Information Table 1 (Supplementary Table S1).
Table 1.
Clinical information and genotype of the Adelaide cohort
| Patient ID | Age at operation | Gender | Diagnosis | Histopathology | Germline ARMC5 mutation status |
|---|---|---|---|---|---|
| III-1 | 69 | M | Cushing's due to PBMAH | PBMAH | Chr16:g.31476121; c.1777C→T; p.(R593W) |
| III-2 | 62 | M | Cushing's due to PBMAH | PBMAH | Chr16:g.31476121; c.1777C→T; p.(R593W) |
| III-3 | 66 | M | Cushing's due to PBMAH | PBMAH | Chr16:g.31476121; c.1777C→T; p.(R593W) |
| CS-01 | 39 | F | Cushing's syndrome | Adrenal adenoma | not tested |
| CS-02 | 48 | F | Cushing's syndrome | Adrenal adenoma | not tested |
| PA-01 | 57 | M | Primary aldosteronism | Adrenal adenoma | not tested |
| PA-02 | 76 | M | Primary aldosteronism | Adrenal hyperplasia - micro- and macronodular | not tested |
| PA-03 | 50 | M | Primary aldosteronism | Adrenal hyperplasia - micro- and macronodular | not tested |
| N-1 | 50 | M | normal adrenal gland a | N/A | not tested |
| N-2 | unknown | M | normal adrenal gland b | N/A | not tested |
For RPB1 expression analysis, PBMAH macronodules were obtained from resected adrenal glands of PBMAH patients with germline ARMC5 mutations. Adrenal gland adenomas and adrenocortical carcinoma tissues not known to have ARMC5 mutations were used as controls. The clinical diagnosis and histopathology of the patients are indicated. aThe normal adrenal gland of patient N-1 was obtained from normal tissues adjacent to the adrenal gland adenoma of patient PA-03. bNormal adrenal gland N-2 was obtained from a patient undergoing nephrectomy for renal cell carcinoma. PBMAH: primary bilateral macronodular adrenal gland hyperplasia. N/A: not applicable.
Immunoprecipitation and Western blotting
For protein-protein interaction experiments, cells were lysed in TNE buffer (50 mM Tris–HCl at pH 7.4, 100 mM NaCl, 0.1 mM EDTA, 1% Triton X100) supplemented with Halt™ Protease Inhibitor Cocktail (ThermoFisher) and Phosphatase Inhibitors (Roche). The extracts were incubated with the corresponding antibodies overnight at 4°C and then incubated with Protein G magnetic beads (Bio-Rad) for another two hours at 4°C. The beads were washed five times with wash buffer (50 mM Tris–HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.2 mM sodium orthovanadate, 1% Triton X100). The bound proteins were eluted by a 2× SDS-loading buffer. For protein ubiquitination experiments, cells or tissues were lysed or homogenized in RIPA buffer (25 mM Tris at pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with Halt™ Protease Inhibitor Cocktail, Phosphatase Inhibitors, and 4 mM of N-ethylmaleimide (Millipore Sigma). The cleared supernatants were incubated with Abs overnight at 4°C, followed by protein G-conjugated magnetic beads (Bio-Rad) for another 2 h at 4°C. The beads were washed five times with RIPA buffer. The bound proteins were eluted by a 2× SDS-loading buffer and then resolved by SDS-PAGE. The proteins were then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% (w/v) milk in TBST (Tris-Buffered Saline, 0.05% Tween 20) and incubated with first Abs for 2 h at room temperature or overnight at 4°C, followed by HRP-conjugated secondary Abs for 1 h at room temperature. The signal was revealed by the Western Lightening pro-ECL (PerkinElmer) and detected with either X-ray film or ChemiDoc Imaging system (Bio-Rad).
Tandem liquid chromatography and mass spectrometry
HEK293 cells were transfected with ARMC5-HA or control plasmid, and after 48 h, the cells were lysed in TNE buffer supplemented with Halt™ Protease Inhibitor Cocktail and Phosphatase Inhibitors. Lysates were centrifuged and immunoprecipitated by anti-HA Ab-conjugated agarose beads (Sigma) at 4°C overnight. The beads were washed five times by wash buffer (50 mM Tris–HCl at pH 7.4, 150 mM NaCl, 1mM EDTA, 0.2 mM sodium orthovanadate, 1% Triton X100), and the precipitated proteins were eluted by HA peptides (GenScript). The eluates were resolved by 4–15% Mini-PROTEAN® TGX™ Precast Protein Gels, and the gels were stained with the silver staining kit (ThermoFisher).
Visible bands in the silver-stained gel were excised and destained in 50% methanol (Sigma-Aldrich). Each band was shrunk in 50% acetonitrile (ACN), reconstituted in 50 mM ammonium bicarbonate with 10 mM Tris (2-carboxyethyl) phosphine hydrochloride (Thermo Fisher Scientific), and vortexed for 1 h at 37°C. Chloroacetamide (Sigma-Aldrich) was added for alkylation to a final concentration of 55 mM. Samples were vortexed for another hour at 37°C. One microgram of trypsin was added, and digestion was performed for 8 h at 37°C. Peptide extraction was conducted with 90% ACN. The extracted peptide samples were dried and solubilized in 5% ACN-0.2% formic acid. The samples were loaded on a homemade C18 pre-column (0.3-mm inside diameter × 5 mm) connected directly to the switching valve. Peptides were separated on a home-made reversed-phase column (150-μm inside diameter × 150 mm) with a 56-min gradient from 10 to 30% ACN-0.2% FA and a 600 nl/min flow rate on an Ultimate 3000 HPLC connected to a Q-Exactive Plus Hybrid Quadrupole-Orbitrap™ mass spectrometer (MS) (Thermo Fisher Scientific). Each full MS spectrum acquired at a resolution of 70 000 was followed by 12 tandem-MS (MS-MS) spectra on the most abundant multiply charged precursor ions. Tandem-MS experiments were performed using collision-induced dissociation at a collision energy of 27%.
The data were processed using PEAKS 8.5 (Bioinformatics Solutions, Waterloo, ON) and a human database. Mass tolerances on precursor and fragment ions were 10 ppm and 0.01 Da, respectively. Variable selected posttranslational modifications were carbamidomethyl (C), oxidation (M), deamidation (NQ), and phosphorylation (STY). The data were analyzed with Scaffold 4.3.0. A protein was categorized as a hit if it met the threshold of 99%, with at least two peptides identified and a false-discovery rate (FDR) of 1% for peptides.
The hits were further filtered for data presentation according to the criteria described in the Result section.
Construction of the E3 3D model
The ARMC5 3D structure was obtained from Alphafold Protein Structure Database (39). The cullin-repeat structure of CUL3 was derived from the crystal structure of the KLHL3-CUL3 complex (Protein Database Band (PDB) 4HXI). The cullin homology domain and C-terminal domain of CUL3 conjugated with RBX1 were extracted from the crystal structure of an RBX1–UBC12∼NEDD8–CUL1–DCN1 complex (PDB 4P5O), in which CUL1 was highly homologous to CUL3. The RPB1 structure was obtained from the crystal structure of the human RNA Pol II complex (PDB 6DRD). The structures of UBE2E1 and UBC were derived from the structures of the TRIM21–UBE2E1 complex (PDB 6FGA) and UbcH5A–UBC complex (PDB 4AP4), respectively. The docking of UBE2E1 on RBX1 was modeled according to the structure of TRIM21–UBE2E1 (PDB 6FGA) by replacing the RING domain of TRIM21 with RBX1. The interaction between ubiquitin UBC and UBE2E1 was based on the UbcH5A-UBC structure (PDB 4AP4) by replacing UbcH5A with UBE2E1. The interactions among RPB1, ARMC5, and RPB1 were modeled according to the results of our deletion studies by positioning the interacting domains close to each other. UCSF Chimera (40) was used to extract and position proteins in the 3D model.
Immunofluorescence
MEFs, ARMC5-HA-transfected HEK293 cells, and SW-13 cells were grown on coverslips in 6-well plates. In some experiments, nuclear export inhibitor leptomycin (20 nM) was added to the culture for the final 2–4 h of culture, as indicated. After 48 hours, the cells were fixed with 4% (w/v) paraformaldehyde for 30 min at room temperature and permeabilized with 0.3% Triton in PBS for 5 min. The cells were then blocked with 5% goat serum in PBS for 1 h at room temperature and reacted with corresponding first Abs overnight at 4°C. The coverslips were washed three times with PBS and incubated with Alexa Fluor 488 goat anti-mouse Ab or Alexa Fluor 555 goat anti-rabbit Ab for 2 h at room temperature. After the third wash, the coverslips were mounted with ProLong Diamond Antifade Mountant containing DAPI (4,6-diamidino-2-phenylindole) (ThermoFisher Scientific). Images were acquired with a Zeiss microscope. For RPB1 signal quantification in MEFs, the corrected total cell fluorescence (CTCF) intensity was evaluated by ImageJ using the following formula: CTCF = integrated density – (area of selected cells × mean fluorescence of background readings).
In vitro ubiquitination assay
HEK293 cells were transfected with plasmids expressing ARMC5-HA or FLAG-RPB1. Forty-eight hours after transfection, the cells were lysed with RIPA buffer. The lysates were immunoprecipitated by anti-HA Ab-conjugated agarose beads or anti-FLAG M2 Ab-conjugated agarose beads. The beads were washed three times with RIPA buffer and then three times with wash buffer (50 mM Tris–HCl at pH 7.4, 300 mM NaCl, 1mM EDTA, 0.2 mM sodium orthovanadate, 1% Triton-X100). The bound proteins were eluted by 10 μg HA peptides (GenScript) or 10 μg FLAG peptides (GenScript) in 200 μl protein preservation buffer (40 mM Tris, 110 mM NaCl, 2.2 mM KCl, 0.04% Tween20, 30% glycerol). ARMC5-HA protein and ARMC5-ΔBTB-HA protein eluates were concentrated by Microcon-30 kDa centrifugal filter devices (Amicon, Millipore). Flag-RPB1 protein eluates were concentrated by Microcon-100 kDa centrifugal filter devices (Amicon, Millipore). The control eluate was extracted from HEK293 cells transfected with empty vectors and underwent the same purification steps. CUL3/Rbx1-GST complexes were obtained from BPS Bioscience.
Preliminary screening for an optimal E2 was performed. Eight different E2s (UBE2N, UBE2C, UBE2L3, UBE2E3, UBE2E1, UBE2D3, UBE2D2 and UBE2D1) were available in the UbcH Enzyme Kit (Boston Biochem) and were tested in in vitro ubiquitination assays, using RPB1-Flag as the substrate. Four of these E2s (i.e. UBE2E3, UBE2E1, UBE2E2 and UBE2D2) were known to interact with CUL3 according to a BioGrid search (Supplementary Figure S1A). The levels of RPB1 ubiquitination profile by these E2s are shown in Supplementary Figure S1B. To avoid E2s that might cause high background, we chose UBE2E1, which generated an intermediate level of the signal, for the final in vitro ubiquitination assay.
The purity of the recombinant proteins used in the in vitro ubiquitination was determined by Coomassie Blue or silver staining (Supplementary Figure S2).
For a typical in vitro ubiquitination reaction, ARMC5-HA, ARMC5-ΔBTB-HA and FLAG-RPB1 (500 ng each, affinity-purified from transfected HEK293 cells) were added to a mixture containing 100 ng of E1 (UBE1), 500 ng of E2 (UBE2E1), 10 μg of His6-ubiquitin (all from BostonBiochem), 260 ng of CUL3/RBX1-GST (BPS Bioscience), and 10 mM ATP in ubiquitination buffer (25 mM Tris–Cl at pH 8.0, 125 mM NaCl, 10 mM MgCl2, 50 μM DTT). The reaction was carried out at 30°C for 90 min and stopped by 20 mM EDTA. The ubiquitinated RPB1 was immunoprecipitated with anti-FLAG Ab plus Protein G conjugated magnetic beads, resolved by 6% SDS-PAGE gel, and immunoblotted with anti-ubiquitin Ab.
RNA-seq
KO and WT adrenal glands were homogenized, and total RNA extracted using an RNeasy kit (Qiagen). Total RNA was quantified using a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, Inc.), and its integrity was assessed on a 2100 Bioanalyzer (Agilent Technologies). rRNA was depleted from 250 ng of total RNA using QIAseq FastSelect (Human 96rxns; Qiagen). cDNA synthesis was performed with the NEBNext RNA First-Strand Synthesis and NEBNext Ultra Directional RNA Second Strand Synthesis Modules (New England BioLabs). The remaining steps of library preparation were performed using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs), according to the manufacturer's instructions. Adapters and PCR primers were purchased from New England BioLabs. Libraries were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) and the Kapa Illumina GA with Revised Primers-SYBR Fast Universal kit (Kapa Biosystems). The average fragment size was determined using a LabChip GX (PerkinElmer).
The libraries were normalized and pooled and then denatured in 0.05 N NaOH and neutralized using HT1 buffer. The pool was loaded at 225 pM on an Illumina NovaSeq S2 lane using the Xp protocol as per the manufacturer's instructions. The run was performed for 2×100 cycles (paired-end mode). A phiX library was used as a control and mixed with libraries at a 1% level. Base-calling was performed with RTA v3. Program bcl2fastq2 v2.20 was then used to demultiplex samples and generate fastq reads.
Reads were trimmed from the 3' end to have a phred score of at least 30. Illumina sequencing adaptors were removed from the reads, and all reads were required to have a length of at least 32 bp. Trimming and clipping were performed using Trimmomatic (41).
Upstream processing of FastQ files was performed as described previously (42), using the ENSEMBL GRCm38 (Mus musculus) release 97 mouse reference genome sequences and annotations. Each readset was aligned using STAR (43), which creates a Binary Alignment Map (.bam) file. Then, all readset BAM files from the same sample were merged into a single global BAM file using Picard.
The pairwise Pearson's correlation values of samples were calculated. The correlation controlled the general transcripts expression consistency between samples. It could check sample mix-up or error in name assignment. Thus, samples belonging to the same design group/condition were expected to show a higher level of correlation.
Gene expression levels were quantified using StringTie. Specifically, the count matrix was extracted from StringTie output using the auxiliary script prepDE.py as provided on the StringTie website. A gene here could be a novel gene with no overlap with a known gene. This resulted in count data for StringTie-assembled gene models.
For this data set, we filtered genes that did not exceed 1 CPM in at least three samples. After filtering, 17,711 genes remained.
Each gene was tested for differential expression between WT and Armc5 KO adrenal glands with EdgeR Likely Ratio Tests. Since the augmented Pol II pool resulting from Armc5 deletion might affect the transcription of all genes in the KO adrenal glands, we set normalization factors for each sample as the ratio between the log2CPM value of Rn7sk of the sample to the average of all log2CPM values of Rn7sk across all samples. Rn7sk is transcribed by Pol III and is thus independent of the putative influence of the Pol II pool size. A similar level of Rn7sk expression in WT and KO adrenal glands is shown in Supplementary Figure S3. This normalization was used instead of using edgeR::calcNormFactors, which uses a trimmed mean of M-value normalization by default.
The heatmaps were constructed using R pheatmap. The volcano plots, pie charts, and bar plots were produced using R v3.6.3. ggplot2. The violin plot was generated by Raincloud Plots (44).
RT-qPCR
Total RNA was extracted by RNeasy kit with on-column DNase A digestion (Qiagen) and reverse-transcribed with SuperScript IV Reverse Transcriptase (Invitrogen). PowerTrack™ SYBR Green Master Mix (ThermoFisher) was used with fast cycling mode (2-min of enzyme activation at 95°C, 40 cycles of 5-s denaturation at 95°C, 30-s annealing, and extension at 60°C) on a 7500 Fast Real-time PCR System (ThermoFisher). Rn7sk was the internal control. Six pairs of mouse biological samples were analyzed for each gene. The ΔΔCT method was applied to the analysis of CT values. The paired two-way Student's t-tests were used for mouse samples and unpaired two-way Student's t-tests for human samples. The primer sequences are found in Supplementary Table S2.
Chromatin immunoprecipitation and sequencing (ChIP-seq)
The adrenal glands from 8- to 12-month-old WT and KO female mice were resected and stored in liquid nitrogen immediately until use. The ChIP method was adapted from Cotney and Noonan's protocol (45). Each gland was homogenized in 200 μl ice-cold PBS buffer with a handheld Polytron homogenizer. The final volume was brought to 1 ml with ice-cold PBS. The homogenized samples were crosslinked with 66.7 μl 16% formaldehyde (1% final) at room temperature for 15 minutes. They were quenched with 107 μl 1.25 M glycine (0.125 M final) at room temperature for another 10 minutes in rotating tubes. The samples were centrifuged, and the pellets were washed twice with ice-cold PBS. The crosslinked pellets were re-homogenized with the Polytron homogenizer and suspended in 300 μl cell lysis buffer (50 mM Tris (pH 8.0), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and incubated on ice for 20 min to release nuclei. The nuclei were harvested by centrifugation, resuspended in 200 μl ChIP sonication buffer (10 mM Tris (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 0.5% SDS), and incubated on ice for 20 min. The nuclei were sonicated with a probe-based sonicator (FB120 with a CL-18 probe; ThermoFisher) at a 20% amplitude setting. The sonication was conducted using 15-s pulses at 15-s intervals for a total of 8 min. The sonicated nuclei were harvested by centrifugation, and then diluted with 800 μl ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris (pH 8.1), 167 mM NaCl) to reach the final SDS concentration of 0.1%. These samples represented sonicated chromatin ready for immunoprecipitation.
To quantify precipitated DNA and assess the size of the fragmented DNA, we used a rapid de-crosslinking protocol (45) by treating 5% of the sonicated nuclei (50 μl/sample) with 10 μg of RNase A for 15 min at 37°C followed by 20 μg of proteinase K for 30 min at 65°C. They were de-crosslinked for 5 min at 95°C. DNA was extracted with the QIAquick PCR Purification Kit (Qiagen). DNA concentration was determined with a Nanodrop 1000 Fluorospectrometer (ThermoFisher). We used electrophoresis to confirm DNA fragment sizes were 200–800 bp in length.
For the remaining 95% chromatin to be used in ChIP-seq, an equal amount of sonicated chromatin, based on their prior DNA measurements, of different samples was incubated with anti-RPB1 N-terminal domain Ab (D8L4Y) (1:100) at 4°C overnight, followed by 40-μl magnetic protein G beads (Bio-Rad) for another 2 h at 4°C. The beads were rinsed with wash buffer (100 mM Tris (pH 8.0), 500 mM LiCl, 1% NP-40, 1% deoxycholic acid) five times and then with TE buffer once. The chromatin was eluted with elution buffer (50 mM Tris (pH 8.0), 10 mM EDTA, 1% SDS) at 65°C for 10 min. The immunoprecipitated chromatins were de-crosslinked at 65°C overnight with NaCl adjusted to 200 mM. The chromatins were then treated with 10 μg RNase A/sample at 37°C for 1 h, followed by 200 μg proteinase K/sample for 2 h at 45°C. DNA was purified with QIAquick PCR Purification kit and quantified by the Bioanalyzer (Agilent).
Libraries were prepared robotically with 0.2–2 ng of fragmented DNA ranging 100–300 bp in length, using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs), as per the manufacturer's recommendations. Adapters and PCR primers were purchased from Integrated DNA Technologies. Size selection was carried out using SparQ beads (Qiagen) prior to PCR amplification (12 cycles). Libraries were quantified using the Kapa Illumina GA with Revised Primers-SYBR Fast Universal kit (Kapa Biosystems). The average size of the fragments was determined using a LabChip GX (PerkinElmer) instrument.
The libraries were normalized and pooled and then denatured in 0.05 N NaOH and neutralized using HT1 buffer. The pool was loaded at 225 pM on an Illumina NovaSeq S4 lane using Xp protocol as per the manufacturer's recommendations. The run was performed for 2×100 cycles (paired-end mode). A phiX library was used as a control and mixed with libraries at 1% level. Each library was sequenced at 25 million reads. Base-calling was performed with RTA v3. Program bcl2fastq2 v2.20 was then used to de-multiplex samples and generate fastq reads.
ChIP-seq reads were first trimmed for adaptor sequences and low-quality score bases using Trimmomatic (41). The resulting reads were mapped to the mouse reference genome (GRCm38) using BWA-MEM (46) in paired-end mode at default parameters. Only reads that had a unique alignment (mapping quality > 20) were retained, and PCR duplicates were marked using Picard tools. Peaks were called and annotated using MACS2 (47) and HOMER (48) software suites, respectively.
To assess differences in Pol II occupancy patterns between WT and KO samples, we obtained ChIP-seq read counts within the following genomic regions using HOMER: the promoter region (from TSS (transcription starting site) –400 bp to TSS +100 bp), gene body (from TSS +100 bp to TES (transcription ending site) –100 bp), region downstream of the gene body (from TES –100 bp to TES +2000 bp), the 5’ untranslated region (5’UTR), intron, 3’UTR, the enhancer region (from TSS –5000 bp to TSS –400 bp), the region from –10 000 bp to TSS, the region from TSS to +10 000 bp, and the intergenic region. Since the RPB1 levels in the KO tissues were elevated, we speculated that there would be more Pol II association with the genes, hence higher RPB1 ChIP signal in the KO promoter regions than in the WT counterparts. Therefore, genes that lacked RPB1 ChIP-seq signal in the promoter region in the KO tissues were filtered out, as these genes were believed to have no signals in WT tissues either. Raw counts were normalized using edgeR’s TMM algorithm (49) and were then transformed to log2 counts per million (log2CPM) using the Voom function implemented in the Limma R package (50).
To construct the global metagene Pol II-binding profile, normalized read counts (Fragments per Kilobase of transcript per Million Mapped reads (FPKM) of a full gene length plus 2000-bp flanks (TSS –2000 bp to TES +2000 bp) were obtained from all the genes that passed the filtering. Both flanks were divided into 20 equal-sized bins of 100 bp each. The gene bodies were scaled to 60 bins for the full gene length. FPKM was calculated from BAM input files using ngs.plot (51) with the following parameters: -G mm10 -R genebody -D ensembl -FL 200 -BOX 0 -SE 1 -VLN 0 -LWD 2 -WD 9. These global metagene Pol II binding profiles were only for visualization of differences in Pol II density, and customarily inferential statistics was not conducted for such profiling.
The peak count versus distance (–10 kb to +10 kb from TSS) profile was generated from 51 equal-sized bins of 400 bp for this region of all the genes that passed filtering. This was meant to give an overall view of the genomic location of all the peaks, and again, inferential statistics were not conducted.
To test for differential Pol II density in WT and KO tissues, we used the R package DESeq2 (52) to analyze the raw counts of the promoter region, gene body, and the region downstream of the gene body. Differential expression analysis of DESeq2 is based on the Negative Binomial (a.k.a. Gamma-Poisson) distribution. Genes with the threshold of 5% FDR were analyzed.
Genome browser tracks were created with the HOMER makeUCSCfile command and bedGraphToBigWig utility from UCSC. Tracks were normalized so that each value represented the read count per base pair per 10 million reads. UCSC Genome Browser (http://genome.ucsc.edu/) was implemented for track visualization.
Patient cohort information
All patients provided written informed consent to the retention of adrenal tissue for research purposes. For the Adelaide and Montreal cohorts, respectively, the study was approved by the Royal Adelaide Hospital Human Research Ethics Committee and the Ethical Committee at Centre hospitalier de l’Université de Montréal (CHUM) (SL05-054).
The Adelaide cohort
PBMAH patients III-1, III-2 and III-3 were three male siblings from the first Australian kindred we previously reported (33,53). Briefly, III-1 presented with advanced Cushing's syndrome due to PBMAH. Despite bilateral adrenalectomy, he died from complications of advanced Cushing's syndrome. His two siblings, III-2 and III-3, self-presented for evaluation and were found to have mild hypercortisolism. Both underwent single adrenalectomy, at the most recent evaluation, remain eucortisolaemic. Their age, gender, and diagnosis are shown in Table 1. Additional clinical and laboratory findings of these three PBMAH patients were detailed previously (53). These PBMAH patients were genotyped by whole-exome sequencing, and ARMC5 mutations were subsequently confirmed by Sanger sequencing (33). They all carried the same heterozygous Chr16:g.31476121; c.1777C→T germline ARMC5 mutation resulting in R593W. For controls, we used adrenal adenomas from patients with ACTH-independent Cushing's (n = 2), a large adrenal incidentaloma (n = 1), hyperplastic adrenal glands from patients with primary aldosteronism (n = 2), and two normal adrenals, one (N-1) from normal adrenal adjacent to the adenoma (patient PA-03, Table 1) and the second (N–2) from a patient undergoing nephrectomy for renal cell carcinoma.
The Montreal cohort
PBMAH patients E35 and E202 were a father and daughter that were described previously (31,54). They had Cushing's syndrome secondary to PBMAH, and their cortisol secretion was beta-adrenergic/vasopressin sensitive. These PBMAH patients were genotyped by whole-exome sequencing, and ARMC5 mutations were subsequently confirmed by Sanger sequencing. They both carried the heterozygous germline pathogenic variant in the ARMC5 gene c.327_328insC, (p.Ala110Argfs*9) (Table 2). Patient E191 was a 47-year-old man with PBMAH co-secreting cortisol and aldosterone with clinical and biochemical Cushing's syndrome and primary aldosteronism. The patient also had a 2.8 cm mass in the pancreatic tail that was resected, and histopathology was compatible with a pancreatic neuroendocrine tumor. This patient carried a heterozygous germline ARMC5 deletion of exons 5–8. The deletion is predicted to prematurely truncate the protein product and cause loss of function. The adrenal gland adenomas from three patients with primary aldosteronism and one patient with adrenocortical carcinoma co-secreting cortisol and androgens were used as controls (Table 2). Two PBMAH patients without ARMC5 mutation were included as additional controls. All the tissue samples were stored at –80°C until use.
Table 2.
Clinical information and genotype of the Montreal cohort
| Patient ID | Age at operation | Gender | Diagnosis | Histopathology | Germline ARMC5 mutation status |
|---|---|---|---|---|---|
| E35 | 56 | M | Cushing's syndrome | PBMAH | c.327_328insC, (p.Ala110Argfs*9) |
| E191 | 37 | M | Cushing's syndrome and primary aldosteronism | PBMAH | Deletion of exons 5 to 8 |
| E202 | 44 | F | Cushing's syndrome | PBMAH | c.327_328insC, (p.Ala110Argfs*9) |
| B148 | 46 | F | Primary aldosteronism | Adrenal adenoma | not tested |
| B193 | 47 | M | Primary aldosteronism | Adrenal adenoma | not tested |
| B17 | 55 | F | Primary aldosteronism | Adrenal adenoma | not tested |
| B183 | 77 | M | Cushing's syndrome and hirsutism | Adrenocortical carcinoma | not tested |
| B206 | 54 | F | Cushing's syndrome and Primary aldosteronism | PBMAH | WT |
| E58 | 33 | F | Cushing's syndrome | PBMAH | WT |
For RPB1 expression analysis, PBMAH nodules were obtained from resected adrenal gland macronodules of PBMAH patients with germline ARMC5 mutations. Adrenal adenomas and adrenocortical carcinoma tissues not known to have ARMC5 mutations were used as controls. Two PBMAH samples with confirmed WT ARMC5 were included as additional controls. The clinical diagnosis and histopathology of the patients are indicated. PBMAH: primary bilateral macronodular adrenal hyperplasia. WT: wild type.
RESULTS
ARMC5 physically interacted with CUL3 and RPB1
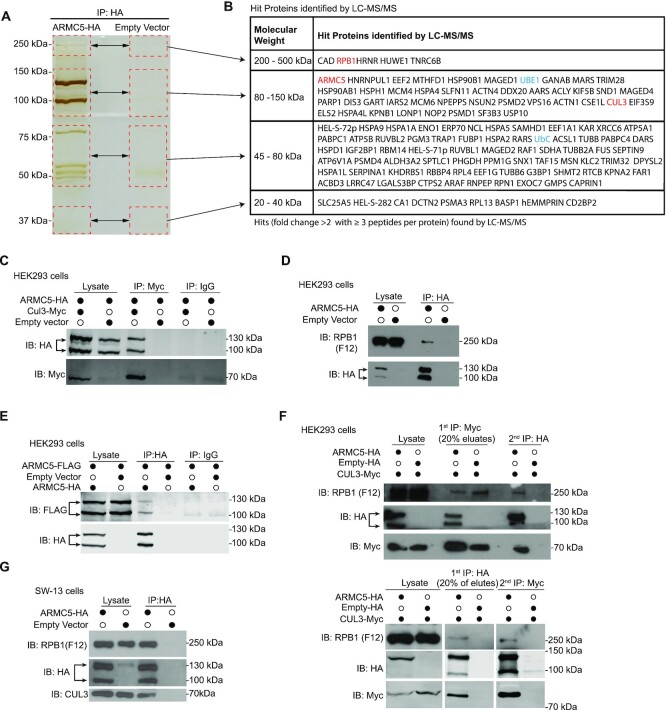
Our Y2H assay showed that CUL3, RPB1 , and ARMC5 itself are potential binding partners of ARMC5 (37). Several additional methods were used to confirm this. HEK293 cells were transfected with a plasmid expressing human ARMC5-HA, and ARMC5 was affinity-precipitated with anti-HA Ab and resolved by SDS-PAGE (Figure 1A). The bands in the gel were analyzed with LC–MS/MS. The experiments were conducted as three biological replicates. Protein hits satisfying both the following conditions in any of the 3 biological replicates are listed in Figure 1B. (i) Three or more peptides corresponding to the protein were detected, and (ii) the number of the peptides in the test sample was >2-fold larger than that in the controls. Two proteins, that is, RPB1 and CUL3 identified in Y2H (37), were also found to associate with ARMC5 in this LC–MS/MS analysis. CUL3 binds with a RING-finger protein RBX1 and forms a multiple-unit RING-finger E3, using a protein containing a BTB domain as its substrate recognition subunit (19). We hypothesized that ARMC5, which harbors a BTB domain at its C-terminus, was the substrate recognition subunit of a novel RPB1-specific multiple-unit RING-finger E3. Consistent with this hypothesis, some components of the ubiquitination system, such as an E1 (UBE) and ubiquitin (UBC) (shown in blue in the list), were also found in ARMC5 co-precipitates. Detailed information about these hits, including the number of peptides representing a given protein and the fold change of the number of these peptides in the test versus control samples, is provided in Supplementary Table S3, in which a less stringent criterion was employed. Proteins were included in this list if they had two or more peptides detected and had a 2-fold higher number of peptides in the test sample than that in the controls. This allows a more comprehensive appreciation of the LC–MS/MS results.
Figure 1.
ARMC5 forms a complex with CUL3, RPB1, and itself. (A) Silver staining of ARMC5 precipitates resolved by SDS-PAGE. Lysates of HEK293 cells transfected with ARMC5-HA-expressing constructs were precipitated with anti-HA Ab. The precipitates were resolved by SDS-PAGE. The regions (rectangles with dashed lines) with visible bands in the test sample and the corresponding positions in the empty vector-transfected lane were excised and were analyzed by LC–MS/MS. Three independent experiments were conducted, and a representative gel with silver staining is shown. (B) Proteins found in the ARMC5 precipitates according to the LC–MS/MS analysis. Proteins met with both the following two conditions in any of the biological replicates (200-500 kDa: duplicates; 80–150 kDa: duplicates; 45–80 kDa: triplicates; 30 kDa: no replicate) were listed. 1) The protein had equal or more than three peptides corresponding to its sequence in the ARMC5-HA transfected sample; 2) the number of the peptides in the ARMC5-HA-transfected sample was more than 2-fold larger than that in the empty vector control. The gel pieces from which the proteins were derived were indicated. (C) ARMC5 interacted with CUL3. HEK293 cells were transfected with plasmids expressing ARMC5-HA and CUL3-Myc. Cell lysates were precipitated with anti-HA Ab and immunoblotted with anti-Myc Ab. (D) ARMC5 interacted with RPB1. HEK293 cells were transfected with plasmids expressing ARMC5-HA. Cell lysates were precipitated with anti-HA Ab and immunoblotted with anti-RPB1 N-terminal Ab (clone F12). (E) ARMC5 interacted with itself. HEK293 cells were transfected plasmids expressing ARMC5-HA and ARMC5-FLAG. Cell lysates were precipitated with anti-HA Ab and immunoblotted with anti-FLAG Ab. (F) ARMC5, CUL3, and RPB1 formed tri-molecule complexes. HEK293 cells were transfected with plasmids expressing ARMC5-HA and CUL3-Myc. In the upper panel, cell lysates were first precipitated with anti-Myc Ab and eluted with Myc peptides. The precipitates were then re-precipitated with anti-HA Ab. The secondary precipitates were blotted with anti-RPB1 N-terminus Ab (clone F12). In the lower panel, the order of precipitation was reversed. The lysates were first precipitated with anti-HA Ab. The precipitates were then precipitated with anti-Myc Ab. (G) ARMC5 interacted with endogenous CUL3 and RPB1 in adrenal cortical carcinoma SW-13 cells. SW-13 cells were transfected with plasmids expressing ARMC5-HA. Cell lysates were precipitated with anti-HA Ab. The precipitates were immunoblotted with anti-CUL3 Ab or anti-RPB1 N-terminus Ab (clone F12). In all the experiments, the lysates were also immunoblotted with Abs against HA, MYC, or FLAG, as applicable, to demonstrate the effectiveness of transfection. Empty vectors were used in transfection as controls. IgG was employed in immunoprecipitation as a control. All the experiments were conducted more than three times, and representative results are shown.
Immunoprecipitation was employed to further prove the interaction among ARMC5, CUL3 and RPB1. CUL3-Myc was found in ARMC5-HA precipitates from HEK293 cells transfected with plasmids expressing ARMC5-HA and CUL3-Myc (Figure 1C), and endogenous RPB1 was present in ARMC5-HA precipitates from HEK293 cells transfected plasmid expressing ARMC5-HA (Figure 1D). This confirms that ARMC5 interacts with CUL3 and RPB1, respectively. ARMC5 always showed as two bands at 130 and 100 kDa. This is due to protease cleavage, as will be further explained in the Discussion. We previously showed in the Y2H analysis that ARMC5 bait interacts with ARMC5 prey (37), suggesting that ARMC5 can dimerize with itself. Indeed, ARMC5-FLAG was found in ARMC5-HA precipitates from HEK293 cells transfected with plasmids expressing ARMC5-FLAG and ARMC5-HA (Figure 1E), indicating that ARMC5 partnered with itself, forming homodimers. Using HEK293 cells transfected with plasmids expressing both ARMC5-HA and CUL3-Myc, we first precipitated CUL3-Myc from the cell lysates and then further precipitated ARMC5-HA from the first-round precipitates. CUL3-Myc, ARMC5-HA, and endogenous RPB1 were all found in the second precipitation (upper panel; Figure 1F), indicating that ARMC5, CUL3 and RPB1 formed a tri-molecule complex. It is to be noted that after the first immunoprecipitation, the endogenous RPB1 could be found in the sample without ARMC5-HA overexpression (lane 4). This is because the endogenous ARMC5 is still present in the cells, and it can bridge the endogenous RPB1 with CUL3-Myc. When we precipitated CUL3-Myc using anti-Myc Ab, the endogenous RPB1 was pulled down via the endogenous ARMC5. We also conducted an experiment using a reverse order in immunoprecipitation, that is, ARMC5-HA was first precipitated by anti-HA Ab, and the precipitates were used for the second round of CUL3-Myc precipitation by anti-Myc Ab. RPB1 was also found in the second round precipitates (lower panel; Figure 1F). This further validates our conclusion that ARMC5, CUL3 and RPB1 form a tri-molecular complex.
To alleviate any artifact caused by excessive protein expression in HEK293 cells, we transfected adrenal cortex carcinoma SW-13 cells with plasmids expressing ARMC5-HA. We detected both endogenous CUL3 and RPB1 in the anti-HA precipitates (Figure 1G), proving that ARMC5 was associated with endogenous CUL3 and RPB1 in these cells. In these experiments, ARMC5 was always detected as two bands of 130 and 100 kDa in size in immunoblotting due to proteolysis. Due to the poor specificity of all the anti-ARMC5 Abs currently available, we were not able to confirm the interactions among endogenous ARMC5, RPB1 and CUL3.
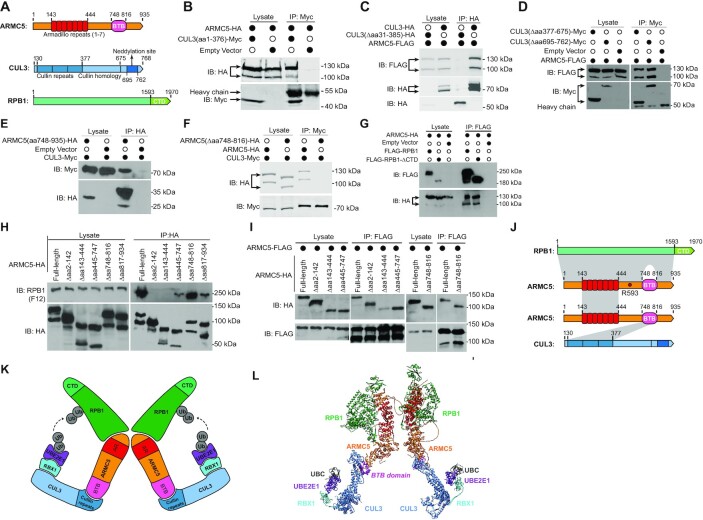
Identification of regions of interaction in ARMC5, CUL3 and RPB1 molecules
Human ARMC5 contains an ARM domain at its N-terminus (aa 143–444) and a BTB domain at its C-terminus (aa 748–816) (Figure 2A). CUL3 has 3 Cullin repeats in its N-terminus (aa 30–377), followed by a Cullin homology domain (aa 378–675). In its C-terminus, there is a neddylation site spanning aa 695–762. We created deletion mutations of both molecules to identify regions of their interaction. A CUL3 deletion mutant containing only the cullin repeats plus the 30-aa N-terminal segment (CUL3(aa 1–376)-Myc; Figure 2B) could still bind to full-length ARMC5-HA (Figure 2B). Conversely, a CUL3 mutant with deletion of the cullin repeats plus its following 9-aa (CUL3(Δaa 31–385)-HA) was no longer bound to full-length ARMC5-FLAG, although the full-length CUL3-HA did (Figure 2C). On the other hand, CUL3 with the cullin homology domain deleted (CUL3 (Δaa 377–675)-Myc) or with the C-terminal neddylation site deleted (CUL3(Δaa 695–762)-Myc) still associated well with ARMC5 (Figure 2D). These deletion studies demonstrate that CUL3 uses its cullin repeats in its N-terminus to interact with ARMC5. A short ARMC5 C-terminal segment containing the BTB domain (ARMC5(aa 748–935)-HA) was sufficient to precipitate down CUL3-Myc (Figure 2E). Further, CUL3-Myc could precipitate the full-length ARMC5-HA but not the mutant ARMC5-HA (ARMC5(Δ748–816)-HA) with the BTB domain deleted (Figure 2F). These data show that the BTB domain in ARMC5 is necessary and sufficient for ARMC5 to associate with the cullin repeats of CUL3. In immunoprecipitated samples, due to higher salt concentration, the proteins often moved more slowly and appeared at a higher position than their counterparts in the lysates, and such retardation was more obvious for molecules of smaller molecular weight and over abundance, such as ARMC5(aa748–935) in lane 3 of panel 2E.
Figure 2.
Identification of the regions of interaction in ARMC5, CUL3, and RPB1 molecules. (A) Schematics of key domains of human ARMC5, CUL3 and RPB1 molecules. (B) The cullin repeats of CUL3 interacted with ARMC5. HEK293 cells were transfected with plasmids expressing CUL3 N-terminal cullin repeats (CUL3(aa1–376)-Myc) and full-length ARMC5-HA. The lysates were precipitated with anti-Myc Ab and blotted with anti-HA and anti-Myc Abs. (C) CUL3 with cullin repeats deleted no longer interacted with ARMC5. HEK293 cells were transfected plasmids expressing either full-length CUL3 or CUL3 with culling repeats deleted (CUL3(Δaa31–385)-HA) and full-length ARMC5-HA. The lysates were precipitated with anti-HA Ab and blotted with anti-FLAG and anti-HA Abs. (D) The CUL3 cullin homology domain and its C-terminal fragment were not necessary for interaction with ARMC5. HEK293 cells were transfected plasmids expressing CUL3 with the cullin homology domain deleted (CUL3(Δaa377–676)-Myc) or with its C-terminal sequence, including the neddylation site deleted (CUL3(Δaa 695–762)-Myc), and full-length ARMC5-FLAG. The lysates were precipitated with anti-Myc Ab and blotted with anti-FLAG and anti-Myc Abs. (E) The ARMC5 C-terminal sequence containing the BTB domain was sufficient to interact with CUL3. HEK293 cells were transfected plasmids expressing CUL3-Myc and the ARMC5 C-terminal sequence containing the BTB domain plus the following 119-aa sequence (ARMC5(aa748–935)-HA). The lysates were precipitated with anti-HA Ab and blotted with anti-HA or anti-Myc Abs. (F) ARMC5 without BTB domain no longer bound to CUL3. HEK293 cells were transfected with plasmids expressing CUL3-Myc and BTB domain-deleted ARMC5-HA (ARMC5(Δaa748–816)-HA). The lysates were precipitated with anti-Myc Ab and blotted with anti-HA or anti-Myc Abs. (G) The CTD of RPB1 was not essential for the association between RPB1 and ARMC5. HEK293 cells were transfected plasmids expressing ARMC5-HA and full-length RPB1 (FLAG-RPB1) or RPB1 with its CTD deleted (FLAG-RPB1-ΔCTD). The lysates were precipitated with anti-FLAG Ab and blotted with anti-HA or anti-FLAG Abs. (H) The N-terminal sequence (aa2–142) before the ARM domain (aa143–444) and the ARM domain of ARMC5 were both needed for RPB1 binding. The sequence after the ARM domain and before the BTB domain, and the sequence after the BTB domain also contributed to RPB1 binding but to a lesser degree. HEK293 cells were transfected plasmids expressing HA-tagged ARMC5 deletion mutants as described. The lysates were precipitated with anti-HA Ab and blotted with anti-RPB1 (clone F12) or anti-HA Abs. (I) ARMC5 interacted with ARMC5 through their ARM domains (positions aa 143-444). FLAG-tagged full-length ARMC5 was transfected into HEK293 cells along with HA-tagged full-length ARMC5 or deletion mutants. The lysates were immunoprecipitated with anti-FLAG Ab and blotted with anti-HA or anti-FLAG Abs. (J) A schematic showing the regions that contributed to the interaction among RPB1, ARMC5, and CUL3. The grey shading between the molecules represents the interaction regions. The lighter shade between RPB1 and ARMC5 indicates a lesser contribution of the regions to the association between these two molecules. The position of ARMC5 R593, which is mutated in Adelaide PBMAH patients, is indicated. (K) A 2D schematic of the novel dimeric RPB1-specific E3. (L) A 3D model of the novel dimeric RPB1-specific E3. In all the experiments, empty vectors were used in transfection as controls. The lysates were also immunoblotted to confirm that the transfected proteins were present. All the experiments were conducted more than three times, and representative results are shown.
We next investigated the interaction between ARMC5 and RPB1. We obtained an RPB1 mutant with its C-terminal repeats deleted (FLAG-RPB1-ΔCTD; Addgene). FLAG-RPB1-ΔCTD could precipitate ARMC5-HA as efficiently as the full-length FLAG-RPB1 (Figure 2G), suggesting that the RPB1 sequence upstream of the CTD was essential for ARMC5 binding. ARMC5 mutants with deletions of the following regions were generated: the N-terminal sequence (ARMC5(Δaa 2–142)-HA) before the ARM domain; the ARM domain (ARMC5(Δaa 143–444)-HA); the sequence between the ARM domain and BTB domain (ARMC5(Δaa 445–747)-HA); the BTB domain (ARMC5(Δaa 748–816)-HA); and the C-terminal sequence after the BTB domain (ARMC5(Δaa 817–935)-HA). These mutants, as well as full-length ARMC5-HA, were expressed in HEK293 cells. ARMC5(Δaa 2–142)-HA and ARMC5(Δaa143–444)-HA could not pull down endogenous RPB1, while the full-length ARMC5-HA and the rest of the mutants could (Figure 2H). This indicates that the ARM domain and the sequence before it at the N-terminus are essential for RPB1 binding. We noticed that ARMC5 mutants with the deletions of the sequence between the ARM domain and BTB domain (ARMC5(Δaa 445–747)-HA) and the sequence after the BTB domain (ARMC5(Δaa 817–935)-HA) were less effective in pulling down RPB1, compared to the full-length ARMC5-HA (Figure 2H). Although the ARMC5 with the BTB domain deleted could pull down RPB1, it was somewhat less effective than the WT ARMC5 (Figure 2H). It is possible that these regions all contribute to RPB1 binding, albeit to a lesser extent.
The essential regions needed for ARMC5 and ARMC5 homologous interaction were assessed by ARMC5 deletion mutants ARMC5(Δaa 2–142)-HA, ARMC5(Δaa 143–444)-HA, ARMC5(Δaa 445–747)-HA, and ARMC5(Δ aa748–816)-HA (Figure 2I). The full-length ARMC5-FLAG could precipitate well the full-length ARMC5-HA and the other HA-tagged deletion mutants, except that it could only weakly precipitate the mutant with the aa 143–444 deletion, which corresponded to the ARM domain, indicating that this domain contributes significantly to the dimeric interaction between two ARMC5 molecules.
It is to be noted that the binding assays, as illustrated in Figures 1 and 2, are qualitative in nature. They are only designed to determine whether the molecules concerned interact with each other, regardless of their post-translational modifications (e.g. phosphorylated versus non-phosphorylated RPB1).
The interacting regions between RPB1 and ARMC5, CUL3 and ARMC5, and ARMC5 and ARMC5 are depicted in Figure 2J.
CUL3 is known to interact with a RING-finger protein RBX1, the enzymatic component of the multiple subunit RING-finger E3s, as previously reported (55). We confirmed this in HEK293 cells (data not shown). This RPB1–ARMC5–CUL3–RBX1 complex has the necessary features of an RPB1-specific multiple subunit RING-finger E3, with ARMC5 as the substrate recognition unit. We later demonstrated that an E2 UBE2E1 was functional for this RPB1-specific E3 according to E2 profiling and in vitro ubiquitination assays. A 2D schematic is depicted in Figure 2K and illustrates the proposed structure of the dimeric RPB1–ARMC5–CUL3–RBX1–UBE2E1–UBC complex based on our results and literature. We extracted X-ray crystallographic 3D information of the components in the complex, that is, RPB1, CUL3, RBX1, UBE2E1, and UBC from the Protein Database (56–59). The 3D structure of ARMC5 has not been resolved but is predicted by AlphaFold (39). A 3D model of this E3 complex was constructed (Figure 2L), providing a better visual perspective. Whilst the 3D structure of each component is reliable, the contour of the complex is speculative.
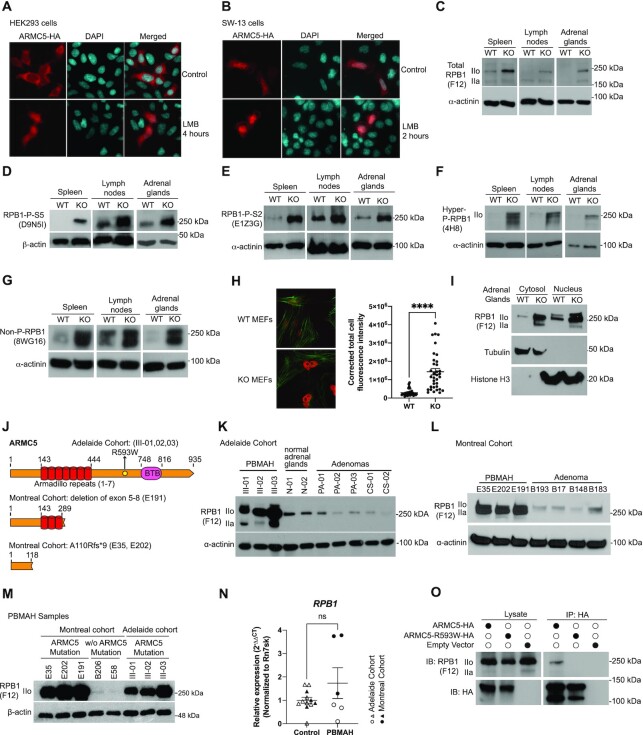
Armc5 KO led to an accumulation of RPB1 in normal organs under a physiological condition and in adrenal glands of PBMAH patients
RPB1 is mainly a nuclear protein. Its E3 should therefore also have a nuclear presence. We previously reported that when ARMC5 was overexpressed in HEK293, ARMC5 signals were found mainly in the cytosol (37). While this still holds true, we found that in the presence of the nuclear export blocker leptomycin B, ARMC5 was easily detectable in the nuclei (Figure 3A). This suggested that ARMC5 did enter the nuclei, but that there was also active shuttling of ARMC5 between the cytosol and nuclei. We also transfected ARMC5-HA-expressing plasmid into human adrenal cortex carcinoma SW-13 cells (Figure 3B). In these cells, ARMC5 was detected both in the cytosol and nuclei in the absence of leptomycin B. However, the presence of leptomycin B enhanced ARMC5 signals in the nuclei. These data indicate that ARMC5 is ubiquitous in the nuclei, but in different cell types, there are different ARMC5 shuttling dynamics between the cytosol and nuclei, resulting in different degrees of distribution of ARMC5 molecules between these two cellular compartments. The detection of intranuclear ARMC5 is dependent on the equilibrium between its import into and export from the nucleus. In SW-13 cells, the ARMC5 import to the nucleus is likely faster than its export, so it could be detected without an export inhibitor. In the HEK293 cells, its export is likely faster than import, so we cannot observe ARMC5 in the nuclei unless the export is blocked.
Figure 3.
ARMC5 KO or mutation led to RPB1 accumulation. (A and B) ARMC5 was presented in both the cytosol and nuclei of HEK293 cells (A) and human adrenal carcinoma SW-13 cells (B). Both types of cells were transfected with plasmids expressing ARMC5-HA. The cells were harvested after 36 h and were stained with anti-HA Ab (pseudo-red) and DAPI (pseudo-cyan). In some cultures, nuclear export inhibitor leptomycin B (LMB; 20 nM) was present for the last 2 or 4 h of culture, as indicated. (C–G) Accumulation of RPB1 in KO tissues. The spleen, lymph node, and adrenal gland protein of KO and WT mice were assessed by immunoblotting for total RPB1 (C); mAb clone F12 against the N-terminal sequence of RPB1), RPB1 with phosphorylated S5 in CTD (D); mAb clone D9N5I), RPB1 with phosphorylated S2 in CTD (E); mAb clone E1Z3G), hyper- and hypo-phosphorylated RPB1 (F); mAb clone 4H8), and non-phosphorylated RPB1 (G): mAb clone 8WG16). β-actin or α-actinin was blotted as a loading control. (H) Elevated RPB1 protein (red) expression in the nuclei of KO MEFs, according to immunofluorescence using rabbit anti-RPB1 Ab (D8L4Y) followed by Alexa Fluor™ 555-conjugated goat anti-rabbit IgG. Filamentous actin (green) was stained with Alexa Fluor™ 488 Phalloidin. Representative micrographs are shown on the left. A bar graph on the right shows the means ± SD of corrected total cell fluorescent intensity (CTCF), which is derived from RPB1 signals of 35 WT and 35 KO MEFs from three independent experiments. ****P < 0.0001 (unpaired two-way Student's t-test). (I). Augmented total RPB1 levels (as determined by F12 mAb) in both the cytosolic and nuclei fractions of KO adrenal glands. Cytosolic tubulin and nuclear histone H3 were used as fraction purity and loading controls. (J) Schematics of ARMC5 mutations in the Adelaide and Montreal cohorts. Patient ID numbers are indicated in the parentheses. (K and L) Elevated RPB1 protein expression in the adrenal macronodules from Adelaide (K) and Montreal (L) PBMAH cohorts with germline ARMC5 mutations. Adrenal adenomas or normal adrenals were employed as controls as indicated. Immunoblotting was performed using mAb (clone F12) against total RPB1 protein. (M) Normal RPB1 protein levels in PBMAH adrenals without ARMC5 mutations. The RPB1 protein levels of adrenal macronodules from two PBMAH patients without ARMC5 mutation (Montreal cohort) were compared to that of six adrenal macronodules from PBMAH patients with germline ARMC5 mutations (Montreal and Adelaide cohorts). (N) RPB1 mRNA levels of the Adelaide and Montreal PBMAH adrenal samples with ARMC5 mutations were similar to those of the controls (adrenal adenomas and normal adrenals). (O) The ARMC5 R539W mutation found in the Adelaide cohort resulted in reduced RPB1 association. HEK293 cells were transfected with plasmids expressing WT ARMC5-HA or ARMC5-R539W-HA. Their association with endogenous RPB1 was detected by immunoblotting after immunoprecipitation.
RPB1 is heavily modified by phosphorylation of the S2 and S5 residues in its C-terminal domain (CTD), which contains multiple 7-aa long repeats (60). RPB1 with different degrees of phosphorylation could be detected by different Abs (61). Total RPB1 (detected by anti-RPB1 N-terminus mAb clone F12, Figure 3C), RPB1 with CTD S5 phosphorylation (detected by mAb clone D9N5I, Figure 3D), RPB1 with CTD S2 phosphorylation (detected by mAb clone E1Z3G, Figure 3E), RPB1 with both high and low phosphorylation of its CTD (detected by mAb 4H8, Figure 3F), and un-phosphorylated RPB1 (detected by mAb clone 8WG16, Figure 3G) were all increased according to immunoblotting in the Armc5 KO lymphoid organs (the spleen and lymph nodes) and adrenal glands, compared to their WT counterparts. Immunofluorescence showed that the nuclear RPB1 level in Armc5 KO mouse embryonic fibroblasts (MEFs) was also augmented (Figure 3H). Such accumulation of RPB1 with different CTD phosphorylation was also observed in all other mouse KO organs tested (i.e. the thymus, liver, kidney, lung, brain, heart, stomach, colon, and small intestine) (Supplementary Figure S4). These results suggest that this E3 acts on RPB1 regardless of its phosphorylation status. As RPB1 phosphorylation is a dynamic process, we cannot exclude the possibility that this E3 only targets RPB1 of a given type of phosphorylation, but due to the consequent RPB1 pool size increase, RPB1 of all the other phosphorylation statuses are affected.
We fractionated the nuclei and cytosolic RPB1 of the KO and WT adrenal glands. The RPB1 levels in both fractions from the KO tissue were elevated (Figure 3I), suggesting that this E3 is active in both these cellular compartments. With that said, since Pol II is assembled in the cytosol and translocated to the nuclei, we cannot completely rule out the possibility that this E3 acts in the cytosol and the accumulated RPB1 in the cytosol spills over to the nuclei.
ARMC5 germline mutations predispose patients to PBMAH. We assessed the RPB1 expression in the adrenal glands of PBMAH patients from the two cohorts, one from Adelaide, Australia, and one from Montreal, Canada, which were described in the Materials and Methods. The ARMC5 mutations of patients in these cohorts are depicted in Figure 3J. In the Adelaide cohort, the three affected siblings all carried the same missense germline C→T point mutation at Chr16:g.31476121, resulting in an R593W mutation in the ARMC5 protein sequence. The R593W mutation was in a region between the ARM domain and BTB domain.
In the Montreal cohort, the heterozygous germline variant in the ARMC5 gene c.327_328insC (p.A110Rfs*9) caused a frameshift starting from the N-terminal region before the ARM domain and resulted in an early truncation and a lack of functional ARMC5 protein (Figure 3J). Another PBMAH patient, E191, from the cohort harbored a heterozygous germline ARMC5 deletion of exons 5–8. This deletion started in the middle of the ARM domain and resulted in a truncation of all the downstream ARMC5 sequences and hence a lack of functional ARMC5.
RPB1 protein levels in the resected PBMAH nodular adrenal tissues with ARMC5 mutations of the Adelaide cohort (Figure 3K) and Montreal cohort (Figure 3L) were all elevated compared to those in the adrenal adenomas and normal adrenal glands. Intriguingly, only the PBMAH samples with ARMC5 mutation had elevated RPB1, while two PBMAH samples without ARMC5 mutation showed RPB1 levels similar to control adenomas (Figure 3M). This indicated that RPB1 accumulation was not a general feature of PBMAH, but rather a result of ARMC5 mutation. RPB1 mRNA expression was similar between PBMAH tissues and controls (Figure 3N), indicating that the upregulation of the RPB1 protein occurred at the post-transcriptional level, likely due to compromised degradation since we found that ARMC5 was the substrate recognition subunit of the novel RPB1-specific E3. We assessed how the ARMC5 mutations found in the patients affected ARMC5 binding to RPB1. Compared to WT ARMC5, ARMC5 with R593W mutation (Adelaide cohort) had a significantly reduced association with the endogenous RPB1 in HEK293 cells (Figure 3O). According to our deletion studies, R593 was located in a region that contributed to the association between RPB1 and ARMC5 (Figure 2H and I). Thus, this mutation resulted in an altering of the interaction between ARMC5 and RPB1. These results highlight the relevance of our in vitro and in vivo data in the KO mice to human pathophysiology and indicate that the novel E3 is indeed essential for maintaining RPB1 homeostasis, hence Pol II pool size, in humans in the absence of massive DNA damage.
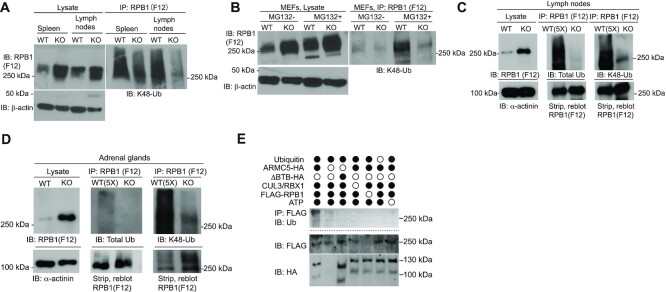
ARMC5-CUL3 was an RPB1-specific E3 according to in vivo and in vitro ubiquitination
The increased RPB1 protein levels in Armc5 KO tissues suggested that ARMC5-CUL3-RBX1 might be an E3 responsible for RPB1 ubiquitination, which is a necessary step to channel RPB1 to the proteasome for degradation. We analyzed the ubiquitination of the endogenous RPB1 in the KO spleen and lymph nodes (Figure 4A) and MEFs (Figure 4B). Although the RPB1 protein levels in these KO tissues were all increased, their K48-linked RPB1 ubiquitination was reduced, indicating that ARMC5 is essential for such RPB1 ubiquitination. When WT MEFs were cultured in the presence of a proteasome inhibitor MG132, their RPB1 ubiquitination was drastically augmented (Figure 4B), suggesting that ubiquitinated RPB1 is usually channeled to the proteasome for degradation. The RPB1 ubiquitination in the KO MEFs was only marginally increased in the presence of MG132 compared to that without the inhibitor, suggesting that in the absence of this putative RPB1-specific E3, RPB1 ubiquitination is very limited, even after the degradation blockage. The slight increase of RPB1 ubiquitination KO MEFs in the presence of MG132 suggests the existence of other RPB1-specific E3(s), which could ubiquitinate RPB1 but to a much lesser extent.
Figure 4.
ARMC5-CUL3-RBX1 as an RPB1-specific E3 based on in vivo and in vitro ubiquitination. (A and B) Reduced K48-linked RPB1 in the KO spleen and lymph nodes (A) and MEFs (B). MEFs were cultured in the absence or presence of proteasome inhibitor MG132 (10 μM) for the last 4 h of culture. β-Actin was blotted for lysate loading control. (C and D) Reduced total and K48-linked RPB1 ubiquitination in KO lymph nodes (C) and adrenal glands (D). Tissue proteins were precipitated with anti-total RPB1 mAb (F12) and immunoblotted with Abs against K48-linked ubiquitin or total ubiquitin, as indicated. α-actinin was blotted for lysate loading control. In (C and D), 5-fold (5×) more WT lysates than the KO counterpart were used as input for immunoprecipitation to detect the weak WT RPB1 ubiquitination signals, using a limited amount of anti-RPB1 Ab during the immunoprecipitation. A similar amount of RPB1 protein in the WT and KO precipitates was shown by immunoblotting. (E) ARMC5-CUL3-RBX1 was a novel RPB1-specific multiple subunit RING-finger E3 according to in vitro ubiquitination assays. Different recombinant proteins, as indicated, were added to the in vitro ubiquitination assay in the presence of E1, E2 (UBE2E1) and ATP. ARMC5 with BTB domain deletion was used as an additional control. The reaction product was immunoprecipitated with anti-FLAG Ab followed by magnetic protein G beads. The immunoprecipitates were blotted with anti-Ub Ab to detect RPB1 ubiquitination. The flow-through of the immunoprecipitation was blotted to confirm the presence of RPB1-FLAG and ARMC5-HA using Abs against these molecules. All the experiments were conducted three times or more, and representative results are shown.
The RPB1 protein level in WT adrenal glands was extremely low. This made the detection of RPB1 ubiquitination in this WT tissue difficult. To circumvent this, we therefore used 5-fold more input protein of the WT tissue than the KO tissue during the immunoprecipitation, along with a limited amount of anti-RPB1 Ab. This allowed us to compare the ubiquitination of a similar amount of precipitated RPB1 proteins in the WT and KO tissues. The validity of this method was first confirmed in lymph nodes because the WT lymph nodes had a reasonable RPB1 signal to be detected for ubiquitination without the equimolar comparison (Figure 4A). The result revealed that on an equimolar RPB1 basis, RPB1 from the KO lymph nodes had drastically lower total ubiquitination as well as K48-linked ubiquitination (Figure 4C), and this result was compatible with that of the equal protein input method. This equal molar RPB1 input method was then applied to the adrenal glands. We showed significantly reduced total and K48-linked ubiquitination of RPB1 in the KO adrenal glands (Figure 4D).
The gold standard to prove E3 activity is the in vitro ubiquitination assay, in which a substrate is ubiquitinated in vitro by a reconstituted ubiquitination enzyme cascade of E1, E3 and E3. The reconstituted ubiquitination enzymatic cascade comprises ARMC5, CUL3, RBX1, E1, E2 (UBE2E1), ATP and WT ubiquitin. This E3 effectively ubiquitinated RPB1 (Figure 4E). A mutant ARMC5 with BTB domain deletion (ARMC5-ΔBTB), which rendered the mutant incapable of binding CUL3, failed to ubiquitinate RPB1 in this system. This finding indicates that ARMC5 is part of a novel RPB1-specific multi-subunit RING-finger E3, and ARMC5 depends on its BTB domain to interact with CUL3 to form a functional E3 complex.
Armc5 KO resulted in increased transcription of a large number of genes in the adrenal glands
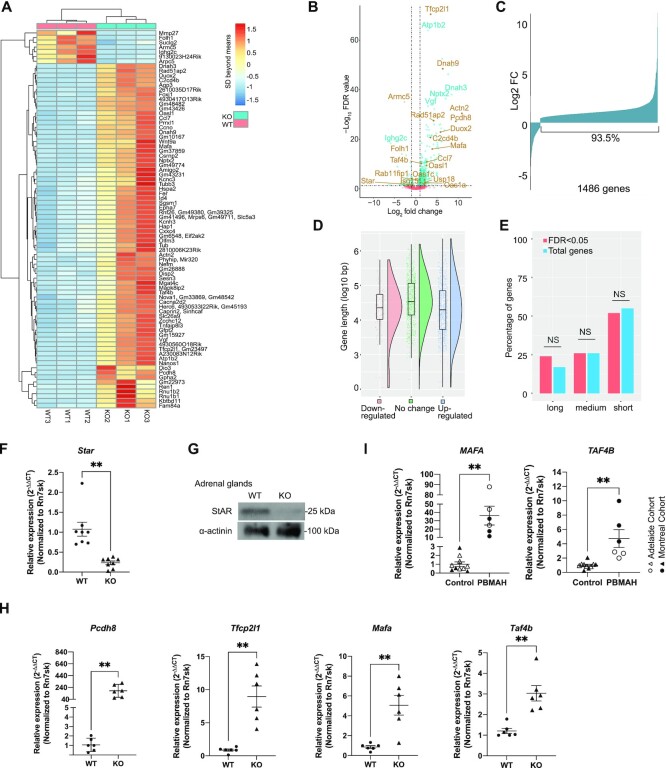
To understand how an enlarged Pol II pool due to ARMC5 deletion affects the transcriptome, we conducted RNA sequencing (RNA-seq) of the WT and KO adrenal glands. The read counts were normalized against Rn7sk RNA, which was transcribed by Pol III and was not subjected to a putative general influence by abnormal levels of Pol II. Indeed, Rn7sk levels in KO and WT adrenal samples had no significant difference (Supplementary Figure S3). A threshold for gene-level significance of < 5% FDR was applied to the paired comparison of RNA-seq results from three KO and three WT adrenal glands. After filtering out nominal genes that were generated by GenPipe but were not presented in the mouse reference genome (GRCm38 release 97), we obtained 1486 genes with significantly different expressions between KO and WT adrenal glands. These genes are listed in Supplementary Table S4, along with their FDRs, fold changes, and raw readcounts. Eighty genes in this list with the lowest FDRs are shown in a heatmap (Figure 5A). A volcano plot illustrates the fold change and FDR of these 1486 genes, with several prominently changed ones annotated (Figure 5B). Armc5 was among the downregulated genes, as expected (Figure 5B and Supplementary Figure S5a). It still had some signal due to transcripts appearing in the undeleted gene body (Supplementary Figure S5b), although these transcripts would not produce any functional proteins due to frameshifts or early stops. The volcano plot shows that there were more up- than down-regulated genes. This is more clearly depicted in a bar graph (Figure 5C). Among the 1486 genes with FDR <0.05, 1389 genes (93.5%) were upregulated. Only a small fraction (97 genes; 6.5%) was downregulated. The gene length of the upregulated, downregulated, and unchanged genes showed no significant difference (Figure 5D). We arbitrarily divided the upregulated genes into short (< 30 kb), medium-sized (≥30 kb and <100 kb), and long (≥100 kb) ones. Most upregulated genes were short. However, their percentage (50.2%) among all the upregulated genes was similar to that of short genes in the genome (55%) (Figure 5E). Therefore, the enlarged Pol II pool does not specifically favor the transcription of short genes.
Figure 5.
RNA-seq of WT and KO adrenal glands. (A) A heatmap of differentially expressed genes. Eighty genes with the lowest FDR were shown. For a given gene, color represents SDs beyond the mean of raw reads in all the six samples (3 KO and 3 WT) tested. (B) A volcano plot of FDR and expression fold changes (FC) of all the genes detected in the KO versus WT adrenal glands according to RNA-seq. Some prominently changed ones are annotated, and dysregulation of genes indicated in brown was confirmed by RT-qPCR. Dashed horizontal and vertical lines represent FDR = 0.05 and log2FC = ±1, respectively. (C) Fold changes of expression for 1486 genes with FDR < 0.05 in the KO versus WT adrenal glands. (D) The length distribution of the genes with significant upregulation or downregulation or without change. The box graphs show the median (solid horizontal line in the box), the 75th percentile (upper part of the box), 25th percentile (lower part of the box), 95th percentile (upper whisker), 5th percentile (lower whisker), and outliers (dots beyond the 95th and 5th percentile) of the gene length of each group. The violin plots illustrate the gene size distribution of each group. (E) The length of the upregulated genes was similar to those in the whole genome. The percentages of short (<30 kb), medium (≥30 kb and <100 kb), and long (≥100 kb) genes of the significantly upregulated genes (FDR < 0.05) and the percentages of genes of these sizes in all the genes in the genome are shown. No significant difference was observed in any length category (P > 0.05; χ2 test). (F) Reduced Star mRNA levels in the KO adrenal glands according to RT-qPCR. (G) Reduced STAR protein levels in the KO adrenal glands according to immunoblotting. A representative blot from three replicates is shown. (H) Elevated mRNA levels of tumor suppressor genes (Pcdh8 and Tfcp2l1) and oncogenes (Mafa and Taf4b) in the mouse KO adrenal glands, according to RT-qPCR. (I). Elevated mRNA levels of oncogenes MAFA and TAF4B in the human PBMAH adrenal gland samples according to RT-qPCR. Genes in (H) and (I) were selected from the ones with significant upregulation in the mouse KO adrenal glands according to RNA-seq. The signal ratios of the test genes versus Rn7sk were presented. Means ± SEM are shown. **P < 0.01; ***P < 0.001 (paired two-way Student's t-tests for mouse samples and unpaired two-way Student's t-test for human samples).
PBMAH patients have inappropriately regulated and increased cortisol levels. The large mass of the nodular adrenals can result in Cushing's syndrome. However, the glucocorticoid biogenesis per cell in the hypertrophic adrenal cortex is reduced (36). STAR is a rate-limiting enzyme in steroidogenesis, regulating cholesterol transfer in the mitochondria (62). RNA-seq revealed that the Star mRNA level was significantly reduced in the KO adrenal gland (Supplementary Table S4). This was confirmed by RT-qPCR (Figure 5F), and reduced STAR protein level in the KO adrenal glands was demonstrated by immunoblotting (Figure 5G). The reduced STAR expression in the Armc5 KO is likely to be a key contributor to the reduced cortisol production by each adrenocortical cell.
Among 1486 differentially expressed genes in the Armc5 WT and KO adrenal gland, we selected those involved in tumorigenesis and subjected them to further RT-qPCR confirmation. The results are presented in Supplementary Figure S6. Four of the confirmed genes are known tumor suppressors (e.g.Pcdh8 and Tfcp2l1) or oncogenes (Mafa and Taf4b), and their validation by RT-qPCR is shown in Figure 5H. The upregulation of two oncogenes at the mRNA level (MAFA and TAF4b) was also validated in the hyperplastic adrenal glands of PBMAH patient samples with ARMC5 mutations (Figure 5I). These findings raise an interesting possibility that these genes are among the effector genes that cause adrenal gland hypertrophy and PBMAH.
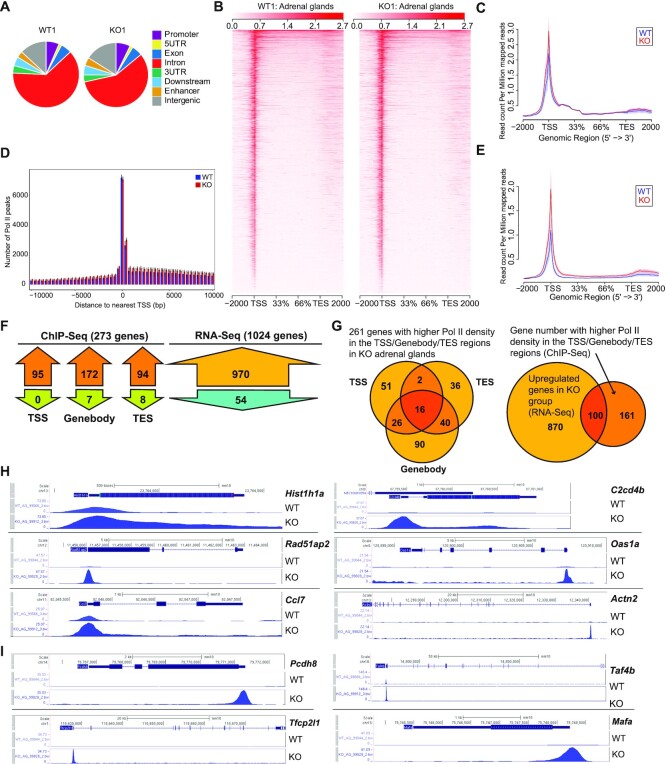
The higher Pol II density in genes of KO adrenal gland cells was not a sign of stalling and did not cause a general decrease in transcription
The accumulation of RPB1 in KO cells raised the question of whether it was part of stalled Pol II due to failed degradation. We conducted an anti-total RPB1 mAb F12 ChIP-seq of the adrenal gland, which was analyzed along with RNA-seq data to address this question. RPB1 signals in ChIP-seq were used customarily as a surrogate marker of Pol II peak density in the genes (11). A total of 12,718 genes showed discernible ChIP-seq signals. The distribution of Pol II peaks in different regions of genes is illustrated in Figure 6A. In both the KO and WT adrenal glands, the introns had the highest peak number, followed by intergenic regions and then the promoter regions. Representative CPM heatmaps for the region from –2000 bp upstream of TSS to +2000 bp downstream of the transcription ending site (TES) of all genes in one pair of WT and KO samples are illustrated in Figure 6B. Within the genes, the highest normalized RPB1 readcounts (readcount per million mapped reads (CPM)) were accumulated near the TSS (Figure 6C). In this metagene analysis (Figure 6C), no visually discernible difference in Pol II peak density was observed between the KO and WT adrenal glands. We also conducted anti-phospho-S2 RPB1 Ab ChIP-seq, which detected Pol II peaks mainly in the gene body and TES, and the results revealed no significant difference between WT and KO adrenal glands either, as expected (data not shown). However, for the mAb F12 ChIP-seq in a fixed region analysis in which the Pol II density of all the genes for the region spanning from –10 kb to +10 kb surrounding the TSS was measured, the KO tissue had a slightly higher Pol II density across this region according to visual inspection (Figure 6D). We also carried out a metagene analysis of the RPB1 CPM distribution of 970 upregulated genes found in KO adrenals according to RNA-seq. As shown in Figure 6E, the KO tissue had higher RPB1 CPM across the TSS, gene-body, and TES regions, supporting our hypothesis that failed RPB1 degradation in the KO tissues does not cause Pol II stalling but rather increases transcription in a subset of genes. We will elaborate on this further later. It is to be noted that such metagene illustration (Figure 6C–E) is not suitable for statistical analysis and is only meant for visual appreciation. The statistical analysis results are presented below.
Figure 6.
RPB1 ChIP-seq of WT and KO adrenal glands. (A) Pol II peak distribution in different gene regions of a representative pair of KO and WT samples. (B) Heatmaps of the normalized readcounts based on data from a representative pair of KO and WT adrenal glands. (C) Means (solid lines) ± SE (shadows) of normalized readcounts (readcounts per million mapped reads) in a metagene analysis for a genomic region from –2 kb of TSS to +2 kb of TES. Data were based on three biological replicates for WT and KO pairs. (D) Pol II peak distribution (mean ± SE) in a fixed region from –10 kb upstream to +10 kb downstream of TSS. (E) Means (solid lines) ± SE (shadows) of normalized readcounts in a metagene analysis (from –2 kb of TSS to +2 kb of TES) of 970 genes that were upregulated in the KO adrenal glands. (F and G) Combined analysis of RNA-seq and ChIP-seq. The numbers of genes with significantly dysregulated Pol II density in different gene regions (TSS, gene body and TES) are shown in the left panel of (F) Please note that one gene could have dysregulation in more than one region. The numbers of genes showing significant up- or down-regulation according to RNA-seq and at the same time with detectable signals in ChIP-seq are shown in the right panel of (F). The locations (i.e. TSS, gene body, and TES) of the significantly different Pol II peak density of 261 genes with increased Pol II density are illustrated in the left panel of (G). The overlap of genes with higher Pol II density and mRNA upregulation is depicted in the right panel of (G). (H) ChIP-seq readcount tracks in the gene regions of six genes with a prominent difference in Pol II density in KO versus WT adrenal glands. (I) ChIP-seq readcount tracks showing increased Pol II density in four genes with upregulated mRNA. The tracks were normalized so that each value was proportional to the readcount per base pair per 10 million reads.
Two hundred and seventy-three of the 12 718 genes that had ChIP-seq signals showed a statistically significant difference (FDR < 0.05) in Pol II density in the KO versus WT adrenal glands (95 genes (all increased) in the TSS region (from TSS –400 bp to TSS +100 bp); 179 genes (172 increased and seven decreased) in the gene body region (from TSS +100 bp to TES –100 bp (transcription ending site)); and 102 genes (94 increased and eight decreased) in the TES region (from TES –100 bp to TES +2000 bp) (left panel, Figure 6F; Suuplmentary Tables S5–S7). Thus, for those 273 genes with FDR <0.05, the majority of them (26) in the KO group presented increased Pol II density. The increased Pol II peak density in different gene regions (i.e. TSS, gene-body, or TES) for the 261 genes with upregulated Pol II peak density are shown in Figure 6G (left panel).
RPB1 ChIP-seq only provided a snapshot of the location of Pol II during the dynamic transcription process. To assess whether these Pol IIs were actively transcribing or stalled, we conducted a combined ChIP-seq and RNA-seq data analysis. In the KO adrenal glands, among the 1486 dysregulated genes according to RNA-seq, 1024 of them (69%) had detectable ChIP-Seq signals, 970 being upregulated and 54 downregulated (right panel, Figure 6F). The upregulated genes tended to have higher Pol II peak density in different regions of the genes, as shown in Figure 6E. The P-values, FCs and FDRs of the ChIP-seq signals of these genes in the TSS, gene body, and TES regions are presented in Suuplmentary Tables S8–S10.
Conversely, among the 261 genes with higher Pol II density, approximately one-third were upregulated (100/261, 38%; right panel, Figure 6G). On the other hand, none of the genes with an increased Pol II density presented decreased mRNA levels (data not shown).
The higher Pol II density of six genes with concomitant mRNA upregulation in the KO adrenal glands is illustrated in Figure 6H. The obviously increased Pol II density in the TSS region (all the six genes), in the gene body (all the genes), and in the TES region (Hist1h1a, Oas1a and Ccl7) could be appreciated by visual inspection.
Four genes (Pddh8, Tfcp2l1, Mafa and Taf4b) with functions related to tumorigenesis had confirmed mRNA upregulation (Figure 5H) accompanied by significantly higher Pol II density in their genes (Figure 6I).
These results suggest that the increased Pol II density favors upregulation of a subset of genes, some of which might be effector genes causing adrenal gland hypertrophy in the KO mice and PBMAH in patients.
This ChIP-seq did not employ control spike-in DNA to normalize inter-sample variation and increase assay sensitivity. As a consequence, this ChIP-seq might not have sufficient sensitivity to detect those genes with low-level differences between the WT and KO samples.
DISCUSSION
In the present study, we show that ARMC5 forms a multiple-subunit E3 ligase complex with CUL3 and RBX1 that targets RPB1, the largest subunit of RNA pol II, for degradation. This E3 was largely responsible for RPB1 ubiquitination in all the tissues tested under physiological conditions. Due to compromised RPB1 ubiquitination and the subsequent degradation via the proteasome pathway, Armc5 deletion caused RPB1 accumulation in all the major organs in KO mice. Similar RPB1 accumulation was also found in the hyperplastic adrenal tissues of PBMAH patients with ARMC5 mutations. Such accumulation likely resulted in an enlarged Pol II pool size. We did not find any evidence of generalized Pol II stalling or a generalized decrease of mRNA transcription in the KO adrenal glands. However, a subset of genes had altered expression, with most being upregulated. It is plausible that some of these affected genes are effectors that cause adrenal hyperplasia in the KO mice and in PBMAH patients with ARMC5 mutations.
ARMC5-CUL3-RBX1 was a primary RPB1-specific E3 under a physiological condition
Several RPB1-specific E3s have been reported in yeast and mammalian cells. Rsp5, an E3 ligase in yeast, polyubiquitinates RPB1 with an S5 hypo-phosphorylated CTD (63,64). Rsp5 also mono-ubiquitinates Rpb1, and then a second E3 Elc1/Cul3 adds polyubiquitin K48-linked chains to Rpb1 (21). NEDD4, a mammalian homolog of yeast Rsp5, cooperates with Elongins/CUL5 and catalyzes polyubiquitination via a two-step reaction (21). However, in the HEK293 cells, the ubiquitination of RPB1 catalyzed by NEDD4 only occurs when they are UV irradiated (20). CRL4-CSA has been suggested as an RPB1-specific E3 in irradiated cells (11). Its E3 activity for RPB1 at K1268 has been proven by in vitro ubiquitination (65). Two RPB1-specific E3 for cell lines without exogenous DNA damage have been reported. pVHL-EloB/EloC-CUL2-RBX1 is specific for RPB1 with a hyperphosphorylated CTD, and its activity is found in unperturbed PC12 cells based on pVHL anti-sense knockdown (26). WWP2, a HECT E3, ubiquitinates the RPB1 CTD domain in the absence of exogenous DNA damage (23). However, this is only proved in vitro in F9 embryonic carcinoma cells, in which WWP2 siRNA knockdown leads to increased total RPB1 as well as RPB1 containing S2 or S5 hyperphosphorylation of the CTD. WWP2 was also shown to ubiquitylate RPB1 in response to DNA breakage in a manner that requires DNA-PK (12). The activity of these RPB1-specific E3s highly depends on RPB1 CTD phosphorylation. WWP2 (23,24) and pVHL (22) KO mice have been generated, but there is no report on the possible RPB1 accumulation in the organs of these mice. In theory, an E3 acting on RPB1 in normal tissues or organs should exist.
The novel ARMC5-CUL3-RBX1 E3 we discovered in this work is a constitutive RPB1-specific E3 under a physiological condition because when its function was compromised by ARMC5 deletion or mutation, there was a considerable accumulation of RPB1 protein in most tissues and organs without exogenously induced DNA damage or stress. The phosphorylation of S2 and S5 in the RPB1 C-terminal domain repeats is related to the location of RPB1 in the genes (66). In the absence of ARMC5-CUL3-RBX1 E3, the degradation of RPB1 with hyper- or hypo-phosphorylated S2 or S5 in the CTD or unphosphorylated RPB1 was all compromised (Figure 3), resulting in significant accumulation of all these RPB1 species. As RPB1 constantly transforms between these different phosphorylation states, we have not determined whether the E3 targets one, a few, or all the forms of RPB1. It is interesting that even in organs where ARMC5 expression is low, e.g., the liver and heart (37), the absence of this E3 still caused drastically increased RPB1 levels. Thus, this E3 likely plays a significant role in maintaining Pol II pool size homeostasis in all the organs and tissues under a physiological condition. We found that the cytosolic RPB1 level in the KO tissue was also increased (Figure 3I), suggesting that this E3 is also involved in degrading misassembled Pol II or misfolded RPB1 under a physiological condition. Due to the vital roles of RPB1 and Pol II in cell biology, other RPB1-specific E3s might provide the much-needed redundancy to allow cells to survive when this major ARMC5–CUL3–RBX1 E3 is dysfunctional or inadequate. However, they cannot fully compensate for the dysfunction of this E3, as evidenced by the accumulation of RPB1 in the KO organs and tissues.
K48-linked ubiquitination ushers proteins to the proteasome for degradation (67). The KO lymphoid organs and adrenal glands showed reduced K48-linked RPB1 ubiquitination (Figure 4A–D). These findings are compatible with augmented RPB1 levels in these tissues.
Using tagged ARMC5 overexpression in HEK293 cells or SW-13 is an effective method of detecting the interaction of ARMC5 with other proteins and identifying interaction regions of ARMC5 with other proteins in a qualitative way. However, this system is not effective at assessing ARMC5’s function on RPB1 levels, which is expected to decrease. Such a decrease was not observed in SW-13 cells (Figure 1G) and was only occasionally found in HEK293 cells at a very moderate degree (Figure 1D and F). This is explained by low plasmid transfection efficiency. The transfection efficiency in SW-13 cells was ∼3%. Therefore, 97% of the SW-13 cells did not have ARMC5 overexpression, and RPB1 levels in these cells were unchanged (Figure 1G). Even if the 3% transfected cells have all their RPB1 destroyed by the overexpressed ARMC5, the 3% decrease of RPB1 in the lysates cannot be detected amongst 97% unchanged background. The transfection efficiency in HEK293 cells was better at about 20–30%. The 70-80% background noise explains why only a slight decrease of RPB1 levels in HEK293 cells overexpressing ARMC5 could occasionally be noticed.
The structure of ARMC5–CUL3–RBX1 E3 and its accessory molecules
Through a series of deletion mutations, we established that ARMC5 utilized its BTB domain to interact with the cullin repeats of CUL3. The ARM domain and the preceding N-terminal sequences were essential for ARMC5’s association with the N-terminal region before the CTD of RPB1. However, the remaining sequences after the ARM domain except the BTB domain in ARMC5 also contributed to RPB1 binding, but to a lesser extent. The critical component, a RING-finger protein RBX1, of this multi-unit RING-finger E3, is well known to bind to CUL3 (68,69). These four molecules formed a basic ensemble. We demonstrated previously by Y2H that ARMC5 is able to homodimerize. We showed that the ARM domain in ARMC5 was critical for its dimerization, according to the deletion study. It is possible that this basic 4-molecule ensemble exists as dimers, as illustrated in Figure 2K, or even exists in polymeric form. We constructed a 3D model of the complex (Figure 3L) based on information extracted from the Protein Database and computer modeling. This model needs to be confirmed and detailed in the future by structural biology studies, especially in the context of this E3 in association with Pol II. We will then have a better understanding of how this dimeric E3 functions and a better answer to the following questions. (i) Does this dimeric E3 (or polymeric E3) target only one RPB1 or different RPB1s in the vicinity simultaneously? (ii) Even more intriguingly, will this dimeric or polymeric E3 simultaneously target other subunits of the Pol II or even other components of the transcription machinery such as those in the pre-initiation complex?
There should be other obligatory components attached to or near this E3 complex for its E3 enzymatic activity, such as E1, E2 and ubiquitin. We indeed found E1 (UBE1) and ubiquitin (UbC) in the ARMC5 immunoprecipitates according to LC–MS/MS analysis (Figure 1B). No E2 was found in the ARMC5 immunoprecipitates, but this was not unexpected, as they might not have survived the immunoprecipitation due to lower affinity to the E3 complex.
In this E3 complex, RBX1 is the catalytic subunit, and CUL3 is the scaffold to bridge the catalytic subunit to the substrate recognition subunit. RBX1, in the presence of E1, E2, ubiquitin and ATP, is likely capable of ubiquitinating anything in its vicinity, including CUL3 and RBX1. Indeed, CUL3-RBX1 auto-ubiquitination has been documented (70). The function of ARMC5 as a substrate recognition subunit is probably to draw a substrate (e.g. RPB1) close enough to the catalytic subunit and retain it there long enough to be ubiquitinated. Since ARMC5 is also near the RBX1 catalytic subunit, it should be ubiquitinated by this E3 as well. Such ARMC5 ubiquitination was recently reported by Cavalcante et al. (71). Whilst ARMC5 could be considered a substrate of RBX1-CUL3, a more accurate characterization of such ARMC5 ubiquitination is auto-ubiquitination of this E3 complex. Enzymes are not usually regarded as a substrate even if it acts on itself because an enzyme needs to work on a different molecule (with a few exceptions such as Lck autophosphorylation) to propagate a signaling cascade or execute a function. Therefore, we envisage that RPB1 but not ARMC5 is a true substrate of this novel ARMC5–CUL3–RBX1 E3.
Each E3 often has more than one substrate (72). Since the ARM domain in an ARM-containing protein can often dock different proteins (73), it follows that ARMC5 may be able to serve as a recognition subunit for several different substrates. Thus, in addition to RPB1, this novel ARMC5–CUL3–RBX1 E3 might have other substrates pending discovery. Potential other substrates could be hits found in our LC–MS/MS analysis of ARMC5 precipitates from HEK293 cells. A total of 164 hits with more than 2-fold presence than that in the controls. Among these, some could be additional substrates recognized by ARMC5. Dysfunction of these additional putative substrates and RPB1 might collectively contribute to Armc5 KO/mutation phenotype in mice and humans.
ARMC5 isoforms and cleavage products
ARMC5 has eight isoforms at the mRNA level (27). Among them, six encode proteins. The dominant isoform ARMC5-201 encodes a peptide of 935 aa in length. This isoform is expressed in most tissues, albeit at different levels. In our study, the cDNA encoding this 935-aa peptide was employed for transfection. The expressed exogenous ARMC5 always appeared as two bands at 130 and 100 kDa. This size is bigger than the calculated size based on mRNA length, probably due to posttranslational modifications. The relative intensity of these two bands varied in different experiments (Figures 1C–E, and 2D). This suggests that the lower band is a protease degradation product rather than a peptide translated from a second ATG downstream in the 935-nt mRNA. Since the HA and FLAG tags are at the C-terminus, according to size calculation, this cleavage site is near the end of the ARM domain (aa 143–444). As expected, when this domain was deleted, the 100-kDa band no longer existed (Figure 2H, the third lane of the left panel). However, with the deletion of aa143–444 or aa445–747, there appeared a new band of about 50 kDa in size, likely due to the exposure of a new protease cleavage site (Figure 2H, third and fourth lanes). These results suggest that in the cells, a fraction of ARMC5 (30–50%) was cleaved in two. The cleaved N-terminal part contained the RPB1-binding site, and the C-terminal part, the CUL3-binding BTB domain. These fragments can no longer serve as an E3 substrate recognition unit, as they cannot bring RPB1 close to CUL3-RBX1. Instead, they might act as negative competitors to interfere with the interaction of the full-size ARMC5 with RPB1 and CUL3.
Decreased RPB1 degradation did not cause Pol II stalling
Transcription by Pol II is a dynamic process. It can frequently pause due to damage of the template DNA, defective Pol II assembly, or stress in growth conditions (74). In quiescent cells at the G0 phase, some Pol IIs are paused at promoter-proximal regions of many genes, and they need to be nudged along to increase the transcription rate to allow cells to enter the G1 phase (75). The paused Pol II will continue its journey once DNA damage is repaired, the quiescent cell status is changed, or the stress is relieved (11–13). If the pause persists, for the transcription to start or to resume, data suggest that the Pol II has to be destroyed by the proteasomes after being ubiquitinated (5,7–13). If the major machinery for Pol II degradation was dysfunctional, as was the case in Armc5 KO or ARMC5 mutations, we expect to see extensive Pol II stalling, and consequently, a general decrease of mRNA transcription. However, this did not happen in the KO adrenal glands.
Among 12 718 genes with detectable anti-total RPB1 ChIP-seq signals, only 261 genes in the KO tissue had a significantly higher Pol II density. A caveat of this ChIP-seq observation is that with our experimental methods, it is likely that only genes with significantly changed Pol II density are detected. With a better normalization approach, we might be able to detect more genes with increased Pol II density. However, it remains true that there was no general transcription-hampering Pol II stalling in the KO tissue, as no gene with higher Pol II density presented lower mRNA levels, and no generally decreased transcription was observed. We also conducted anti-P-S2 ChIP-seq, which mainly detects RPB1 in the genebody. No obvious Pol II stalling was detected (Supplementary Figure S7) either, similar to that found using anti-total RPB1 ChIP-seq.
There are two possible non-mutually exclusive explanations for such an unexpected result. Most of our knowledge related to Pol II stalling and the role of the proteasome system in resolving such stalling was derived from experiments where significant DNA damage was induced (74) or from experiments conducted in vitro (76,77). It is possible that the ubiquitination/proteasome system is not required at all for removing the stalled Pol II in vivo. This is the case in yeast (78,79). Recently, two groups reported that K1268 ubiquitination is responsible for UV-induced RPB1 degradation by the proteasome (11,13). In HEK293 cells with RPB1 K1628R mutation, in spite of failed RPB1 degradation after UV irradiation, Pol II still comes off from the damaged DNA sites, and computer modeling suggests it recycles normally (11). This is consistent with our finding that there was no apparent Pol II stalling in the absence of this major RPB1-specific ARMC5–CUL3–RBX1 E3. The primary function of this novel E3 is probably to maintain the homeostasis of the Pol II pool size and to degrade misfolded or misassembled RPB1. Another possible explanation is that under a physiological condition, Pol II stalling is an insignificant event. Hence, the task of removing the stalled Pol II, although vital, is light. E3 redundancy is in place, as manifested by the existence of several other RPB1-specific E3s (11–12,21,23,26). Although most of the other E3s play a minor role without massive DNA damage, in the absence of ARMC5, they are probably sufficient to carry out the light-duty of removing the stalled Pol II.
Another surprising observation is that the accumulation of RPB1 was correlated to significantly increased mRNA levels of a large number of genes (1,389) in the adrenal glands, while only a small number of genes (97) had reduced mRNA levels (Figure 5C). Such a heavily skewed expression pattern has rarely been seen in RNA-seq datasets with other gene deletions or mutations. Although the steady-state mRNA level is determined by the balance of mRNA transcription and degradation, in most cases, it reflects the rate of mRNA transcription. It is, therefore, reasonable to assume that in the KO adrenal glands, the heavily biased expression pattern towards the augmentation for a subset of genes was due to a generally increased transcription rate. The increased transcription is probably due to a bigger Pol II pool in the KO cells. In supporting this hypothesis, Vidakovic et al. reported that the K1268R mutation of RPB1 renders it incapable of being degraded by the ubiquitination/proteasome system in UV irradiated HEK293 (13). Consequently, there is an enlarged Pol II pool in cells after UV irradiation, resulting in increased transcription of >1600 genes but decreased transcription of fewer than 400. Thus, both our results and the results of Vidakovic et al. support the notion that a larger Pol II pool favors enhanced transcription, at least for a subgroup of genes in certain tissues (13).
A larger Pol II pool does not affect all the genes, unlike we would expect intuitively. Instead, only a subgroup of genes, 1,389 out of 18,500 expressed genes in the adrenal glands, showed increased transcription. Vidakovic et al. reported that a large Pol II pool size preferably increases the transcription of short genes after UV irradiation (13). Although in our study, 50.2% of the genes in the upregulated group are short ones, this percentage was not significantly different from that of short genes in the entire genome, in which short genes are predominant. Therefore, gene length is not a factor for the Pol II pool size to influence transcription under an unperturbed condition.
In the KO adrenal glands, among 261 genes with increased Pol II density, 100 (38%) were upregulated at the mRNA level (Figure 6G). What is the mechanism for a larger Pol II pool to upregulate some genes? For the genes with an increase in both mRNA levels and Pol II density, they must have a particular gene sequence to enable more active transcription due to a larger Pol II pool. A possible location of such sequences is the TSS region, where most Pol IIs reside according to ChIP-seq (Figure 6B). About 24% of human genes have a TATA-like element as the TSS in the region (80). Others might have multiple noncanonical Pol II binding sites for transcription initiation (80,81). It is possible that under a condition of excessive Pol IIs, genes with multiple TSSs have an increased transcription rate since they can dock multiple Pol IIs. It will be interesting to identify these Pol II pool size-sensitive noncanonical Pol II binding motifs (transcription initiation sites) in the promoter region. Of course, such augmentation of transcription will likely be subjected to further tissue-specific regulation. Different tissue types had different numbers and different subsets of upregulated genes under the influence of a larger Pol II. For example, in the Armc5 KO mouse embryonic fibroblasts, similar to the KO adrenal, more than 1,000 genes were upregulated, but most of them were different from those upregulated in the adrenal (data not shown). Another example is the upregulated genes in HEK293 cells with RPB1 K1268R mutation (13). The 1,600 upregulated genes, in that case, were mostly different from the 1,389 upregulated ones we observed in the Armc5 KO adrenal glands.
It is more difficult to understand why a larger Pol II pool causes the downregulation of some genes. It is possible that some of the downregulated genes lack other means to remove the stalled Pol II, which exists functionally but is not detected as significant in ChIP-seq after multiple-testing correction. It is also possible that some of these downregulated genes are indirectly influenced by the larger Pol II pool via upregulated ones.
The biological consequence of the Pol II pool size has not attracted much attention until recently (11,13). The increased Pol II pool size could cause abnormal regulation of a subset of genes, and the consequence is often detrimental, as demonstrated in the RPB1 K1268R knock-in mice (11), which have a Cockayne syndrome-like manifestation. These mice and our Armc5 KO mice share some common pathology, such as a significant degree of prenatal lethality, defective neural development (our KO mice had a high incidence of neural tube defects; data not shown), and dwarfism. Based on their cellular studies of the effect of the enlarged Pol II pool caused by RPB K1268R mutation, Vidakovic et al. suggested that “the Pol II pool size might contribute significantly to genome instability disorders (e.g., Cockayne syndrome, Fanconi anemia, Blooms syndrome, and Huntington's disease), and perhaps even to the regulation of cell-type-specific transcription programs in normal cells.” (13). Indeed, our current work proved the biological importance of the Pol II pool size in normal cells and whole animals/humans without irradiation.
ARMC5 mutations and diseases
A general phenotype of the Armc5 KO cells was reduced proliferation. This was observed in T cells (37), MEFs, neural tube cells, and neural progenitor cells (data not shown). The generally reduced proliferation might also be a contributing factor to dwarfism. How do we reconcile this phenotype with adrenal hyperplasia and meningiomas found in PBMAH patients with ARMC5 mutation? It is possible that some upregulated genes caused by the larger Pol II pool are oncogenes. However, such an oncogenic effect is counteracted by reduced cell proliferation, which is caused by another set of anti-proliferation genes (detailed examples will be given later). As a result of these two opposing effects, ARMC5 mutations lead to very slow-growing hyperplasia (adrenal hyperplasia in mice and PBMAH in humans) or tumors (meningiomas in humans). Indeed, both PBMAH and meningiomas take decades to develop and are often detected after 50–60 years of age (28,82).
Our studies of two cohorts from Adelaide, Australia, and Montreal, Canada, were able to show that PBMAH patients with ARMC5 mutations had abnormally high RPB1 levels in their hyperplastic adrenal nodules. This has demonstrated that our findings of ARMC5-CUL3-RBX1 as a novel RPB1-specific E3 are relevant to PBMAH. The three Australian PBMAH kindred had a missense mutation of R593W caused by a point mutation at C1777T in the ARMC5 coding sequence. The mutation is located in a region between the ARM domain and BTB domain. According to our deletion studies (Figure 2H), this region contributes to the association between RPB1 and ARMC5. Based on the 3D structure of ARMC5 predicted by AlphaFold, R593 is located at a hinge region of the ARMC5 molecule. We speculate that this mutation might alter the angulation of ARMC5 and consequently cause a weaker binding between ARMC5 and RPB1. Such reduced interaction was proven in vitro using ARMC5 R593W mutants according to immunoprecipitation (Figure 3O). As a consequence, the E3 became less potent, resulting in inadequate RPB1 ubiquitination followed by RPB1 accumulation. Among many point mutations that occur naturally in ARMC5, only the critical ones will render pathological manifestations, hence becoming clinically apparent. Some PBMAH patients of other cohorts have mutations in the ARMC5 BTB domain (83), which, based on our deletion experiment, is critical for binding with CUL3. It is likely that any critical mutations affecting the interaction between the subunits of this RPB1-ARMC5-CUL3-RBX1 complex will result in compromised RPB1 ubiquitination, which in turn causes RPB1 accumulation and an abnormally larger Pol II pool.
It is known that the PBMAH patients with germline mutations also develop somatic ARMC5 mutations in the hyperplastic adrenal tissues (30). Such biallelic mutations generate a condition in their adrenals akin to the biallelic ARMC5 KO in mice, leading to severely reduced ARMC5 levels, which in turn result in RPB1 accumulation in the adrenals. The monoallelic germline mutations in the patients probably can still produce enough ARMC5 and hence do not cause an obvious phenotype, like heterozygous KO mice, until the second allele is compromised somatically later in life. Since the somatic mutations occur randomly and it takes time to affect critical nucleotides by chance, this can also explain the late onset of the disease.
The adrenal hyperplasia in the KO mice and PBMAH patients is likely caused by increased cortical cell proliferation or reduced apoptosis, or both. We conducted gene ontogeny analysis of genes with significantly different transcription accompanied by upregulated Pol II peak density and validated their dysregulated transcription by RT-qPCR. Some confirmed upregulated genes seem to function as tumor suppressors and are anti-proliferative. For example, Pcdh8 encodes a membrane protein belonging to the cadherin superfamily and is a candidate tumor suppressor in breast cancer (84). Tfcp2l1 encodes a transcription factor that is known to maintain the pluripotency of embryonic stem cells (85). It positively regulates CRYAB protein expression, and its downregulation is associated with thyroid carcinomas (86). They were both highly upregulated (∼200-fold and ∼6-fold, respectively) in KO adrenal glands.
On the other hand, some confirmed upregulated genes can act as oncogenes. Mafa is a case in point. It is a large-Maf family member and has a strong transforming capability in fibroblasts (87). However, its oncogenic activity depends on the cell context, and under some circumstances, it counteracts the oncogenic function of the Ras/Raf/MEK pathway activation (88). This gene was highly upregulated in KO adrenals (5-fold) and greatly so in human PBMAH adrenal nodules (40-fold). TAF4b also belongs to this oncogene category. Its protein is a subunit of the transcription factor IID (TFIID) (89), which is essential for the initiation of transcription by Pol II (90). TAF4b is known to activate anti-apoptotic genes and can thus promote cell survival (91). It was upregulated 3- and 5-fold in the KO mouse adrenal gland and human PBMAH adrenal glands, respectively.
It is possible that the collective effects of these multiple dysregulated genes but not of a single gene resulted in the phenotype observed in the KO mice and patients with ARMC5 mutations. The upregulation of tumor suppressor and anti-proliferative genes such as Pcdh8 and Tfcp2l1 curbs the growth of cells and tumors while elevated levels of oncogenes such as Mafa and Taf4b trigger oncogenesis. The end results depend on the equilibrium of these two opposing forces in different tissues and often lead to gradual hyperplasia or tumors such as PBMAH and meningiomas. The upregulation of the anti-proliferative genes (Pcdh8 and Tfcp2l1 or similar ones) can also explain the generally reduced growth rate of many types of KO cells, such as lymphocytes (37), MEFs, and neural progenitor cells (data not shown). There is no prior literature related to this hypothesis, and further validation is warranted.
Since ARMC5 mutation is associated with PBMAH and meningioma, can it be characterized as a tumor suppressor gene? To be defined as an oncogene or tumor suppressor gene, it should affect cell transformation, proliferation, or death in most cell types and have a clear, immediate mechanism leading to such phenotypes. ARMC5 does not fulfill these criteria. The abnormally large Pol II pool size after ARMC5 deletion or mutation might influence some real effector genes (e.g. upregulating certain oncogenes or tumor suppressor genes), causing cell type-dependent indirect effects, be they anti-proliferative or pro-proliferative, and anti-apoptosis or pro-apoptosis. Thus, despite the eventual cell growth or death caused by ARMC5 KO/mutation, it is inappropriate to categorize ARMC5 as either a tumor suppressor gene or an oncogene per se, for the same reason that we do not consider ARMC5’s downstream target RPB1 as a tumor suppressor or oncogene even Pol II transcribes every tumor suppressor gene and oncogene.
Although ARMC5 mutation is associated with PBMAH risks, only about 25% of PBMAH patients have ARMC5 mutations. What causes PBMAH in the remaining 75% of patients? We found that only in PBMAH patients with ARMC5 mutations the RPB1 protein levels in their adrenal gland nodules were elevated. This finding confirms that ARMC5 mutation results in RPB1 accumulation. It also supports the heterogeneous nature of PBMAH pathogenesis. We propose that for PBMAH patients with ARMC5 mutations, the enlarged Pol II pool size is the cause, although not a direct one, but this hypothesis is yet to be validated. The real culprits might be the downstream effector genes that are dysregulated by the enlarged Pol II pool. Some of these effector genes are upregulated oncogenes such as MAFA and TAF4B (Figure 5I). For those PBMAH patients without ARMC5 mutations, although they have a normal Pol II pool size, their PBMAH might be caused by the mutations of the same effector genes (e.g. MAFA or TAF4B, as alluded above). Thus, precisely identifying the Pol II downstream effector genes in the PBMAH patients with ARMC5 mutations might help identify risk genes for those PBMAH patients without ARMC5 mutations.
Three of our coauthors previously conducted Affymetrix microarray analysis to discover differentially expressed genes in three PBMAH versus two normal adrenal gland tissues (92). The complete dataset has now been deposited in the Gene Expression Omnibus of NCBI (accession #GSE171558). We compared the mouse RNA-seq and human microarray datasets and found 43 genes that were commonly upregulated in the mouse KO and human PBMAH adrenal gland tissues and four genes that were commonly downregulated (Supplementary Table S11). The oncogene TAF4b was among the upregulated ones found in both studies. It will be interesting to investigate TAF4b and other commonly regulated genes for their roles in PBMAH pathogenesis. There are apparent profile differences between the mouse RNA-seq and human microarray data. This is not unexpected due to the following reasons: (a) the sensitivity and specificity of RNA-seq and microarray are different; (b) in the KO mouse tissue, all the cells have bi-allelic Armc5 deletion, while all the PBMAH tissues carry a monoallelic R593W point mutation; (c) in the mouse samples, the whole adrenal gland, including the medulla, was used. Armc5 in all the cell types in the gland was deleted. The human samples were derived from macronodules of the adrenal gland cortex, but they also contained non-malignant cells (e.g. cells in the blood vessel and fibroblasts) without ARMC5 mutations. In both datasets, there were false-positive and false-negative genes. Mafa is one such example. This gene was significantly upregulated in mouse RNA-seq. This prompted us to assess its expression in the PBMAH samples. The RT-qPCR results showed that it was also highly upregulated (25–90-fold) (Figure 5I) in the same three Adelaide PBMAH samples used in the microarray, which did not identify it as significant and d) there is also a species difference.
ARMC5 mutations cause PBMAH accompanied by reduced cortisol biogenesis per cell, although due to the massive adrenal gland hyperplasia, the patients have varying degrees of biochemical hypercortisolism. Among the small number of downregulated genes, Star had particular relevance to the reduced per cell cortisol biogenesis. The protein coded by this gene is a transport protein for cholesterol within the mitochondria and is the rate-limiting enzyme for the biogenesis of glucocorticoids (93). Thus, Star might be one of the effector genes downstream of the enlarged Pol II for cortisol biogenesis. The significantly reduced Star mRNA and protein levels in the KO adrenal gland likely contribute to the reduced cortisol production per cell. Mechanisms by which the large Pol II pool-caused suppression of Star transcription need to be elucidated. There was no significant increase in Pol II density in the Star gene in the KO adrenal glands, as it is absent from the list of genes with significantly different Pol II densities (Supplementary Table S5-7). Possibly the Star downregulation is an indirect effect mediated by some upregulated genes. We cannot, however, exclude the possibility that for this gene, some minimal degree of Pol II stalling occurs, and this gene lacks a degradation-independent Pol II recycling mechanism as alluded to before, resulting in depressed transcription.
The enlarged Pol II pool due to ARMC5 mutation might cause other diseases, some of which are subtle and can only be revealed if carefully examined. The Armc5 KO phenotype in mice can serve as a guide in searching for the cause of such human diseases.
In summary, we discovered a novel and major RPB1-specific E3 that functioned under a physiological condition. The dysfunction of this E3 led to an enlarged Pol II pool, which rendered dysregulation of a subset of genes. Some of these genes might be effectors causing adrenal gland hyperplasia in the KO mice and PBMAH in humans.
DATA AVAILABILITY
The mouse RNA-seq dataset and ChIP-seq dataset have been deposited to the Gene Expression Omnibus of NCBI (accession #GSE169263 (Token: yjchiquobjebnyl) and #GSE169578 (Token: wxyzicsyvrynvqb), respectively). The human PBMAH microarray dataset has also been deposited to the Gene Expression Omnibus of NCBI (accession #GSE171558).
Supplementary Material
ACKNOWLEDGEMENTS
Proteomics analyses were performed by the Center for Advanced Proteomics Analyses, a Node of the Canadian Genomics Innovation Network that is supported by the Canadian Government through Genome Canada. RNA-seq and ChIP-Seq services were provided by the Génome Québec Innovation Centre, Montreal. RNA-seq and ChIP-Seq data analyses were assisted by Canadian Centre for Computational Genomics, Montreal. The authors thank the professionals in these platforms for providing these essential services.
Author contributions: H.L. and J.W. conceived and designed the experiments. L.L., I.B., L.G., X.H., W.S., B.H., M.T., Y.H., J.P., B.C., D.T., H.S. and A.L. performed the experiments. L.L., I.B., L.G., X.H., H.L. and J.W. analyzed the data. L.L., I.B., L.G., A.L., H.L. and J.W. wrote the manuscript.
Notes
Present address: Wei Shi, Department of Neonatology, the Children's Hospital, Zhejiang University School of Medicine, Hangzhou 310003, China.
Present address: Yan Hu, Department of Anesthesiology, State University of New York Downstate Health Sciences University, 450 Clarkson Avenue, Brooklyn, NY 11203-2098, USA.
Contributor Information
Linjiang Lao, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Isabelle Bourdeau, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada; Endocrinology Division, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Lucia Gagliardi, Adelaide Medical School, University of Adelaide, Adelaide, SA, 5000, Australia; Endocrine and Metabolic Unit, Royal Adelaide Hospital, Adelaide, SA, 5000, Australia; Department of Genetics and Molecular Pathology, SA Pathology, Adelaide, SA, 5006, Australia; Endocrine and Diabetes Unit, Queen Elizabeth Hospital, Adelaide, SA, 5011, Australia.
Xiao He, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Wei Shi, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Bingbing Hao, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China.
Minjia Tan, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China.
Yan Hu, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Junzheng Peng, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Benoit Coulombe, Department of Translational Proteomics, Institut de Recherches Cliniques de Montréal, Montréal, Québec, Canada; Department of Biochemistry and Molecular Medicine, Université de Montréal, Montreal, Quebec, Canada.
David J Torpy, Adelaide Medical School, University of Adelaide, Adelaide, SA, 5000, Australia; Endocrine and Metabolic Unit, Royal Adelaide Hospital, Adelaide, SA, 5000, Australia.
Hamish S Scott, Adelaide Medical School, University of Adelaide, Adelaide, SA, 5000, Australia; Department of Genetics and Molecular Pathology, SA Pathology, Adelaide, SA, 5006, Australia; Centre for Cancer Biology, an alliance between SA Pathology and the University of South Australia, Adelaide, SA, 5001, Australia; UniSA Clinical and Health Sciences, University of South Australia, Adelaide, SA, 5001, Australia.
Andre Lacroix, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada; Endocrinology Division, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Hongyu Luo, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
Jiangping Wu, Centre de recherché, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada; Nephrology Division, Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Québec H2X 0A9, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
J.-Louis Lévesque Foundation; Canadian Institutes of Health Research [PJT180284, in part]; Arthritis Society of Canada [SOG-21-0158]; Natural Science and Engineering Research Council of Canada [PGPIN-2017-04790 to J.W.]. Funding for open access charge: FRQS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Villicana C., Cruz G., Zurita M.. The basal transcription machinery as a target for cancer therapy. Cancer Cell Int. 2014; 14:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young R.A. RNA polymerase II. Annu. Rev. Biochem. 1991; 60:689–715. [DOI] [PubMed] [Google Scholar]
- 3. Edenberg E.R., Downey M., Toczyski D.. Polymerase stalling during replication, transcription and translation. Curr. Biol. 2014; 24:R445–R452. [DOI] [PubMed] [Google Scholar]
- 4. Noe Gonzalez M., Blears D., Svejstrup J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2020; 22:3–21. [DOI] [PubMed] [Google Scholar]
- 5. van den Heuvel D., van der Weegen Y., Boer D.E.C., Ogi T., Luijsterburg M.S.. Transcription-coupled DNA repair: from mechanism to human disorder. Trends Cell Biol. 2021; 31:359–371. [DOI] [PubMed] [Google Scholar]
- 6. Lans H., Hoeijmakers J.H.J., Vermeulen W., Marteijn J.A.. The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 2019; 20:766–784. [DOI] [PubMed] [Google Scholar]
- 7. Karakasili E., Burkert-Kautzsch C., Kieser A., Strasser K.. Degradation of DNA damage-independently stalled RNA polymerase II is independent of the E3 ligase Elc1. Nucleic Acids Res. 2014; 42:10503–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gillette T.G., Gonzalez F., Delahodde A., Johnston S.A., Kodadek T.. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svejstrup J.Q. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 2003; 116:447–451. [DOI] [PubMed] [Google Scholar]
- 10. Noe Gonzalez M., Blears D., Svejstrup J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2021; 22:3–21. [DOI] [PubMed] [Google Scholar]
- 11. Nakazawa Y., Hara Y., Oka Y., Komine O., van den Heuvel D., Guo C., Daigaku Y., Isono M., He Y., Shimada M.et al.. Ubiquitination of DNA damage-stalled RNAPII promotes transcription-coupled repair. Cell. 2020; 180:1228–1244. [DOI] [PubMed] [Google Scholar]
- 12. Caron P., Pankotai T., Wiegant W.W., Tollenaere M.A.X., Furst A., Bonhomme C., Helfricht A., de Groot A., Pastink A., Vertegaal A.C.O.et al.. WWP2 ubiquitylates RNA polymerase II for DNA-PK-dependent transcription arrest and repair at DNA breaks. Genes Dev. 2019; 33:684–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tufegdzic Vidakovic A., Mitter R., Kelly G.P., Neumann M., Harreman M., Rodriguez-Martinez M., Herlihy A., Weems J.C., Boeing S., Encheva V.et al.. Regulation of the RNAPII pool is integral to the DNA damage response. Cell. 2020; 180:1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iconomou M., Saunders D.N.. Systematic approaches to identify E3 ligase substrates. Biochem. J. 2016; 473:4083–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schulman B.A., Harper J.W.. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009; 10:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart M.D., Ritterhoff T., Klevit R.E., Brzovic P.S.. E2 enzymes: more than just middle men. Cell Res. 2016; 26:423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uchida C., Kitagawa M.. RING-, HECT-, and RBR-type E3 ubiquitin ligases: involvement in human cancer. Curr. Cancer Drug Targets. 2016; 16:157–174. [DOI] [PubMed] [Google Scholar]
- 18. Chew E.H., Hagen T.. Substrate-mediated regulation of cullin neddylation. J. Biol. Chem. 2007; 282:17032–17040. [DOI] [PubMed] [Google Scholar]
- 19. Andérica-Romero A.C., González-Herrera I.G., Santamaría A., Pedraza-Chaverri J.. Cullin 3 as a novel target in diverse pathologies. Redox. Biol. 2013; 1:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anindya R., Aygün O., Svejstrup J.Q.. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell. 2007; 28:386–397. [DOI] [PubMed] [Google Scholar]
- 21. Harreman M., Taschner M., Sigurdsson S., Anindya R., Reid J., Somesh B., Kong S.E., Banks C.A., Conaway R.C., Conaway J.W.et al.. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl Acad. Sci. U.S.A. 2009; 106:20705–20710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurihara T., Kubota Y., Ozawa Y., Takubo K., Noda K., Simon M.C., Johnson R.S., Suematsu M., Tsubota K., Ishida S.et al.. von Hippel-Lindau protein regulates transition from the fetal to the adult circulatory system in retina. Development. 2010; 137:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Zhang Z., Wang B., Zhang J., Zhao Y., Jin Y.. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol. Cell. Biol. 2007; 27:5296–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mokuda S.et al.. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat. Commun. 2019; 10:2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caron P., Pankotai T., Wiegant W.W., Tollenaere M.A.X., Furst A., Bonhomme C., Helfricht A., Groot A.d., Pastink A., Vertegaal A.C.O.et al.. WWP2 ubiquitylates RNA polymerase II for DNA-PK-dependent transcription arrest and repair at DNA breaks. Genes Dev. 2019; 33:684–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuznetsova A.V., Meller J., Schnell P.O., Nash J.A., Ignacak M.L., Sanchez Y., Conaway J.W., Conaway R.C., Czyzyk-Krzeska M.F.. von Hippel–Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl Acad. Sci. U.S.A. 2003; 100:2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berthon A., Faucz F., Bertherat J., Stratakis C.A.. Analysis of ARMC5 expression in human tissues. Mol. Cell. Endocrinol. 2017; 441:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Venanzi A., Alencar G.A., Bourdeau I., Fragoso M.C., Lacroix A.. Primary bilateral macronodular adrenal hyperplasia. Curr Opin Endocrinol Diabetes Obes. 2014; 21:177–184. [DOI] [PubMed] [Google Scholar]
- 29. Kyo C., Usui T., Kosugi R., Torii M., Yonemoto T., Ogawa T., Kotani M., Tamura N., Yamamoto Y., Katabami T.et al.. ARMC5 alterations in primary macronodular adrenal hyperplasia (PMAH) and the clinical state of variant carriers. J Endocr Soc. 2019; 3:1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Assié G., Libé R., Espiard S., Rizk-Rabin M., Guimier A., Luscap W., Barreau O., Lefèvre L., Sibony M., Guignat L.et al.. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing's syndrome. N. Engl. J. Med. 2013; 369:2105–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bourdeau I., Oble S., Magne F., Levesque I., Caceres-Gorriti K.Y., Nolet S., Awadalla P., Tremblay J., Hamet P., Fragoso M.C.et al.. ARMC5 mutations in a large French-Canadian family with cortisol-secreting beta-adrenergic/vasopressin responsive bilateral macronodular adrenal hyperplasia. Eur. J. Endocrinol. 2016; 174:85–96. [DOI] [PubMed] [Google Scholar]
- 32. Elbelt U., Trovato A., Kloth M., Gentz E., Finke R., Spranger J., Galas D., Weber S., Wolf C., Konig K.et al.. Molecular and clinical evidence for an ARMC5 tumor syndrome: concurrent inactivating germline and somatic mutations are associated with both primary macronodular adrenal hyperplasia and meningioma. J. Clin. Endocrinol. Metab. 2015; 100:E119–E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gagliardi L., Schreiber A.W., Hahn C.N., Feng J., Cranston T., Boon H., Hotu C., Oftedal B.E., Cutfield R., Adelson D.L.et al.. ARMC5 mutations are common in familial bilateral macronodular adrenal hyperplasia. J. Clin. Endocrinol. Metab. 2014; 99:E1784–E1792. [DOI] [PubMed] [Google Scholar]
- 34. Bourdeau I., Antonini S.R., Lacroix A., Kirschner L.S., Matyakhina L., Lorang D., Libutti S.K., Stratakis C.A.. Gene array analysis of macronodular adrenal hyperplasia confirms clinical heterogeneity and identifies several candidate genes as molecular mediators. Oncogene. 2004; 23:1575–1585. [DOI] [PubMed] [Google Scholar]
- 35. Antonini S.R., Baldacchino V., Tremblay J., Hamet P., Lacroix A.. Expression of ACTH receptor pathway genes in glucose-dependent insulinotrophic peptide (GIP)-dependent Cushing's syndrome. Clin. Endocrinol. (Oxf). 2006; 64:29–36. [DOI] [PubMed] [Google Scholar]
- 36. Cavalcante I.P., Nishi M., Zerbini M.C.N., Almeida M.Q., Brondani V.B., Botelho M., Tanno F.Y., Srougi V., Chambo J.L., Mendonca B.B.et al.. The role of ARMC5 in human cell cultures from nodules of primary macronodular adrenocortical hyperplasia (PMAH). Mol. Cell. Endocrinol. 2018; 460:36–46. [DOI] [PubMed] [Google Scholar]
- 37. Hu Y., Lao L., Mao J., Jin W., Luo H., Charpentier T., Qi S., Peng J., Hu B., Marcinkiewicz M.M.et al.. Armc5 deletion causes developmental defects and compromises T-cell immune responses. Nat. Commun. 2017; 8:13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berthon A., Faucz F.R., Espiard S., Drougat L., Bertherat J., Stratakis C.A.. Age-dependent effects of Armc5 haploinsufficiency on adrenocortical function. Hum. Mol. Genet. 2017; 26:3495–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 41. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bourgey M., Dali R., Eveleigh R., Chen K.C., Letourneau L., Fillon J., Michaud M., Caron M., Sandoval J., Lefebvre F.et al.. GenPipes: an open-source framework for distributed and scalable genomic analyses. Gigascience. 2019; 8:giz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Allen M., Poggiali D., Whitaker K., Marshall T.R., Kievit R.A.. Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res. 2019; 4:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cotney J.L., Noonan J.P.. Chromatin Immunoprecipitation with Fixed Animal Tissues and Preparation for High-Throughput Sequencing. Cold Spring Harb. Protoc. 2015; 2015:pdb.prot084848. [DOI] [PubMed] [Google Scholar]
- 46. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids. Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gagliardi L., Hotu C., Casey G., Braund W.J., Ling K.H., Dodd T., Manavis J., Devitt P.G., Cutfield R., Rudzki Z.et al.. Familial vasopressin-sensitive ACTH-independent macronodular adrenal hyperplasia (VPs-AIMAH): clinical studies of three kindreds. Clin. Endocrinol. (Oxf). 2009; 70:883–891. [DOI] [PubMed] [Google Scholar]
- 54. Lacroix A., Tremblay J., Rousseau G., Bouvier M., Hamet P.. Propranolol therapy for ectopic beta-adrenergic receptors in adrenal Cushing's syndrome. N. Engl. J. Med. 1997; 337:1429–1434. [DOI] [PubMed] [Google Scholar]
- 55. Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., Feng S., Bostick M., Callis J., Hellmann H.et al.. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. 2005; 17:1180–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anandapadamanaban M., Kyriakidis N.C., Csizmok V., Wallenhammar A., Espinosa A.C., Ahlner A., Round A.R., Trewhella J., Moche M., Wahren-Herlenius M.et al.. E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2. J. Biol. Chem. 2019; 294:11404–11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jishage M., Yu X., Shi Y., Ganesan S.J., Chen W.Y., Sali A., Chait B.T., Asturias F.J., Roeder R.G.. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat. Struct. Mol. Biol. 2018; 25:859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Plechanovova A., Jaffray E.G., Tatham M.H., Naismith J.H., Hay R.T.. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012; 489:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scott D.C., Sviderskiy V.O., Monda J.K., Lydeard J.R., Cho S.E., Harper J.W., Schulman B.A.. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014; 157:1671–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Egloff S., Murphy S.. Cracking the RNA polymerase II CTD code. Trends Genet. 2008; 24:280–288. [DOI] [PubMed] [Google Scholar]
- 61. Stock J.K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A.G., Pombo A.. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007; 9:1428–1435. [DOI] [PubMed] [Google Scholar]
- 62. Walker J.J., Spiga F., Gupta R., Zhao Z., Lightman S.L., Terry J.R.. Rapid intra-adrenal feedback regulation of glucocorticoid synthesis. J. R. Soc. Interface. 2015; 12:20140875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Somesh B.P., Reid J., Liu W.F., Sogaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005; 121:913–923. [DOI] [PubMed] [Google Scholar]
- 64. Somesh B.P., Sigurdsson S., Saeki H., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007; 129:57–68. [DOI] [PubMed] [Google Scholar]
- 65. Kokic G., Wagner F.R., Chernev A., Urlaub H., Cramer P.. Structural basis of human transcription-DNA repair coupling. Nature. 2021; 598:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Phatnani H.P., Greenleaf A.L.. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006; 20:2922–2936. [DOI] [PubMed] [Google Scholar]
- 67. Saeki Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017; 161:113–124. [DOI] [PubMed] [Google Scholar]
- 68. Lee D.F., Kuo H.P., Liu M., Chou C.K., Xia W., Du Y., Shen J., Chen C.T., Huo L., Hsu M.C.et al.. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell. 2009; 36:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Petroski M.D., Deshaies R.J.. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005; 6:9–20. [DOI] [PubMed] [Google Scholar]
- 70. Bosu D.R., Kipreos E.T.. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008; 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cavalcante I.P., Vaczlavik A., Drougat L., Lotfi C.F.P., Perlemoine K., Ribes C., Rizk-Rabin M., Clauser E., Fragoso M., Bertherat J.et al.. Cullin 3 targets the tumor suppressor gene ARMC5 for ubiquitination and degradation. Endocr. Relat. Cancer. 2020; 27:221–230. [DOI] [PubMed] [Google Scholar]
- 72. Nalepa G., Rolfe M., Harper J.W.. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 2006; 5:596–613. [DOI] [PubMed] [Google Scholar]
- 73. Tewari R., Bailes E., Bunting K.A., Coates J.C.. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010; 20:470–481. [DOI] [PubMed] [Google Scholar]
- 74. Rothenberg E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014; 32:283–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gala H.P., Saha D., Venugopal N., Aloysius A., Dhawan J.. RNA polymerase II pausing regulates a quiescence-dependent transcriptional program, priming cells for cell cycle reentry. 2018; bioRxiv doi:19 January 2018, preprint: not peer reviewed 10.1101/250910. [DOI]
- 76. Selby C.P., Sancar A.. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M.A., Chong J., Hare A.A., Dervan P.B., DiMaio F., Leschziner A.E.et al.. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017; 551:653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lommel L., Bucheli M.E., Sweder K.S.. Transcription-coupled repair in yeast is independent from ubiquitylation of RNA pol II: implications for Cockayne's syndrome. Proc. Natl Acad. Sci. U.S.A. 2000; 97:9088–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Woudstra E.C., Gilbert C., Fellows J., Jansen L., Brouwer J., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002; 415:929–933. [DOI] [PubMed] [Google Scholar]
- 80. Yang C., Bolotin E., Jiang T., Sladek F.M., Martinez E.. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007; 389:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vo Ngoc L., Wang Y.L., Kassavetis G.A., Kadonaga J.T.. The punctilious RNA polymerase II core promoter. Genes Dev. 2017; 31:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ostrom Q.T., McCulloh C., Chen Y., Devine K., Wolinsky Y., Davitkov P., Robbins S., Cherukuri R., Patel A., Gupta R.et al.. Family history of cancer in benign brain tumor subtypes versus gliomas. Front. Oncol. 2012; 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Albiger N.M., Regazzo D., Rubin B., Ferrara A.M., Rizzati S., Taschin E., Ceccato F., Arnaldi G., Pecori Giraldi F., Stigliano A.et al.. A multicenter experience on the prevalence of ARMC5 mutations in patients with primary bilateral macronodular adrenal hyperplasia: from genetic characterization to clinical phenotype. Endocrine. 2017; 55:959–968. [DOI] [PubMed] [Google Scholar]
- 84. Yu J.S., Koujak S., Nagase S., Li C.M., Su T., Wang X., Keniry M., Memeo L., Rojtman A., Mansukhani M.et al.. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008; 27:4657–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sun H., You Y., Guo M., Wang X., Zhang Y., Ye S.. Tfcp2l1 safeguards the maintenance of human embryonic stem cell self-renewal. J. Cell. Physiol. 2018; 233:6944–6951. [DOI] [PubMed] [Google Scholar]
- 86. Smallridge R.C., Marlow L.A., Copland J.A.. Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr. Relat. Cancer. 2009; 16:17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nishizawa M., Kataoka K., Vogt P.K.. MafA has strong cell transforming ability but is a weak transactivator. Oncogene. 2003; 22:7882–7890. [DOI] [PubMed] [Google Scholar]
- 88. Pouponnot C., Sii-Felice K., Hmitou I., Rocques N., Lecoin L., Druillennec S., Felder-Schmittbuhl M.P., Eychène A.. Cell context reveals a dual role for Maf in oncogenesis. Oncogene. 2006; 25:1299–1310. [DOI] [PubMed] [Google Scholar]
- 89. Dikstein R., Zhou S., Tjian R.. Human TAFII105 is a cell type–specific TFIID subunit related to hTAFII130. Cell. 1996; 87:137–146. [DOI] [PubMed] [Google Scholar]
- 90. Louder R.K., He Y., Lopez-Blanco J.R., Fang J., Chacon P., Nogales E.. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016; 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yamit-Hezi A., Dikstein R.. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 1998; 17:5161–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gagliardi L., Ling K.-H., Kok C.H., Carolan J., Brautigan P., Kenyon R., D’Andrea R.J., Van der Hoek M., Hahn C.N., Torpy D.J.et al.. Genome-wide gene expression profiling identifies overlap with malignant adrenocortical tumours and novel mechanisms of inefficient steroidogenesis in familial ACTH-independent macronodular adrenal hyperplasia. Endocr. Relat. Cancer. 2012; 19:L19–L23. [DOI] [PubMed] [Google Scholar]
- 93. Kallen C.B., Billheimer J.T., Summers S.A., Stayrook S.E., Lewis M., Strauss J.F. III. Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein. J. Biol. Chem. 1998; 273:26285–26288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse RNA-seq dataset and ChIP-seq dataset have been deposited to the Gene Expression Omnibus of NCBI (accession #GSE169263 (Token: yjchiquobjebnyl) and #GSE169578 (Token: wxyzicsyvrynvqb), respectively). The human PBMAH microarray dataset has also been deposited to the Gene Expression Omnibus of NCBI (accession #GSE171558).