Abstract
Heterogenous nuclear ribonucleoproteins (hnRNPs) are abundant proteins implicated in various steps of RNA processing that assemble on nuclear RNA into larger complexes termed 40S hnRNP particles. Despite their initial discovery 55 years ago, our understanding of these intriguing macromolecular assemblies remains limited. Here, we report the biochemical purification of native 40S hnRNP particles and the determination of their complete protein composition by label-free quantitative mass spectrometry, identifying A-group and C-group hnRNPs as the major protein constituents. Isolated 40S hnRNP particles dissociate upon RNA digestion and can be reconstituted in vitro on defined RNAs in the presence of the individual protein components, demonstrating a scaffolding role for RNA in nucleating particle formation. Finally, we revealed their nanometer scale, condensate-like nature, promoted by intrinsically disordered regions of A-group hnRNPs. Collectively, we identify nuclear 40S hnRNP particles as novel dynamic biomolecular condensates.
INTRODUCTION
Eukaryotic cell compartmentalization evolved to increase the efficiency of cellular pathways by enriching required components in a confined space. This subcellular up-concentration governing cellular activities can occur either within membranous or membrane-less organelles (1). Many pathways, especially those related to RNA metabolism, take place in membrane-less macromolecular assemblies or organelles, also known as biomolecular condensates. Nuclear processes are often compartmentalized in biomolecular condensates (2), of which nucleoli (3), Cajal bodies (4), paraspeckles (5) and nuclear speckles (6) are best known. Both RNA and RNA binding proteins (RBPs) can undergo a phenomenon called liquid–liquid phase separation (LLPS) (7), which has recently been widely implicated in promoting formation of such biomolecular condensates (8).
RBPs undergoing phase separation often contain condensate-promoting intrinsically disordered region (IDR) and tethering domains required for binding to RNA. Examples include the IDRs of heterogeneous nuclear ribonucleoproteins (hnRNPs) A1 and A2B1 that undergo LLPS by forming in vitro micrometer size droplets (9,10). HnRNPs are a large group of abundant nuclear proteins that coat the pre-mRNA and play a major role in transcription, splicing, 3′ end processing, export and RNA degradation (11–14). Six hnRNPs form the core of 40S hnRNP particles that package nuclear RNA and were named as A1, A2, B1, B2, C1 and C2 (15). C1 and C2 are isoforms that form (C1)3C2 tetramers (16,17). Likewise, A2 and B1 are isoforms (18), and A2/B1 as well as A1/B2 also assemble with a 3:1 ratio in the context of the 40S hnRNP (19,20). 40S hnRNP particles were initially isolated from rat liver nuclear extracts and shown to sediment as a major peak at 40S on sucrose density gradients, (21). This peak is composed mainly of single spherical particles with a diameter of about 20 nm (22), initially referred to as ‘informofers’ (23) and later termed ‘40S hnRNP particles’ (24). Interestingly, addition of RNAse inhibitors re-distributed the 40S peak to a series of distinct peaks of higher density, suggesting a beads-on-a-string-like arrangement of 40S hnRNP particles on RNA (25). Consistent with this idea, the lengths of the RNA fragments extracted from the higher density peaks also increased in increments of about 700 nucleotides per additional 40S hnRNP particle (24,25). Moreover, in RNase treated nuclear extracts, hnRNP particles were reconstituted upon addition of in vitro transcribed RNA of about 700 nucleotides (26). Because hnRNPs are of comparable abundance as histones and the overall architecture of 40S hnRNP particles is reminiscent to nucleosomes, it was proposed that 40S hnRNP particles represent ‘ribonucleosomes’ that might play important roles in RNA compaction and in regulating RNA processing (27).
In contrast to these early reports indicating that the 40S hnRNP particles might constitute the functional entities, most of our current knowledge about the different functions of hnRNPs is based on studies of individual hnRNP proteins, and it remains largely unknown how individual hnRNPs interact with each other and with RNA to form hnRNP particles. To address this, we employed and optimized protocols for affinity purification of native 40S hnRNP particles from human embryonic kidney (HEK) 293 cells and identified their protein composition by label-free quantitative mass spectrometry (MS). In line with the early papers, we show that the integrity of the particles depends on co-purified, short RNA fragments, and that particles dissociated by treatment with micrococcal nuclease (MNase) can re-assemble on in vitro transcribed RNA into 40S particles. Moreover, we reconstituted hnRNP-like particles from purified components and showed that their size and C to A-group hnRNPs stoichiometries are almost identical to the particles purified from cells. Furthermore, we show a combined role of the RNA recognition motifs (RRMs) of A-group hnRNPs and IDRs for the formation of higher-order structures, pointing at IDR-driven phase separation as a possible mechanism of the particle assembly. Finally, we map a part of the IDR to the M9 nuclear localization signal (28) and suggest it to act as a possible interaction interface for the 40S hnRNP particle biogenesis.
MATERIALS AND METHODS
Cell lines
HEK293 Flip-In T-REx cells (Invitrogen #R78007) were cultured as a monolayer in DMEM/F-12 medium (Gibco #32500043) supplemented with 10% FCS, 100 U/ml penicillin and 100 μg/ml streptomycin (37°C, 5% CO2, humidified incubator). HnRNP C and hnRNP A1 constructs including alternatively spliced introns were gene synthesized (General Biosystems) and cloned into pcDNA5/FRT/TO 3xFLAG (C) plasmid (29).
Cells stably expressing hnRNP C-3xFLAG, or 3xFLAG-hnRNP A1 were established using pcDNA5/FRT/TO vector-based protocol accordingly to manufacturer's instructions.
Immunoprecipitation (IP)
For IP, dox-induced HEK293 Flip-In T-REx cells were grown on 15 cm dish till full confluency (4 × 107). After harvesting, the cell pellet was resuspended in the lysis buffer (90 mM NaCl, 0.5% Triton X-100, 50 mM HEPES pH 7.4, protease inhibitor cocktail and 250 U of RNAsin (NxGen Rnase inhibitor, 50 000 U, Lucigen 30281-2) and sonicated 3 × 5 s (Vibra-Cell Processor, Sonics). Next, the lysate was clarified by centrifugation (16 100 × rcf, 10 min, 4°C) and incubated with anti-FLAG beads (M2 antibodies coupled to Dynabeads M-270 Epoxy) for 1 hour with rotation at 4°C (cold room). The beads were washed in the lysis buffer and proteins eluted either in presence of 3xFLAG peptide (1 mg/ml), or loading buffer (1.5× LDS, 50 mM DTT).
Protein electrophoresis and silver staining
Protein samples were prepared in 1.5× LDS loading buffer (Invitrogen #NP0008) supplemented with 50 mM DTT and heated up at 75°C prior to loading. The samples were loaded onto 4–12% Bis–Tris gel (Invitrogen #WG1401BOX) and run in 1× MOPS running buffer at 200 V. After electrophoresis, the gel was fixed and stained with silver nitrate.
Purification of 40S hnRNP particles
HEK293 Flip-In T-REx cells expressing hnRNP C-3xFLAG (or 3xFLAG-hnRNP A1) were grown in cell factories (Nunc #140410) and 24 h before harvesting, the medium was supplemented with 1 μg/ml doxycycline to induce expression of tagged protein. Cells were harvested by trypsinization, aliquoted into 2 g pellets, frozen and stored at −80°C. Immunoprecipitation of tagged hnRNPs was performed as above. Natively eluted proteins (3xFLAG peptide elution) were loaded onto 15–35% glycerol gradient and centrifuged for 16 h at 26 400 rpm, 4°C (SW 41 Ti rotor, Beckman Coulter). Fractions were collected using the piston gradient fractionator (BioComp Instruments) and analyzed by protein electrophoresis and silver staining. For GraFix gradients, the heavy solution was supplemented with 0.2% glutaraldehyde (Sigma-Aldrich #G5882-10X1ML). Fractions were collected using the piston gradient fractionator (speed: 0.2, distance: 3.71, number of fractions: 24; BioComp Instruments), and cross-linking was quenched with 50 mM Tris 7.5, and analyzed by protein electrophoresis (26-well 4–12% Bis-Tris gel, Invitrogen #WG1403BOX) followed by silver staining.
In vitro transcription and purification of RNA
To generate the 717 nucleotides long human β-globin (HBB) RNA, the corresponding DNA fragment was synthesized by GenScript, adding GGGATC to the 5′ end to facilitate efficient transcription initiation, and inserted in the pTx1 vector. For run-off in vitro transcription, the plasmid was linearized with EcoRV.
For producing the 984 nucleotides long RPL35A RNA, a fragment of RPL35A intron 3 was PCR-amplified from genomic DNA of HEK293 Flip-In T-REx cells using the following primers: Fwd_HindIII 5′-TATATAAAGCTTGCATTTTGTTAATGCCTTAAAATTATGGG-3′ and Rev_XhoI 5′-ATATATCTCGAGAGACAATAGGATTTGCTCTTGTTGTC-3′. The resulting PCR product was cleaved with HindIII and XhoI and inserted into the corresponding restriction sites of pcDNA3.1(+).
In vitro run-off transcription reactions using T7 RNA polymerase were performed first in small-scale (50 μl; to determine optimal MgCl2 concentrations) and then in large-scale (10 ml) following a standard protocol. After 3 h incubation at 37°C, transcripts were purified by denaturing anion-exchange chromatography using HPLC. An aliquot of each sample was analyzed for correct length and purity by denaturing PAGE. The in vitro synthetized RNAs desalted by three rounds of butanol precipitation, lyophilised, resuspended in water and stored at −80°C. See Supplemental Information for the sequences of the HBB and RPL35A RNAs.
Protein expression in bacteria and purification
DNA encoding full-length human hnRNP A1 (AA 1−320) or UP1 (AA 1–196), with an N-terminal His6-tag and a TEV cleavage site, were cloned into pET28a between the BamHI and XhoI restriction sites (30). DNA encoding full-length human hnRNP A2 (AA 1–341), with an N-terminal His6-tag, a TEV cleavage site and a GB1-tag, was cloned into pET24a between EcoRI and XhoI restriction sites by BioCat GmbH. All constructs were expressed in BL21(DE3) codon-plus (RIL) cells. The cells were grown in LB media at 37°C to OD600 ∼0.6 and induced by adding a final concentration of 0.5 mM isopropyl-b-d-thiogalactopyranoside. After induction, cells were grown for 6 h at 30°C and harvested by centrifugation. Cell pellets were resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 1 M NaCl, 30 mM imidazole, 0.5 mM DTT) and lysed by sonication. Lysate was centrifuged for 45 min at 17 000 rpm at 4°C. Supernatant was loaded on a gravity flow Ni-NTA column, His-hnRNP A1 was eluted with an imidazole gradient. The fractions containing protein were pooled and dialysed against lysis buffer for 19 h at room temperature. During this time, His-TEV protease was added to the pooled sample at a 1:100 (w/w) protease:hnRNP A1 ratio. The dialysed product was loaded on a gravity flow Ni-NTA column to remove His-TEV protease and cleaved His6-tag from hnRNP A1 sample. Pure protein was dialysed against 40S reconstitution buffer (50 mM HEPES 7.4, 150 mM NaCl, 1 mM DTT) and concentrated to required concentrations with a Vivaspin 10 000 MWCO (Sartorius Stedim Biotech).
In vitro reconstitution of 40S hnRNP particles
Recombinantly expressed hnRNP A1 and A2B1 were mixed with in vitro transcribed RNA and incubated 30 min on ice prior to loading on a glycerol gradient. When added, hnRNP C tetramer was purified from 2 g of cells. To obtain a prep free of other proteins, the mixture was treated with RNase A before elution.
Western blot
After electrophoresis, proteins were transferred onto a nitrocellulose membrane (Invitrogen #IB23001) using the iBlot 2 Dry Blotting System (Invitrogen #IB21001). Membrane was first blocked in 5% powdered milk /TBS-T for 1 h at RT then incubated with the primary antibodies overnight at 4°C. After washing with TBS-T, the membrane was incubated with the secondary antibodies for 1 h at room temperature. After washing with TBS-T, the membrane was dried and scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence
HEK293 Flip-In T-REx cells were seeded into the poly-d-lysine-coated chamber slide (SPL Life Sciencies #30104) at 105 cells per chamber in growth medium supplemented with 1 μg/ml doxycycline for protein induction. After 24 h, cells were fixed (4% formaldehyde in PBS) (sigma #) and permeabilized (0.5% Triton X-100 and 6% FCS in PBS). Incubation with primary antibodies (1:500, anti-FLAG M2 in 0.1% Triton X-100 and 6% FCS in PBS) was performed for 1 h at room temperature on a rocking table. Next, the cells were washed 3× with PBS and incubated with the secondary antibodies (Alexa Fluor 594, or 488, chicken anti-mouse in 0.1% Triton X-100 and 6% FCS in PBS) as above. After washing, cells were stained with DAPI, then dried and mounted using the mounting medium (Vector Laboratories, Vectashield H-1000-10). Images of slides were taken using the Leica microscope.
PLA assay
Proximity ligation assay was performed using the Duolink (Sigma-Aldrich #DUO92008) kit accordingly to manufacturer's instruction.
Mass spectrometry
For the analysis of hnRNP C-3xFLAG interactome, IP was performed in triplicate for RNase A-treated and untreated conditions. Eluates were run 1 cm into 4–12% Bis–Tris gel, stained with blue silver (31) and excised as vertical, 3 × 8 mm gel bands. Excised bands were further cut into smaller cubes and proteins were trypsinized using an in-gel digestion protocol (32). Digests were analyzed on a QExactive instrument with identical conditions as described elsewhere (33). Mass spectrometry data was interpreted with MaxQuant against the human SwissProt protein sequence database (version 2017_12) with carbamidomethylation of cysteine set as static modification and protein N-terminal acetylation, oxidation of methionine, and deamidation of asparagine and glutamine set as variable modification. All other settings and statistical evaluation of differential LFQ intensities were as previously described (33).
For label free quantitative mass spectrometry, proteins from fraction 9 were precipitated overnight at −80°C with 1 volume of acetone and 0.1 volume of 100% (w/v) TCA. Precipitated material was pelleted by centrifugation and washed 3 times with cold acetone. Samples were dried in SpeedVac, reconstituted in 8 M Urea, 100 mM Tris pH8 and double digested with LysC and trypsin as described elsewhere (34). The digests were analyzed by liquid chromatography (LC)–MS/MS (PROXEON coupled to a QExactive HF mass spectrometer, ThermoFisher Scientific) with three injections of 5 μl digests. Peptides were trapped on a μPrecolumn C18 PepMap100 (5 μm, 100 Å, 300 μm × 5 mm, ThermoFisher Scientific, Reinach, Switzerland) and separated by backflush on a C18 column (5 μm, 100 Å, 75 μm × 15 cm, C18) by applying a 90-min gradient of 5% acetonitrile to 40% in water, 0.1% formic acid, at a flow rate of 350 nl/min. The Full Scan method was set with resolution at 60 000 with an automatic gain control (AGC) target of 1E06 and maximum ion injection time of 50 ms. The data-dependent method for precursor ion fragmentation was applied with the following settings: resolution 15 000, AGC of 1E05, maximum ion time of 110 milliseconds, mass window 1.6 m/z, collision energy 28, under fill ratio 1%, charge exclusion of unassigned and 1+ ions, and peptide match preferred, respectively.
The samples were processed by the four search engines Comet (35), Xtandem (36), MSGF (37) and Myrimatch (38) against the manually curated Swiss-Prot (39) database [release 2017_12 for 20180201_MD (Figure 1E), release 2018_07 for 20180724_MD (Figures 2D and 3E), and release 2020_03 for 20200901_MD (Figure 4E)] concatenated to reversed decoy sequences. Search parameters were set to high resolution parameters, namely 10 ppm and 20 ppm for the peptide and fragment mass tolerance, respectively; cleavage rule was set to strict trypsin, allowing for 3 missed cleavages; allowed modifications were fixed carbamidomethylation of cysteines, variable oxidation of methionines and acetylation of protein N-termini, plus deamidation of asparagines and glutamines for the 20180720_MD samples. Each search was followed by the application of the PeptideProphet (40) tool from the Transproteomics pipeline (41), followed by the application of iprophet (42) from TPP in order to combine the search results, which were filtered at the false discovery rate of 0.01; furthermore, the identification was only accepted if at least two of the search engines agreed on the identification. Protein inference was performed with ProteinProphet (43) from TPP. For those protein groups accepted by a false discovery rate filter of 0.01, a Normalized Spectral Abundance Factor (44) was calculated based on the peptide to spectrum match count; shared peptides were accounted for by the method of Zhang et al. (45). The per gene aggregation was performed based on the protein prophet report, so that a PSM sum per gene is calculated from the weighted sums of all the PSMs attributed to the ID group, and the NSAF calculated based on the length of the longest protein of the ID group. The results were plotted using the Prism 9 software.
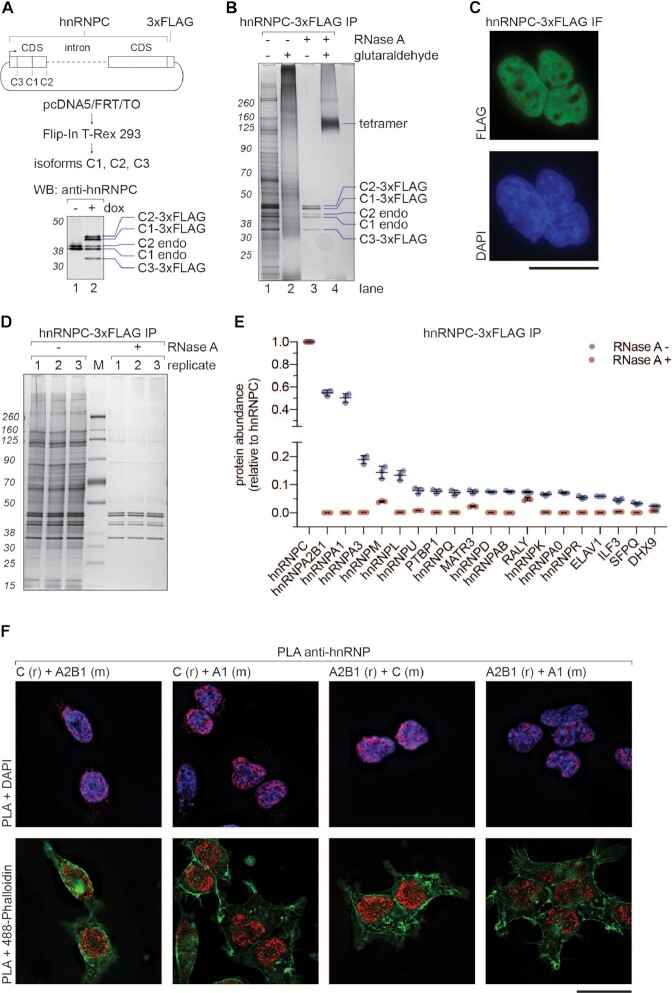
Figure 1.
HnRNP C tetramer associates with A-group hnRNPs. (A) Upper panel: schematic representation of the construct used for doxycycline-inducible expression of hnRNP C-3xFLAG in HEK293 Flip-In T-Rex cells. Lower panel: Western blot analysis of the whole cell extract from cells expressing hnRNP C-3xFLAG. (B) SDS-PAGE/silver staining analysis of hnRNP C-3xFLAG IPs. Samples were treated with RNase A before elution (lanes 3 and 4). Glutaraldehyde crosslinking (final 0.2%) was applied after native elution (3xFLAG peptide) to probe for higher order assemblies (lanes 2 and 4). (C) Immunofluorescence analysis of hnRNP C-3xFLAG using anti-FLAG and Alexa Fluor 488 antibodies. DAPI stain was used as a nuclear marker. Scale bar represents 25 μm. (D) SDS-PAGE/silver staining analysis of eluates from hnRNP C-3xFLAG IPs performed in triplicate with (+) and without (−) RNase A treatment. (E) MS analysis of samples from (D). Column chart displaying abundance of selected proteins (top 20 hits) from hnRNP C-3xFLAG eluates. Y axis shows relative abundance values normalized to the bait protein. Note disruption of the axis to reveal intensities of plotted factors. Error bars represent standard deviation triplicates. (F) Proximity ligation assay. Capital letters represent used anti-hnRNP antibodies, whereas (r), or (m) animals they were raised in (r for rabbit and m for mouse). DAPI and 488-Phalloidin were included as nuclear and cytoplasmic markers, respectively. Scale bar represents 25 μm.
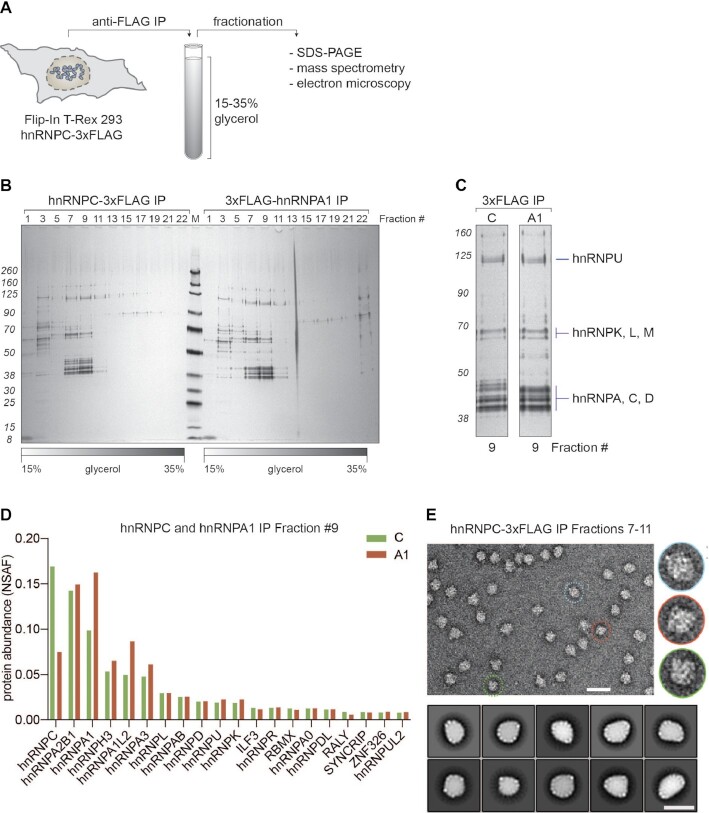
Figure 2.
Biochemical characterization of 40S hnRNP particles. (A) Schematic representation of the workflow established to purify 40S hnRNP particles from cells expressing hnRNP C-3xFLAG (or other hnRNP particle components). Native eluates are fractionated on a glycerol gradient. Collected fractions are analyzed using SDS-PAGE, mass spectrometry, or electron microscopy. (B) SDS-PAGE/silver staining analysis of 15–35% glycerol gradient fractions from hnRNP C-3xFLAG (left) and 3xFLAG-hnRNP A1 (right) purifications. Every other fraction was loaded (odd numbers). (C) Fractions 9 with labeled protein bands from gradient analysis displayed in (B). (D) MS analysis of fractions 9 from gradients displayed in B. Plotted are top 20 the most abundant proteins from both purifications (hnRNP C-3xFLAG in green, 3xFLAG-hnRNP A1 in red), sorted based on their NSAF (normalized spectral abundance factor) protein abundance values. (E) Negative stain EM analysis of endogenous 40S hnRNP particles. A representative electron micrograph (scale bar: 50 nm) and magnified views of individual particles (right, inset) are shown. Lower panel: 2D classification of endogenous 40S hnRNP particles. Class averages are sorted by occupancy, top 10 classes of 25 are shown (scale bar: 25 nm).
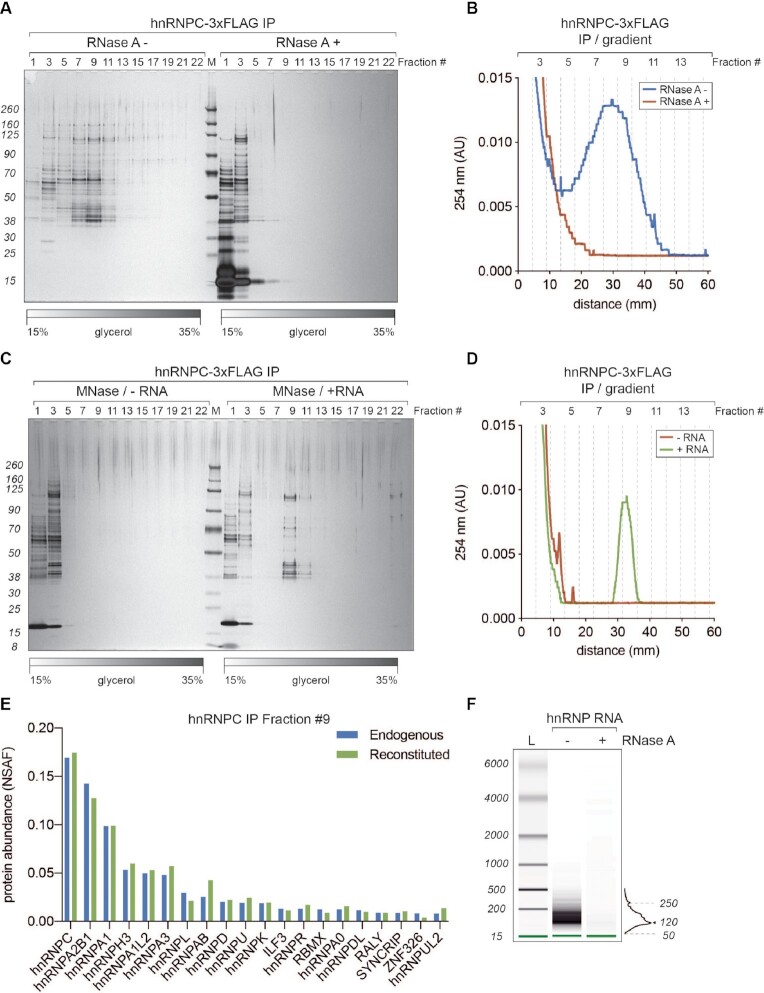
Figure 3.
40S hnRNP particles are dynamic, RNA-dependent assemblies. (A) SDS-PAGE/silver staining analysis of 15–35% glycerol gradient fractions from hnRNP C-3xFLAG purifications without (RNase A–) and with (RNase A+) treatment after the elution. (B) UV A254 nm profile of two gradients displayed in A. Blue trace corresponds to RNase A– and red to RNase A+ condition, respectively. (C) SDS-PAGE/silver staining analysis of 15–35% glycerol gradient fractions from hnRNP C-3xFLAG purifications where eluates were treated with micrococcal nuclease (MNase) to remove associated RNA. Before loading on a gradient, MNase-digested samples were either incubated with an in vitro transcribed fragment of HBB pre-mRNA (+ RNA) or loaded directly (–RNA). (D) UV A254 nm profile of the gradients displayed in (C). Green trace corresponds to +RNA and red to –RNA condition, respectively. (E) MS analysis of fractions 9 from gradients displayed in C (+RNA) and 2B (hnRNP C-3xFLAG IP). Plotted are top 20 the most abundant proteins from both purifications (hnRNP C-3xFLAG in blue and reconstituted in green, respectively). Hits are sorted based on their NSAF (normalized spectral abundance factor) values. (F) RNA length analysis (BioAnalyzer system) of samples extracted from gradient hnRNP fractions displayed in (A).
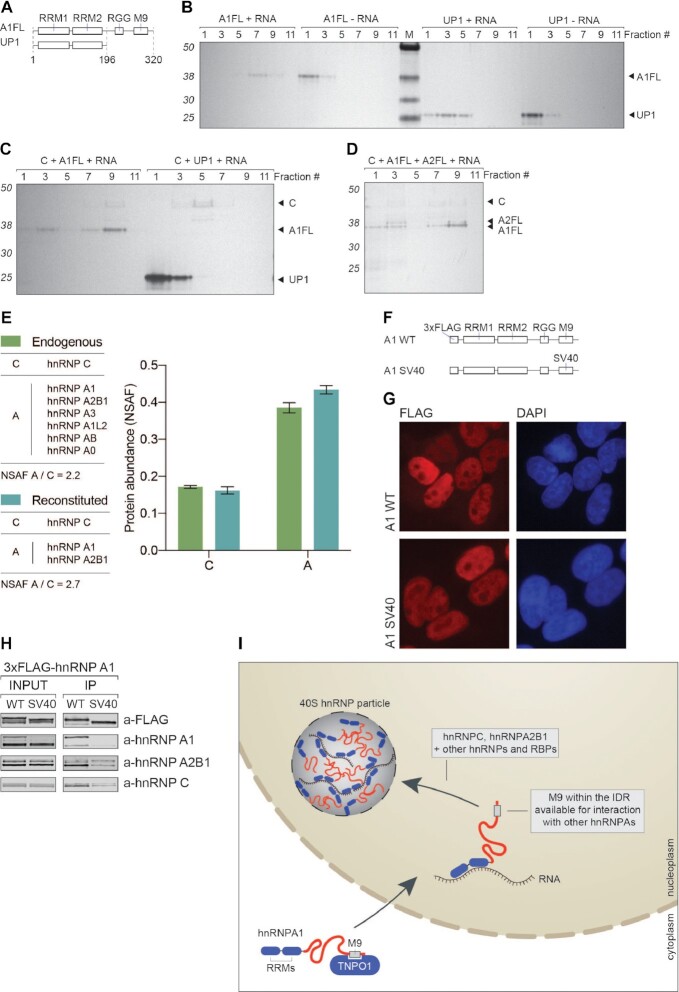
Figure 4.
IDRs of A-group hnRNPs promote assembly of 40S hnRNP particles. (A) Domain organization of hnRNP A1 full length (A1FL) and UP1 proteins. (B) SDS-PAGE/silver stain analysis of 15–35% glycerol gradient fractions. Mixtures of hnRNP A1 full length (A1FL), or UP1 protein and in vitro transcribed fragment of RPL35A pre-RNA were incubated for 30 min on ice prior to loading on gradients. (C) SDS-PAGE/silver stain analysis of 15–35% glycerol gradient fractions. Mixtures of hnRNP C tetramer (C), hnRNP A1 full length (A1FL), or UP1 proteins and in vitro transcribed fragment of RPL35A pre-RNA were incubated for 30 min on ice prior to loading on gradients. (D) SDS-PAGE/silver stain analysis of 15–35% glycerol gradient fractions. Mixtures of hnRNP C tetramer (C), hnRNP A1 full length (A1FL), hnRNP A2B1 full length (A2FL) proteins and in vitro transcribed fragment of RPL35A pre-RNA. (E) MS analysis and comparison of protein abundance in fraction 9 from 4D (reconstituted) and 3E (endogenous). In both cases C refers to the abundance of hnRNP C tetramer and A to the abundance of listed A-group hnRNP proteins. (F) Schematic representation of 3xFLAG-hnRNP A1 expression constructs. In A1 SV40 construct, the M9 region was exchanged for 3x SV40 NLS sequence. (G) Immunofluorescence analysis of A1 WT and SV40 3xFLAG-hnRNP A1 proteins (FLAG in red). DAPI was used as a nuclear marker. Scale bar represents 25 μm. (H) Co-IP analysis of 3xFLAG-hnRNP A1 WT and SV40 proteins. Western blot using antibodies against listed proteins was used to assess the effects of exchanging M9 region for 3x SV40 NLS sequence. (I) Schematic representation of our current understanding of 40S hnRNP particle formation. After translation, hnRNP A1 protein becomes a cargo for the TNPO1 recognizing its M9 region. After translocation to nucleus, hnRNP A1 is free to bind to RNA and its IDR becomes accessible for interactions with other hnRNPs, such as hnRNP C and hnRNP A2B1.
Electron microscopy specimen preparation
For single-particle analysis, fractions 7–11 were re-immunoprecipitated to remove the glycerol, using anti-FLAG beads, and eluted in 50 ul using 0.125 mg/ml 3xFLAG peptide and otherwise identical buffer conditions.
Negative staining EM, data collection and analysis
Samples for negative staining (46) were prepared on Quantifoil R2/2 holey carbon grids covered with a continues carbon foil. Before staining, the grids were glow discharged with negative polarity, 25 mA for 45 s. 5 μl of sample was deposited on the grid, incubated between 30 s and 10 min (depending on the concentration of the sample), before excess sample was removed using Whatman paper and the grid was washed with distilled water twice for 30 s. After blotting, the grid was twice floated on 1% uranyl acetate solution for 30 s and then dried under an incandescent lamp. Negative staining micrographs of the hnRNPC IP eluate (Supplementary Figure S1D) were recorded on a TFS Morgagni 269 microscope operated at 100 kV. The negative staining analysis of gradient-purified hnRNP particles was performed on a Thermo Fisher Scientific Tecnai F20 electron microscope (200 keV) using a Falcon II camera at the ETH ScopeM facility. Micrographs were recorded at a magnification of 62000x, with a pixel size of 1.7 Å/pixel at an electron dose of 110–140 e/Å2. Number of micrographs, particles, for each sample are listed in Supplementary Table S1. Defocus was determined using gctf (47). Particles were manually selected and used to train a neural network in crYOLO 1.7.6 (48), which was then used to automatically pick the particles. Accuracy of particle picks was confirmed by visual inspection. Particles were binned and extracted and subsequently subjected to 2D classification in Relion 3.1 (49,50). Particles of samples of endogenous 40S, A1FL + RNA, and UP1 + RNA were classified into 25 classes. For comparison, particles of endogenous and reconstituted samples were intitially classified into 100 classes. After removal of junk classes, 5000 particles were selected randomly from the remaining particles and subjected to a further round of 2D classification into 25 classes.
Cryo-EM data collection
Endogenous hnRNP complexes stabilized using the GraFix method were vitrified for Cryo-EM using the Thermo Fisher Scientific Vitrobot mark IV plunge freezer, on Quantifoil Cu R2/2 grids covered with 1 nm continuous carbon support film. 3.6 μl sample were applied to the glow discharged grids and plunge-frozen after an incubation time of 10 s using a blotting time of 3 s, at 95% humidity and at 4°C. Data were collected with a Thermo Fisher Scientific Titan Krios microscope operating at 300 keV, equipped with a GIF-Quantum energy filter (used slit width of 20 eV). Images were recorded on a K3 camera operating in counting mode (105k magnification, pixel size 0.84 Å/pix), with an exposure time of 1.8 s and a total dose of 82 e/A2. A target defocus range of −1.8 μm to −3 μm was used to collect 7382 micrographs. 5732 micrographs were selected for processing based on power spectra quality judged by visual inspection, quality of the movie-alignments and the CTF determinations. Defocus was determined using gctf (47). From representative micrographs of each sample, particles were manually selected and used to train a neural network in crYOLO 1.7.6 (48), which was then used to automatically pick 171 698 particles, which were subjected to 2D classification into 100 classes in Relion 3.1 (49,50).
RESULTS
Affinity purification of hnRNP C reveals its association with A-group hnRNPs
HnRNP C protein, together with hnRNP A1 and A2B1, was initially identified as a core component of hnRNP particles (15) and suggested to nucleate its formation upon binding to RNA (51). Available methods for purification of endogenous hnRNP C protein are based either on anion exchange chromatography (16) or immunoprecipitation (IP) using a monoclonal antibody (52), none of which allows for its native elution. To affinity-purify hnRNP C, we therefore established HEK293 Flip-In T-REx cells stably expressing hnRNP C-3xFLAG and employed anti-FLAG antibody coupled beads for IP (Figure 1A) (29). Importantly, we included the endogenous hnRNP C intron 3–4 (HNRNPC-230 ENST00000557201.5) that undergoes alternative splicing, leading to the generation of isoforms C1, C2 and C3, as confirmed by western blot (Figure 1A). Since endogenous hnRNP C forms a tetramer (16,17), we performed IP of hnRNP C-3xFLAG followed by RNase A treatment and glutaraldehyde crosslinking to test whether the tagged version of the protein also forms tetramers (Figure 1B). The eluate contained many proteins, most of which co-purified in an RNA-dependent manner (Figure 1B, compare lanes 1 and 3). RNase A-treated hnRNP C-3xFLAG co-immunoprecipated 3xFLAG-tagged isoforms C1, C2 and C3 but also untagged, endogenous isoforms C1 and C2 (Figure 1B, lane 3), indicating that tagging does not affect the oligomerization properties of hnRNP C protein and also allows for an interaction between endogenous and tagged proteins. Upon combining RNase A-treatment with glutaraldehyde crosslinking, all hnRNP C bands collapsed into one, corresponding in size to an hnRNP C tetramer (Figure 1B, lane 4), providing further support for their existence. We obtained virtually identical results using N-terminally tagged protein (Supplementary Figure S1A). Based on the design and stoichiometry of isoforms expressed from our construct in comparison with the endogenous proteins, we conclude that the cellular abundance of isoforms C1, C2 and the less well characterized C3 is determined by alternative splicing events of the introns 3–4, more specifically by alternative 5′ splice site selection. According to our results, it is plausible that the cellular abundance of hnRNP C isoforms reflects their stoichiometry within the tetramer, however, with our approach we could not directly address that. Notably, we did not find any evidence for A-group hnRNPs to form stable tetramers (Supplementary Figures S1B and C), in contrast to a previous report (20).
Intracellular localization of 3xFLAG-tagged protein is, as endogenous hnRNP C (52), also restricted mainly to the nucleoplasm (Figure 1C). To gain further insights into the interactome of hnRNP C-3xFLAG, we next performed IP in the presence or absence of RNAse A, followed by MS analysis. As shown in Figure 1D, most of the co-purified proteins associate with hnRNP C in an RNA-dependent manner, which was confirmed by the MS analysis (Figure 1E) and indicates that hnRNP C tetramers are part of larger RNP-like assemblies. Importantly, we identified the A-group hnRNPs A2B1 and A1 as the most abundant interaction partners of hnRNP C (Figure 1E), apart from other hnRNPs and RBPs that co-purified with lower abundance.
Negative staining electron microscopy (EM) of this IP sample showed particles of different sizes and shapes (Supplementary Figure S1D). A large proportion of them are spherical particles of ∼20 nm diameter (see encircled in red and blue) resembling the previously reported 40S hnRNP particles (22,26), while smaller particles (see encircled in yellow) could represent partially dissociated substructure of 40S particles or unrelated hnRNP C-containing complexes.
Since all previous studies reporting on 40S hnRNP particles were performed on extracted nuclei and none have addressed their existence in vivo, we applied proximity ligation assays (PLA) to test if the major 40S protein constituents co-localize inside cells. PLA results in a punctate signal if two proteins localize within 40 nm distance. We tested different combinations of antibodies against hnRNP C, A2B1 and A1 (Figure 1F) and all conditions resulted in the detection of puncta in the nucleoplasm, indicating the presence of nuclear assemblies composed of hnRNP C, A2B1 and A1. Notably, their concentration and intensity appeared higher in the vicinity of the inner nuclear membrane, which is in line with previous results based on the immunofluorescent labeling of hnRNP C and confocal microscopy (53).
Biochemical characterization of 40S hnRNP particles
Given the heterogeneity of particle sizes in the IP samples (Supplementary Figure S1D), we next aimed at enriching our samples specifically for 40S hnRNP particles. To this end, we fractionated the IP eluates on a linear glycerol gradient, separated the fractions by SDS-PAGE followed by silver staining, performed MS analysis and electron microscopy (Figure 2A) (54). To maximally preserve the 40S hnRNP particle integrity, cells were lysed by sonication at 90 mM NaCl (Barnett S.F. et al., 1990) and in the presence of RNAse inhibitors. Using this workflow, we were able to isolate 40S hnRNP particles from cells expressing either hnRNP C-3xFLAG or 3xFLAG-hnRNP A1 (Figure 2B). In both cases, we observed a strong enrichment of particles, revealed by the characteristic pattern of hnRNP protein bands migrating around 40, 60 and 120 kDa, in fractions 7–11, with a peak around fraction 9. This result is consistent with published IPs of hnRNP C using a monoclonal 4F4 antibody (52,55), indicating the robustness of our approach. Based on the relatively broad distribution of the particles over five fractions, we hypothesize that native 40S hnRNP particles are heterogeneous assemblies with variable density, mass and/or shape. This observation is consistent with previous studies using sucrose gradient sedimentation to fractionate HeLa nuclear extracts (15).
To gain information regarding protein abundance in the 40S hnRNP particles and compare the particles purified using two different bait proteins, we analyzed fractions 9 from both gradients (Figure 2C). Interestingly, label-free quantitative MS analysis revealed an almost identical protein composition and quantity between the two purifications, except for hnRNP C and A1, which were overrepresented in the respective purification in which they served as the bait protein (Figure 2D). This analysis indicated that the core of 40S hnRNP particles is composed predominantly of hnRNPs of the C and A families and identified hnRNP H3, A1L2, L, D, U and K as additional components present at lower stoichiometries. As estimated by MS analysis, the cellular abundance of A-group hnRNPs reflects their stoichiometry within the hnRNP particle (Supplementary Figure S2A and B). Additionally, partial knockdown of hnRNP A2B1, but not of A1, slightly changed the migration of the hnRNP particle in the gradient towards the lighter fractions (Supplementary Figure S2C–E), confirming that hnRNP A2B1 is a component of the 40S particle.
For assessing the overall size and shape of the 40S hnRNP particles, IP eluates were continuously cross-linked with glutaraldehyde during gradient centrifugation following the GraFix protocol (56) and factions 7–11 were analyzed by negative staining EM. Remarkably, the spherical particles look indistinguishable to those initially reported (Figure 2E) (22): They appear as globular macromolecular assemblies with a diameter of ∼22 nm. This was validated by cryo-EM with 40S hnRNP particles embedded in vitreous ice (Supplementary Figure S2F). However, extensive 2D-classification trials did not align (Figure 2E and Supplementary Figure S2F), reflecting strong heterogeneity at this level of single-particle analysis. Similarly, 3D refinements failed to converge to a single structural solution, indicating a continuum of structural states and a high level of intrinsic heterogeneity and/or flexibility of these particles. Collectively, the EM analyses suggest that 40S hnRNP particles do not exhibit a rigid structure but constitute heterogenous and rather amorphous assemblies. Yet, we found the relatively homogenous size and overall shape of the sample unexpected, especially since hnRNP A-group proteins, which are major constituents of the particle, are known to phase separate in vitro into large (often tens on micrometers) droplets (9,10).
40S hnRNP particles are dynamic, RNA-dependent assemblies
HnRNP particles were initially shown to bind and form on pre-mRNA (55). More recently, the CLIP-Seq analyses of hnRNP C, A1 and A2B1 proteins revealed the intronic sequences of pre-mRNAs as their primary binding sites (57–60). Based on the available literature, we hypothesized that intronic RNA plays an important role in 40S hnRNP particle formation. Furthermore, following our observation on the RNA-dependence of the hnRNP C-3xFLAG interactome, we decided to include RNase A treatment of the eluate in our 40S hnRNP particle isolation workflow (Figure 3A). The protein pattern visible throughout the gradient (untreated sample) collapsed into the top fractions upon addition of RNase A. This effect was confirmed using the UV 254 nm readout during the gradient fractionation procedure (Figure 3B), highlighting the requirement of RNA for the particle integrity.
Next, we decided to test whether the particle can re-assemble on in vitro transcribed RNA. Eluates from the hnRNP C-3xFLAG IP were treated with MNase to degrade associated endogenous RNAs. After inhibiting MNase with EGTA, we added a 717 nucleotides long fragment of in vitro transcribed human β-globin pre-mRNA spanning exon 1, intron 1 and exon 2 [analogous to the previously used mouse β-globin RNA (26); see Supplemental Information], and fractionated this mixture on a glycerol gradient. As seen by SDS-PAGE followed by silver staining and UV 254 nm readout, the 40S hnRNP particle re-assembled spontaneously, with a peak signal in fraction 9 (Figure 3C and D). This finding led us to conclude that RNA nucleates the formation of 40S hnRNP particles, by acting as a scaffold recruiting hnRNPs and allowing for higher-order assembly of RNAs and proteins. To confirm that, we analyzed gradient fraction number 9 by MS and compared it to the endogenous particle (Figure 3E). Interestingly, the composition and protein abundance of the reconstituted particle turned out to be almost identical to its endogenous counterpart.
Since our workflow of isolating 40S particles preserves the RNA scaffold (Figure 3A), we analyzed RNA fragments isolated from the hnRNP C-3xFLAG gradient fractions (Figure 3F). Material extracted from fractions 7–11 contains RNA species in a size range of 50–250 nucleotides, with a peak around 120 nucleotides. Notably, the same size range of RNA fragments was shown to hold together the hnRNP particles upon partial RNAse degradation (55). Taken together, our data points towards a very dynamic nature of 40S hnRNP particles. These nuclear protein-RNA assemblies strictly depend on the integrity of the RNA to which they bind, and when the RNA scaffold is degraded, hnRNPs most likely become free to associate with new binding target.
40S hnRNP particles are a novel class of nuclear condensates
Biomolecular condensates are defined by the propensity of their components to locally up-concentrate certain factors to fulfill their function (61). IDRs of hnRNP A1 and A2B1 have recently been shown to undergo LLPS (9,10). Their biophysical disposition to form droplets in vitro together with their RNA-binding capabilities (62) may drive the assembly of 40S hnRNP particles. Knowing the relative abundance of the 40S hnRNP particle components (Figures 2D and 3E), we decided to test their role in particle formation using proteins purified either from HEK293 cells (hnRNP C tetramer) or proteins recombinantly expressed and purified from bacteria (hnRNP A1 and A2B1). We first focused on full-length hnRNP A1 protein (A1FL) and its UP1 variant missing the IDR (Figure 4A) (63). Their migration on a glycerol gradient was assessed in the presence of an in vitro transcribed 1000 nucleotides intronic sequence (a fragment of RPL35A pre-mRNA; see Supplemental Information). Interestingly, A1FL alone, in the absence of RNA (A1FL – RNA), was unable to form higher-order structures and did not enter the gradient (Figure 4B), whereas upon addition of the RNA (A1FL + RNA), A1FL formed complexes migrating deeper into the gradient (fraction 7). UP1 on its own (UP1 – RNA), similarly to A1FL, did not enter the gradient. When mixed with the RNA (UP1 + RNA), it entered the gradient but migrated mostly till fraction 3. Fractions 7 (A1FL + RNA) and 3 (UP1 + RNA) were additionally analyzed with GraFix cross-linking followed by negative stain electron microscopy (Supplementary Figure S3A). We were able to detect bigger particles only in the case of the A1FL but not with the IDR-missing UP1. These observations suggest that UP1 can bind the RNA via its two RRMs, but only A1FL is able to form higher-order structures upon binding to RNA. This points towards a specific role of the IDRs in the formation of higher-order hnRNP-like particles. Intriguingly, when supplementing the above-mentioned mixtures with hnRNP C tetramer, we observed the formation of 40S hnRNP-like particles similar in migration to endogenous 40S hnRNP particles (with a peak in fraction 9) only in presence of A1FL (C + A1FL + RNA) but not with UP1 (C + UP1 + RNA) (Figure 4C). We conclude that formation of 40S hnRNP particles is promoted by hnRNP C tetramers and A-group hnRNPs upon binding to the RNA scaffold.
Next, we sought to reconstitute the 40S hnRNP particle from purified packaging core components. We mixed recombinantly expressed full-length hnRNP A1 (A1FL), A2B1 (A2FL), hnRNP C tetramers (C) isolated from HEK293 cells, and in vitro transcribed RPL35A RNA. The mixture was then fractionated on a glycerol gradient (Figure 4D). Strikingly, the migration pattern of the formed complex resembled that of the endogenous particle (with a peak in fraction 9) and the ratio between hnRNP C and hnRNP A-group proteins was almost identical, as determined by label-free quantitative MS (Figure 4E and Supplementary Figure S3B). The in vitro reconstituted particles were also compared to their endogenous counterparts with negative stain electron microscopy using cross-linked samples (Supplementary Figure S3C) and they both appeared to be very similar in size and their overall globular shape, pointing towards their dynamic and universal nature to assemble spontaneously in the presence of RNA. The particles, although globular and of an estimated size of around 20 nm, are still heterogenous in nature, therefore obtaining a high-resolution cryo-EM structure at this stage was not feasible. Collectively, we demonstrate that the IDRs of A-group hnRNPs are required for the particle formation, since the particle cannot form with UP1, suggesting that 40S particle formation involves LLPS. We also demonstrate that RNA plays a scaffolding role and together with the low-complexity domains of hnRNP A-group proteins contributes to the assembly of the 40S hnRNP particle.
The M9 NLS of A-group hnRNPs is crucial for interactions within the 40S hnRNP particle
The M9 region present within the IDR of hnRNP A1 was initially identified as a non-canonical nuclear localization signal (NLS) (28) and shown to be recognized by Transportin-1 (TNPO1, also known as Karyopherinβ2) for its import to the nucleus. In the nucleoplasm, Ran GTPase displaces TNPO1 from the IDR of hnRNP A1 (64), rendering the protein available to bind its RNA substrates and interact with other proteins. TNPO1 was also shown to prevent LLPS of FUS by binding to its M9 NLS (65). We therefore reasoned that the M9 region possibly plays an important role in mediating interactions between A-group hnRNPs in the context of the 40S hnRNP particle formation. Additional evidence on the importance of the M9 region in IDR-mediated interactions has recently been provided from cryo-EM analysis of hnRNP A1 amyloid fibrils (66). To test the proposed role of the M9 region in mediating interactions between major components of the 40S hnRNP particle, we established HEK293 Flip-In T-Rex cells stably expressing 3xFLAG-tagged hnRNP A1 WT (A1 WT), or an A1 variant in which the M9 region was replaced by 3x SV40 NLS (A1 SV40, Figure 4F). We confirmed by immunofluorescence microscopy that both constructs localized to the nucleoplasm as expected (Figure 4G). hnRNP A1 WT co-purified endogenous hnRNP A1, A2B1 and C (Figure 4H). Intriguingly, their levels were significantly reduced when A1 SV40 was used as bait protein (Figure 4H). We conclude that the M9 region is not only responsible for the correct nuclear localization of hnRNP A1 protein but also for its interaction with other hnRNP proteins and thereby for the formation of the 40S hnRNP particle as depicted in our model (Figure 4I).
DISCUSSION
Our affinity purification of hnRNP particles led to the rediscovery of the ribonucleosome, a hnRNP particle sedimenting around 40S that has been first described 55 years ago (21). Using state-of-the-art label-free mass spectrometry analysis allowed us to identify the protein composition of these 40S hnRNP particles (Figure 1). 40S hnRNP particles are mainly composed of hnRNP A and C proteins and the particles can be affinity purified by expressing either tagged hnRNP C or hnRNP A1 (Figure 2). Despite of their overall defined size and shape, structural heterogeneity prevented the determination of a high-resolution structure of the 40S hnRNP particle by single particle cryo-EM approaches.
In line with the early papers, we showed that the integrity and the assembly of the 40S hnRNP particle depends on RNA: the affinity-purified particles dissolved upon digestion of the RNA and spontaneously re-assembled in vitro upon addition of an in vitro transcribed RNA (Figure 3). Moreover, hnRNP particles with a size and stoichiometric ratio between hnRNP A and C proteins that is indistinguishable from the affinity-purified 40S hnRNP particles could also be reconstituted from purified components (Figure 4A–E). We further demonstrated that the IDR of hnRNP A1 proteins is required for the in vitro assembly of these particles, suggesting that the assembly of the 40S hnRNP particles might occur through an IDR-driven phase separation mechanism. Corroborating the crucial role of the hnRNP A1 IDR for the formation of 40S hnRNP particles, we finally revealed that the M9 NLS, which resides within the IDR of hnRNPA1, is necessary for the in vivo interaction of hnRNP A1 with hnRNP C, hnRNP A2B1, and itself (Figure 4G–H).
Collectively, the results presented herein indicate that the 40S hnRNP particles represent a novel example of nanometer scale, nuclear biomolecular condensates forming upon hnRNPs binding to target RNA. The plausible function of the 40S hnRNP particle could be analogous to the chromatin compaction, suggested to also be LLPS-driven and mediated by multivalent nucleosome interactions (67). RNA compaction by 40S hnRNP particles could be a prerequisite for RNA processing in the crowded, nuclear environment. Indeed, IDR-mediated multivalent hnRNP assemblies were shown to control alternative splicing (68,69), supporting this idea. Follow-up studies will address the cellular functions of the 40S hnRNP particles and the transcriptome-wide identification of the 40S hnRNP particle binding sites.
DATA AVAILABILITY
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD031677.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Christian Kroun Damgaard (Aarhus University) for the pcDNA5/FRT/TO 3xFLAG(N) plasmid. We would like to thank Miroslav Peterek from the ETH ScopeM facility in Zürich, Switzerland, where EM experiments were conducted, and Georg Dorn for valuable advice and help. We are also grateful to Evan Karousis for his valuable comments on the manuscript.
Author contributions: E.D., M.D., F.H.T.A and O.M. conceived the project; M.D., E.D., M.E.P., S.C., A.C., J.R., P.A., conducted the experiments; S.B., M.H. and A.C.U produced and interpreted the mass spectrometry results; M.D., E.D., J.R., P.A., D.B., J.N., F.H.T.A and O.M. analyzed and interpreted the results; M.D., O.M. and E.D. wrote the manuscript, with contributions by all other co-authors; F.H.T.A. and O.M. provided funding for the project.
Notes
Present address: Michal Domanski, Lonza AG, Visp, Switzerland.
Present address: Emil Dedic, Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10C, 8000 Aarhus C, Denmark.
Present address: Sébastien Campagne, ARNA laboratory, INSERM U1212, University of Bordeaux, 2, rue Robert Escarpit, 33600 Pessac, France.
Contributor Information
Michal Domanski, Department of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Freiestrasse 3, 3012 Bern, Switzerland.
Emil Dedic, Institute of Biochemistry, Department of Biology, ETH Zürich, Hönggerbergring 64, 8093 Zürich, Switzerland.
Maria Escura Pérez, Institute of Biochemistry, Department of Biology, ETH Zürich, Hönggerbergring 64, 8093 Zürich, Switzerland.
Antoine Cléry, Institute of Biochemistry, Department of Biology, ETH Zürich, Hönggerbergring 64, 8093 Zürich, Switzerland.
Sébastien Campagne, Institute of Biochemistry, Department of Biology, ETH Zürich, Hönggerbergring 64, 8093 Zürich, Switzerland.
Anne-Christine Uldry, Proteomics & Mass Spectrometry Core Facility, Department for BioMedical Research (DBMR), University of Bern, Murtenstrasse 28, 3008 Bern, Switzerland.
Sophie Braga, Proteomics & Mass Spectrometry Core Facility, Department for BioMedical Research (DBMR), University of Bern, Murtenstrasse 28, 3008 Bern, Switzerland.
Manfred Heller, Proteomics & Mass Spectrometry Core Facility, Department for BioMedical Research (DBMR), University of Bern, Murtenstrasse 28, 3008 Bern, Switzerland.
Julius Rabl, Cryo-EM Knowledge Hub, ScopeM, Otto Stern Weg 3, ETH Zürich, 8093 Zürich, Switzerland.
Pavel Afanasyev, Cryo-EM Knowledge Hub, ScopeM, Otto Stern Weg 3, ETH Zürich, 8093 Zürich, Switzerland.
Daniel Boehringer, Cryo-EM Knowledge Hub, ScopeM, Otto Stern Weg 3, ETH Zürich, 8093 Zürich, Switzerland.
Jiří Nováček, Cryo-Electron Microscopy and Tomography Core Facility, Central European Institute of Technology, Masaryk University, Kamenice 753/5, 625 00 Brno, Czech Republic.
Frédéric T Allain, Institute of Biochemistry, Department of Biology, ETH Zürich, Hönggerbergring 64, 8093 Zürich, Switzerland.
Oliver Mühlemann, Department of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Freiestrasse 3, 3012 Bern, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Center of Competence in Research (NCCR) on RNA & Disease funded by the Swiss National Science Foundation (SNSF); SNSF [310030B-182831 to O.M., 310030B_189379 and 31003A_170130 to F.H.T.A.]; Canton of Bern (University intramural funding to O.M.). Funding for open access charge: SNSF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Alberti S., Hyman A.A.. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021; 22:196–213. [DOI] [PubMed] [Google Scholar]
- 2. Sabari B.R., Dall’Agnese A., Young R.A.. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 2020; 45:961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lafontaine D.L.J., Riback J.A., Bascetin R., Brangwynne C.P.. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021; 22:165–182. [DOI] [PubMed] [Google Scholar]
- 4. Machyna M., Heyn P., Neugebauer K.M.. Cajal bodies: where form meets function. Wiley Interdiscip. Rev. RNA. 2013; 4:17–34. [DOI] [PubMed] [Google Scholar]
- 5. Fox A.H., Nakagawa S., Hirose T., Bond C.S.. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018; 43:124–135. [DOI] [PubMed] [Google Scholar]
- 6. Ilik I.A., Aktas T.. Nuclear speckles: dynamic hubs of gene expression regulation. FEBS J. 2021; 10.1111/febs.16117. [DOI] [PubMed] [Google Scholar]
- 7. Razin S.V., Gavrilov A.A.. The role of liquid–liquid phase separation in the compartmentalization of cell nucleus and spatial genome organization. Biochemistry (Mosc). 2020; 85:643–650. [DOI] [PubMed] [Google Scholar]
- 8. Tauber D., Tauber G., Parker R.. Mechanisms and regulation of RNA condensation in RNP granule formation. Trends Biochem. Sci. 2020; 45:764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P.. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015; 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D.et al.. A molecular grammar governing the driving forces for phase separation of Prion-like RNA binding proteins. Cell. 2018; 174:688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke J.P., Thibault P.A., Salapa H.E., Levin M.C.. A comprehensive analysis of the role of hnRNP A1 function and dysfunction in the pathogenesis of neurodegenerative disease. Front. Mol. Biosci. 2021; 8:659610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 13. Geissler R., Simkin A., Floss D., Patel R., Fogarty E.A., Scheller J., Grimson A.. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev. 2016; 30:1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geuens T., Bouhy D., Timmerman V.. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016; 135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beyer A.L., Christensen M.E., Walker B.W., LeStourgeon W.M.. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977; 11:127–138. [DOI] [PubMed] [Google Scholar]
- 16. Barnett S.F., Friedman D.L., LeStourgeon W.M.. The C proteins of hela 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol. Cell. Biol. 1989; 9:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rech J.E., LeStourgeon W.M., Flicker P.F.. Ultrastructural morphology of the hnRNP C protein tetramer. J. Struct. Biol. 1995; 114:77–83. [DOI] [PubMed] [Google Scholar]
- 18. Burd C.G., Swanson M.S., Gorlach M., Dreyfuss G.. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:9788–9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barnett S.F., Northington S.J., LeStourgeon W.M.. Isolation and in vitro assembly of nuclear ribonucleoprotein particles and purification of core particle proteins. Methods Enzymol. 1990; 181:293–307. [DOI] [PubMed] [Google Scholar]
- 20. Lothstein L., Arenstorf H.P., Chung S.Y., Walker B.W., Wooley J.C., LeStourgeon W.M.. General organization of protein in HeLa 40S nuclear ribonucleoprotein particles. J. Cell Biol. 1985; 100:1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samarina O.P., Krichevskaya A.A., Georgiev G.P.. Nuclear ribonucleoprotein particles containing messenger ribonucleic acid. Nature. 1966; 210:1319–1322. [DOI] [PubMed] [Google Scholar]
- 22. Samarina O.P., Lukanidin E.M., Georgiev G.P.. On the structural organization of the nuclear complexes containing messenger RNA. Biochim. Biophys. Acta. 1967; 142:561–564. [DOI] [PubMed] [Google Scholar]
- 23. Lukanidin E.M., Zalmanzon E.S., Komaromi L., Samarina O.P., Georgiev G.P.. Structure and function of informofers. Nat. New Biol. 1972; 238:193–197. [DOI] [PubMed] [Google Scholar]
- 24. Samarina O.P. hnRNP particles. Bioessays. 1996; 18:595–601. [Google Scholar]
- 25. Samarina O.P., Lukanidin E.M., Molnar J., Georgiev G.P.. Structural organization of nuclear complexes containing DNA-like RNA. J. Mol. Biol. 1968; 33:251–263. [DOI] [PubMed] [Google Scholar]
- 26. Conway G., Wooley J., Bibring T., LeStourgeon W.M.. Ribonucleoproteins package 700 nucleotides of pre-mRNA into a repeating array of regular particles. Mol. Cell. Biol. 1988; 8:2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chung S.Y., Wooley J.. Set of novel, conserved proteins fold pre-messenger RNA into ribonucleosomes. Proteins. 1986; 1:195–210. [DOI] [PubMed] [Google Scholar]
- 28. Siomi H., Dreyfuss G.. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995; 129:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Domanski M., Molloy K., Jiang H., Chait B.T., Rout M.P., Jensen T.H., LaCava J.. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. BioTechniques. 2012; 0:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barraud P., Allain F.H.. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: how the relative orientation between RRMs influences the nucleic acid binding topology. J. Biomol. NMR. 2013; 55:119–138. [DOI] [PubMed] [Google Scholar]
- 31. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G.M., Carnemolla B., Orecchia P., Zardi L., Righetti P.G.. Blue silver: a very sensitive colloidal coomassie G-250 staining for proteome analysis. Electrophoresis. 2004; 25:1327–1333. [DOI] [PubMed] [Google Scholar]
- 32. Gunasekera K., Wuthrich D., Braga-Lagache S., Heller M., Ochsenreiter T.. Proteome remodelling during development from blood to insect-form trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012; 13:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winzer P., Muller J., Imhof D., Ritler D., Uldry A.C., Braga-Lagache S., Heller M., Ojo K.K., Van Voorhis W.C., Ortega-Mora L.M.et al.. Neospora caninum: differential proteome of multinucleated complexes induced by the bumped kinase inhibitor BKI-1294. Microorganisms. 2020; 8:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braga-Lagache S., Buchs N., Iacovache M.I., Zuber B., Jackson C.B., Heller M.. Robust Label-free, quantitative profiling of circulating plasma microparticle (MP) associated proteins. Mol. Cell. Proteomics. 2016; 15:3640–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eng J.K., Hoopmann M.R., Jahan T.A., Egertson J.D., Noble W.S., MacCoss M.J.. A deeper look into Comet–implementation and features. J. Am. Soc. Mass. Spectrom. 2015; 26:1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Craig R., Beavis R.C.. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003; 17:2310–2316. [DOI] [PubMed] [Google Scholar]
- 37. Kim S., Pevzner P.A.. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014; 5:5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tabb D.L., Fernando C.G., Chambers M.C.. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007; 6:654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi H., Ghosh D., Nesvizhskii A.I.. Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. J. Proteome Res. 2008; 7:286–292. [DOI] [PubMed] [Google Scholar]
- 41. Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B.et al.. A guided tour of the trans-proteomic pipeline. Proteomics. 2010; 10:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shteynberg D., Deutsch E.W., Lam H., Eng J.K., Sun Z., Tasman N., Mendoza L., Moritz R.L., Aebersold R., Nesvizhskii A.I.. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics. 2011; 10:M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nesvizhskii A.I., Aebersold R.. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 2005; 4:1419–1440. [DOI] [PubMed] [Google Scholar]
- 44. Zybailov B.L., Florens L., Washburn M.P.. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol. Biosyst. 2007; 3:354–360. [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y., Wen Z., Washburn M.P., Florens L.. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010; 82:2272–2281. [DOI] [PubMed] [Google Scholar]
- 46. Ohi M., Li Y., Cheng Y., Walz T.. Negative staining and image classification - Powerful Tools in modern electron microscopy. Biol. Proced. Online. 2004; 6:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D.et al.. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019; 2:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zivanov J., Nakane T., Scheres S.H.W.. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020; 7:253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang M., Rech J.E., Northington S.J., Flicker P.F., Mayeda A., Krainer A.R., LeStourgeon W.M.. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1994; 14:518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi Y.D., Dreyfuss G.. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 1984; 99:1997–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mattern K.A., van der Kraan I., Schul W., de Jong L., van Driel R.. Spatial organization of four hnRNP proteins in relation to sites of transcription, to nuclear speckles, and to each other in interphase nuclei and nuclear matrices of hela cells. Exp. Cell. Res. 1999; 246:461–470. [DOI] [PubMed] [Google Scholar]
- 54. Domanski M., Upla P., Rice W.J., Molloy K.R., Ketaren N.E., Stokes D.L., Jensen T.H., Rout M.P., LaCava J.. Purification and analysis of endogenous human RNA exosome complexes. 2016; 22:1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Choi Y.D., Dreyfuss G.. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:7471–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010; 481:109–126. [DOI] [PubMed] [Google Scholar]
- 57. Bruun G.H., Doktor T.K., Borch-Jensen J., Masuda A., Krainer A.R., Ohno K., Andresen B.S.. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 2016; 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li M., Zhuang Y., Batra R., Thomas J.D., Li M., Nutter C.A., Scotti M.M., Carter H.A., Wang Z.J., Huang X.S.et al.. HNRNPA1-induced spliceopathy in a transgenic mouse model of myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:5472–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nguyen E.D., Balas M.M., Griffin A.M., Roberts J.T., Johnson A.M.. Global profiling of hnRNP A2/B1-RNA binding on chromatin highlights LncRNA interactions. RNA Biol. 2018; 15:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wiedner H.J., Giudice J.. It's not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 2021; 28:465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jain N., Lin H.C., Morgan C.E., Harris M.E., Tolbert B.S.. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu R.M., Jokhan L., Cheng X., Mayeda A., Krainer A.R.. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997; 5:559–570. [DOI] [PubMed] [Google Scholar]
- 64. Lee B.J., Cansizoglu A.E., Suel K.E., Louis T.H., Zhang Z., Chook Y.M.. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006; 126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.D., Simons M., Niessing D., Madl T.et al.. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018; 173:706–719. [DOI] [PubMed] [Google Scholar]
- 66. Sun Y., Zhao K., Xia W., Feng G., Gu J., Ma Y., Gui X., Zhang X., Fang Y., Sun B.et al.. The nuclear localization sequence mediates hnRNPA1 amyloid fibril formation revealed by cryoEM structure. Nat. Commun. 2020; 11:6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Farr S.E., Woods E.J., Joseph J.A., Garaizar A., Collepardo-Guevara R.. Nucleosome plasticity is a critical element of chromatin liquid–liquid phase separation and multivalent nucleosome interactions. Nat. Commun. 2021; 12:2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gueroussov S., Weatheritt R.J., O’Hanlon D., Lin Z.Y., Narula A., Gingras A.C., Blencowe B.J.. Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell. 2017; 170:324–339. [DOI] [PubMed] [Google Scholar]
- 69. Ule J., Blencowe B.J.. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell. 2019; 76:329–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD031677.