Abstract
Translational readthrough (TR) occurs when the ribosome decodes a stop codon as a sense codon, resulting in two protein isoforms synthesized from the same mRNA. TR has been identified in several eukaryotic organisms; however, its biological significance and mechanism remain unclear. Here, we quantify TR of several candidate genes in Drosophila melanogaster and characterize the regulation of TR in the large Maf transcription factor Traffic jam (Tj). Using CRISPR/Cas9-generated mutant flies, we show that the TR-generated Tj isoform is expressed in a subset of neural cells of the central nervous system and is excluded from the somatic cells of gonads. Control of TR in Tj is critical for preservation of neuronal integrity and maintenance of reproductive health. The tissue-specific distribution of a release factor splice variant, eRF1H, plays a critical role in modulating differential TR of leaky stop codon contexts. Fine-tuning of gene regulatory functions of transcription factors by TR provides a potential mechanism for cell-specific regulation of gene expression.
Graphical Abstract
Graphical Abstract.
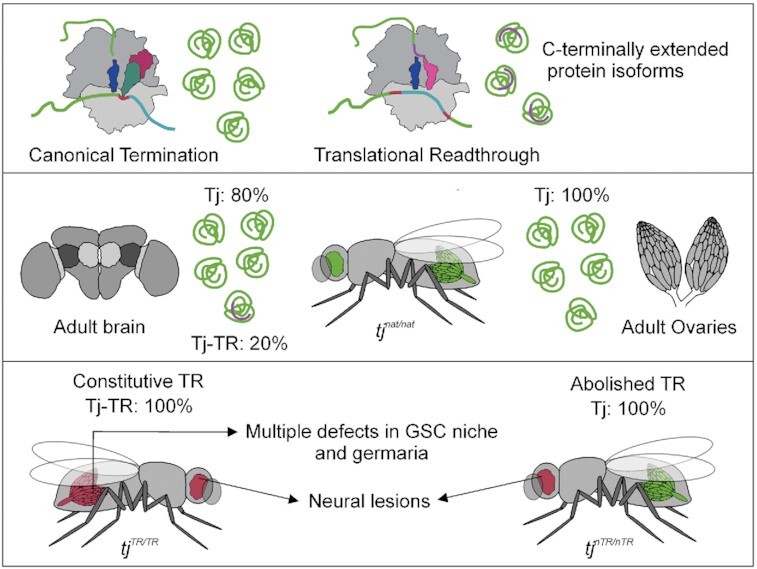
Tissue-specific regulation of translational readthrough in Tj is crucial for preservation of neuronal integrity and maintenance of reproductive health.
INTRODUCTION
Eukaryotes employ several mechanisms to enlarge the coding capacity of their genomes, such as alternative splicing, alternative polyadenylation, frameshifting and alternative initiation of translation (1–4). Translational readthrough (TR) is yet another strategy to increase the diversity of the proteome by supplying C-terminally extended protein isoforms with potentially altered physiological functions. Studies over the last decade have revealed the occurrence of TR in several eukaryotic systems (5–8). The TR efficiency depends on the stop codon and its sequence context and is lowest on the UAA and highest on the UGA stop codon (5,9–11). A cytidine (C) at position +4 enhances TR (nucleotide numbering starting with the first nucleotide of the stop codon), whereas nucleotides +4 to +9 modulate TR in a number of viral and eukaryotic genes (5,9,12–14).
The prevalence of TR varies between organisms. It is widely employed by viruses to expand the coding potential of their small genomes (15–18). Several cases of TR were described in mammalian cells, yeast, and mosquito (6,8,19–21). Analysis of the stop codon contexts of 12 Drosophila species and ribosome profiling studies suggested potential TR in several hundred Drosophila genes (7,8,19). The majority of TR candidate genes identified in Drosophila have regulatory roles, suggesting that appending a functional C-terminal extension may confer conditional advantage to protein function. Computational analysis of TR protein isoforms indicates that these are mostly long, modular proteins with intrinsically disordered C-termini of low sequence complexity (22,23). The lack of a structurally ordered C-terminus might provide conformational pliability that allows the TR extensions to perform functions without distorting the native protein.
Of several hundred candidate genes that can undergo TR in Drosophila, only a few have been validated experimentally. One of the early examples of a gene undergoing TR is kelch, which encodes a short native protein and a longer extended TR protein (24). The TR efficiency of kelch changes through development, approaching a 1:1 ratio of the two isoforms during metamorphosis. The Kelch-TR isoform is expressed in a tissue-specific manner and is particularly enriched in the larval and adult central nervous system (CNS) and imaginal discs (25,26), but its function is not known. Another example is the headcase (hdc) gene; the TR isoform is necessary for Drosophila tracheal development (27). Further examples are Synapsin (Syn), nonsense alleles of embryonic lethal abnormal vision (elav), and wingless (wg) (28–30). Recently, tissue-specific TR was identified in a transcription factor Ventral veins lacking/Drifter (Vvl/Dfr). TR in Vvl/Dfr is necessary for regulation of several downstream genes and is crucial for larval development (31). For the majority of the candidates, the efficiency and the biological relevance of TR are not known.
Here, we tested the TR efficiency for several candidate genes in Drosophila. We then selected one gene, traffic jam (tj) that shows high TR efficiency, to explore the expression and biological significance of the TR protein isoform in vivo. tj encodes a large Maf transcription factor that regulates gonad morphogenesis, including stem cell niche specification during ovarian development (32–34) and collective cell migration during oogenesis (35). Tj has extensive sequence similarity with its mammalian orthologs c-Maf and MafB, which modulate tissue-specific gene expression and cell differentiation by binding to the regulatory regions of target genes and by interacting with other transcription factors. Like all Maf factors, Tj has a leucine-zipper domain, a DNA-binding basic domain, and a Maf-specific extended homology domain located at the C-terminal region of the factor (36). Tj is expressed in the somatic cells of the gonads where it plays a crucial role in regulating the expression of several adhesion molecules such as Fasciclin 3 (Fas3), DE-Cadherin (DE-Cad) and Neurotactin (Nrt) (33,35–37). Disruption of tj function causes impaired interaction between germ cells and somatic cells, which eventually leads to defective gonad development and sterility (36). Apart from somatic gonadal cells, Tj is expressed in the embryonic and larval CNS, adult heads as well as adult fat bodies (36,38). A recent study also identified Tj as one of the transcription factors that fine-tune the molecular and morphological features of the developing neurons, contributing to remarkable glutamatergic neuronal cell-type diversity in the adult brain (39).
To explore the biological significance of TR in tj, we utilized CRISPR/Cas9 genome editing to create mutant fly lines that exhibit different levels of TR in tj. Using immunohistochemistry, we show that the expression of the longer TR isoform of Tj is tissue-specific and is tightly translationally controlled. The expression of the Tj-TR isoform is important for the cells of the nervous system but is detrimental for the correct development of the gonads. Furthermore, we show that there exist considerable tissue-specific differences in the relative abundance of various protein factors that are directly or indirectly involved in translation termination. In particular, we identify a strong link between the tissue-specific distribution of eRF1H, a splice variant of eukaryotic release factor eRF1, and its capacity to enhance readthrough of leaky stop codon contexts. Strict control of TR in a transcription factor provides a yet unidentified link between transcription and translation, which may be particularly relevant for the nervous cells that are under pressure to rapidly adapt to external stimuli and conditions.
MATERIALS AND METHODS
Plasmid construction
Vector templates and/or inserts for molecular cloning were amplified by PCR using Phusion High-Fidelity PCR master mix (New England Biolabs). Desired PCR products were purified using Nucleospin® Gel and PCR clean up kit. Point mutations were introduced into plasmid vectors using the Agilent QuikChange II site-directed mutagenesis protocol. Insertional and deletion mutagenesis were performed using a blunt-end ligation method. Phosphorylation and ligation reactions were performed using T4 Polynucleotide kinase and T4 ligase, and the ligated products were transformed into competent cells. Molecular cloning of TR test cassettes into psiCHECK™-2 vector was achieved by isothermal assembly (Gibson assembly) (40). Insert sequences for Gibson assembly were amplified from gDNA or cDNA using primers containing 18 bp overhangs that overlap with the blunt ends of PCR-amplified, linearized vectors. Gibson assembly was performed by incubating the purified inserts and 100 ng linearized vectors in a molar ratio of 3:1 with ‘in-house’ prepared Gibson assembly mix for 1 h at 50°C in a total volume of 15 μl. 1 μl of end product was transformed into competent cells. Escherichia coli NovaBlue Singles™ Competent cells-Novagen endA1 hsdR17 (rK12– mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac F′(proA+B+ lacIqZΔM15::Tn1)(TetR) were used for all cloning and plasmid construction purposes. All primers were ordered from Eurofins Scientific.
Dual Luciferase reporter assay
The commercial psiCHECK™-2 (Promega) vector was modified by deleting the Rluc poly(A) signal and the firefly luciferase (Fluc) herpes simplex virus thymidine kinase (HSV TK) promoter using blunt-end ligation, such that both the reporters are transcribed as a single monocistronic mRNA controlled by an SV40 promoter. A self-cleaving P2A sequence (66 bp) was inserted between the two reporters using two-step insertional blunt-end ligation mutagenesis. The start codon of Fluc was deleted using blunt-end ligation mutagenesis. The primers used for vector modification are listed in Supplementary Table S4. TR test cassettes (105 bp) from the candidate genes containing the leaky stop codons, flanked by 51 bp on either side, were amplified from w1118 cDNA using primers with 18 bp overhangs and inserted into modified linearized vector using Gibson assembly. UGA to UUC and UAAA point mutations were introduced into each construct using blunt-end ligation mutagenesis. The 39 bp Tobacco mosaic virus (TMV) TR motif was inserted via blunt-end ligation. The primers for cloning of TR motifs and point mutations are listed in Supplementary Table S5. Transfections were performed in triplicates in Drosophila S2R+ cells (Drosophila Genomics Resource Center). S2R + cells were cultured in 25 cm2 flasks at 25°C in a CO2 incubator in Schneider's Gibco® Drosophila medium (ThermoFisher), supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transfections were performed when the cell confluency reached approximately 70% using an Effectene® Transfection Kit (Qiagen). 100 ng of dual reporter vector constructs were used for transfections in 96-well plates. The activities of Renilla (Rluc) and Firefly (Fluc) luciferases were measured 72 h after transfection using Beetle juice and Renilla Glow Juice (PJK GmbH). Measurements were performed in a luminometer with a delay time of 2 s and an integration time of 5 s. The absolute RLuc values of the test (UGA/UAG/UAA), UAAA and UUC constructs for each gene cassette tested were comparable. Background luminescence (obtained from cell lysates prepared from S2 cells transfected with empty transfection mixes) was subtracted from the raw readouts of the luminescence signals. The ratio of Fluc:Rluc signal was calculated for each construct containing the native and the UAA-A stop codon context in their TR motif, as well as for the corresponding constructs where the stop codon is mutated to UUC. To calculate TR efficiency of test constructs with native and UAA-A stop codon contexts, their respective Fluc:Rluc values were divided with Fluc:Rluc values of constructs containing a UUC codon, which serve as positive controls. Non-paired two-tailed Student's t-test was used to analyze the results. For co-transfection assays, 100 ng of ubiquitous expression vector was used along with 100 ng of TMV dual reporter constructs. pUb expression vectors were constructed by using a modified pUWG vector (DGRC #1284) The modified pUGW vector with gateway cassette and mini white marker removed, was a kind gift from Herbert Jäckle, Max Planck Institute for Biophysical Chemistry, Göttingen. The genes of interest were cloned into pUb expression vectors using Gibson assembly. The primers used for cloning are listed in Supplementary Table S9. The co-transfected S2 cells were visualized using Olympus LV200 Microscope with objective 20× XAPO NA 0.8. Bioluminescence was measured with an exposure time of 1 min.
Drosophila handling and maintenance
Fly stocks were maintained on standard food with yeast, cornmeal, and agar in a controlled environment with constant temperature of 25°C, constant humidity, and 12 h–12 h light–dark cycle, unless otherwise stated. For control animals, we collected the progeny of w1118 (Bloomington Drosophila Stock Center, BDSC #5905) males crossed to OregonR (BDSC #5) virgin females.
CRISPR/Cas9 design
CRISPR/Cas9-based genome editing was employed to create three different genetic mutants of D. melanogaster that harbor mutations in and around the stop codon of the traffic jam (tj) gene (sequence location 2L:19 64 267 to 19 467 758). The CRISPR target finder tool (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) was used to find optimal protospacer adjacent motif (PAM) sites on the tj gene that flank the TR region between the first and the second stop codon of the tj ORF. The proximal PAM site was 5′ AGAGCTTT|GGCTATCGCCGC CGG 3′ and the distal PAM site was 5′ ACACAATG|TATAAGGTAAAT TGG 3′, where the NGG motifs are highlighted in bold. The 20 bp proximal and distal PAM regions were introduced upstream of gRNA scaffold into two separate pU6-BbsI-chiRNA vectors (Addgene #45946) (41) via blunt-end ligation-mediated insertional mutagenesis using primer pairs PK241_F/PK243_R and PK242_F/PK243_R respectively (Supplementary Table S6).
pHD-DsRed vector (Addgene #51434) carrying the homology arm 1 (HA1), the Template for Recombination (TfR), and the homology arm 2 (HA2) were generated in subsequent steps using Gibson assembly. HA1 (1100 bp) + TfR (250 bp) was amplified from gDNA obtained from w1118 as a single fragment and inserted upstream of loxP-DsRed-SV40poly(A)-loxP sequence. HA2 (1144 bp) was amplified and inserted immediately downstream of this sequence. The QuikChange mutagenesis protocol was used to introduce synonymous mutations into the proximal PAM sequence that borders HA1 and TfR, in order to prevent Cas9 from cleaving the vector once injected into the embryos. UGA to UUC mutation was then introduced in the TfR at the tj stop codon by QuikChange mutagenesis and 3xUAA was inserted downstream of the tj stop codon by blunt-end ligation method. 3xFlag was inserted upstream of the second stop codon by Gibson assembly. These cloning steps were performed in a pHD-DsRed vector in which the loxP1 site had been deleted in order to avoid complications associated with redundant primer binding sites. Finally, the loxP1 site was reinserted. Additionally, the dispensable phage pC31 attP site was removed from the pHD-DsRed vector during PCR amplification. The primers used for Gibson assembly, point mutations, and blunt-end ligation cloning are listed in Supplementary Table S6. Due to the introduction of an independent SV40 transcription termination signal in the TfR, the biogenesis of tj-derived piRNAs in the CRISPR-derived recombinants is inhibited. To overcome this limitation, the loxP-flanked DsRed-SV40 poly(A) marker cassette was removed by Cre-Lox recombination, which restored the native tj 3′ UTR in tj-TR mutants (tjmut). The introduction of the desired mutations was verified via sequencing of a genomic DNA-derived amplicon. DsRed deletion was confirmed via screening of eyes for negative fluorescence as well as by sequencing. The genome engineering services offered by BestGene Inc. Chino Hills, CA, USA were used for CRISPR injections. The fly strain used for injection had the following genotype: y1w1118; attP2(nos-cas9)/TM6C, Sb Tb.
DsRed-positive CRISPR mutants were crossed with Sco/CyO, Dfd-YFP (Sco/CDY) balancer lines (BDSC #8578) to obtain CDY-balanced mutant lines for second chromosome. tjmut(+DsRed)/CDY lines were crossed with Sco/Cre lines in order to achieve Cre recombinase-mediated removal of the DsRed marker. The progenies, tjmut(±DsRed)/Cre, were back-crossed with Sco/CDY balancer lines to obtain DsRed-deleted, tjmut/CDY flies that served as stocks. DsRed deletion was confirmed by screening individual balanced flies for the absence of DsRed. The tjmut/tjmut obtained by back-crossing of tjmut/CDY flies were used for experimental purposes. gDNA was extracted from the homozygous mutant flies. Using it as template, the genomic region flanking PAM sites was amplified using primers PK277_F and PK278_R and sequenced using primers PK277_F and PK279_F to confirm the introduced mutations (Supplementary Table S6). w1118 flies were used as wild-type controls as they have the closest genetic background to the mutants.
Immunohistochemistry
Immunofluorescent studies were performed using standard procedures (König and Shcherbata, 2013). Primary antibodies used were: guinea pig anti-Tj (1:10 000; a kind gift from Dorothea Godt, University of Toronto, Canada), rabbit anti-Vasa (1:5000; a gift from Herbert Jäckle, MPI-bpc, Göttingen), mouse anti-Engrailed (1:20; Developmental Studies Hybridoma Bank, DSHB), mouse anti-Fas3 (1:20; DSHB), rat anti-DE-Cadherin (1:20; DSHB), rabbit anti-mCherry (1:200; Abcam), and mouse anti-Flag (1:500; Sigma Aldrich). The following secondary antibodies were used: goat anti-guinea pig Alexa 647 (1:500; Life Technologies, A-21450), goat anti-mouse Alexa 488 (1:500; Molecular Probes) and goat anti-rabbit Alexa 568 (1:500; Molecular Probes). DAPI (Sigma) was used to stain the nuclei. Samples were imaged using Zeiss LSM 700. ImageJ and CorelDrawX8 were used to make Figures.
Histology of Drosophila brains
For analysis of adult brain morphology, 7 μm paraffin-embedded sections were cut from fly heads. To prepare Drosophila brain sections, the fly heads were immobilized in collars in the required orientation and fixed in Carnoy fixative solution (6:3:1 = ethanol:chloroform:acetic acid) at 4°C overnight. Tissue dehydration and embedding in paraffin was performed as described previously (42). Histological sections were prepared using a Hyrax M25 (Zeiss) microtome and stained with hematoxylin and eosin (H&E) as described previously (43). All chemicals for these procedures were obtained from Sigma Aldrich.
Gene expression analysis
Standard miniprep protocol for D. melanogaster genomic DNA extraction was followed as described in Huang et al. (44). Total RNA was extracted from heads and ovaries of 3–4 days old adult flies of each genotype using TRIzol™ reagent (Jena Bioscience) following manufacturer's protocol. Extracted RNA was quantified and treated with DNaseI (2 units per μg of RNA). Total cDNA was prepared using random primers with High Capacity Reverse Transcriptase (ThermoFisher) following manufacturer's instructions. 20 μl reverse transcription (RT) reactions were set up for 1 μg RNA template. RT-qPCR was performed using Fast SYBR® Green PCR kit (Applied Bioystems™). Each reaction was performed in 15 μl volume using 20 ng cDNA template and 200 nM primers. All reactions were performed in triplicates. Control reactions were set up for each target gene using non-RT templates. The primer sequences used for each transcript quantification were designed using the Drosophila RNAi Screening Center (DRSC) FlyPrimerBank and are listed in Supplementary Table S7 (45). The qPCR reaction conditions used were according to manufacturer's instructions. αTub84B was used as endogenous control and tjnat/nat flies were used as control samples. The analysis of the acquired threshold cycle (CT) values was performed using StepOne Software. CT value denotes the fractional cycle number at which the fluorescence signal for each test sample passes a defined threshold. Average CT values from three technical replicates of respective genes were subtracted from that of the αTub84B control to obtain ΔCT. These ΔCT values for each gene were normalized again by subtracting the ΔCT of the control sample from the ΔCT of the test sample. The ΔΔCT values thus obtained were used to calculate gene expression levels by using the formula: relative quantification (RQ) = 2−ΔΔCT. Non-paired two-tailed Student's t-test was used for calculating p-values. For quantification of individual cellular tRNAs, a modified stem-loop qPCR method was used as published by (46). Stem-loop reverse transcription primers were used for individual tRNAs (Supplementary Table S8). A separate stem–loop primer was also designed for 18S rRNA, which is used as an internal control. cDNA was prepared using SuperScript III™ Reverse Transcriptase following manufacturer's protocol. The amplification for individual tRNAs using RT-qPCR was performed using tRNA-specific forward primers and a universal reverse primer (Supplementary Table S8). RNA sequencing services were provided by Transcriptome and Genome Analysis Laboratory (TAL), Göttingen.
Stress induction and fly survival assays
Stress induction experiments were carried out with 5–6 days old flies. Approx. 50 flies were used for each condition. Flies were starved by placing them in empty vials with a glass fiber disk soaked in water. Acute heat stress was induced by incubating the flies at 36°C for 3 h. Oxidative stress was induced by placing flies in vials with a glass fiber disk soaked in 5% sucrose and 20 mM paraquat solution for 12 h. For sleep deprivation, vials containing flies were shaken at 300 rpm under constant light source for 24 h. To study ageing, flies were reared on standard food vials for 3 weeks, with food changed every week. The method for lysate preparation from stressed fly heads is described in the following section. The survivability rate of flies upon oxidative stress was recorded by observing fly activity for 3 days upon feeding them 5% sucrose, 20 mM paraquat solution as mentioned above. Twenty flies were taken for each genotype and the study was performed in five biological replicates.
Quantitative western blot
Whole tissue lysates were prepared from ovaries and heads using RIPA buffer (Sigma Aldrich), and the protein concentration was measured using Pierce BCA Protein Assay Kit (ThermoFisher). Equal amounts of total protein extract were loaded as samples for western blot. Instead of using housekeeping genes as loading controls, we utilized the total protein normalization (TNP) method (Revert™700 Total Protein Stain Li-Cor). The TNP staining gives a linear signal range, which is especially relevant for low-abundance proteins such as Tj. Anti-DYKDDDDK-HRP antibodies (Miltenyi Biotec) were used to immunostain Tj-TR-3xFlag bands. HRP signal was developed using Chemiluminescent substrates (ThermoFisher) and imaged in the Odyssey Fc Imaging System. The anti-Flag chemiluminescent signal from each tissue derived from tjnat/nat mutants was normalized to the respective TNP signal and compared to the anti-Flag/TNP signal from tissues derived from tjTR/TR mutants to calculate TR efficiency of Tj.
Quantification and statistical analysis
For luciferase assays, error bars show the standard deviation obtained from at least three technical replicates (n = 3). For constructs with stop codon context of UGAC, three independent biological replicates were performed. Statistical significance was determined by two-tailed unpaired Student's t-test wherein P-values of <0.05 were considered significant in all cases. RNAfold web server was used for prediction of secondary structures on mRNA sequences. All western blots were performed using head samples from three biological replicates for each mutant fly line. Two-way tables and Chi-square testes were used to determine significant differences in the phenotypes between the mutants. Statistical analyses were performed with Graphpad prism 8. RT-qPCR studies were performed from cDNA obtained from three independent biological samples. Three technical replicates were used for each biological sample. StepOne Software v2.3 was used to perform comparative CT (ΔΔCT) quantification. The data were normalized against average ΔCT of the housekeeping gene αTub84B. Data values from tjnat/nat sample were used as endogenous control. P-values are calculated using two-tailed unpaired Student's t-test with Welch's correction of standard deviation from ΔΔCT values of tjnat/nat and tjTR/TR samples.
For quantification of neural lesions, CBs and OLs were analyzed independently, and the percentage of CBs or OLs exhibiting neural lesions, out of the total brains analyzed, was calculated for each genotype. For immunostained brains, z-stack images were acquired through the brains using a Zeiss LSM700 confocal microscope. Examination of all slices was taken into account to identify lesions, since they are found to occur throughout both CBs and OLs of analyzed brains. For H&E-stained and sectioned brains, multiple sections for each brain were viewed and assessed for presence of lesions.
For quantification of niche, germarium, and ovariole phenotypes, we used immunofluorescent staining for common markers to discern the presence of mutant phenotypes. Ovaries from each genotype were stained, and then ovarioles were separated from one another on slides prior to imaging. Each ovariole, corresponding germarium, and germline stem cell (GSC) niche was examined, and each was counted independently as either normal or exhibiting mutant phenotypes. En staining was used to evaluate GSC niches, as cap cells are countable with this marker. Vasa staining was used to assess the health and differentiation of the germline. Fas3 staining was used to visualize the follicular epithelium and to monitor the process of germline encapsulation in the posterior end of germaria. Finally, DAPI combined with all other stains was used to evaluate the overall shape and size of germaria; in addition, condensed and fragmented DAPI staining was used as an indicator of cell death in egg chambers. Two-way tables and chi-square tests were used to determine significant differences in the phenotypes between the mutants.
RESULTS
Quantification of TR in candidate genes in Drosophila
Phylogenetic analysis identified >300 TR candidates in Drosophila (7), the majority of which have not been experimentally tested. We narrowed down the list of potential candidates to 11 genes that perform important functions during fly development (Supplementary Table S1). Their functions are well-characterized and associated with traceable phenotypes. Among the selected genes, klumpfuss (klu), doublesex (dsx), traffic jam (tj), seven up (svp), chronologically inappropriate morphogenesis (chinmo), fruitless (fru) and broad (br) encode transcription factors or transcriptional regulators; atypical protein kinase C (aPKC), and discs large 1 (dlg1) encode protein kinases involved in cell signaling; wishful thinking (wit) encodes a signaling receptor; and Kinesin heavy chain-73 (Khc-73) encodes a motor protein that regulates cell polarity. The lengths of the expected TR extensions range from 11 to 236 amino acids (Supplementary Table S1). Out of the candidate genes selected, wit mRNA has a strong propensity to form a secondary structure within the 3′ UTR region accommodated by the reporter test cassette as predicted by RNAfold (7).
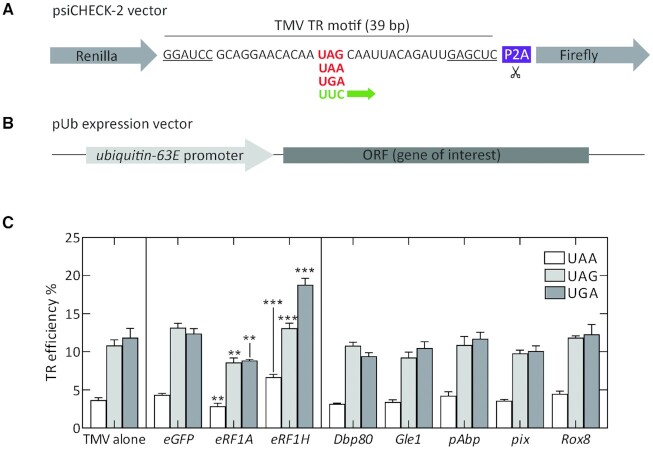
To validate the TR of the candidate genes, we constructed dual luciferase reporters for the 11 candidate genes and tested their expression in S2 cell lines (Figure 1A). We generated a psiCHECK™-2 based dual luciferase vector in which the Renilla and Firefly luciferase genes are fused into a single ORF, separated by the test cassette for each gene and a self-cleaving P2A sequence. For each gene construct, we generated a positive control by mutating the native stop codons to the UUC sense codon, resulting in constitutive Firefly synthesis. Additionally, we mutated the extended tetranucleotide stop codon sequence to UAA-A to obtain control constructs with a highly efficient translation termination context. αTubulin 84B (αTub84B), which is not a TR candidate, was used as a negative control. The constructs were introduced into S2 cells, and TR efficiency was calculated from the activity of Renilla and Firefly reporters. TR efficiency of the control αTub84B sequence is 0.35% (Figure 1B), similar to a basal TR frequency of 0.02–1.4% reported for yeast and mammalian cell lines (15,47–51). Mutating the native tetranucleotide termination signal of αTub84B from UAA-G to UGA-C does not increase TR values, indicating that the 105 bp αTub84B test cassette represents a robust sequence with efficient termination independent of the immediate stop codon context.
Figure 1.
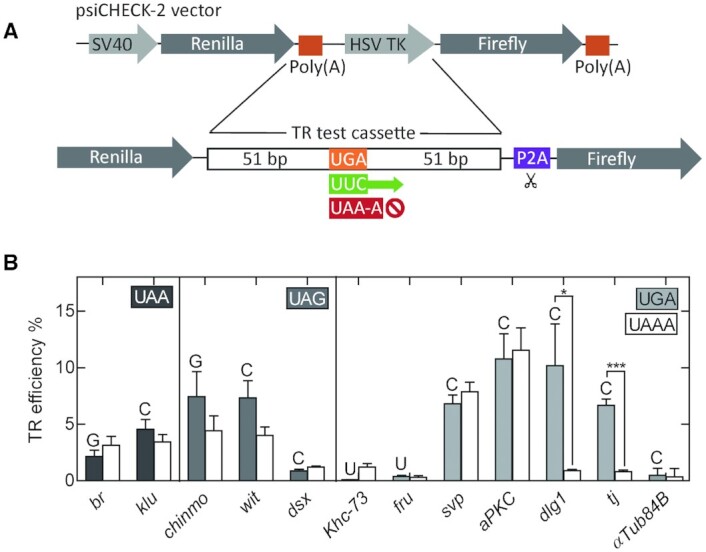
Quantification of TR in putative candidate genes using dual luciferase reporter assay in S2 cells. (A) Construct design for dual luciferase reporter vectors. (B) TR efficiencies of putative candidate genes with UAA, UAG, and UGA stop codons determined by dual luciferase assay in S2 cells. The +4 nucleotide for each gene is indicated by the letter above each bar. White bars represent TR efficiencies for corresponding genes upon mutating the native tetranucleotide termination signal to UAA-A. αTub84B represents a negative control. The error bars indicate the SD from three biological replicates. P-values are calculated using two-tailed unpaired Student's t-test from datasets generated from three biological replicates, each with three technical replicates (*P< 0.05, ***P< 0.0005).
Among the candidate genes selected, dsx, Khc73, and fru show basal TR levels, i.e. they do not undergo TR at the conditions tested (Figure 1B). The TR efficiency of br and klu, which harbor a UAA stop codon followed by G or C, is independent of the nucleotide at +4 position, suggesting that the moderate TR may be induced by elements beyond the stop codon. For genes containing a UAG stop codon, chinmo and wit, the TR efficiency is reduced by about a half by mutating nucleotides +3 and +4, suggesting that TR depends not only on the extended stop codon, but also on an external signal(s). The four genes with the UGA-C stop signal, svp, aPKC, dlg1 and tj, exhibit the highest TR efficiencies, ranging from about 7% to 11%. Mutating the tetranucleotide signal to UAA-A in dlg1 and tj abolishes TR efficiency, indicating that the immediate nucleotide context is the only requirement to drive TR in these genes. TR in svp and aPKC is unaffected upon mutating the tetranucleotide termination signal to UAA-A.
Tissue-specific TR in tj during embryogenesis and in adult Drosophila
To study the physiological relevance of TR in Drosophila, we have chosen tj as a model TR gene. The leaky UGA-C tetranucleotide is sufficient to induce TR in tj (Figure 1B). Because the gene encoding tj lacks introns, genetic manipulation of the gene is relatively simple and avoids the complications of working with multiple splice isoforms. The ORF of tj is 509 codons-long; the TR extension would append an additional 44 amino acids, generating a larger protein that we call the Tj-TR isoform.
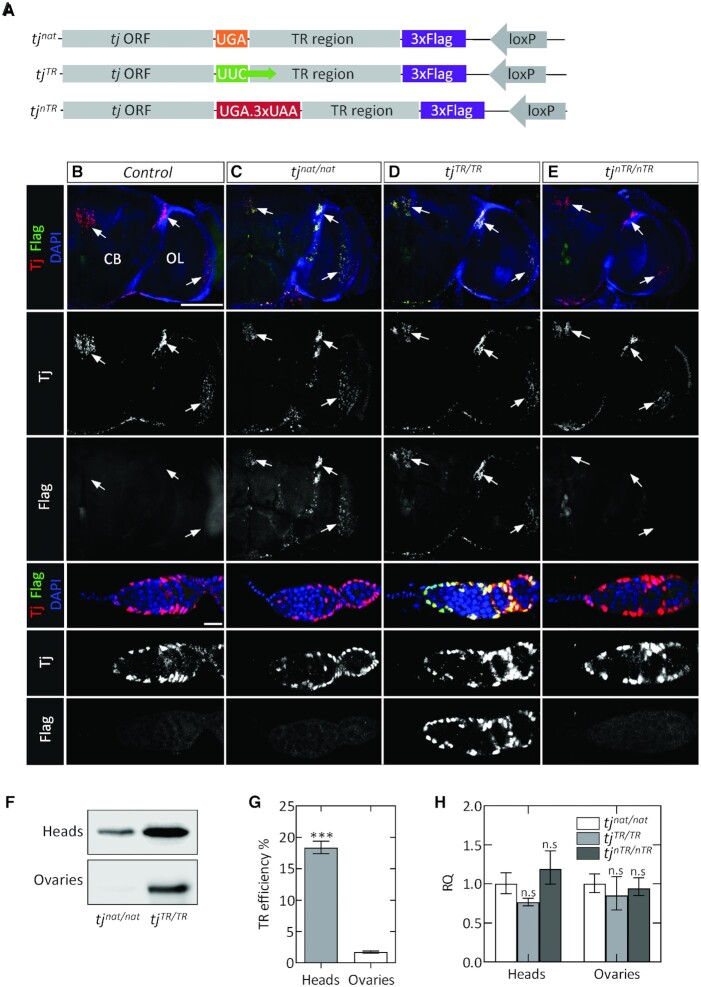
To study TR in Drosophila, we created three mutant fly lines that harbor mutations at the termination sequence of tj using CRISPR/Cas9-based genome editing (Supplementary Figure S1). The mutants were designed to code for a Flag epitope tag downstream of the TR extension (Figure 2A). The first mutation, tjnat, does not alter the primary stop signal that terminates the tj ORF; TR in this mutant is expected to occur at the same frequency as in the native tj. The tjTR mutation replaces the primary tj stop codon with a UUC sense codon, such that the mutant flies undergo constitutive TR and produce only the Flag-tagged TR isoform. Finally, the tjnTR mutation introduces multiple stop codons after the primary tj stop codon, which leads to complete abolition of TR. Homozygous flies for each of the three genomic mutations are viable and do not exhibit obvious phenotypes.
Figure 2.
Tissue-specific expression of the Tj-TR isoform in Drosophila. (A) Sequence map of chromosomal modifications introduced in the tj locus by CRISPR/Cas9 genome editing. (B–E) Adult brains and ovaries of tj-TR mutants; each column depicts brains (upper panels) and germaria (lower panels) of the indicated genotypes. For both organs, Tj and Flag are shown in single channels below the merged images. (B) Control tissues express no Flag. Endogenous Tj is expressed in the brain, especially in three distinct clusters (arrows) and in several types of somatic cells in the germarium. (C) tjnat/nat brains express Tj in a pattern indistinguishable from control. In addition, brains express the Flag-tagged TR isoform, demonstrating the occurrence of TR in neural cells. In germaria, Tj appears as in control, but no Flag staining is detectable. (D) Overlapping pattern of Tj and Flag staining in tjTR/TR brains and germaria, consistent with constitutive induction of TR in Tj. (E) Abrogation of TR in Tj observed in tjnTR/nTR brains and germaria, evidenced by the lack of Flag stain. Scale bars: 100 μm for brains; 10 μm for germaria. (F) Western blot with anti-Flag antibodies showing the relative abundance of Flag-tagged Tj-TR isoform in adult tissues from tjnat/nat and tjTR/TR mutants. (G) TR efficiency estimated in adult heads and ovaries using quantitative western blot using total protein normalization. P-values were calculated using unpaired Student's t-test obtained from three biological replicates (*P< 0.05, **P< 0.005, ***P< 0.0005). (H) RT-qPCR analysis of tj transcripts using cDNA prepared from adult heads of tjnat/nat, tjTR/TR and tjnTR/nTR mutants. Error bars represent the upper and lower limit of RQ defined by the standard deviation of ΔΔCT from three biological replicates. The data were normalized against average ΔCT of the housekeeping gene αTub84B. Two-tailed unpaired Student's t-tests were performed from the ΔΔCT values of tjnat/nat vs tjTR/TR and tjnat/nat versus tjnTR/nTR (n.s., not significantly different from control).
To detect the expression of Tj and Tj-TR isoforms during embryonic development, we stained stage 15–17 embryos with antibodies specific to Tj and Flag. Anti-Tj recognizes both the Tj and Tj-TR isoforms while anti-Flag detects only Tj-TR. In controls and in all tj-TR mutant embryos, Tj is expressed in a subset of neural cells in the ventral nerve cord (VNC) and brain as well as in the somatic gonadal precursors (SGPs) of the embryonic ovary (Supplementary Figure S2). The Tj-TR isoform is expressed in the VNC but not in the ovaries of tjnat/nat embryos, suggesting that TR in tj is regulated in a tissue-specific manner during embryogenesis (Supplementary Figure S2B). tjTR/TR mutants constitutively express the Tj-TR isoform in the VNC as well as gonads, while tjnTR/nTR mutants only express the native Tj (Supplementary Figure S2C and S2D). Furthermore, the Tj and Tj-TR isoforms in tjTR/TR embryonic SGPs remain strictly nuclear (Supplementary Figure S2C), indicating that the TR extension does not affect the nuclear localization of Tj.
Next, we sought to determine whether tissue-specific expression of the Tj-TR isoform persists until adulthood. First, we inspected adult brains. The brain is composed of two bilateral hemispheres, each of which is divided into a medial compartment termed the central brain (CB) and a lateral optic lobe (OL), which receives primary visual input from photoreceptor neurons. Marking tj-expressing cells with membrane-bound RFP (tj > mRFP) allows the visualization of neuronal processes and demonstrates that the brain contains many Tj-positive neurons, especially dispersed throughout the OL; in addition, several prominent clusters of Tj-expressing cells exist, particularly in the junction of the lobula and the CB and in some neurons of the pars intercerebralis (PI) (Supplementary Figure S3A). Next, we examined brains from control as well as all three tj-TR mutants (Figure 2B–E). The pattern and number of Tj-expressing cells do not appear to be different in any of these genotypes. Interestingly, the cells expressing Tj in the brains of tjnat/nat flies also express the Tj-TR isoform, indicating that the nervous tissue-specific regulation of TR in tj is maintained in adults (Figure 2C). As expected, in tjTR/TR brains, Flag expression exhibits complete overlap with Tj (Figure 2D), while Flag expression is absent from tjnTR/nTR brains (Figure 2E).
Next, we examined adult ovaries for the existence of tj-TR. Each ovary in Drosophila is composed of about nineteen parallel egg production units called ovarioles. At the anterior tip of each ovariole is a structure called the germarium that houses the germline stem cells (GSCs) as well as several somatic cell subtypes that express Tj. We stained control germaria as well as germaria from the three tj-TR mutants with antibodies against Tj and Flag (Figure 2, bottom rows). We did not observe differences in the expression pattern of native Tj protein in any of the mutants. As expected, Flag staining is undetectable in control and tjnTR/nTR germaria, and Flag overlaps precisely with Tj in tjTR/TR germaria. However, in tjnat/nat germaria, in contrast to the pattern in adult brains in which Flag and Tj overlap, Flag staining was undetectable (Figure 2B, compare Flag in brain and germarium). These results demonstrate that the tissue-specific regulation of TR in tj exists during adulthood as in embryogenesis.
To estimate the TR efficiency in adult tissues, we utilized quantitative western blot and probed the abundance of Flag-tagged Tj-TR isoform in tissue lysates from tjnat/nat and tjTR/TR adult flies using anti-Flag antibodies. We estimated a TR efficiency of ∼20% in adult heads, whereas adult ovaries exhibited only basal levels of TR (Figure 2F, G). The potential differences in tj transcript levels between the mutants were ruled out by RT-qPCR experiments (Figure 2H). Given that the Tj staining is very similar in both tjTR/TR and tjnTR/nTR mutant lines (Figure 2 and Supplementary Figure S2), the TR extension does not seem to trigger selective degradation of the protein. Thus, TR is regulated on a translational level and is not caused by tissue-specific differences in mRNA levels or protein stability or by altered cellular localization of the two Tj isoforms.
Effect of TR induction in adult brains
Because Tj-TR is expressed in the nervous system (Figure 2 and Supplementary Figure S2), we analyzed the effect of differential TR in adult brains. We also included brains from tj hypomorphic flies (tjhypo) to compare with the tj-TR mutants. The hypomorphic flies bore the tjPL3 allele, a genetic null, in trans to tj-Gal4, which itself is reported to be a weak hypomorphic allele (34). We stained brains with an antibody against the cell adhesion molecule DE-Cadherin (Cad), ubiquitous expression of which allows visualization of the neuropils of the brain (Figure 3A). In addition, we prepared brain sections stained with hematoxylin and eosin (H&E) for histological evaluation. Using these methods, we found evidence of neural lesions in tjhypo brains as well as in both tjTR/TRand tjnTR/nTR brains (Figure 3 and Supplementary Figure S3). In immunofluorescence images, these lesions are visible either as regions in which Cad staining is cleared, indicating the absence of neural cells or projections (Figure 3A–E). In H&E sections, lesions are visible as stain-free clearings (Supplementary Figure S3B). We quantified brain lesions in the CB (Figure 3F) and in the OL (Figure 3G). The incidence of these lesions in both compartments is significantly greater in brains of tjhypo, tjTR/TR and tjnTR/nTR flies than in tjnat, which exhibits lesions at a rate similar to control brains (Figure 3F and G). These results show that in the CNS, regulation of Tj and Tj-TR expression is crucial for neuroprotection.
Figure 3.
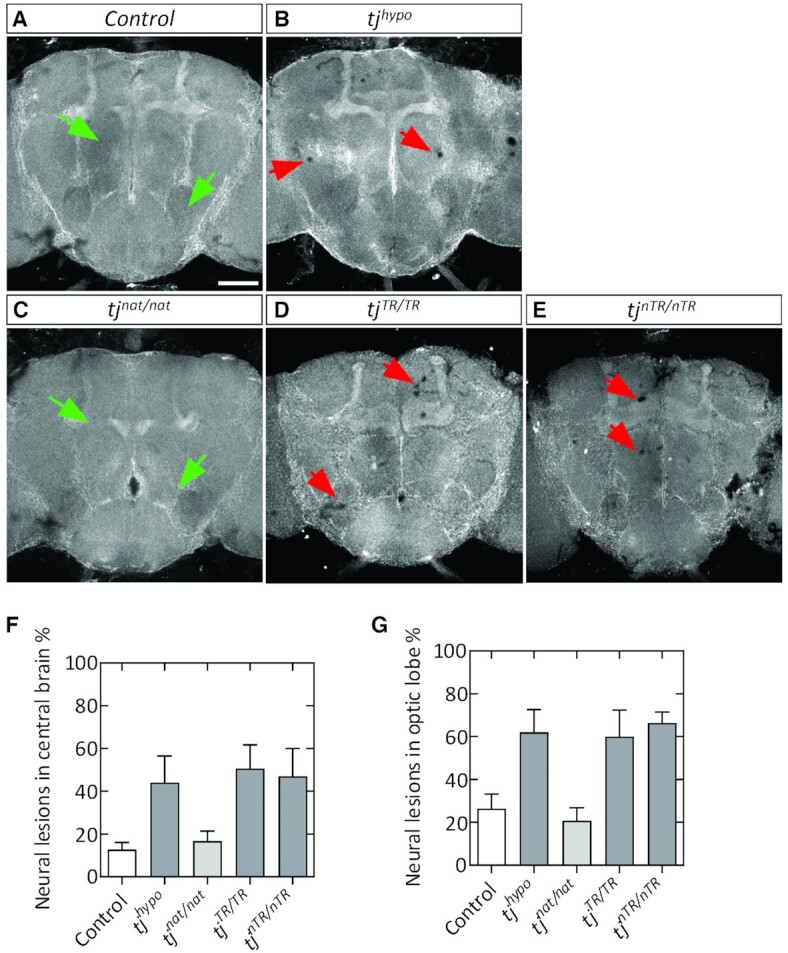
Neural lesions in adult brains caused by perturbation of proper regulation of tj-TR. (A–E) Young (3–4 day old) adult brains stained with DE-Cadherin (Cad). (A, B) Cad staining usually appears homogeneous in control brains (A; e.g. green arrowheads) and is disrupted by instances of sporadic lesions in tjhypo brains (B; red arrowheads). (C–E) tj-TR mutant brain phenotypes. tjnat/nat brains appear similar to control brains (C). Lesions occur sporadically throughout tjTR/TR (D) and tjnTR/nTR (E) brains, similar to tjhypo. Scale bar: 100 μm. (F, G) Quantifications show the percentage of CBs (F) and OLs (G) that exhibit neural lesions, out of the total brains analyzed. At least three biological replicates were performed. n ≥ 42 CBs and n ≥ 74 OLs per genotype. Student's t-test was used to determine significance (*P < 0.05; n.s., not significantly different from control).
Effect of TR induction in ovaries
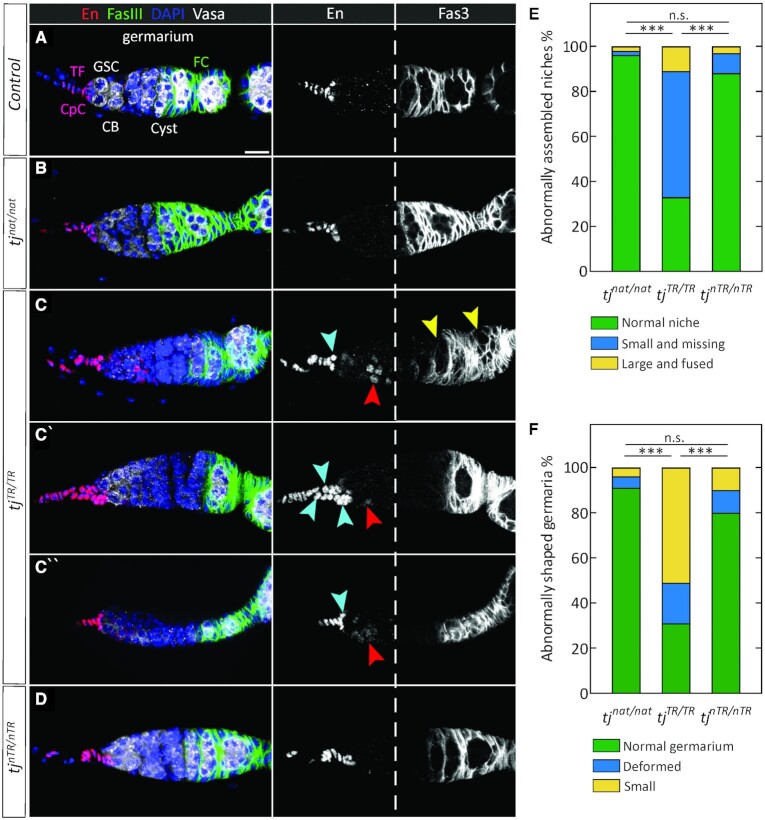
As Tj plays an important role in gonad development, we examined how TR affects Tj function in adult ovaries, specifically focusing on germaria, the structures that house the GSCs and GSC niches. The GSC niche comprises an anterior stack of disc-shaped terminal filament cells (TFCs) and a cluster of 6–7 cap cells (CpCs) that make direct contact with GSCs (Figure 4A) (52). GSCs divide asymmetrically to generate cystoblasts that further divide to yield a 16-cell germline cluster. These clusters become enveloped by somatic follicle cells to form cysts that bud off from the posterior end of the germarium. Germline cells can be identified based on expression of Vasa, an RNA-binding protein. TFCs and CpCs express the transcription factor Engrailed (En). Tj is expressed in CpCs but not in the TFCs (34,36). Tj is also expressed in some other somatic cells, including the follicle cells that surround the germline cysts, as well as escort cells (ECs), which are interspersed with and comprise a differentiation niche for germline cells in the germarium (Figure 4) (34,36). The cell adhesion molecule Fas3 is expressed in the follicle cells in early egg chambers as well (53).
Figure 4.
Defects in adult germaria due to constitutive TR in Tj. (A) Control germarium. Engrailed (En) is expressed in TFCs and CpCs. Fas3 is present in follicle cells, which are present in the posterior half of the germarium (right panel from the white dashed line) and in the follicular epithelia surrounding the early-stage egg chambers. Shown are some of the relevant cell types (CB, cystoblast; CpC, cap cell; FC, follicle cell; GSC, germline stem cell; TF, terminal filament). Also indicated is a single Cyst, a 16-cell germline cluster surrounded by FCs. Scale bar: 10 μm. (B) Wild-type-like morphology of tjnat/nat germaria. (C–C’’) Diverse phenotypes observed in tjTR/TR niches and germaria. Some niches appear small (blue arrowheads, C and C″), some are enlarged, appearing to be composed of multiple niches that have fused (multiple blue arrowheads, C′) while some are indiscernible or absent. Similarly, a majority of the germaria are small or deformed (C″). Ectopic expression of En is observed in a subset of escort cells in all tjTR/TR germaria (red arrowheads). In some germaria, the Fas3-positive follicle cells fail to encapsulate the germline cells (yellow arrowheads). (D) Organization of tjnTR/nTR germaria. The GSC niche and germaria of tjnTR/nTR ovaries appear normal and are indistinguishable from control or tjnat/nat. (E) Quantification of niche phenotypes described above and observed in (B–D). (F) Quantification of defects in germaria observed in (B–D, n > 120 per genotype). Two-way tables and chi-square tests were used to determine any significant differences in the distribution of phenotypes between the mutants (***P < 10–4; n.s., not significantly different).
Using En as a marker for TFCs and CpCs, it is possible to distinguish between them based on the disc shape and stacked organization of the TFCs and the clustered, round, germline-adjacent nature of CpCs. Thus, we used En staining and determined that the GSC niches appear indistinguishable from control in the tjnat/nat and tjnTR/nTR flies (Figure 4), a result consistent with our observation that TR is absent in ovaries. In contrast, constitutive TR in tj produces diverse defects (Figure 4C–C″ and Supplementary Figure S4). Whereas tjnat/nat and tjnTR/nTR germaria nearly always have normal terminal filaments and a cluster of about six CpCs (Figure 4B and D), tjTR/TR niches are often small or absent (Figure 4C and C″, blue arrowheads). In addition, a small number of mutant niches appear enlarged, perhaps resulting from two or more adjacent niches that have fused (Figure 4C′, blue arrowheads; quantified in Figure 4E and Supplementary Table S2).
Germaria in the tjTR/TR mutants are frequently small or exhibit deformities (Figure 4C″ and Supplementary Figure S4; quantified in Figure 4F and Supplementary Table S2); these phenotypes are similar to those in tjhypo ovarioles (Supplementary Figure S4B and S4C). While in tjnat/nat and tjnTR/nTR, En was restricted to the TFCs and CpCs, we found that 100% of tjTR/TR germaria exhibit ectopic En expression in escort cells, albeit more weakly than in the GSC niche cells (Figure 4C-C″, red arrowheads). These results are in agreement with previous observations (54), in which somatic knockdown of tj resulted in derepression of En in anterior ECs, which form the differentiation niche regulating the efficiency of GSC progeny differentiation (55). Since En is an activator of dpp expression (56), its presence in ECs can disrupt the differentiation niche and lead to a failure of the germline to develop properly. Thus, our data suggest that the Tj-TR protein behaves as a hypomorphic mutant in the somatic cells of the germarium.
In the posterior part of the germarium, germline becomes encapsulated by follicle cells, which are derived from two follicle stem cells (FSCs), located about halfway along the A-P axis of the germarium. Fas3 is expressed in early follicle cells and can be used to visualize the morphology and organization of the follicular epithelium (Figure 4A). Using this marker, we frequently saw tjTR/TR germaria in which follicle cells fail to surround the germline (Figure 4C, yellow arrowheads), whereas in control, tjnat/nat and tjnTR/nTR germaria, germline cysts are consistently encapsulated by Fas3-positive cells (Figure 4A, B and D). Germline left unprotected by the follicular epithelium is known to be susceptible to cell death; indeed, tjTR/TR ovarioles have a strong increase in the number of dying egg chambers (Supplementary Figure S4D and Supplementary Table S2).
TR modulates selective gene regulatory properties of Tj
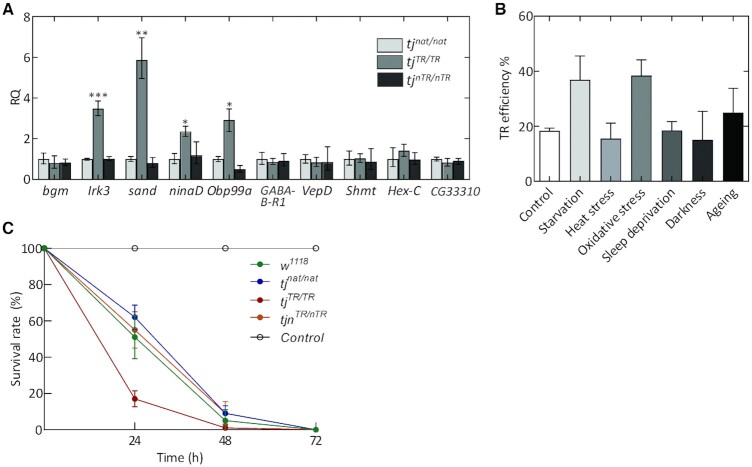
Because the Tj-TR isoform is specific to neural cells of the CNS, we studied how induction or disruption of TR affects the expression of the known target genes regulated by Tj. We performed RT-qPCR with adult head tissues from all three tj-TR mutants. The expression levels of the VGlut, Rh6, melt and wts, which are regulated by Tj (36,39,57), did not change significantly (Supplementary Figure S5). High-throughput RNA sequencing (RNA-seq) on adult brain samples from the three tj-TR mutants identified genes dysregulated in tjTR/TR or tjnTR/nTR brain samples. Included are genes that act in metabolic processes, stress responses, signaling pathways, and mitochondria (Supplementary Table S3, Supplementary Figure S6). We then validated the selective dysregulation of several candidate target genes identified by RNA-seq using RT-qPCR analysis. Genes that exhibit marked upregulation upon TR induction in tj include Inwardly rectifying potassium channel 3 (Irk3), sandman (sand), neither activation nor afterpotential D (ninaD) and Odorant-binding protein 99a (Obp99a) (Figure 5A). These genes represent potential transcriptional targets that might be positively regulated by the Tj-TR isoform. This suggests that the Tj-TR isoform affects CNS functions related to perception of external stimuli (such as light and olfactory molecules) and homeostatic cellular response to such stimuli. The observation that the expression levels of these genes in tjnTR/nTR mutants were comparable to tjnat/nat suggests that the effects are due to the overexpression of the Tj-TR isoform in tjTR/TR flies, whereas a moderate Tj-TR expression in controlled lab conditions do not upregulate these genes. It is also possible that an effect could also be masked by using whole heads instead of isolated brains.
Figure 5.
Effect of TR in Tj on the transcriptome profile in adult heads. (A) RT-qPCR analysis of genes dysregulated in tj-TR mutants using cDNA prepared from adult heads. Error bars represent the upper and lower limits of RQ values defined by the standard deviation of ΔΔCT. The data were normalized against average ΔCT of the housekeeping gene αTub84B. P-values were calculated using two-tailed unpaired Student's t-test with Welch's correction of standard deviation from ΔΔCT values of tjnat/nat and tjTR/TR samples, obtained from three biological replicates (*P< 0.05, **P< 0.005, ***P< 0.0005). (B) Effect of different stress conditions on TR efficiencies in Tj. Head lysates from stressed tjnat/nat and tjTR/TRmutant flies were used to perform western blot. Total protein normalization was used for determining TR efficiencies. Error bars represent SD from three biological replicates. (C) Survival rate of wild-type (w1118), tjnat/nat, tjTR/TR and tjnTR/nTR mutants upon stress induction via paraquat ingestion (20 mM paraquat in 3% sucrose). Control group represents w1118 flies that were not exposed to paraquat. Error bars represent SD from five biological replicates.
Because several genes upregulated in tjTR/TR mutants are largely involved in homeostatic functions, we tested whether the TR efficiency is affected by different stress conditions. We subjected tjnat/nat and tjTR/TR mutants to several stressors such as starvation, heat stress, oxidative stress, sleep deprivation, and constant exposure to darkness and measured TR efficiency in adult heads using quantitative western blot. We also aged the adult flies for two weeks to test whether TR efficiency changes with chronological ageing. While heat stress, sleep deprivation, and darkness do not have an effect on TR in Tj, starvation and oxidative stress enhance the TR efficiency to ∼40%; ageing has only a modest effect (Figure 5B and Supplementary Figure S5B). Thus, the expression level of Tj-TR can be fine-tuned at certain stress conditions.
In order to understand the physiological relevance of these observations, we tested the effect of stress induction on the survival rates of 4–5 days old wild-type and mutant flies (Figure 5C). We induced oxidative stress via paraquat ingestion and recorded the viability of flies over three days. Interestingly, the survival rates of the tjnTR/nTRmutants was comparable to that of tjnat/nat and wild-type flies, while the tjTR/TR mutants exhibited reduced survival rates. Although we did not observe any susceptibility in tjnTR/nTRflies, we cannot rule out the possibility that TR in Tj might aid in internal cellular responses that do not manifest in organismal survivability. On the other hand, the importance of a proper regulation of TR is further corroborated by the observation that tjTR/TR mutants have decreased viability when subject to acute oxidative stress.
The tissue-specific repertoire of factors involved in translation termination
Tissue-specific differences in TR can be attributed to factors that can potentially upregulate global TR at leaky stop codons or trans factors that drive gene-specific TR by binding to target mRNA elements outside the stop codon (20). In particular, differences in the relative abundance, activity, and modifications of eukaryotic release factors (eRF1 and eRF3) and tRNAs that can recognize stop codons can play a role in modulating tissue-specific TR (58–60). As our luciferase data suggest that TR in tj is solely determined by the tetranucleotide termination signal, and is independent of extended mRNA element (Figure 1B), the most likely tissue-specific regulators are those interacting with the stop codon directly.
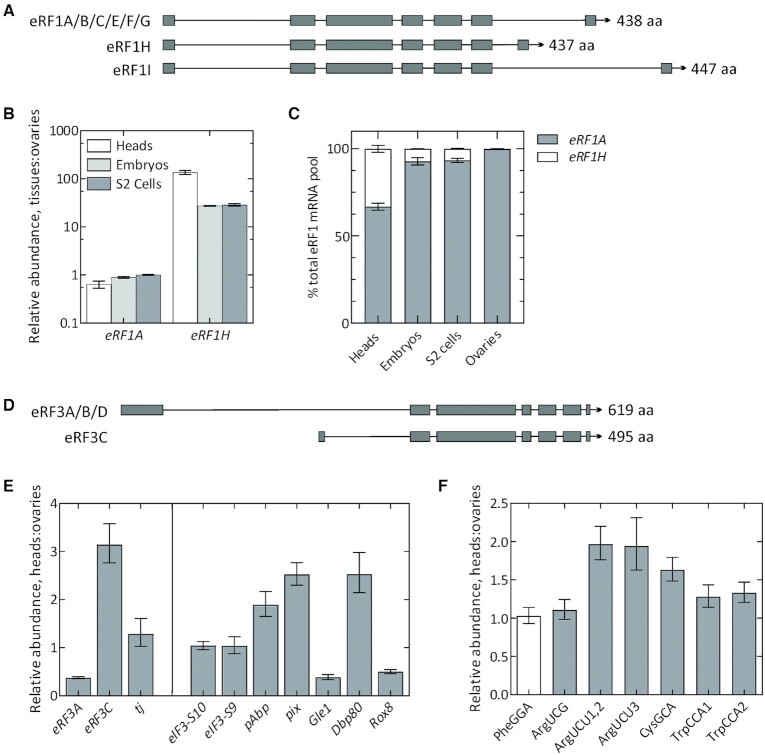
To test this hypothesis, we first quantified the relative levels of eukaryotic release factors, eRF1 and eRF3. eRF1 in Drosophila has 8 annotated transcript isoforms with at least 3 different predicted protein isoforms (Figure 6A). These protein isoforms have a conserved core but possess differentially spliced C-termini. Isoforms A, B, C, E, F and G possess the same C-terminus and give rise to a 438 aa long protein product (henceforth, referred to as eRF1A). eRF1H and eRF1I are unique isoforms that are 437 aa and 447 aa in length, respectively. We performed RT-qPCR-based relative quantification of each of these factors and their isoforms using isoform-specific primer pairs. eRF1A, which is the major eRF1 isoform, is expressed at similar levels across different tissues tested, although its relative abundance is slightly lower in heads. Interestingly, the eRF1H isoform is significantly more abundant in heads (∼130-fold), as well as in embryos and S2 cells (∼30-fold), compared to ovaries (Figure 6B). The relative expression levels of eRF1H across different tissue types correlates with the tissue-specific readthrough efficiencies (Figures 1B and 2G) We note that the eRF1 transcript pool in ovaries is comprised almost entirely of eRF1A, whereas eRF1H constitutes 33%, 7.3% and 6.7% of the total eRF1 transcript pool in heads, embryos, and S2 cells, respectively (Figure 6C). We excluded eRF1I from our analyses as it comprises <0.001% of total eRF1 transcript pool.
Figure 6.
Tissue-specific differences in factors directly involved in leaky termination. Genome map depicting the ORFs of different isoforms of eRF1 (A) and eRF3 (D). (B) RT-qPCR analysis of eRF1 isoforms across different tissue types. For simplicity, isoforms A, B, C, E, F and G are collectively depicted as eRF1A. Note the logarithmic scale for relative abundance. (C) Relative composition of total eRF1 mRNA pool across different tissue types. (E) RT-qPCR analysis of full-length eRF3 (A, B, D), depicted as eRF3A; N-terminally truncated isoform eRF3C, and tj (left panel); and other accessory factors implicated in termination fidelity (right panel): eIF3 subunits S10 and S9, pAbp, pix, Gle1, Dbp80 and Rox8 transcripts using cDNA prepared from adult fly heads and ovaries. CT values for each transcript were normalized against the respective CT values for αTub84B. (F) RT-qPCR analysis of ArgUCG, ArgUCU, CysGCA and TrpCCA isoacceptor tRNAs that are near-cognate to UGA stop codon. PheGGA serves as a control tRNA which is non-cognate to UGA. For cases where individual isodecoders are quantified using separate primer pairs, the isodecoder identity is indicated by the number at the end of the tRNA. CT values for each transcript were normalized against the respective CT values for 18S rRNA. The ΔCT values obtained from each test transcript were then compared between the tissues to derive ΔΔCT. Error bars represent the upper and lower limit of RQ defined by the standard deviation of ΔΔCT from three biological replicates.
eRF3 has 4 annotated transcript isoforms (Figure 6D): isoforms A, B and D give rise to full-length eRF3 (619 amino acids) and have a high degree of sequence similarity (henceforth referred to as eRF3A), whereas isoform C possesses an N-terminal truncation of 124 aa. In the case of eRF3, the full-length isoform, eRF3A, was relatively less abundant in heads, and the N-terminally truncated eRF3C isoform was three times more abundant in heads compared to the ovaries (Figure 6E). However, the eRF3C isoform comprises only ∼0.2% of total eRF3 pool in heads and ∼0.02% in ovaries.
Additionally, we also tested the relative abundance of Drosophila orthologs of several factors that are directly or indirectly implicated in eukaryotic translation termination fidelity and ribosome recycling. eIF3 has been reported to increase TR by interfering with eRF1 decoding of stop codon at the third/wobble position (61). Rli1/ABCE1 facilitates recruitment of eRFs to the ribosome and promotes termination (62,63). Dbp5/DDX19 stabilizes termination complex and prevents premature dissociation of eRFs (62,64), while Gle1 functions together with Dbp5 to regulate termination (65). Pub1 and poly(A) binding protein (PABP) are known to interact with eRFs and stimulate termination efficiency (66,67). Upon quantification, we did not find any difference in the relative abundance of eIF3 (S10 and S9, orthologous to eIF3a and eIF3b) between heads and ovaries. However, pAbp (PABP), pix (Rli1/ABCE1) and Dbp80 (Dbp5/DDX19) were found to be 2–3 times more abundant in heads while Gle1 and Rox8 (Pub1) were relatively less abundant in heads (Figure 6E).
Another potential mechanism for tissue-specific TR regulation might be the relative abundance of near-cognate tRNAs (nc-tRNAs) that can read stop codons due to wobble interactions with the nucleotide at the 1st or 3rd codon position. Differences in the relative abundance of tRNAs have been linked to modulation of recoding events such as frameshifting (68) and TR (58,59,69). Specifically, Trp-, Cys- and Arg-specific tRNAs can be inserted at UGA to stimulate TR (58,70–72). We performed relative quantification of four nc-tRNA isoacceptors that can potentially be incorporated at UGA: Arg-tRNA with the anticodon 5′-UCG-3′ (ArgUCG) (x4) and 5′-UCU-3′ (ArgUCU) (x3), Cys-tRNA with the anticodon 5′-GCA-3′ (CysGCA) (x4) and Trp-tRNA with the anticodon 5′-CCA-3′ (TrpCCA) (x2). In Drosophila, each tRNA isoacceptor has several isodecoders, i.e. tRNAs that have different sequences but decode the same codon with varied gene copy numbers; the number of isodecoders for each tRNA isoacceptor is indicated in brackets. The isodecoders for ArgUCU and TrpCCA vary considerably in sequence conservation, which necessitates the use of separate primer pairs for quantification. The isodecoder identity is indicated by a number following the isoacceptor name (e.g.: TrpCCA1, ArgUCU1,2). Upon quantification, we found that the levels of all three ArgUCU isodecoders were two times more abundant in heads compared to ovaries, while CysGCA and TrpCCA were modestly overexpressed in heads. ArgUCG tRNAs did not show any difference in relative abundance. As a control, we used the non-cognate PheGGA tRNA, which is present in the same amounts in heads and ovaries (Figure 6F). Together, these data indicate the existence of considerable differences in the tissue-specific repertoire of eRF1 and eRF3 isoforms, accessory protein factors involved in termination fidelity, and specific nc-tRNAs that can read the UGA stop codon.
Role of eRF1H isoform in modulating TR
In order to test the effects of different protein factors on termination fidelity, we designed a dual luciferase reporter construct for expression in S2 cells. We used a well-established standard TR motif from TMV harboring a UAG stop codon, which has been extensively characterized for TR in different eukaryotic systems. The TMV motif undergoes efficient TR at all three stop codons with values ranging from 10% to 80% (73–76). We inserted the 39 bp TMV TR motif between two luciferase genes to obtain a psiCHECK2-based dual luciferase reporter vector that is similar in construction to the reporter vectors depicted in Figure 1A. We also created mutant variants of the TMV reporter that sample all three stop codons and also a positive UUC control that undergoes constitutive Fluc synthesis (Figure 7A). Upon transfecting S2 cells with the TMV TR constructs, we obtained TR values of approximately 4%, 11% and 12% for UAA, UAG and UGA stop codons respectively (Figure 7C). Next, we created ubiquitous expression vectors that harbor the ORF of different protein factors under the control of a ubiquitin-63E promoter. By calculating the TR efficiency of the TMV TR motif upon co-transfecting S2 cells with pUb expression vectors, we were able to estimate the effects of different protein factors on TR efficiency. We found that despite having subtle tissue-specific differences in their abundance, factors Dbp80, Gle1, pAbp, pix and Rox8 did not alter TR efficiencies at any of the three stop codons. eRF1A overexpression slightly reduced TR at all three stop codons. In contrast, the eRF1H isoform promoted TR at all three stop codons, with the most prominent effect being observed for the UGA stop codon, with TR value reaching 20% (Figure 7C, Supplementary Figure S7). Given the ability of the eRF1H isoform to promote TR of leaky stop codon contexts and its overrepresentation in head tissues, TR modulation by a splice variant of translation termination factor might provide an attractive explanation for the reason behind elevated incidence of TR of leaky genes in nervous tissues.
Figure 7.
Effect of different protein factors on TR modulation of TMV motif in S2 cells. (A) Construct design for a dual luciferase reporter vector containing TMV TR motif. (B) pUb expression vector design containing ORF for the gene of interest. (C) TR efficiencies of TMV reporter constructs when transfected into S2 cells alone (left panel) or when co-transfected with pUb expression vector containing the ORF for various protein factors as indicated. pUb constructs expressing eGFP were used as control. The error bars indicate the SD from four to five biological replicates. P-values are calculated using two-tailed unpaired Student's t-test. (**P< 0.005, ***P< 0.0005).
DISCUSSION
TR generates proteins with extended C-termini that can change protein functions and help in adaptation and evolution. A growing body of evidence from bioinformatics and ribosome profiling data indicates that TR is utilized by higher eukaryotes such as Drosophila, mosquitos and mammals, but the functions of the extended protein isoforms in an organism remain largely unexplored (7,8,19,21). Of the 11 candidate Drosophila genes tested here, several do not undergo TR in S2 cells, which may indicate that some of the predicted candidates are either false positives or do not undergo TR in this particular cell type. High levels of TR in genes ending with UGA-C, such as svp, aPKC, dlg1 and tj, are consistent with the notion that UGA-C is the leakiest stop codon context (5,9,48,77,78). The UGA-C sequence can act as the major determinant of TR, as observed with dlg1 and tj. However, in other cases, such as aPKC, a regulatory sequence outside of the stop codon context must drive TR. The TR stimulatory elements outside the stop codon context can be difficult to predict bioinformatically, but can be of potential use as a synthetic biology tool to induce programmed TR in specific tissues.
Using TR in the tj gene as a model, we show that Tj-TR is absent in most tissues, including embryonic and adult gonads, but is selectively expressed in the embryonic CNS and adult brain. The different expression levels are controlled at the translation level, which is surprising given that the UGA-C termination context appears to be necessary and sufficient for TR in tj. Tissue-specific regulation of TR was first reported for the Drosophila gene kelch (25). Subsequently, ribosome profiling revealed differential ribosomal footprints in several other genes that show significant TR in samples derived from early embryos and S2 cells (8). More recently, several studies in Drosophila and mice have reported the highest levels of TR in tissues of the CNS, mainly neurons (26,79–81). These observations, combined with significant overrepresentation of neuronal genes in TR candidates (7,19) support the idea that elevated TR in susceptible genes might be an idiosyncratic feature of neuronal tissues, although the mechanism of such TR regulation remains unexplored.
Towards understanding the physiological basis for tissue-specific TR, we found appreciable differences in the expression levels of key factors that are directly or indirectly involved in the process of termination. First, we observed that a specific splice variant of eRF1 (eRF1H) constitutes a substantial portion of the total eRF1 transcript pool – particularly in those tissues that exhibit considerable TR – and is absent in ovaries where we only observe basal TR. It is interesting to note that alternative splicing occurs at a high frequency in the nervous system, allowing neuronal cells to expand their transcriptomic diversity (82–84). Consequently, eRF1H is most abundant in head tissues (33% of total eRF1 pool) where we observe the highest level of TR (20%) in tj. We substantiated this correlation by overexpressing the eRF1H isoform in S2 cells, which led to enhanced TR of the TMV TR motif with an effect that was particularly pronounced for the UGA stop codon. While the depletion of eRF1 has been previously shown to enhance TR at all three stop codons (85–87), there are no studies addressing the role of eRF1 isoforms in modulating termination fidelity. eRF1H differs from the major eRF1A isoform in its extreme C terminus (amino acid residues 422–437). The C-terminal domain of eRF1 directly interacts with domain 3 of eRF3 via hydrophobic patches in helices α8, α11 and strand β10 (88,89). The structure of the C-terminal residues of eRF1 is disordered and not resolved in the available structures from S. cerevisiae (residues 422–437), S. pombe (residues 427–433) and mammalian pre-termination complexes (residues 420–437) (88–91). However, biochemical studies show that the C-terminal residues of eRF1 are required for high-affinity interaction between eRF1 and eRF3 in yeast (92), wherein even a small deletion of 4–6 amino acids form the C terminus results in decreased interaction between eRF1 and eRF3 (93). It is thus likely that the eRF1H isoform has reduced affinity towards eRF3 and acts as a negative inhibitor thereby affecting termination fidelity.
Another attractive explanation for tissue-specific TR is the difference in the relative abundance of TR-prone suppressor tRNAs that can compete with release factors by wobble base pairing at the first or third position of the leaky stop codon (58,94). Our data indeed support this idea, as we found subtle enrichment in heads compared to ovaries of specific tRNA isodecoders that are near-cognate to UGA. Apart from these core players that directly interact with the stop codon, we also found tissue-specific differences in the abundance of Drosophila orthologs of protein factors that interact with the translation machinery, such as PABP, Rli1/ABCE1, Gle1, Dbp5/DDX19 and Pub1. However, our dual reporter assays did not show any strong correlation between the overexpression of these factors with changes in TR efficiencies. Taking into account all of these findings, it is likely that the selective elevation of TR of leaky stop contexts in neuronal tissues is caused by differences in the cellular repertoire of factors involved in translation termination, with eRF1H isoform being one of the prime candidates. Such tissue-specific differences are unlikely to disrupt the homeostatic cellular proteome, as UAA is the preferred stop codon in highly expressed and housekeeping genes (95). Additionally, the presence of secondary in-frame stop codons within a few positions downstream of the primary stop codon or appendage of a peptide sequence that destabilizes the TR-extended protein mitigates any dominant-negative effects that can be attributed to erroneously extended TR genes (96,97). Thus, selective upregulation of TR in genes with leaky termination contexts serves to enrich the diversity of the neuronal proteome.
The tight control of TR is biologically important, as constitutive induction of TR in tj results in several defective phenotypes in adult ovaries, such as abnormally assembled niches, ectopic expression of En, deformed germaria, and defective encapsulation of germline cysts by follicular epithelia. Several of these phenotypes are reminiscent of previously characterized hypomorphic tj variants (34,54). Since the expression level of Tj in tjTR/TR ovaries is comparable to that in tjnat/nat ovaries, the observed phenotypes can be attributed to altered transcriptional activity caused by the Tj-TR isoform in soma-specific gene regulatory functions during gonad development.
In adult brains of tjTR/TR and tjnTR/nTR mutants, constitutive induction or abolition of TR in tj gives rise to neural phenotypes in the OL and the CB. The phenotypes manifest in the formation of sporadic lesions in both mutants with comparable frequencies. These results suggest that the maintenance of a distinct ratio between the Tj and Tj-TR isoforms is crucial for the preservation of neuronal structure in the brain (Figure 8). While the lesions that arise due to genetic dysregulation caused by the forced presence of either Tj or the Tj-TR isoform have a similar appearance, at the level of our histological analysis it is not possible to determine whether they have an identical nature or result from the same cellular dysfunctions. Indeed, many disparate genetic defects manifest as neurodegeneration and neural lesions similar in appearance to those that we report here (98–101). It is apparent that neural gene regulation must be tightly controlled, and perturbations of this regulation commonly result in neural lesions.
Figure 8.
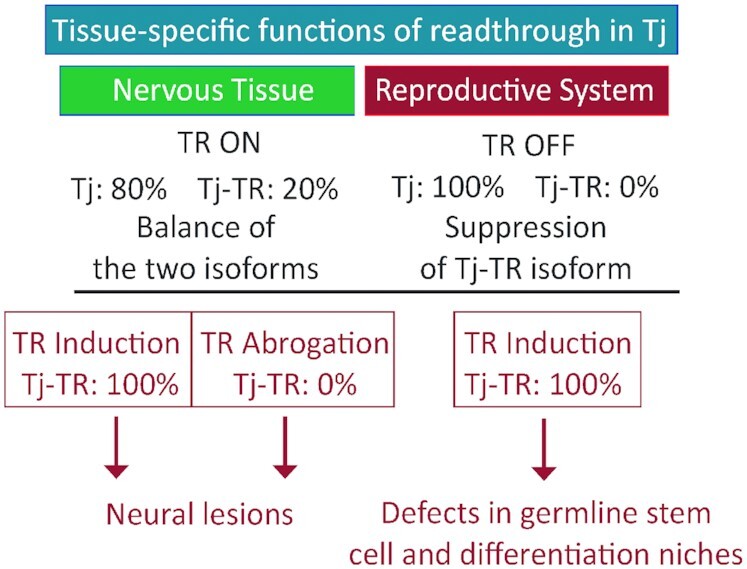
Scheme depicting the importance of proper regulation of Tj-TR. In nervous tissue, TR of tj transcript results in a mixture of Tj and Tj-TR isoforms. Abrogation of either isoform results in neural lesions, while a properly regulated balance of isoforms is key to brain health. In the reproductive system, preventing Tj-TR is crucial for reproductive capacity.
The results of the RT-qPCR and RNA-seq experiments begin to uncover a network of potentially complex interactions modulated by Tj-TR. Constitutive expression of Tj-TR leads to upregulation of a number of genes such as irk3, sand, ninaD and Obp99a, suggesting that the Tj-TR isoform can act as activator of these targets. The genes that are dysegulated upon constitutive induction of TR in tj are mostly involved in CNS functions related to perception of external stimuli such as photo stimuli and odorants and regulation of bodily responses towards such stimuli.
Because the function of Tj in the nervous system is not well-understood, the exact mechanism of regulation of Tj function by TR is not clear at present. Given its proximity to the DNA-binding leucine-zipper motif in the C-terminal region of the protein, it is possible that the TR motif alters the DNA or protein-binding properties of native Tj. Alternatively, as bZIP transcription factors are known to form homo/heterodimers, the TR motif could also affect the dimerization potential of Tj, leading to alteration of its gene regulatory functions (102,103). The structurally disordered C-termini generated by TR (22,23) may ensure the accessibility for new interactors or affect liquid-liquid phase transitions involving transcription factors in the nucleus (104,105), thereby affecting the native protein function.
The nervous system is under constant pressure to modulate the neuronal proteomic diversity in response to environmental cues and stress conditions. Indeed, regulatory processes that occur at a post-transcriptional level such as adenosine-to-inosine editing and alternative splicing are known to be prevalent in neurons (83,106). Given the selective enrichment of neuron-specific genes in the phylogenetically predicted list of TR candidates in Drosophila (7) and increasing evidence of neuron-specific TR, regulation of the neuronal proteome at the translational level via TR represents a novel mechanism of fine-tuning neuronal gene expression. In our study, we present one such case of tissue-specific TR in Tj, wherein the expression of Tj-TR is not only upregulated upon exposure to selective conditions such as heat and oxidative stress, but also plays a crucial role in shaping the neuronal transcriptome via regulation of selective genes. It would be interesting to study the precise mechanism of TR regulation by selective stressors and the role of TR isoforms in the pleiotropic regulatory cascades that mediate stress responses. Exploring the repertoire of TR-generated proteins and their functions will significantly augment our understanding of the neuronal proteomic diversity and its implications on physiological processes.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Serge Birman for insights on adult brain morphology, Jasmin Rehman and Ibrahim Ömer Cicek for help with CRISPR/Cas9 mutant design, Tobias Boothe for help with microscopy, Anna Pfeifer, Olaf Geintzer, Sandra Kappler, Christina Kothe, Vanessa Herold, Franziska Hummel, Tessa Hübner, and Michael Zimmermann for expert technical assistance. We thank Melissa Harrison, Kate O’Connor-Giles and Jill Wildonger for gifting us pU6-BbsI-chiRNA and pHD-DsRed vectors. We thank Clara Moch and Jean-Rene Huynh for pUWG vector and Herbert Jäckle for modifications in pUWG. We thank Zhaohui Wang and Dorothea Godt for anti-Tj antibodies. The work was supported by a grant of the Deutsche Forschungsgemeinschaft (SFB860 and Leibniz Prize to M.V.R.) and VolkswagenStiftung (Az. 97750 to H.R.S.).
Notes
Present address: Cristina Maracci, Department of Life and Environmental Science, Polytechnic University of Marche, 60131 Ancona, Italy.
Contributor Information
Prajwal Karki, Department of Physical Biochemistry, Max Planck Institute for Biophysical Chemistry, 37077 Goettingen, Germany.
Travis D Carney, Gene Expression and Signaling Group, Institute of Cell Biochemistry, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.
Cristina Maracci, Department of Physical Biochemistry, Max Planck Institute for Biophysical Chemistry, 37077 Goettingen, Germany.
Andriy S Yatsenko, Gene Expression and Signaling Group, Institute of Cell Biochemistry, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.
Halyna R Shcherbata, Gene Expression and Signaling Group, Institute of Cell Biochemistry, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.
Marina V Rodnina, Department of Physical Biochemistry, Max Planck Institute for Biophysical Chemistry, 37077 Goettingen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB860 and Leibniz Prize to M.V.R.]; Volkswagen Stiftung [Az. 97750 to H.R.S.]. Funding for open access charge: MPG.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kornblihtt A.R., Schor I.E., Allo M., Dujardin G., Petrillo E., Munoz M.J.. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013; 14:153–165. [DOI] [PubMed] [Google Scholar]
- 2. Kim E., Magen A., Ast G.. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007; 35:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tian B., Manley J.L.. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017; 18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.C., Vagner S.. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 2003; 95:169–178. [DOI] [PubMed] [Google Scholar]
- 5. Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F.. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014; 42:8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Namy O., Duchateau-Nguyen G., Hatin I., Hermann-Le Denmat S., Termier M., Rousset J.P. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2003; 31:2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jungreis I., Lin M.F., Spokony R., Chan C.S., Negre N., Victorsen A., White K.P., Kellis M.. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011; 21:2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cridge A.G., Crowe-McAuliffe C., Mathew S.F., Tate W.P.. Eukaryotic translational termination efficiency is influenced by the 3' nucleotides within the ribosomal mRNA channel. Nucleic Acids Res. 2018; 46:1927–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manuvakhova M., Keeling K., Bedwell D.M.. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000; 6:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howard M.T., Shirts B.H., Petros L.M., Flanigan K.M., Gesteland R.F., Atkins J.F.. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 2000; 48:164–169. [PubMed] [Google Scholar]
- 12. Beier H., Grimm M.. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001; 29:4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Urban C., Zerfass K., Fingerhut C., Beier H.. UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Res. 1996; 24:3424–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrell L., Melcher U., Atkins J.F.. Predominance of six different hexanucleotide recoding signals 3' of read-through stop codons. Nucleic Acids Res. 2002; 30:2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Firth A.E., Wills N.M., Gesteland R.F., Atkins J.F.. Stimulation of stop codon readthrough: frequent presence of an extended 3' RNA structural element. Nucleic Acids Res. 2011; 39:6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pelham H.R. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978; 272:469–471. [DOI] [PubMed] [Google Scholar]
- 17. Felsenstein K.M., Goff S.P.. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J. Virol. 1988; 62:2179–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hofstetter H., Monstein H.J., Weissmann C.. The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta. 1974; 374:238–251. [DOI] [PubMed] [Google Scholar]
- 19. Jungreis I., Chan C.S., Waterhouse R.M., Fields G., Lin M.F., Kellis M.. Evolutionary dynamics of abundant stop codon readthrough. Mol. Biol. Evol. 2016; 33:3108–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eswarappa S.M., Potdar A.A., Koch W.J., Fan Y., Vasu K., Lindner D., Willard B., Graham L.M., DiCorleto P.E., Fox P.L.. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014; 157:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams I., Richardson J., Starkey A., Stansfield I.. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004; 32:6605–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pancsa R., Macossay-Castillo M., Kosol S., Tompa P.. Computational analysis of translational readthrough proteins in Drosophila and yeast reveals parallels to alternative splicing. Sci. Rep. 2016; 6:32142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kleppe A.S., Bornberg-Bauer E.. Robustness by intrinsically disordered C-termini and translational readthrough. Nucleic Acids Res. 2018; 46:10184–10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue F., Cooley L.. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993; 72:681–693. [DOI] [PubMed] [Google Scholar]
- 25. Robinson D.N., Cooley L.. Examination of the function of two kelch proteins generated by stop codon suppression. Development. 1997; 124:1405–1417. [DOI] [PubMed] [Google Scholar]
- 26. Hudson A.M., Szabo N.L., Loughran G., Wills N.M., Atkins J.F., Cooley L.. Tissue-specific dynamic codon redefinition in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2012793118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steneberg P., Samakovlis C.. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001; 2:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klagges B.R., Heimbeck G., Godenschwege T.A., Hofbauer A., Pflugfelder G.O., Reifegerste R., Reisch D., Schaupp M., Buchner S., Buchner E.. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J. Neurosci. 1996; 16:3154–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samson M.L., Lisbin M.J., White K.. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995; 141:1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chao A.T., Dierick H.A., Addy T.M., Bejsovec A.. Mutations in eukaryotic release factors 1 and 3 act as general nonsense suppressors in Drosophila. Genetics. 2003; 165:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y., Lindberg B.G., Esfahani S.S., Tang X., Piazza S., Engstrom Y.. Stop codon readthrough alters the activity of a POU/Oct transcription factor during Drosophila development. BMC Biol. 2021; 19:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wingert L., DiNardo S.. Traffic jam functions in a branched pathway from Notch activation to niche cell fate. Development. 2015; 142:2268–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai C.M., Lin K.Y., Kao S.H., Chen Y.N., Huang F., Hsu H.J.. Hedgehog signaling establishes precursors for germline stem cell niches by regulating cell adhesion. J. Cell Biol. 2017; 216:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Panchal T., Chen X., Alchits E., Oh Y., Poon J., Kouptsova J., Laski F.A., Godt D. Specification and spatial arrangement of cells in the germline stem cell niche of the Drosophila ovary depend on the Maf transcription factor Traffic jam. PLos Genet. 2017; 13:e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gunawan F., Arandjelovic M., Godt D. The Maf factor traffic jam both enables and inhibits collective cell migration in Drosophila oogenesis. Development. 2013; 140:2808–2817. [DOI] [PubMed] [Google Scholar]
- 36. Li M.A., Alls J.D., Avancini R.M., Koo K., Godt D. The large Maf factor traffic jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol. 2003; 5:994–1000. [DOI] [PubMed] [Google Scholar]
- 37. Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M.C.. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009; 461:1296–1299. [DOI] [PubMed] [Google Scholar]
- 38. Gelbart W., Emmert D. Flybase high throughput expression pattern data. FlyBase Analysis. 2013; (29 October 2013, date last accessed)https://flybase.org/reports/FBrf0221009.html. [Google Scholar]
- 39. Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C.. Phenotypic convergence distinct transcription factors regulate common terminal features. Cell. 2018; 174:622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 41. Gratz S.J., Cummings A.M., Nguyen J.N., Hamm D.C., Donohue L.K., Harrison M.M., Wildonger J., O’Connor-Giles K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013; 194:1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kucherenko M.M., Marrone A.K., Rishko V.M., Yatsenko A.S., Klepzig A., Shcherbata H.R.. Paraffin-embedded and frozen sections of Drosophila adult muscles. J. Vis. Exp. 2010; 46:e2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shcherbata H.R., Yatsenko A.S., Patterson L., Sood V.D., Nudel U., Yaffe D., Baker D., Ruohola-Baker H.. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007; 26:481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang A.M., Rehm E.J., Rubin G.M.. Quick preparation of genomic DNA from Drosophila. Cold Spring Harb. Protoc. 2009; 2009:pdb.prot5198. [DOI] [PubMed] [Google Scholar]
- 45. Hu Y., Sopko R., Foos M., Kelley C., Flockhart I., Ammeux N., Wang X., Perkins L., Perrimon N., Mohr S.E.. FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda). 2013; 3:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan Makhtar W.R., Browne G., Karountzos A., Stevens C., Alghamdi Y., Bottrill A.R., Mistry S., Smith E., Bushel M., Pringle J.H.et al.. Short stretches of rare codons regulate translation of the transcription factor ZEB2 in cancer cells. Oncogene. 2017; 36:6640–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fearon K., McClendon V., Bonetti B., Bedwell D.M.. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J. Biol. Chem. 1994; 269:17802–17808. [PubMed] [Google Scholar]
- 48. Bonetti B., Fu L., Moon J., Bedwell D.M.. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995; 251:334–345. [DOI] [PubMed] [Google Scholar]
- 49. Namy O., Duchateau-Nguyen G., Rousset J.P.. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol. 2002; 43:641–652. [DOI] [PubMed] [Google Scholar]
- 50. Keeling K.M., Lanier J., Du M., Salas-Marco J., Gao L., Kaenjak-Angeletti A., Bedwell D.M.. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Napthine S., Yek C., Powell M.L., Brown T.D., Brierley I.. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA. 2012; 18:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yatsenko A.S., Shcherbata H.R.. Stereotypical architecture of the stem cell niche is spatiotemporally established by miR-125-dependent coordination of Notch and steroid signaling. Development. 2018; 145:dev159178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bai J., Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002; 129:5377–5388. [DOI] [PubMed] [Google Scholar]
- 54. Li M., Hu X., Zhang S., Ho M.S., Wu G., Zhang L.. Traffic jam regulates the function of the ovarian germline stem cell progeny differentiation niche during pre-adult stage in Drosophila. Sci. Rep. 2019; 9:10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Konig A., Shcherbata H.R.. Soma influences GSC progeny differentiation via the cell adhesion-mediated steroid-let-7-Wingless signaling cascade that regulates chromatin dynamics. Biol Open. 2015; 4:285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eliazer S., Palacios V., Wang Z., Kollipara R.K., Kittler R., Buszczak M.. Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLos Genet. 2014; 10:e1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jukam D., Xie B., Rister J., Terrell D., Charlton-Perkins M., Pistillo D., Gebelein B., Desplan C., Cook T.. Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science. 2013; 342:1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blanchet S., Cornu D., Argentini M., Namy O.. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beznoskova P., Gunisova S., Valasek L.S.. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016; 22:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chauvin C., Salhi S., Le Goff C., Viranaicken W., Diop D., Jean-Jean O.. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 2005; 25:5801–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Beznoskova P., Wagner S., Jansen M.E., Haar T., Valasek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beissel C., Neumann B., Uhse S., Hampe I., Karki P., Krebber H.. Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3. Nucleic Acids Res. 2019; 47:4798–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shoemaker C.J., Green R.. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mikhailova T., Shuvalova E., Ivanov A., Susorov D., Shuvalov A., Kolosov P.M., Alkalaeva E.. RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes. Nucleic Acids Res. 2017; 45:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R.. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008; 134:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ivanov A., Mikhailova T., Eliseev B., Yeramala L., Sokolova E., Susorov D., Shuvalov A., Schaffitzel C., Alkalaeva E.. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016; 44:7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Urakov V.N., Mitkevich O.V., Safenkova I.V., Ter-Avanesyan M.D.. Ribosome-bound Pub1 modulates stop codon decoding during translation termination in yeast. FEBS J. 2017; 284:1914–1930. [DOI] [PubMed] [Google Scholar]
- 68. Korniy N., Goyal A., Hoffmann M., Samatova E., Peske F., Pohlmann S., Rodnina M.V.. Modulation of HIV-1 Gag/Gag-Pol frameshifting by tRNA abundance. Nucleic Acids Res. 2019; 47:5210–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Roy B., Leszyk J.D., Mangus D.A., Jacobson A.. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Feng Y.X., Copeland T.D., Oroszlan S., Rein A., Levin J.G.. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:8860–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Urban C., Beier H.. Cysteine tRNAs of plant origin as novel UGA suppressors. Nucleic Acids Res. 1995; 23:4591–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zerfass K., Beier H.. The leaky UGA termination codon of tobacco rattle virus RNA is suppressed by tobacco chloroplast and cytoplasmic tRNAs(Trp) with CmCA anticodon. EMBO J. 1992; 11:4167–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beier H., Barciszewska M., Krupp G., Mitnacht R., Gross H.J.. UAG readthrough during TMV RNA translation: isolation and sequence of two tRNAs with suppressor activity from tobacco plants. EMBO J. 1984; 3:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Namy O., Hatin I., Rousset J.P.. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001; 2:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lao N.T., Maloney A.P., Atkins J.F., Kavanagh T.A.. Versatile dual reporter gene systems for investigating stop codon readthrough in plants. PLoS One. 2009; 4:e7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Valle R.P., Drugeon G., Devignes-Morch M.D., Legocki A.B., Haenni A.L.. Codon context effect in virus translational readthrough. A study in vitro of the determinants of TMV and Mo-MuLV amber suppression. FEBS Lett. 1992; 306:133–139. [DOI] [PubMed] [Google Scholar]
- 77. Cassan M., Rousset J.P.. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol. 2001; 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P.. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen Y., Sun T., Bi Z., Ni J.Q., Pastor-Pareja J.C., Javid B.. Premature termination codon readthrough in Drosophila varies in a developmental and tissue-specific manner. Sci. Rep. 2020; 10:8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sapkota D., Lake A.M., Yang W., Yang C., Wesseling H., Guise A., Uncu C., Dalal J.S., Kraft A.W., Lee J.M.et al.. Cell-type-specific profiling of alternative translation identifies regulated protein isoform variation in the mouse brain. Cell Rep. 2019; 26:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Palazzo C., Abbrescia P., Valente O., Nicchia G.P., Banitalebi S., Amiry-Moghaddam M., Trojano M., Frigeri A.. Tissue distribution of the readthrough isoform of AQP4 reveals a dual role of AQP4ex limited to CNS. Int. J. Mol. Sci. 2020; 21:1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Raj B., Blencowe B.J.. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015; 87:14–27. [DOI] [PubMed] [Google Scholar]
- 83. Su C.H., D D., Tarn W.Y. Alternative splicing in neurogenesis and brain development. Front Mol Biosci. 2018; 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Venables J.P., Tazi J., Juge F.. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res. 2012; 40:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Janzen D.M., Geballe A.P.. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 2004; 32:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Carnes J., Jacobson M., Leinwand L., Yarus M.. Stop codon suppression via inhibition of eRF1 expression. RNA. 2003; 9:648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Csibra E., Brierley I., Irigoyen N.. Modulation of stop codon read-through efficiency and its effect on the replication of murine leukemia virus. J. Virol. 2014; 88:10364–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cheng Z., Saito K., Pisarev A.V., Wada M., Pisareva V.P., Pestova T.V., Gajda M., Round A., Kong C., Lim M.et al.. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009; 23:1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. des Georges A., Hashem Y., Unbehaun A., Grassucci R.A., Taylor D., Hellen C.U., Pestova T.V., Frank J.. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic Acids Res. 2014; 42:3409–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000; 100:311–321. [DOI] [PubMed] [Google Scholar]
- 91. Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O., Green R., Becker T., Beckmann R.. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014; 8:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ito K., Ebihara K., Nakamura Y.. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998; 4:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Eurwilaichitr L., Graves F.M., Stansfield I., Tuite M.F.. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999; 32:485–496. [DOI] [PubMed] [Google Scholar]
- 94. Beznoskova P., Bidou L., Namy O., Valasek L.S.. Increased expression of tryptophan and tyrosine tRNAs elevates stop codon readthrough of reporter systems in human cell lines. Nucleic Acids Res. 2021; 49:5202–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Trotta E. Selective forces and mutational biases drive stop codon usage in the human genome: a comparison with sense codon usage. BMC Genomics. 2016; 17:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fleming I., Cavalcanti A.R.O.. Selection for tandem stop codons in ciliate species with reassigned stop codons. PLoS One. 2019; 14:e0225804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Arribere J.A., Cenik E.S., Jain N., Hess G.T., Lee C.H., Bassik M.C., Fire A.Z.. Translation readthrough mitigation. Nature. 2016; 534:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cao Y., Chtarbanova S., Petersen A.J., Ganetzky B.. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E1752–E1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Loewen C.A., Ganetzky B.. Mito-nuclear interactions affecting lifespan and neurodegeneration in a Drosophila model of Leigh syndrome. Genetics. 2018; 208:1535–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Melentev P.A., Ryabova E.V., Surina N.V., Zhmujdina D.R., Komissarov A.E., Ivanova E.A., Boltneva N.P., Makhaeva G.F., Sliusarenko M.I., Yatsenko A.S.et al.. Loss of swiss cheese in neurons contributes to neurodegeneration with mitochondria abnormalities, reactive oxygen species acceleration and accumulation of lipid droplets in Drosophila brain. Int. J. Mol. Sci. 2021; 22:8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cauchi R.J., van den Heuvel M.. The fly as a model for neurodegenerative diseases: Is it worth the jump. Neurodegener Dis. 2006; 3:338–356. [DOI] [PubMed] [Google Scholar]
- 102. Kataoka K., Noda M., Nishizawa M.. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 1994; 14:700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kurokawa H., Motohashi H., Sueno S., Kimura M., Takagawa H., Kanno Y., Yamamoto M., Tanaka T.. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell. Biol. 2009; 29:6232–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Peng A., Weber S.C. Evidence for and against liquid-liquid phase separation in the nucleus. Noncoding RNA. 2019; 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Owen I., Shewmaker F.. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int. J. Mol. Sci. 2019; 20:5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Behm M., Ohman M.. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 2016; 32:165–175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.