Abstract
Schlafen-5 (SLFN5) is an interferon-induced protein of the Schlafen family, which are involved in immune responses and oncogenesis. To date, little is known regarding its anti-HIV-1 function. Here, the authors report that overexpression of SLFN5 inhibits HIV-1 replication and reduces viral mRNA levels, whereas depletion of endogenous SLFN5 promotes HIV-1 replication. Moreover, they show that SLFN5 markedly decreases the transcriptional activity of HIV-1 long terminal repeat (LTR) via binding to two sequences in the U5-R region, which consequently represses the recruitment of RNA polymerase II to the transcription initiation site. Mutagenesis studies show the importance of nuclear localization and the N-terminal 1–570 amino acids fragment in the inhibition of HIV-1. Further mechanistic studies demonstrate that SLFN5 interacts with components of the PRC2 complex, G9a and Histone H3, thereby promoting H3K27me2 and H3K27me3 modification leading to silencing HIV-1 transcription. In concert with this, they find that SLFN5 blocks the activation of latent HIV-1. Altogether, their findings demonstrate that SLFN5 is a transcriptional repressor of HIV-1 through epigenetic modulation and a potential determinant of HIV-1 latency.
INTRODUCTION
Host restriction factors (RFs) are host proteins that potently inhibit a group of viruses. They are often constitutively expressed at various levels in the absence of viral infection and are generally inducible by interferons (IFN). An arsenal of RFs has been discovered against human immunodeficiency viruses type 1 (HIV-1) by targeting multiple steps of the HIV-1 replication including capsid uncoating (Trim5α) (1), reverse transcription (Apobec3G) (2,3), nuclear import and integration (MxB) (4–6), viral budding and release (Tetherin) (7), and the production of infectious virus particles (SERINC3/5) (8,9). Several studies have indicated that the Schlafen (SLFN) family play a critical role in immune regulation and antiviral responses (10–12). More specifically, several members of the SLFN protein family have been implicated in inhibiting HIV-1 replication. For example, SLFN11 and SLFN13 have been reported to restrict HIV-1 by blocking HIV-1 protein synthesis (13) and depleting tRNA/rRNA in an endonucleolytic activity-dependent manner (14). However, antiviral activities of other SLFN members remain to be determined.
To date, 10 SLFN members in mice and six in humans have been identified, which are categorized into three subgroups (11,15). SLFN5 belongs to the third subgroup and contains a specific N-terminal AAA-domain, a SWADL domain that is unique to SLFN II and SLFN III subfamilies, and a C-terminal extension that is homologous to superfamily I RNA helicases. Notably, SLFN5 was reported to harbor a helix-turn-helix domain (COG2865) in its N-terminus, which is predicted to have DNA binding activity and is thought to be involved in transcription modulation (16). Unlike other SLFN family members, SLFN5 is highly sensitive to type I interferon induction, which suggests its potential role in innate immunity. Various functional roles of SLFN5 in the regulation of tumorigenesis have been documented in several studies (17–24). For example, SLFN5 has been reported to display important antineoplastic effects in renal cell carcinoma cells (22) and malignant melanoma cells (21), decrease the mobility and invasiveness of malignant renal cells carcinoma cells by downregulating the expression of matrix metalloproteinase genes (22). Additionally, SLFN5 has been shown to be an important protective factor against breast cancer (20,23) and lung adenocarcinoma (25) by orchestrating apoptosis via regulating PTEN transcription and the downstream AKT pathway. On the contrary, SLFN5 can promote the growth and invasion of glioblastoma cells by inhibiting transcription of the driving signal transducer and activator of transcription 1 (STAT1) (17). These studies have highlighted that SLFN5 may regulate tumorigenesis by acting as a transcriptional repressor for specific genes in a cell-type dependent manner.
Transcription from the HIV-1 promoter is initiated by binding of RNA polymerase II (RNAP II) to the U3 region of 5′ long terminal repeat (5′LTR). 5′LTR-driven HIV-1 gene expression is regulated by multiple host factors. Lack of sufficient transcription factors or presence of repressive histone markers can lead to transcriptionally silent proviruses in CD4+ T cells (26). HIV-1 latency, which involves transcriptional silencing of HIV-1 proviral DNA, has been shown to be controlled by multiple mechanisms. For example, histone modification by methyltransferases (27), such as G9A, EZH2 and Polycomb Repressive Complex 2 (PRC2), keep the HIV-1 promoter in a heterochromatic state resulting in latency (28–30). Heterochromatin-associated histone markers including H3K9me3 (30), H3K27me3 (28) and H4K20me1 (31) have been reported to suppress HIV-1 transcription and impede proviral activation.
In the present study, we found that HIV-1 replication was significantly increased with depletion of SLFN5 while overexpression of SLFN5 substantially suppressed HIV-1. We further found that SLFN5 significantly decreased basal and Tat-transactivated transcription of HIV-1 LTR. Chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA) demonstrated the binding of SLFN5 to the U5-R region, which decreases the recruitment of RNA Pol II to the transcription initiation site. Notably, SLFN5 was found to have a strong interaction with histone H3 as well as with the PRC2 complex, RBBP7, EZH1 and G9a. Depletion of EZH1 or G9a by small interfering RNA or inhibition with specific inhibitors led to a partial or almost complete loss of the anti-HIV-1 effects of SLFN5. Finally, we found that SLFN5 suppressed the reactivation of HIV latency by JQ-1 in two latently infected cell models. Our findings demonstrate that SLFN5 is a novel transcriptional repressor of HIV-1 and maintains HIV latency through epigenetic modulation.
MATERIALS AND METHODS
Molecular cloning and cell lines
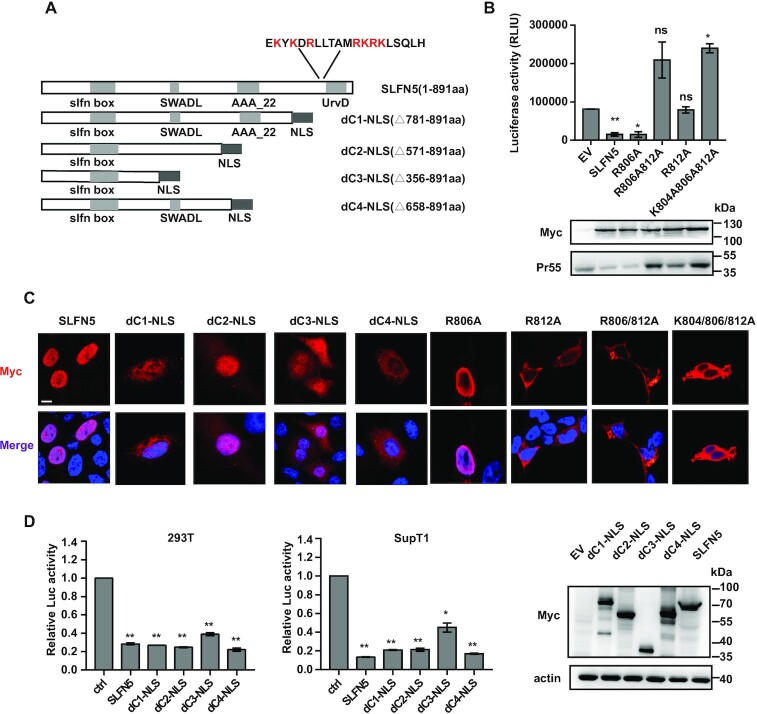
cDNA of SLFN5 constructs including full-length, N-terminal and C-terminal truncations (dN1150–891, dN2335–891, dN3450–891, dN4660–891, dC11–780, dC21–570 and dC31–335) were individually cloned into a pcDNA4-Myc vector. The NLS of SV40 T antigen was fused into the C-terminus of the truncated variants including dC11–780, dC21–570, dC31–335 and dC41–657 and inserted into a pDNA4-Myc vector. Details of all constructs are illustrated in Figure 3A and Supplementary Figure S2a. NLS-deficient mutants (R806A, R806A812A, K804A806A812A) were generated by site-directed mutagenesis using Stratagene's QuickChange site-directed mutagenesis protocol. The commercial pGL3-basic vector (Promega) was used to construct vectors with firefly luciferase reporter gene downstream of LTRs from different HIV strains (ZM246F, ZM247F, Indie, NL4-3, HIV-2) and promoters of SV40 and GAPDH. Two recombinant LTRs (R1 and R2) were constructed by swapping 1–530nt (R1) or 1–300nt of LTR (R2) derived from 89.6 strain with the counterparts of HIV-2 LTR by overlap PCRs and the recombinant fragments were inserted into pGL3-basic vector.
Figure 3.
The N-terminal domain and the NLS allow SLFN5 to inhibit HIV-1. (A) Schematic representation of SLFN5 NLS and truncated mutants fused in-frame to NLS of SV40 T antigen. Light grey rectangles represent the domains of SLFN5 and dark grey rectangles represent NLS of SV40 T antigen. (B) HEK293T cells were transfected with wild type SLFN5-Myc or its NLS mutants, NL4-3luc.R-E- and VSV-G DNA. Levels of infectious viruses in the supernatants were determined by infecting SupT1 cells followed by measuring luciferase activity. Levels of SLFN5 and its mutants in cell lysates were measured by immunoblot. (C) Confocal microscopy of subcellular localization of SLFN5 and its mutants in transfected HeLa cells. The nuclei were stained with DAPI. Scale bar, 10 μm. (D) Effect of SLFN5 NLS-fused mutants on HIV-1 production. HEK293T cells were transfected with wild type SLFN5-Myc or SLFN5 truncated mutants (containing SV40 NLS), NL4-3luc.R-E- and VSV-G. 48 h post-transfection, cells were harvested and luciferase activity was measured (left panel). Viruses in the supernatants were used to infect SupT1 cells. 48 h later, luciferase activity in SupT1 cells was measured (middle panel). Whole cell extracts were analysed by immunoblotting with α-Myc and α-actin antibodies (right panel). Data represent the mean ± SD of three independent experiments. n.s. non-significant, **P < 0.01, *P < 0.05.
Unless otherwise specified, HeLa and HEK293T cells were individually maintained at 37°C in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco). SupT1 cells were maintained in RPMI 1640 medium with 10% FBS. HeLa SLFN5 knockdown cell lines were generated by transfection of commercial SLFN5 shRNAs (Sigma, Mission shRNA library) and a scramble shRNA (negative control). Transfected HeLa cells were then grown in 2 μg/ml puromycin and split 1:5 once the cell density reached 80–90% confluence. Cells were grown over two successive passages with RPMI 1640. Clones that survived were selected and expanded. Knockdown of SLFN5 protein for each cell clone was confirmed by immunoblotting using α-SLFN5 and α-actin antibodies (Sigma-Aldrich).
The knock-out oligonucleotides containing the target sequence of guide RNA (gRNA) in human SLFN5 gene (TAG AAG CCC TCA AGC TCG TA) were annealed and inserted into lentiviral vector LentiCRISPR-v2 vector (Addgene). LentiCRISPR-v2 vector expressing the gRNA/Cas9 was transfected into HEK293T cells with packaging plasmid (pMD2.G and pSPAX2) using Lipofectamine 2000 transfection reagent according to the manufacturer's instructions (Thermo Fisher, USA). At 48h post-transfection, the supernatant was collected. 5 × 105 SupT1 cells were infected with the filtered supernatant and the cell culture media was replaced with fresh media containing 2 μg/ml puromycin after 48 h. Two weeks later, puromycin-resistant cells were expanded. Knockout of SLFN5 in SupT1 cells was confirmed by immunoblotting using an α-SLFN5 antibody and gene sequencing. The positive cell population was collected and subjected to single clone selection.
Subcellular localization analysis
HEK293T cells and HeLa cells were grown on round coverslips and transfected with full-length SLFN5 with a C-terminal Myc tag, N-terminal or C-terminal truncations or NLS-deficient mutants using Lipofectamine 2000 (Thermo Scientific). After 24 h, the cells were fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS, pH 7.4) at room temperature for 10 min and washed with PBS containing 1% glycine three times. Fixed cells were then permeabilized in 0.4% TritonX-100 followed by incubation with α-SLFN5 (Abcam #121537) and PE-conjugated α-rabbit antibodies. The cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) for nuclear staining and mounted in antifade mounting medium (Thermo Scientific). Cells were viewed with FV1000 fluorescent microscope (Olympus).
Measurement of HIV-1 virus production
To measure HIV production, pNL4-3-R-E-Luc vector and pCMV-VSV-G packaging vector were transfected into HEK293T cells together with an empty pcDNA4 vector, pcDNA4-SLFN5, or the corresponding SLFN5 truncations and mutants using Lipofectamine 2000 (Thermo Scientific) according to the manufacturer's instructions. At 6 h post-transfection, the culture media was replaced with fresh media. At 48 h post-transfection, the supernatants were collected. The amount of infectious virus particles in the supernatants were determined by using the supernatant to infect 1 × 105 SupT1 cells per well for 6 h and then replaced with fresh media. At 48 h post-infection, the SupT1 cells were examined for luciferase activity using the Luciferase Assay Kit (Promega). We also measured total p24 level using an HIV-1 p24 ELISA Kit (Clontech) according to the manufacturer's instructions.
Quantitative PCR (qPCR) of viral RNA
HIV viral RNA (vRNA) in the cytoplasm was prepared using the RLN buffer. Briefly, cells were lysed with RLN buffer (50 mM Tris–Cl (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) for 5 min on ice and then centrifuged at 300 × g for 2 min at 4°C. 1 ml of TRIzol® Reagent (Thermo Scientific) was added to the collected supernatants following the manufacturer's instructions. Viral protein and DNA were removed by centrifugation at 4°C for 15 min at 12 000 × g. vRNA was extracted from the upper aqueous phase and subsequently reverse transcribed using PrimeScript RT reagent Kit (TaKaRa). The concentrations of total vRNA and unspliced vRNA were individually determined by qPCR with ABI7300 Real-Time System using SYBR Premix ExTaq Kit (TaKaRa). One cycle of denaturation (95°C for 10 min) was performed, followed by 40 cycles of amplification (95°C for 15 s, 55°C for 30 s and 72°C for 30 s). Total vRNA, unspliced or partially spliced RNA were measured by quantification of HIV Gag-Pol, Vpu and Rev mRNA respectively. The primers used are as follows:
Gag-FP: 5′TCAGACAGGATCAGAAGAAC3′; Gag-RP: 5′ACGCGTCCTGAAGCTTATG3′; Vpu-FP: 5′GCAACCTATAATAGTAGCA3′; Vpu-RP: 5′TCTTCTGCTCTTTCTATTAG3′; Rev-FP: 5′CTATCAAAGCAACCCACCTC3′; Rev-RP: 5′CAAGAGTAAGTCTCTCAAGC3′; GAPDH-FP: 5′GTCCACTGGCGTCTTCACCA3′; GAPDH-RP: 5′GTGGCAGTGATGGCATGGAC3′.
Chromatin immunoprecipitation (ChIP)
HEK293T cells were transfected as indicated in the figures. After 48 h, cells were harvested for cross-linked ChIP using EZ-Magna ChIP™ G Chromatin Immunoprecipitation Kit (Millipore). qPCR was performed using primer pairs targeting U3 (LTR1, LTR2), R-U5 region, Gag, Pol, Vpr and Env coding sequences respectively. The primers used were as follows: LTR1-FP: 5′ACTTCCCTGATTGGCAGAACT3′ LTR1-RP: 5′CTACTTGCTCTGGTTCAACTGG3′; LTR2-FP: 5′ACTGACCTTTGGATGGTGCTT3′ LTR2-RP: 5′AAAGCTCGATGTCAGCAGTCT3′; R-U5-FP: 5′TCTGGCTAACTAGGGAACCCA3′; R-U5-RP: 5′CTGACTAAAAGGGTCTGAGG3′; Gag-FP: 5′GAACAGGGACTTGAAAGCGA3′; Gag-RP: 5′TTTTTGGCGTACTCACCAGTC3′; POL-FP: 5′CCAGGAGCGACACTAGAAGA3′ POL-RP: 5′TTTCCACATTTCCAACAGCCC3′; Vpr-FP: 5′ACAGAGGGAGCCATACAATGA3′ Vpr-RP: 5′GTCGAGTAACGCCTATTCTGC3′; Env-FP: 5′TATCGTTTCAGACCCACCTCC3′; Env-RP: 5′TATTTGAGGGCTTCCCACCC3′. An aliquot of chromatin was amplified in parallel, and the value obtained for immunoprecipitation was normalized using the value for chromatin (% input).
Co-immunoprecipitation (Co-IP)
1 × 107 HEK293T cells overexpressing SLFN5-Myc were collected and lysed in 1 ml of Non-idet P 40 (NP40) buffer supplemented with 10 μl protease inhibitor cocktail (Topscience). SLFN5 was immunoprecipitated by α-Myc antibody (CST #8146) and protein A/G agarose beads (Beyotime #P2019). Beads were washed three times with 1 ml NP40 buffer and subsequently incubated with 50 μl 1× protein sample loading buffer at 100°C for 10 min to remove bound proteins. The elutes were analyzed by immunoblotting using the indicated antibodies.
Immunofluorescence microscopy
HEK293 cells were seeded in a four-chamber slide 1 day prior to transfection with plasmid DNA expressing Myc-tagged SLFN5 and different Myc-tagged SLFN5 truncated mutants. The cells were fixed with 4% paraformaldehyde (in 1X PBS) for 10 min at room temperature and subsequently permeabilized with 0.1% Triton X-100 for 10 min at room temperature. The cells were then stained for 2 h at room temperature with an α-Myc antibody (1:500 dilution, Rabbit) or α-Myc antibody (1:1000 dilution, Mouse). After washing with 1× PBS, cells were incubated with Alexa Fluor 488-conjugated secondary α-rabbit or α-mouse antibody (1:2000 dilution; Invitrogen). Nuclei were stained with DAPI (1 μg/ml in 1× PBS). The images were recorded using the Zeiss Pascal laser scanning confocal microscope.
Flow cytometry
Jurkat E4 and 2D10 cells were washed twice with 1× PBS, and resuspended in fixation/permeabilization solution (B&D) for 20 min at 4°C. After washing twice in Perm/Wash buffer (B&D), cells were resuspended in Perm/Wash buffer for 15 min at 4°C and analyzed by flow cytometry on a FACS Calibur in the Flow Cytometry Core Lab in the Cancer Hospital Chinese Academy of Medical Sciences. Data analysis was performed using FlowJo version 7.6. GFP-expressing cells were sorted using the defined gate, and the percentage of GFP-positive cells was determined.
Electrophoretic mobility shift assay (EMSA)
EMSA was conducted using a chemiluminescent EMSA kit (Beyotime) according to the manufacturer's instructions using the following biotin 3′-end labelled duplex DNA oligonucleotides: 5′-CTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCA-3′. Briefly, 4 μg nuclear extracts obtained from SLFN5-expressing HEK293T cells were preincubated with 5× binding buffer with or without 1 μl competitor probes for 10 min. 2 μl annealed probes (10 nM) were then added and the mixture was incubated for 20 min. 1 μl 10× loading buffer was finally added before loading on a 7% non-denaturingpolyacrylamide gel electrophoresis (PAGE) gel. The gel was separated for 60 min (100 V; 10 mA) and then transferred onto nitrocellulose membranes (380 mA) for 60 min. The membranes were cross-linked under UV irradiation and detected by chemiluminescence.
ChIP-sequencing (ChIP-seq)
We generated ChIP-seq samples from SupT1 cells by using the Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (Merck Millipore). Briefly, ChIP-seq was conducted with E-GENE Tech Co. Ltd. First, the cultured cells were collected in a centrifuge tube and then washed with 1× PBS. After centrifugation and removing the supernatant, 1% formaldehyde in PBS was used to cross-link DNA and proteins for 10 min at 37°C. Next, 0.125 M of glycine was used to stop the crosslinking reaction. The cells were then washed with pre-cooled PBS containing 0.5% bovine serum and by PBS supplemented with protease inhibitor compound (PIC). The cells were collected by centrifugation at 850 rpm for 3 min after each wash. The cells were subsequently resuspended in 200 μl ice-cold lysates buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 7.5 plus PIC) and then thawed on ice for 10 min. The cell lysate was sonicated using Bioruptor Pico to generate chromatin fragments of size range from 100 to 800 bp. One-tenth of the sonicated chromatin sample was separated for input control. The remaining chromatin was immunoprecipitated in ChIP dilution buffer (1% Triton-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 7.5) with 4 μg of α-SLFN5 antibody (ab121537, Abcam) that was pre-incubated with protein A/G magnetic beads (Invitrogen; 10003D). The immunoprecipitation reaction was incubated overnight at 4°C and the beads were washed twice with each of the following buffers at 4°C: RIPA buffer (10 mM Tris–Cl, 1 mM EDTA, 0.1% SDS, 0.1% Na-deoxycholate, 0.1% Triton X-100); RIPA buffer plus 0.3 M NaCl; LiCl buffer (0.25 M LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and TE buffer. The captured DNA and input control sample were reverse cross-linked by eluting the beads with elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C for 2 h, respectively. The eluted DNA was purified by phenol-chloroform extraction and precipitated with ethanol and glycogen. The obtained DNA was subjected to library preparation and sequenced on Illumina/novo-seq sequencing platform.
RESULTS
SLFN5 inhibits HIV-1 replication and expression of viral proteins
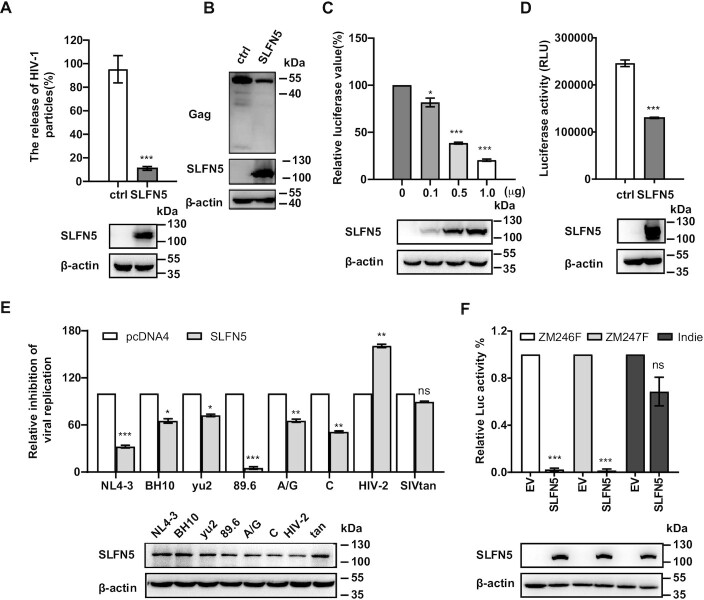
In mammals, viral infections often induce type I interferon production, which plays a pivotal role in host innate immunity. SLFN5 is inducible by interferon, but its biological relevance is not well understood. SLFN5 has been reported to be constitutively expressed in immune cells and peripheral blood mononuclear cells (PBMCs) (16). Upon infection of PBMCs from three healthy donors with VSVG-pseudotyped NL4-3 reporter virus, SLFN5 expression was upregulated (Supplementary Figure S1a). To gain functional insight into the potential antiviral activity of SLFN5, we transfected HEK293T cells with HIV-1 DNA NL4-3luc.R-E- and VSV-G along with a vector expressing SLFN5 or a control vector (pcDNA4). The level of VSVG psedotyped NL4-3luc.R-E- derived viruses (HIVNL4-3luc) in culture supernatants was determined by infecting SupT1 cells followed by measuring luciferase activity. Indeed, SLFN5 potently inhibited HIVNL4-3luc production (Figure 1A). In agreement with these data, SLFN overexpression decreased cellular p55Gag production (Figure 1B). In contrast, the level of β-actin was unaffected, indicating that the reduction of viral protein is not due to a global shutdown of protein synthesis. Co-transfection with NL4-3luc.R-E- and VSV-G with different doses of SLFN5-Myc resulted in a dose-dependent decrease in HIV-1 production, further demonstrating the anti-HIV-1 activity of SLFN5 (Figure 1C). To examine whether SLFN5 inhibits HIV-1 infection, HEK293T cells were transfected with SLFN5 and then infected with VSVG-pseudotyped HIV-1 reporter virus. The results showed a 2-fold reduction of luciferase activity in HIV-1 infected SLFN5-expressing cells (Figure 1D). We next tested SLFN5 against a number of different HIV-1 strains including Yu2, BH10, 89.6, A/G, C, as well as HIV-2 and SIVtan. The HIV-1 89.6 strain was inhibited the most by SLFN5 as opposed to 2 fold enhancement of HIV-2 and little inhibition of SIVtan, indicating the selectivity of SLFN5 antiviral function (Figure 1E). Since 80% of heterosexual HIV-1 infections are established by transmitted/founder (T/F) HIV-1 (32–35), we next tested the antiviral effects of SLFN5 on two T/F reporter viruses ZM246Fluc and ZM247Fluc as well as a chronic subtype C HIV-1 reporter virus Indieluc which containing a nonsense mutation in the viral Env gene and having the firefly luciferase gene inserted into the Nef locus (36,37). SLFN5 decreased the production of both TF HIV-1 by over 10-fold, while it inhibited much less the chronic HIV-1 isolate Indieluc (Figure 1F).
Figure 1.
Exogenous SLFN5 inhibits HIV-1 replication and Gag expression. (A, B) HEK293T cells were transfected with NL4-3luc.R-E-, VSV-G and SLFN5 or a control vector (ctrl) DNA. 48 h post-transfection, levels of viral particles in the supernatants were determined by p24 ELISA (A). Amounts of viral Gag protein and SLFN5 in cell lysates were determined by immunoblot (B). (C) HEK293T cells were transfected with 1 μg NL4-3luc.R-E-, VSV-G and increasing amounts of SLFN5 DNA (0.1, 0.5 or 1 μg). 48 h post-transfection, levels of infectious viruses in the supernatant were determined by infecting SupT1 cells and measuring luciferase activity. (D) HEK293T cells were transfected with 0.8 μg SLFN5 or a control vector. 24 h later, cells were infected with VSV-G pseudotyped NL4-3luc.R-E- reporter viruses. Luciferase activity was measured 48 h after infection. (E) SupT1 cells were first transduced to express SLFN5, followed by infection of wild type HIV-1 strains NL4-3, BH10, yu2, 89.6, A/G, C or HIV-2 and SIVtan. Levels of infectious viruses in the supernatants were determined by infection the TZM-bl cells. (F) HEK293T cells were transfected with 1 μg of reporter viral DNA ZM246Fluc, ZM247Fluc or Indieluc, VSV-G and SLFN5. 48 h post-transfection, levels of viruses in the supernatants were determined by infecting SupT1 cells and measuring luciferase activity. Molecular weight markers in kDa are indicated in immunoblots. Data represent the mean ± SD of three independent experiments. P-values were calculated using a standard Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
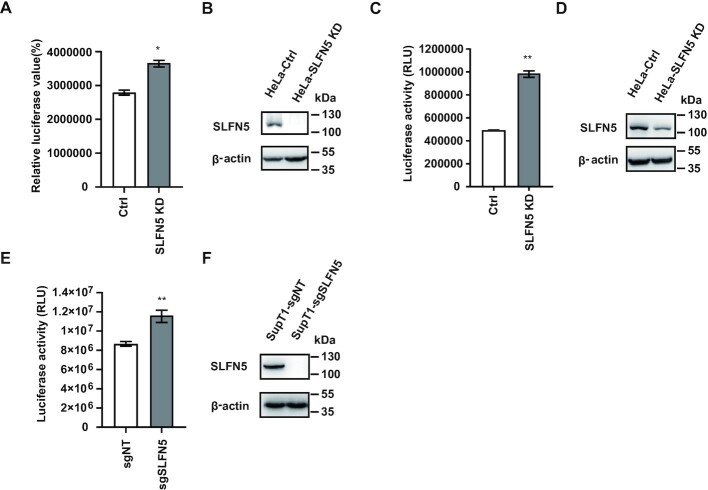
To determine whether endogenous SLFN5 has anti-HIV-1 activity, we used SLFN5 short hairpin RNA (shRNA) to generate stable HeLa cell lines that are deleted of SLFN5 expression (HeLa-SLFN5 KD). We also generated control cell lines that stably express a scramble shRNA (HeLa-Ctrl). We then infected HeLa-SLFN5 KD and HeLa-Ctrl with HIV-1NL4-3luc reporter virus. In SLFN5 knockdown cells, a 20% increase in luciferase activity was detected compared to that of the control cells (Figure 2A, B), and IFN inhibition of HIV-1 infection was alleviated by 50% in SLFN5 knockdown cells (Figure 2C, D). To exclude that SLFN5 blocks the cell cycle of HeLa cells, the proliferation profiles of HeLa SLFN5 KD cell line were compared with that of HeLa-Ctrl cells during a course of 84 h by an incuCyte live cell analysis system, little difference in cellular proliferation was observed between HeLa SLFN5 KD cells and their parental cells (Supplementary Figure S1b). In addition, SLFN5 did not show an influence on HeLa cells by transient transfection with increasing amounts of SLFN5 measured using a CCK8 assay (Supplementary Figure S1c). To assess HIV-1 inhibition by SLFN5 in CD4+ T cells, we generated stable SLFN5-knockout SupT1 cell lines (SupT1-sgSLFN5). Upon infecting SupT1-sgSLFN5 cells and control cells (SupT1-sgNT) with the HIV-1NL4-3luc reporter virus, the luciferase activity was 30% higher in SupT1-sgSLFN5 cells compared to SupT1-sgNT cells (Figure 2E, F).
Figure 2.
Endogenous SLFN5 inhibits HIV-1 replication. (A, B) HeLa-Ctrl and HeLa-SLFN5 KD cells were infected with VSV-G pseudotyped NL4-3luc.R-E- reporter viruses. 48 h post-infection, luciferase activity in HeLa cells was measured (A), and the expression of SLFN5 in HeLa-Ctrl (Ctrl) and HeLa-SLFN5 KD (SLFN5 KD) cell lines was determined by immunoblot (B). (C, D) HeLa-Ctrl and HeLa-SLFN5 KD cells were treated with 103 U/ml IFN-α/β and infected with VSV-G pseudotyped NL4-3luc.R-E- reporter viruses. 48 h post-infection, luciferase activity in HeLa cells was measured (C), and expression of SLFN5 was determined by immunoblot (D). (E, F) Control cells (SupT1-sgNT) and SLFN5-deficient (SupT1-sgSLFN5) cells were infected with VSV-G pseudotyped NL4-3luc.R-E- reporter viruses. 48h post-infection, luciferase activity in SupT1 cells was measured (E). Expression of SLFN5 in SupT1-sgNT and SupT1-sgSLFN5 cell lines was determined by immunoblot (F). Data represent the mean ± SD of three independent experiments. P-values were calculated using a standard Student's t-test. *P < 0.05, **P < 0.01.
N-terminal domain and nuclear localization are required for the anti-HIV-1 activity of SLFN5
To define the domains involved in the anti-HIV-1 activity of SLFN5, we constructed seven SLFN5 truncations in either the N-terminus or C-terminus, designated as SLFN5-dC1 (1–780aa), SLFN5-dC2 (1–570aa), SLFN5-dC3 (1–335aa), SLFN5-dN1 (150–891aa), SLFN5-dN2 (335–891aa), SLFN5-dN3 (450–891aa) and SLFN5-dN4 (660–891aa) (Supplementary Figure S2a). Surprisingly, none of the constructs showed the anti-HIV-1 activity observed with the wild type SLFN5 (Supplementary Figure S2b–d). Notably, all C-terminal truncations were distributed in the cytoplasm whereas the wild type SLFN5 was localized to the nucleus (Supplementary Figure S2e). These results led us to investigate whether the nuclear localization is required for the antiviral activity of SLFN5. Accordingly, we locate a putative nuclear localization signal (NLS) to amino acid positions 801–820 (EKYKDRLLTAMRKRKLSQLH) using a NucPred (http://www.sbc.su.se/∼maccallr/nucpred/) (Figure 3A). By mutating the lysine and arginine residues in the predicted NLS, we found that substitution of lysine 812 to alanine abolished both the anti-HIV-1 activity (Figure 3B) and the nuclear localization of SLFN5 (Figure 3C). The same results were observed in double or triple mutants in which lysine 806 and/or arginine 804 were mutated in addition to lysine 812 (Figure 3B), thereby demonstrating the importance of the nuclear localization in SLFN5 inhibition of HIV-1.
The N-terminal domain has been implicated in conferring putative transcriptional activity in several studies (16,21). While the loss of antiviral activity by the three C-terminal truncation constructs was likely due to the failure of nuclear localization (Supplementary Figure S2e), this does not explain the loss of antiviral activity observed with the N-terminal truncation constructs. To better understand how the N-terminal domain contributes to the anti-HIV-1 activity of SLFN5, SV40 T NLS was attached to a series of SLFN5 N-terminal fragments including dC1, dC2, dC3 and a new N-terminal fragment (1–657aa) designated as dC4 (Figure 3A). We confirmed that the heterologous NLS was functional by rendering nuclear localization of these SLFN5 fragments (Figure 3C). The SV40 NLS-containing SLFN5 fragments were transfected into HEK293T cells together with NL4-3luc.R-E- and VSV-G to produce pseudotyped viruses. These viruses were used to infect SupT1 cells. The results showed that dC1-NLS, dC2-NLS and dC4-NLS inhibited HIV-1 production as strongly as the full-length SLFN5, while dC3-NLS displayed partial anti-HIV-1 activity (Figure 3D). Overall, these data demonstrate the importance of N-terminal 1–570aa fragment for the anti-HIV-1 activity of SLFN5.
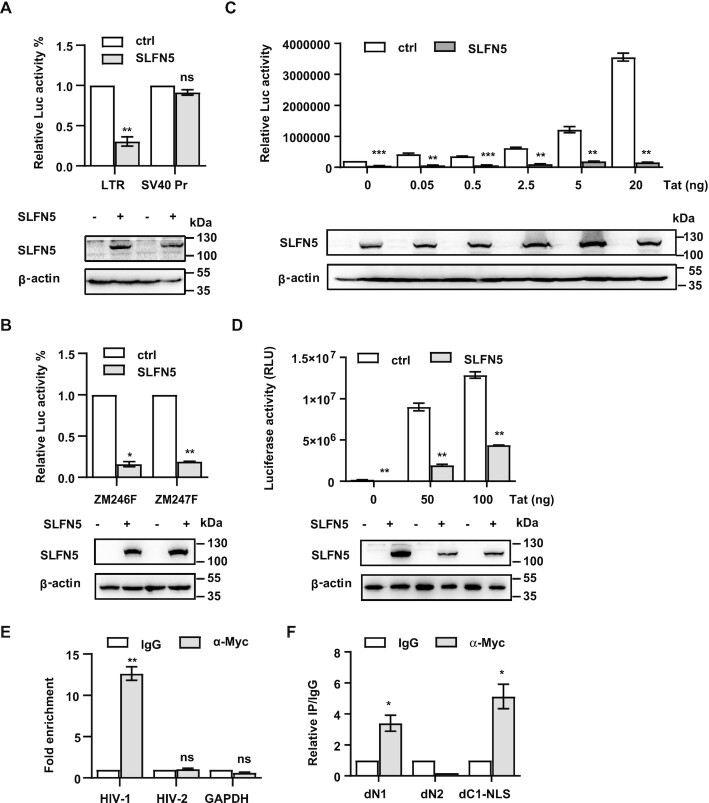
SLFN5 binds to HIV-1 LTR and represses LTR-driven gene expression
Given that the nuclear localization of SLFN5 is critical for its anti-HIV-1 activity, we sought to determine whether SLFN5 represses HIV-1 transcription. We used a firefly luciferase reporter construct pGL3-LTR, which contains the HIV-1 LTR sequence inserted upstream of a firefly reporter gene, to determine the effect of SLFN5 on LTR-driven transcription. We transfected this reporter plasmid into HEK293T cells together with SLFN5 or a control vector to test whether SLFN5 suppresses the LTR promoter. SLFN5 repressed HIV-1 LTR activity by about 70% (Figure 4A). Importantly, SLFN5 reduced LTR-driven transcription of TF viruses by over 90% (Figure 4B), which is in agreement with the one cycle infection assay (Figure 1F). We also investigated whether SLFN5 affects Tat-transactivation of HIV-1 LTR promoter. Different doses of Tat were co-transfected into HEK293T cells along with SLFN5 and pGL3-LTR. SLFN5 repressed Tat-induced LTR transcription by over 10-fold in the presence of less than 20 ng of Tat (Figure 4C). However, SLFN5 suppression of Tat-transactivation of HIV-1 LTR was partially lost when Tat concentration exceeded 20 ng (Figure 4D), suggesting a saturation effect of Tat on SLFN5 suppression.
Figure 4.
SLFN5 inhibits basal and Tat-transactivated HIV-1 transcription by specifically binding to the LTR via its N-terminal domain. (A) HEK293T cells were transfected with 500 ng of either NL4-3 LTR-luc (LTR) or SV40 promoter-Luc (SV40 Pr) reporter DNA, and SLFN5 DNA or a control vector (ctrl). 48h later, luciferase activity was measured. Expression of SLFN5 was detected by immunoblot. (B) HEK293T cells were transfected with 500 ng pZM246F-LTR-Luc (ZM246F) or pZM247F-LTR-Luc (ZM247F) and SLFN5 DNA or a control vector. Luciferase activity was measured after 48 h. Expression of SLFN5 was detected by immunoblot. (C) HEK293T cells were transfected with 0.5 μg NL4-3 LTR-luc and 0.5 μg SLFN5 with increasing doses of Tat DNA (0, 0.05, 0.5, 2.5, 5 and 20 ng). 48 h post-transfection, cells were lysed and luciferase activity was measured. Expression of SLFN5 was detected by immunoblot with α-Myc antibodies. (D) HEK293T cells were transfected with 0.5 μg LTR-luc and 0.5 μg SLFN5 with higher doses of Tat DNA (0, 50 and 100 ng). 48 h post-transfection, cells were lysed and luciferase activity was measured. Expression of SLFN5 were detected by immunoblot with α-Myc antibodies (E) 293T cells were transfected with NL4-3luc.R-E-, HIV-2 LTR or GAPDH promoter DNA, or SLFN5 DNA. The cells were subject to ChIP assay using an α-Myc antibody or mouse IgG as a negative control. Primer sets targeting HIV-1 U5-R, HIV-2 LTR or GAPDH promoter were used for qPCR. (F) HEK293T cells were transfected with NL4-3luc.R-E- and SLFN5-dN1, dN2 or dC1-NLS, and were subject to CHIP assay. Primers targeting U5-R were used for qPCR. Results shown are the average of three independent experiments. n.s. non-significant, **P < 0.01, *P < 0.05, ***P < 0.001.
Next we investigated whether SLFN5 affects splicing, nuclear export or stability of vRNA. HIV-1 transcripts are classified into three groups: full-length unspliced transcripts which serve as genomic RNA or Gag-Pol polyprotein mRNA, incompletely spliced RNA (Env, Vpu, Vpr, Vif), and completely spliced RNA (Tat, Rev, Nef) (38). Both full-length and incompletely spliced transcript harbor rev response element (RRE), which recruit Rev to facilitate its nuclear export (39). To assess a possible alteration of vRNA splicing pattern elicited by SLFN5, we determined the levels of Gag, Vpu and Rev RNA to measure full-length, incompletely and completely spliced transcripts in SLFN5-overexpressing HEK293T cells by qRT-PCR. There were no significant differences in the relative ratio of the three differentially spliced transcripts between SLFN5-overexpressing cells and cells transfected with a control vector (Supplementary Figure S3a). We also measured vRNA in the cytoplasmic fraction, as the nuclear export of unspliced RNA is a hallmark of HIV-1 vRNA processing. Total cellular RNA and RNA of cytoplasmic fraction were extracted separately, and transcripts of Gag, Vpu and Rev were examined in both fractions. We found no variations for levels of transcripts in both cytoplasmic and total mRNA between cells overexpressing SLFN5 and control vector-transfected cells, suggesting that SLFN5 does not affect the nuclear export of vRNA (Supplementary Figure S3b). This prompted us to investigate whether SLFN5 affects vRNA stability. HEK293T cells were transfected with NL4-3luc.R-E- along with SLFN5-Myc or control vector. At 24 h post-transfection, cells were treated with actinomycin D (actD), a transcription inhibitor to halt new mRNA synthesis. Cells were harvested at 0, 4, 8, 12, 16, 24 and 28 h after actD treatment and total RNA was extracted. Then Gag mRNA was quantified by qRT-PCR. Gag mRNA were not affected by SLFN5, indicating no effect of SLFN5 on the stability of viral mRNA (Supplementary Figure S3c). In addition, SLFN5 did not affect the stability of Gag determined by measuring its expression level after cycloheximide (CHX) treatment at different time points (Supplementary Figure S3d). We also sought to determine whether SLFN5 inhibits HIV-1 replication cycle at the reverse transcription or integration stage. Using Efavirenz (EFV, viral reverse transcriptase inhibitor) or Raltegravir (Ral, viral integrase inhibitor) as positive controls, HeLa cells were transfected with SLFN5 or a control vector, and challenged with HIV-1 reporter virus 24 h later. We quantified early reverse transcription product (U5-R) and integration product (2-LTR and Alu-LTR) in SLFN5-expressing cells or control cells, and in cells treated with EFV or RAL. Untreated control cells produced the same levels of both early reverse transcription products and integration products as the SLFN5-expressing cells (Supplementary Figure S3e), which suggests that the SLFN5 does not target the early stage of the HIV-1 replication cycle.
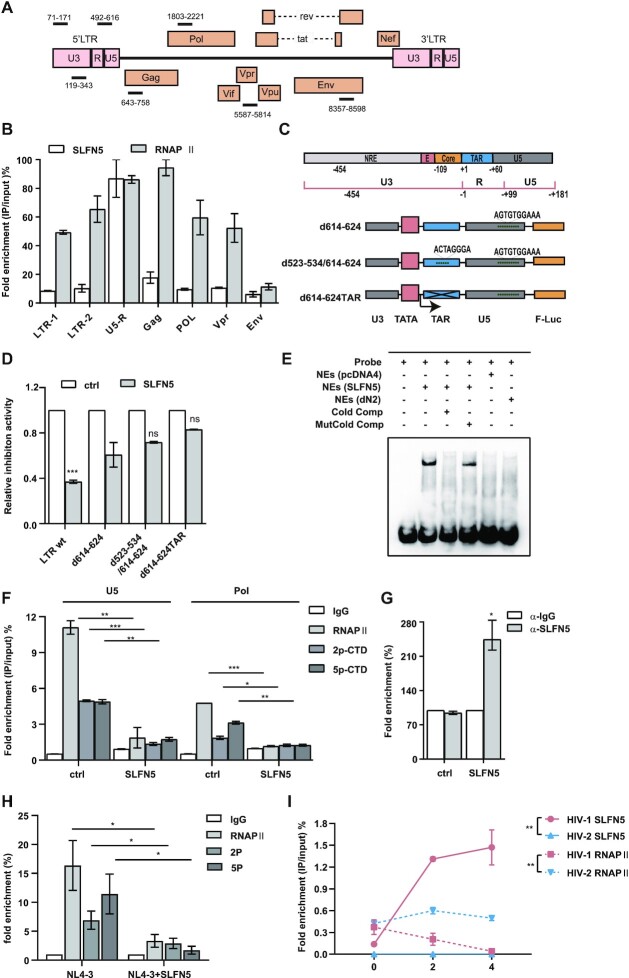
SLFN5 binds to HIV-1 U5-R region by its N-terminal domain
Since we found that SLFN5 inhibited HIV-1 transcription, we sought to determine which LTR sequences were necessary for the inhibitory effects of SLFN5. HIV-1 5′ LTR has a region at nt –454 to –109 known as the Negative Regulatory Element (NRE) (40) that downregulates LTR-directed gene expression. To determine whether NRE mediates SLFN5 inhibition, we transfected HEK293T cells with SLFN5 and either an F-luc reporter driven by parental LTR or a mutant lacking NRE (LTRdNRE), and observed similar inhibition of both reporters by SLFN5, indicating that NRE is not involved in SLFN5 inhibition (Supplementary Figure S4a). Moreover, HIV-1 LTR promoter contains two NFκB binding sites and three SP1 sites. The Tat-activated element (TAR) in the R region acts as the binding site for Tat and is essential for Tat transactivation (40). Thus, we investigated whether NFκB binding sites, SP1 binding sites and TAR are required for SLFN5 inhibition by transfecting HEK293T cells with either a WT LTR or mutated LTRs lacking NFκB (pGL3-LTR-dNF-κB), SP1 binding sites (pGL3-LTR-dSP1) or TAR element (pGL3-LTR-dTAR) along with SLFN5 or a control vector. SLFN5 reduced luciferase level driven by LTRdTAR by about 50%, which is less robust than WT LTR. In contrast, SLFN5 reduced luciferase level driven by LTR-dNF-κB and LTR-dSP1 at a similar level as WT LTR (Supplementary Figure S4b). Since HIV-2 is resistant whereas the 89.6 strain is susceptible to SLFN5 inhibition, two recombinant LTRs were constructed by swapping 1–530nt (R1) or 1–300nt of LTR (R2) derived from 89.6 strain with the counterparts of HIV-2 LTR. As expected, SLFN5 did not inhibit HIV-2 LTR, while SLFN5 inhibited the two recombinant LTR irrespectively of Tat activation (Supplementary Figure S4c).
It was reported that SLFN5 contains a DNA-binding domain in its N-terminus, which raises the possibility that SLFN5 represses the HIV LTR promoter via direct binding. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) assay using HEK293T cells transfected with SLFN5-Myc and NL4-3luc.R-E- DNA. SLFN5-Myc was immunoprecipitated with an α-Myc antibody and the possibly associated HIV-1 LTR was detected by qPCR (Figure 4E). The results showed a 13-fold enrichment of HIV-1 LTR in the Myc antibody IP samples compared to the IP performed with non-specific IgG control, suggesting a specific binding of SLFN5 to HIV-1 LTR. In contrast, no enrichment was detected for HIV-2 LTR or GAPDH promoter (Figure 4E). To evaluate whether the N-terminal COG2685 domain of SLFN5 was directly involved in LTR binding, we performed a ChIP assay of HEK293T cells that were co-transfected with NL4-3luc.R-E- and either dN1, dC1-NLS, or dN2 which lack the COG2865 domain. dN1 and dC1-NLS maintained strong binding with LTR whereas dN2, which lacks an assumed transcriptional regulatory domain, lost LTR binding (Figure 4F).
The interaction of SLFN5 with the HIV-1 LTR prompted us to determine which sequences in the LTR are recognized by SLFN5. Seven DNA fragments covering U3 (LTR1, LTR2), R-U5 region, Gag, Pol, Vpr and Env coding sequences (Figure 5A) were detected in ChIP-qPCR. While binding of Pol II was strongly enriched for the Gag region, SLFN5 binding to the U5-R region was specifically enriched (Figure 5B). Since HIV-1 U5-R region might not be the sole DNA element bound by SLFN5, we performed ChIP-seq in SupT1 cells to comprehensively identify SLFN5-bound DNA sequences. 28 binding sites for SLFN5 predicted by Homer software were listed in Supplementary Table S1, two of which resemble the sequence between 614–624 nt located in the U3 region (LTR614–624) and 523–534nt (LTR523–534) located in the R region of the LTR, of which the predicted binding of LTR614–624 showed high confidence level (Figure 5C). Thus, we investigated whether 614–624 nt and 523–534 nt bind to SLFN5. Three Luciferase reporter constructs were generated by inserting LTR lacking 614–624 nt (d614–624), LTR lacking 614–624 nt and 523–534 nt (d614–624/523–534) and LTR lacking both TAR and 614–624 nt (d614–624TAR) into a pGL3 vector. HEK293T cells were transfected with WT LTR, LTR d614–624, d614–624/523–534 or d614–624TAR in the presence or absence of SLFN5. SLFN5 lost its inhibitory effect on luciferase level completely with d614–624TAR, but only partially with d614–624/523–534 and d614–624 (Figure 5D). Collectively, the data suggest that 614–624 nt of the LTR and TAR are essential for the suppression by SLFN5. To further corroborate SLFN5 binding to LTR614–624, we performed a gel electrophoresis mobility shift assay (EMSA) and detected the association of SLFN5 with LTR614–624 using SLFN5-expressing HEK293T nuclear extracts (lane 2), but not using nuclear extracts of control vector-transfected cells (lane 5) or dN2-transfected cells (lane 6) (Figure 5E).
Figure 5.
SLFN5 inhibits HIV-1 LTR transcription by binding to the U5-R region and decreases the recruitment of RNAP II. (A) Schematic representation of the HIV-1 genome. The black lines indicate the position of each pair of primers used for ChIP-qPCR experiments in (B). (B) HEK293T cells were transfected with NL4-3luc.R-E- and SLFN5, then subject to ChIP assay. Different primer sets spanning HIV-1 genome were used for qPCR. Results are the average of two independent assays ± SD. (C) Illustration of HIV-1 LTR and its indicated mutants. The 5′LTR is divided into the U3, R and U5 regions. The U3 region is further divided into negative response element (NRE) (grey), enhancer (E) (rose pink) and basal/core promoter (core) (orange). R region contains trans-acting response element (TAR) (blue). The mutated LTR reporter constructs (d614–624, d614–624/523–534 and d614–624TAR) with the predicted SLFN5 binding sites deleted were shown below. (TATA TATA box; F-Luc; firefly luciferase). (D) HEK293T cells were transiently transfected with pGL3-LTR, pGL3-d614–624, pGL3-d614–624/523–534 or pGL3-d614–624TAR together with SLFN5 or a control vector. Luciferase activity was measured 48 h later. (E) EMSA assay was performed using SLFN5-expressing nuclear extracts (SLFN5-NEs, lane 2), control nuclear extracts (pcDNA4-NEs lane 5) and dN2-expressing nuclear extracts (dN2, lane 6). Cold comp indicates cold competition probe. Mutcold comp indicates mutated cold competition probe. NEs indicates nuclear extracts. (F) HEK293T cells were transfected with NL4-3luc.R-E- and either SLFN5 DNA or a control vector, then subject to ChIP assay using a phospho-Rpb1 CTD rabbit mAb (α-RNAPII), phospho-Rpb1 CTD (Ser2) rabbit mAb (α-2p-CTD), phospho-Rpb1 CTD (Ser5) rabbit mAb (α-5p-CTD), or an IgG control. qPCR was performed using primer sets to amplify U5-R and Pol, respectively. (G, H) VSV-G-pseudotyped NL4-3luc.R-E- reporter viruses were used to infect HeLa cells followed by ChIP assay at 2 dpi using an α-SLFN5 antibody (G) or α-RNAPII, α-2p-CTD and α-5p-CTD. (H). (I) HEK293T cells were transfected with 0, 2 or 4 μg of SLFN5-Myc and 4 μg of NL4-3luc.R-E- or HIV-2 LTR. 48 h post-transfection, cells were subject to ChIP assay using α-RNAPII, α-SLFN5 antibody or IgG control. qPCR was performed using primer sets to amplify HIV-1 U5-R and HIV-2 TAR, respectively. Results shown are the average of three independent experiments. n.s. non-significant, ** P < 0.01, * P < 0.05, *** P < 0.001.
RNA Pol II binding to the HIV-1 LTR is reduced by SLFN5
Transcription of HIV-1 RNA starts with recruitment of RNA polymerase II to the transcription initiation site where it interacts with the transcription initiation complex and is phosphorylated at serine 5 of C-terminal domain (CTD) by CDK7 (41) and associated Cyclin H. Shortly after transcription initiation, RNA Pol II pauses at the tertiary structure of the tat-transactivating response element (TAR). HIV-1 accessory protein Tat recruits the P-TEFb complex comprising of CDK9 and cyclinT1, and promotes additional serine 2 phosphorylation in the CTD of RNA Pol II which becomes more efficient in transcription elongation (42). To identify the stage of transcription targeted by SLFN5, we performed ChIP assay to measure LTR binding by total Pol II, serine 2 phosphorylated Pol II (2p-CTD) and serine 5 phosphorylated Pol II (5p-CTD). The results showed that SLFN5 decreased binding Pol II to the LTR by ∼80% (Figure 5F). Similar results were observed for 5p-CTD or 2p-CTD, which suggests that SLFN5 significantly reduces the initiation step of HIV-1 transcription. To determine whether SLFN5 inhibits the recruitment of RNA Pol II in the context of HIV-1 infection, HeLa cells were infected with HIV-1NL4-3luc reporter virus. ChIP-qPCR showed that SLFN5 associated with the LTR DNA during HIV-1 infection (Figure 5G) and impeded the binding of total Pol II as well as Ser-2 and Ser-5 phosphorylated Pol II to LTR (Figure 5H). To further examine whether SLFN5 binding to LTR reduces LTR-associated RNA Pol II, different amounts of SLFN5 were transfected into HEK293T cells with NL4-3luc.R-E- DNA. HIV-2 LTR was tested as a negative control. The interactions of SLFN5 and RNA Pol II with the LTR were examined by ChIP and PCR using α-SLFN5 or RNA Pol II antibodies. The results showed that increased binding of SLFN5 to LTR led to decreased association of RNA Pol II with LTR (Figure 5I), In addition, similar results were obtained by using Ser-2 and Ser-5 phosphorylated RNA Pol II (Supplementary Figure S6). Collectively, these data demonstrate that SLFN5 suppresses HIV-1 LTR transcription by blocking the recruitment of RNA Pol II.
SLFN5 increases repressive epigenetic marks at U5-R by recruiting histone methyltransferase
To further investigate how SLFN5 suppresses HIV-1 LTR transcription, we examined SLFN5-associated proteins by mass spectrometry (Supplementary Table S2), and detected RBBP7, one component of the PRC2 complex, and histone proteins, which suggests that SLFN5 may modulate chromatin modification. We next performed co-IP and immunoblots, and confirmed SLFN5 binding with RBBP7 (Figure 6A) and histone H3 (Figure 6B). With this, we examined whether SLFN5 changes the repressive chromatin marks in HIV-1 LTR. Results of ChIP-qPCR experiments using either H3K27me2 or H3K27me3 antibodies showed that SLFN5 increased H3K27me2 and H3K27me3 marks in HIV-1 R-U5 region by 1.5-fold and 2-fold, respectively, compared to the controls (Figure 6C and D). To further determine which methyltransferase was involved in this process, we examined six histone methyltransferases that are known to associate the H3K27me modification, including Suv39H1/2, G9a/GLP and EZH1/2, by transfecting HEK293T cells with NL4-3luc.R-E- and SLFN5 along with siRNAs targeting each of these methyltransferases. EZH1 siRNA and G9a siRNA abolished SLFN5 inhibition of HIV-1 LTR-directed luciferase expression (Figure 6E). To corroborate this observation, G9a inhibitor BRD4770 and EZH1 inhibitor UNC1999 were utilized to treat HEK293T cells that were transfected with SLFN5 and NL4-3luc.R-E-. The DOT1L inhibitor EPZ004777 was included as a negative control. BRD4770 inhibition of G9a eliminated SLFN5 inhibition of HIV-1 LTR, UNC1999 inhibition of EZH1 partially antagonized SLFN5 inhibition (Figure 6F). Interestingly, SLFN5 increased the expression of G9a (Figure 6F, lower panel). Next, we asked whether SLFN5 interacts with G9a and EZH1. Results of co-IP showed the association of SLFN5 with both EZH1 and G9a, but not with dN2 which lacks the binding domain for LTR. Treatment with 20 μM BRD4770 impaired the associations between SLFN5 with EZH1 or G9a (Figure 6G). Taken together, these data demonstrate that SLFN5 deposits repressive epigenetic markers on HIV-1 LTR by recruiting histone methyltransferase.
Figure 6.
SLFN5 represses HIV-1 transcription by depositing of H3K27me2 and H3K27me3 marks and interacting with histone H3 and modifying complex. (A) Co-IP of RBBP7 and Myc-tagged SLFN5 protein of HEK293T cells followed by immunoblot. HEK293T cells were transfected with SLFN5-Myc, 48h later, half of the cell lysis was immunoprecipitated by α-Myc antibody and the association with RBBP7 was detected by α-RBBP7 antibody. In a reciprocal Co-IP, the other half of the lysis was immunoprecipitated by α-RBBP7 antibody and the association with SLFN5-Myc was detected by α-Myc antibody. (B) Co-IP of histone H3 and Myc-tagged SLFN5 protein followed by immunoblot. HEK293T cells were transfected with SLFN5-Myc, 48h later, the cell lysis was immunoprecipitated by α-Myc antibody and the immunoprecipitates were detected by α-histone H3 antibody (left panel). In a reciprocal Co-IP, the cell lysis was immunoprecipitated by α-histone H3 antibody and the immunoprecipitates were detected by α-Myc antibody (right panel). (C) ChIP with α-H3K27me2 antibodies was performed. H3K27me2 associated with the U3 (left panel) and R-U5 regions (right panel) is shown relative to the background signal in IgG negative control ChIPs. (D) ChIP with α-H3K27me3 antibodies was performed. H3K27me3 associated with the U3 (left panel) and R-U5 (right panel) regions is shown relative to the background signal in IgG negative control ChIPs. (E) HEK293T cells were transfected with the indicated siRNA, NL4-3luc.R-E- and either SLFN5 DNA or a control vector. 48h post-transfection, luciferase activity was measured (upper panel) and siRNA knockdown was analysed by immunoblot (lower panel). (F) HEK293T cells were transfected with NL4-3luc.R-E-, and either SLFN5 DNA or a control vector, followed by treatment of BRD4770 (20 μM), UNC1999 (450 nM) or EPZ004777 (4 nM). 5 h post-transfection. 48 h later, luciferase activity was measured. Levels of G9a were determined by immunoblot. (G) Co-IP of EZH1, G9a, H3K27me2 and H3K27me3 with SLFN5 or dN2 using α-Myc antibody in the presence or absence of BRD4770 (20 μM). Results shown are the average of three independent experiments. n.s. non-significant, ** P < 0.01, * P < 0.05, *** P < 0.001.
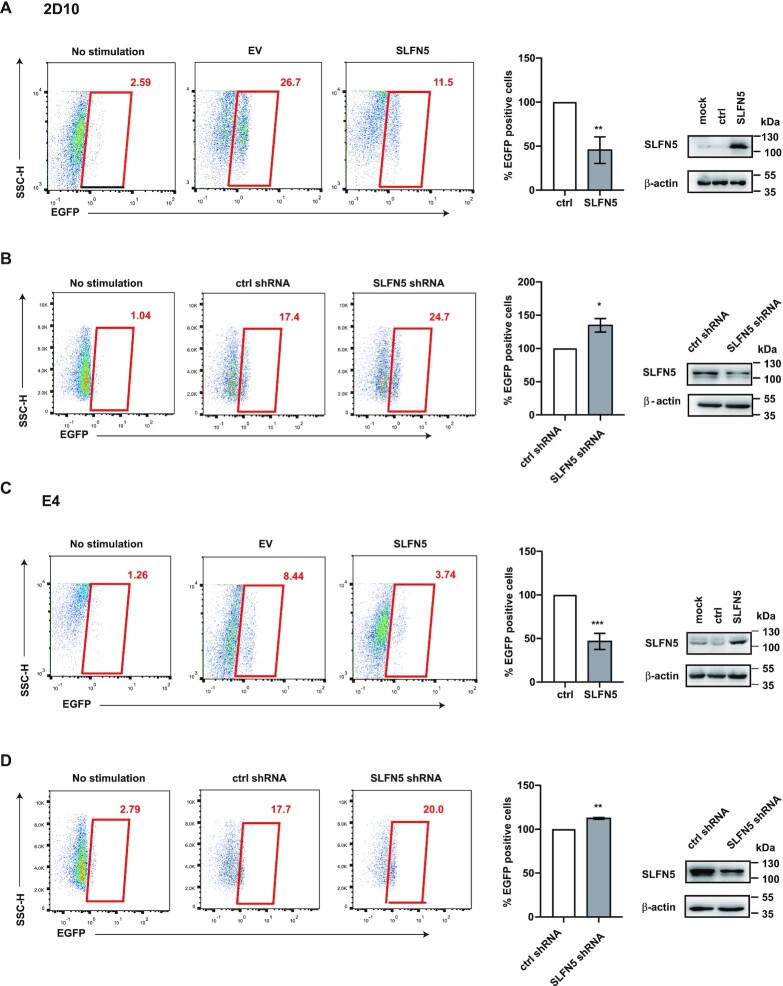
SLFN5 impedes the activation of HIV latency by SAHA and JQ-1
Since pharmacological inhibition of G9a or EZH2 has been shown to reactivate latent HIV-1(27,30), we thus examined whether SLFN5 has a role in HIV-1 latency. Two Jurkat cell-derived HIV-1 latency models (2D10 and E4) (43) were used, which both harbor one silent EGFP reporter provirus. We treated these two cell models with latency reversal agents (LRAs) JQ-1 followed by transfection with SLFN5 or a control vector. JQ1 treatment increased EGFP + cells to 26.7% in control vector transfected 2D10 cells, and this number dropped to 11.5% in SLFN5-transfected 2D10 cells (Figure 7A). With respects to E4 cells, the number dropped from 8.4% to 3.7% (Figure 7C). We further knocked down the endogenous SLFN5 using shRNA, and observed the increase of JQ1-activated EGFP + cells from 17.4% to 24.7% in 2D10 cells (Figure 7B) and from 17.7% to 20% in E4 cells (Figure 7D). Moreover, we excluded that JQ1 treatment affected the expression level of G9a, EZH1, histone H3 and RBBP7 (Supplementary Figure S5). These data suggest that SLFN5 contributes to the maintenance of HIV-1 latency.
Figure 7.
SLFN5 impairs activation of latent proviruses. (A, C) HIV-1 latently infected 2D10 (A) and E4 cells (C) were stimulated by JQ-1 (1 μM) for 48 h, followed by electroporation of SLFN5-Myc DNA or pcDNA4 as a negative control. EGFP + cells were scored by flow cytometry. SLFN5-Myc expression was analysed by immunoblot. (B, D) HIV-1 latently infected 2D10 (B) and E4 cells (D) were stimulated by JQ-1 (1 μM) for 48 h, followed by electroporation of SLFN5 shRNA or ctrl shRNA as a negative control. Levels of endogenous SLFN5 were examined by immunoblot. Results shown are the average of three independent experiments. ** P < 0.01, * P < 0.05.
DISCUSSION
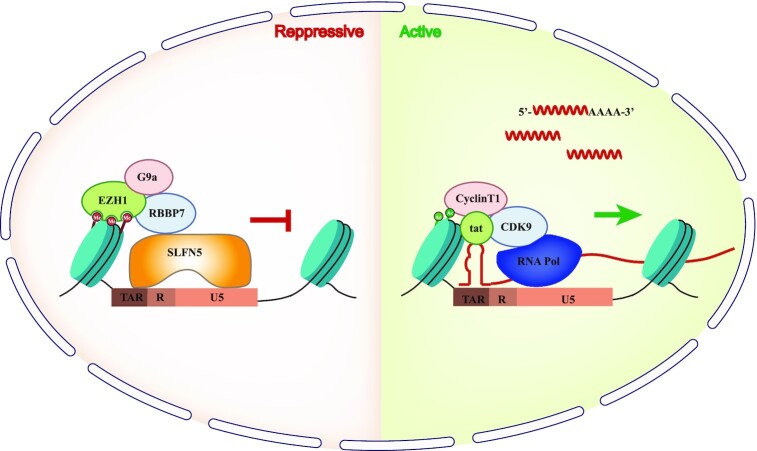
In this study, we have found that SLFN5 potently represses HIV-1 LTR-directed transcription and inhibits different HIV-1 strains to various degrees. Mechanistic studies demonstrated that SLFN5 targets two noncontiguous but closely linked sequences in HIV-1 LTR, LTR614–624 and TAR, and deters the binding of RNA Pol II to the LTR promoter. We further discovered that SLFN5 does so by depositing H3K27 repressive markers on HIV-1 LTR through recruiting the G9a and PRC2 histone methyltransferase complex (Figure 8).
Figure 8.
A model to illustrate how SLFN5 inhibits HIV-1 LTR transcription. SLFN5 binds to two sequences in the R and U5 regions through its N-terminal domain. Binding of SLFN5 to HIV-1 LTR increases H3 repressive marks at lysine 27 which are deposited by G9a and EZH1, thereby preventing the recruitment of RNA Pol II and suppressing HIV-1 transcription. Once latent provirus infection is established, SLFN5 prevents reactivation of the provirus by latency reversal agents.
SLFN5 carries the N-terminal ‘slfn box’ which shares homology with the signature domain COG2865 found in transcription regulators and helicases (16,21). We were able to show that the N-terminal domain is essential for the anti-HIV-1 activity of SLFN5, whereas RNA-modeling domains are dispensable, by testing a series of SLFN5 deletion mutants. Interestingly, fusion of an exogenous NLS into N-terminal fragments of SLFN5 restored the nuclear localization and anti-HIV-1 activity, suggesting that N-terminal 1–570 aa sequence and the NLS are essential for the anti-HIV-1 function of SLFN5.
The role of SLFN5 in transcription regulation has been implicated in several earlier studies. The reported function of SLFN5 in cell proliferation, anchorage-independent growth and tumorigenesis may also be attributed to the regulation of specific gene expression. This function of SLFN5 has also been reported on other SLFN family members. For instance, SLFN9 was shown to co-localize with phosphorylated RNA Pol (15). We now show that SLFN5 is a novel transcriptional repressor for the HIV-1 promoter. Specifically, SLFN5 binds to the R-U5 region and interferes with the recruitment of RNA Pol II, thus blocking the initiation of HIV-1 transcription. By this repressor function SLFN5 also regulates cellular genes, since in our ChIP-seq assay, we found that SLFN5 binds to the promoter region of ZEB1 gene, in agreement with two previous reports showing that ZEB1 is negatively regulated by SLFN5 in breast cancer cells (20,23).
The PRC2 complex regulates chromatin compaction by catalyzing the methylation of histone H3 at lysine 27 through its histone methyltransferases EZH1 and EZH2 (44). Several studies have reported that histone lysine methyltransferase, including EZH2 and G9a, play a prominent role in the regulation of HIV-1 provirus silencing (27,30). An interaction between G9a and PRC2 was recently reported (45). Overexpression of G9a increases both H3K9 and H3K27 methylation, reduces E-cadherin expression, and induces epithelial-mesenchymal transition in PANC-1 pancreatic cancer cells. Our data demonstrate that G9a and EZH1 interact with SLFN5 and are involved in SLFN5-mediated suppression of HIV-1 LTR transcription. Interestingly, EZH2 is dispensable for SLFN5 inhibition of HIV-1, suggesting that SLFN5 inhibits HIV-1 gene expression by a mechanism that is distinct from the previously reported EZH2-dependent ones. In agreement with this, a recent study reported E-cadherin was down-regulated in SLFN5-overexpressing A549 cells, possibly in a similar epigenetic way mediated by G9a (46).
It is worthy to note that G9a inhibitor is sufficient to eliminate SLFN5′s function. This suggests deposition of repressive marks by PRC2 and G9a is required for the action of SLFN5. In another hand, SLFN5 binds with the sequence in R and U5 sequence, and deletion of the binding sequences is able to abolish the inhibitory activity of SLFN5 on HIV-1 LTR. The two processes might act sequentially to achieve a transcriptional repression state. It is possible that SLFN5 binds with specific sequences of HIV-1 LTR and impedes the occupancy of RNA polymerase II, which triggers the recruitment of PRC2 and G9a, resulting in a latency state by an as yet undetermined mechanism. Alternatively, SLFN5 recruits G9a and PRC2 complex to add repressive marks, resulting in the failure of binding of RNA Pol II to LTR. In both scenarios, the epigenetic mechanism plays a dominant role in this process the details of which warrant further investigations. Importantly, Schlafen family play an essential role in T cell differentiation and development (16), it is also intriguing to study whether Schlafen family proteins and PRC2 complex as well as G9a act in synergy for T cell lineage commitment in future.
One interesting observation is that different HIV-1 strains, HIV-2 and SIV exhibited markedly different sensitivity to SLFN5 inhibition. One possible explanation is that sequence variations in the LTR promoters of these lentiviruses affect the binding of SLFN5. We noted that T/F viruses, which establish mucous HIV-1 infection, are more potently inhibited by SLFN5 compared to chronic HIV-1 strains. This suggests a role of SLFN5 in preventing HIV-1 acquisition by restricting the infection of T/F HIV-1. Different viruses may adapt different strategies to antagonize SLFN5 inhibition. A recent study reported that the ICP0 protein, an E3 ubiquitin ligase encoded by herpes simplex virus 1 (HSV-1), causes the ubiquitination and degradation of SLFN5, thus confer refraction to SLFN5 restriction (47). It remains to be determined whether HIV-1 uses a similar mechanism to evade SLFN5 inhibition.
Our data showed that SLFN5 inhibits both basal LTR transcription as well as Tat transactivation. Although SLFN5 binds to the TAR sequence in the HIV-1 LTR, Tat binding to TAR is not affected in SLFN5-overexpressing cells, this is likely because SLFN5 binds to TAR DNA whereas Tat binds to the TAR RNA structure. Nonetheless, SLFN5 inhibits Tat-transactivated HIV-1 LTR transcription to a much greater degree than inhibits HIV-1 LTR basal transcription, suggesting a direct impact of SLFN5 on Tat function. We also noted that SLFN5 inhibition was partially lost when more Tat plasmid DNA was used in transfection, which indicates SLFN5 inhibition can be saturated.
We further showed that SLFN5 is able to assist the maintenance of latent HIV-1. This interesting role of SLFN5 should depend on its levels in HIV-1 reservoir cells including resting memory T cells. Given its interferon inducible nature, at the initial stage of HIV-1 infection when host cells respond and produce interferon, the stimulated SLFN5 may operate with other cellular factors including the epigenetic machineries to silence the integrated HIV-1 DNA and stop HIV-1 production and spread to new target cells. As a result, HIV-1 latency is created. In this context, SLFN5 may have a role at the early stage of HIV-1 infection to promote the establishment of latent HIV-1. Theoretically, targeting SLFN5 may help delay or diminish HIV-1 latency, and increase the efficiency of HIV-1 latency reversal agents such as JQ1.
Several SLFN family members have now been reported to inhibit HIV-1. SLFN11 was first found to suppress HIV-1 protein synthesis by a mechanism of codon-bias discrimination (13). Recently, SLFN13 was shown to restrict HIV-1 replication by acting as a tRNA/rRNA-targeting endoribonuclease (14). Here, we identified SLFN5 as a novel transcription repressor suppressing HIV-1 gene expression through depositing histone suppression markers on viral LTR DNA. It is worthwhile testing whether SLFN5, SLFN11 and SLFN13 inhibit other types of retroviruses, whether other members of the SLFN members also bear anti-retrovirus function, and finally, whether the expansion of the SLFN protein family is a response to the many retrovirus assaults in the long history of human evolution.
DATA AVAILABILITY
All data are available from the corresponding author upon request. The flow cytometry data have been deposited in FlowRepository (ID: FR-FCM-Z4VV, URL: https://flowrepository.org/id/RvFrcYXoyPzUboxFpHLeYMVAFzwRptjjKUSB4gZzN. cQFLEsk9TTKR5twPb36xYRw). Raw ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE197700.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Microbial Resource Center (No. NMRC-2020–3) and the CAMS Collection Center of Pathogenic Microorganisms (CAMS-CCPM-A) for providing valuable reagents.
Author contributions: S.C., J.D. and X.L. designed the study; J.D. and S.W. performed the main part of the experiment; Z.W., S.C. and J.Z. performed the antiviral testing; Z.L. supervised the in vitro binding experiment; L.M. and J.W. contributed to the data analysis; J.D. prepared the manuscript; M.S., C.L. and S.C. revised the manuscript; S.C., X.L. and F.G. conceived and supervised the project; all authors approved the final version of the manuscript.
Contributor Information
Jiwei Ding, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Shujie Wang, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Zhen Wang, Lady Davis Institute for Medical Research and McGill AIDS Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Shumin Chen, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Jianyuan Zhao, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Magan Solomon, Lady Davis Institute for Medical Research and McGill AIDS Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Zhenlong Liu, Lady Davis Institute for Medical Research and McGill AIDS Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Fei Guo, Institute of Pathogen Biology, Chinese Academy of Medical Science, Beijing, China.
Ling Ma, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Jiajia Wen, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Xiaoyu Li, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China.
Chen Liang, Lady Davis Institute for Medical Research and McGill AIDS Centre, Jewish General Hospital, Montreal, Quebec, Canada.
Shan Cen, Institute of Medicinal Biotechnology, Chinese Academy of Medical Science, Beijing, China; CAMS Key Laboratory of Antiviral Drug Research, Chinese Academy of Medical Science, Beijing, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [81973220, 31870164]; Fundamental Research Funds for the Central Universities [33320200046]; CAMS Innovation Fund for Medical Sciences [2021-I2M-1-038, 2021-I2M-1-030]. Funding for open access charge: Natural Science Foundation of China [81973220, 31870164]; Fundamental Research Funds for the Central Universities [33320200046]; CAMS Innovation Fund for Medical Sciences [2021-I2M-1-038, 2021-I2M-1-030].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kaiser S.M., Malik H.S., Emerman M.. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007; 316:1756–1758. [DOI] [PubMed] [Google Scholar]
- 2. Bishop K.N., Holmes R.K., Sheehy A.M., Malim M.H.. APOBEC-mediated editing of viral RNA. Science. 2004; 305:645. [DOI] [PubMed] [Google Scholar]
- 3. Lecossier D., Bouchonnet F., Clavel F., Hance A.J.. Hypermutation of HIV-1 DNA in the absence of the vif protein. Science. 2003; 300:1112. [DOI] [PubMed] [Google Scholar]
- 4. Goujon C., Moncorgé O., Bauby H., Doyle T., Ward C.C., Schaller T., Hué S., Barclay W.S., Schulz R., Malim M.H.. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013; 502:559–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C.. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013; 14:398–410. [DOI] [PubMed] [Google Scholar]
- 6. Kane M., Yadav S.S., Bitzegeio J., Kutluay S.B., Zang T., Wilson S.J., Schoggins J.W., Rice C.M., Yamashita M., Hatziioannou T.et al.. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013; 502:563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perez-Caballero D., Zang T., Ebrahimi A., McNatt M.W., Gregory D.A., Johnson M.C., Bieniasz P.D.. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009; 139:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Usami Y., Wu Y., Gottlinger H.G.. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by nef. Nature. 2015; 526:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S.L., McCauley S.M., Nowosielska A., Antonarakis S.E., Luban J.et al.. HIV-1 nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015; 526:212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mavrommatis E., Fish E.N., Platanias L.C.. The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 2013; 33:206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu F., Zhou P., Wang Q., Zhang M., Li D.. The schlafen family: complex roles in different cell types and virus replication. Cell Biol. Int. 2018; 42:2–8. [DOI] [PubMed] [Google Scholar]
- 12. Katsoulidis E., Carayol N., Woodard J., Konieczna I., Majchrzak-Kita B., Jordan A., Sassano A., Eklund E.A., Fish E.N., Platanias L.C.. Role of schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J. Biol. Chem. 2009; 284:25051–25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li M., Kao E., Gao X., Sandig H., Limmer K., Pavon-Eternod M., Jones T.E., Landry S., Pan T., Weitzman M.D.et al.. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012; 491:125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J.Y., Deng X.Y., Li Y.S., Ma X.C., Feng J.X., Yu B., Chen Y., Luo Y.L., Wang X., Chen M.L.et al.. Structure of schlafen13 reveals a new class of tRNA/rRNA- targeting Rnase engaged in translational control. Nat. Commun. 2018; 9:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neumann B., Zhao L., Murphy K., Gonda T.J.. Subcellular localization of the schlafen protein family. Biochem. Biophys. Res. Commun. 2008; 370:62–66. [DOI] [PubMed] [Google Scholar]
- 16. Geserick P., Kaiser F., Klemm U., Kaufmann S.H., Zerrahn J.. Modulation of t cell development and activation by novel members of the schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004; 16:1535–1548. [DOI] [PubMed] [Google Scholar]
- 17. Arslan A.D., Sassano A., Saleiro D., Lisowski P., Kosciuczuk E.M., Fischietti M., Eckerdt F., Fish E.N., Platanias L.C.. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene. 2017; 36:6006–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Companioni Napoles O., Tsao A.C., Sanz-Anquela J.M., Sala N., Bonet C., Pardo M.L., Ding L., Simo O., Saqui-Salces M., Blanco V.P.et al.. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J. Gastroenterol. 2017; 52:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fischietti M., Eckerdt F., Blyth G.T., Arslan A.D., Mati W.M., Oku C.V., Perez R.E., Lee-Chang C., Kosciuczuk E.M., Saleiro D.et al.. Schlafen 5 as a novel therapeutic target in pancreatic ductal adenocarcinoma. Oncogene. 2021; 40:3273–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu X., Wan G., Yang Y., Liu Y., Yang X., Zheng Y., Jiang L., Zhang P., Liu D., Zhao W.et al.. SLFN5 influences proliferation and apoptosis by upregulating PTEN transcription via ZEB1 and inhibits the purine metabolic pathway in breast cancer. Am. J. Cancer Res. 2020; 10:2832–2850. [PMC free article] [PubMed] [Google Scholar]
- 21. Katsoulidis E., Mavrommatis E., Woodard J., Shields M.A., Sassano A., Carayol N., Sawicki K.T., Munshi H.G., Platanias L.C.. Role of interferon {alpha} (IFN{alpha})-inducible schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J. Biol. Chem. 2010; 285:40333–40341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sassano A., Mavrommatis E., Arslan A.D., Kroczynska B., Beauchamp E.M., Khuon S., Chew T.L., Green K.J., Munshi H.G., Verma A.K.et al.. Human schlafen 5 (SLFN5) is a regulator of motility and invasiveness of renal cell carcinoma cells. Mol. Cell. Biol. 2015; 35:2684–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wan G., Zhu J., Gu X., Yang Y., Liu Y., Wang Z., Zhao Y., Wu H., Huang G., Lu C.. Human schlafen 5 regulates reversible epithelial and mesenchymal transitions in breast cancer by suppression of ZEB1 transcription. Br. J. Cancer. 2020; 123:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez R.S., Salji M.J., Rushworth L., Ntala C., Rodriguez Blanco G., Hedley A., Clark W., Peixoto P., Hervouet E., Renaude E.et al.. SLFN5 regulates LAT1-Mediated mTOR activation in castration-resistant prostate cancer. Cancer Res. 2021; 81:3664–3678. [DOI] [PubMed] [Google Scholar]
- 25. Gu X., Zhou L., Chen L., Pan H., Zhao R., Guang W., Wan G., Zhang P., Liu D., Deng L.L.et al.. Human schlafen 5 inhibits proliferation and promotes apoptosis in lung adenocarcinoma via the PTEN/PI3K/AKT/mTOR pathway. Biomed. Res. Int. 2021; 2021:6628682. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Shang Hong-tao, Ying Yu-shu, Tao-Wu Qiu-liZhang, Liang Fu-jun. Progress and challenges in the use of latent HIV-1 reactivating agents. Acta. Pharm. Sin. 2015; 36:908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen K., Das B., Dobrowolski C., Karn J.. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. Mbio. 2017; 8:e00133-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Friedman J., Cho W.K., Chu C.K., Keedy K.S., Archin N.M., Margolis D.M., Karn J.. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of zeste 2. J. Virol. 2011; 85:9078–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. du Chene I., Basyuk E., Lin Y.L., Triboulet R., Knezevich A., Chable-Bessia C., Mettling C., Baillat V., Reynes J., Corbeau P.et al.. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007; 26:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imai K., Togami H., Okamoto T.. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 2010; 285:16538–16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boehm D., Jeng M., Camus G., Gramatica A., Schwarzer R., Johnson J.R., Hull P.A., Montano M., Sakane N., Pagans S.et al.. SMYD2-Mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe. 2017; 21:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H.et al.. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008; 105:7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haaland R.E., Hawkins P.A., Salazar-Gonzalez J., Johnson A., Tichacek A., Karita E., Manigart O., Mulenga J., Keele B.F., Shaw G.M.et al.. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype a and c HIV-1. PLoS Pathog. 2009; 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abrahams M.R., Anderson J.A., Giorgi E.E., Seoighe C., Mlisana K., Ping L.H., Athreya G.S., Treurnicht F.K., Keele B.F., Wood N.et al.. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype c reveals a non-poisson distribution of transmitted variants. J. Virol. 2009; 83:3556–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y., Li N., Zhang T., Huang X., Cai F., Vandergrift N., Xin R., Meng Z., Zhang X., Jiang C.et al.. Comprehensive characterization of the transmitted/founder env genes from a single MSM cohort in china. J. Acquir. Immune Defic. Syndromes. 2015; 69:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding J.W., Zhao J.Y., Mi Z.Y., Wei T., Cen S.. [The sensitivity of transmitted/founder HIV-1 to anti-HIV drugs]. Yao Xue Xue Bao. 2016; 51:367–372. [PubMed] [Google Scholar]
- 37. Salazar-Gonzalez J.F., Salazar M.G., Keele B.F., Learn G.H., Giorgi E.E., Li H., Decker J.M., Wang S., Baalwa J., Kraus M.H.et al.. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 2009; 206:1273–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukumori T., Kagawa S., Iida S., Oshima Y., Akari H., Koyama A.H., Adachi A.. Rev-dependent expression of three species of HIV-1 mRNAs (review). Int. J. Mol. Med. 1999; 3:297–302. [DOI] [PubMed] [Google Scholar]
- 39. Pollard V.W., Malim M.H.. The HIV-1 rev protein. Annu. Rev. Microbiol. 1998; 52:491–532. [DOI] [PubMed] [Google Scholar]
- 40. Ne E., Palstra R.J., Mahmoudi T.. Transcription: insights from the HIV-1 promoter. Int. Rev. Cell Mol. Biol. 2018; 335:191–243. [DOI] [PubMed] [Google Scholar]
- 41. Kim M., Vasiljeva L., Rando O.J., Zhelkovsky A., Moore C., Buratowski S.. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006; 24:723–734. [DOI] [PubMed] [Google Scholar]
- 42. Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014; 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pearson R., Kim Y.K., Hokello J., Lassen K., Friedman J., Tyagi M., Karn J.. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008; 82:12291–12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D.. Ezh1 and ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008; 32:503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pan M.-R., Hsu M.-C., Chen L.-T., Hung W.-C.. G9a orchestrates PCL3 and KDM7A to promote histone H3K27 methylation. Scientific Rep. 2015; 5:18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo L., Liu Z., Tang X.. Overexpression of SLFN5 induced the epithelial-mesenchymal transition in human lung cancer cell line A549 through β-catenin/Snail/E-cadherin pathway. Eur. J. Pharmacol. 2019; 862:172630. [DOI] [PubMed] [Google Scholar]
- 47. Kim E.T., Dybas J.M., Kulej K., Reyes E.D., Price A.M., Akhtar L.N., Orr A., Garcia B.A., Boutell C., Weitzman M.D.. Comparative proteomics identifies schlafen 5 (SLFN5) as a herpes simplex virus restriction factor that suppresses viral transcription. Nat. Microbiol. 2021; 6:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon request. The flow cytometry data have been deposited in FlowRepository (ID: FR-FCM-Z4VV, URL: https://flowrepository.org/id/RvFrcYXoyPzUboxFpHLeYMVAFzwRptjjKUSB4gZzN. cQFLEsk9TTKR5twPb36xYRw). Raw ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE197700.