Abstract
The chromosome dimer resolution machinery of bacteria is generally composed of two tyrosine recombinases, XerC and XerD. They resolve chromosome dimers by adding a crossover between sister copies of a specific site, dif. The reaction depends on a cell division protein, FtsK, which activates XerD by protein-protein interactions. The toxin-linked cryptic satellite phage (TLCΦ) of Vibrio cholerae, which participates in the emergence of cholera epidemic strains, carries a dif-like attachment site (attP). TLCΦ exploits the Xer machinery to integrate into the dif site of its host chromosomes. The TLCΦ integration reaction escapes the control of FtsK because TLCΦ encodes for its own XerD-activation factor, XafT. Additionally, TLCΦ attP is a poor substrate for XerD binding, in apparent contradiction with the high integration efficiency of the phage. Here, we present a sequencing-based methodology to analyse the integration and excision efficiency of thousands of synthetic mini-TLCΦ plasmids with differing attP sites in vivo. This methodology is applicable to the fine-grained analyses of DNA transactions on a wider scale. In addition, we compared the efficiency with which XafT and the XerD-activation domain of FtsK drive recombination reactions in vitro. Our results suggest that XafT not only activates XerD-catalysis but also helps form and/or stabilize synaptic complexes between imperfect Xer recombination sites.
INTRODUCTION
Repair by homologous recombination can lead to the formation of chromosome dimers when the chromosomes are circular, as is generally the case in bacteria and archaea. Chromosome dimers physically impede the segregation of genetic information. They are resolved by the addition of a crossover between the sister copies of a specific chromosomal locus, dif, by a highly conserved chromosomally encoded tyrosine recombination (Xer) machinery (1).
Many mobile elements such as plasmids exploit the high conservation of the Xer machinery (1). Indeed, Xer was initially characterized as a multicopy plasmid dimer resolvase (2,3). Integrative Mobile Elements exploiting Xer (IMEX) have also been reported (4,5). Plasmids and IMEX participate in the acquisition of antibiotic resistance and pathogenicity factors. In particular, the continuous emergence of new pathogenic forms of Vibrio cholerae is linked to cycles of integration and excision of several IMEX (6–10). Foremost among those are the Cholera ToXin phage, CTXΦ, which harbours the genes responsible for the deadly diarrhoea associated with cholera (11), and a Toxin-Linked Cryptic satellite phage, TLCΦ (12). It has been proposed that prior integration of TLCΦ is necessary to facilitate the integration of CTXΦ into the dif site of the larger of the two chromosomes of its host, chr1 (13). One or more TLCΦ-integration events preceded the integration of CTXΦ in most clinical strains (14–18). For instance, two and three TLCΦ-integration events took place before the integration of CTXΦ on the chr1 of the N16961 clinical isolate from the on-going seventh cholera pandemic and the O395 clinical isolate from the sixth cholera pandemic, respectively (Figure 1A). In both cases, a first copy of TLCΦ integrated into difA, the chr1 dif site of the clinical isolate environmental ancestor, which led to the formation of a new dif site, dif1, into which a second copy of TLCΦ integrated, prior to the integration of CTXΦ (Figure 1A and B).
Figure 1.
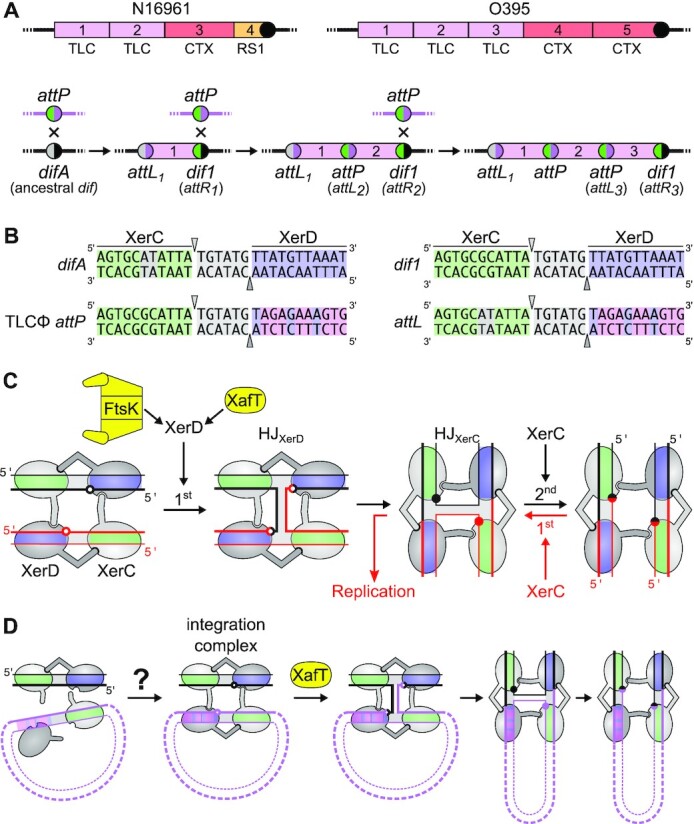
Xer recombination. (A) Schematic representation of the IMEX arrays found on chr1 of a V. cholerae strain from the 7th (N16961) and the 6th (O395) pandemics (top) and of the successive integrations of TLCΦ leading to the formation of those arrays (bottom). The arrays result from successive lysogenic conversion events, a new dif site being generated on the right side of each newly-integrated IMEX. Pink, red, orange blocks: TLCΦ, CTXΦ and RS1 prophages; Coloured disks: difA (grey/black), TLCΦ attP (green/pink), TLCΦ attL1 (grey/pink), dif1 (green/black) and current dif site of the strains (black disk). Numbers indicate the order of integration of the phage, which can be directly deduced from its position relative to the current dif site. (B) V. cholerae difA and dif1, TLCΦ attP and attL1 sequences. Green: XerC-binding site; Light grey: central region and non-canonical bp of difA; Blue: XerD-binding site; Pink: non-canonical bp of attP. The top and bottoms are cleaved by XerC and XerD, respectively. The positions of cleavage are indicated by triangles. (C) Xer recombination pathways. XerC and XerD are shown in light and dark grey, respectively. The central region, XerC and XerD-arms of the recombination sites are shaded in grey, green and blue, respectively. Black arrows: conventional recombination pathway; Red arrows: non-conventional asymmetric recombination pathway. The XerD and XerC cleavage points are depicted by empty and filled disks. (D) TLCΦ integration. Black and pink lines: V. cholerae chr1 and TLCΦ DNA, respectively. Question mark: the low affinity of XerD for the XerD-arm of TLCΦ attP poses questions about the possible formation of integration complexes.
In V. cholerae, as in most bacteria, the Xer machinery is composed of two closely related recombinases, XerC and XerD (Supplementary Figure S1A). The dif sites are composed of an 11 base pair (bp) XerC-binding arm and an 11 bp XerD-binding arm, which are separated by a short 6 bp central region (Figure 1B and Supplementary Figure S1B). The XerC- and XerD-arms are partial inverted repeats. XerC and XerD each catalyse the cleavage and transfer of a specific pair of DNA sequences within the tetrameric nucleoprotein complex (Figure 1B and C). To resolve chromosome dimers, XerCD follow the conventional recombination pathway of tyrosine recombinases. A first pair of DNA strands is cleaved and transferred leading to the formation of a Holliday junction (HJ) intermediate that is subsequently resolved into a crossover by the exchange of the second pair of strands (Figure 1C). During chromosome dimer resolution, the exchange of the first pair of strands is catalysed by XerD and the second by XerC (Figure 1C, (19,20)). This process is under the control of a DNA translocase anchored in the cell division septum, FtsK. The C-terminal γ-domain of FtsK activates XerD by protein-protein interactions (Figure 1C, (19,21–23)). Hence, Xer recombination is normally restricted to the time of cell division and to synaptic complexes located in the proximity of the cell division apparatus (24–28). Many IMEX including CTXΦ integrate via a non-conventional FtsK-independent recombination pathway which is initiated by XerC-mediated strand exchanges. The resulting HJ is resolved into fully integrated CTXΦ by DNA replication (Figure 1C, (7,8,29)). Dimers of the ColE1 multicopy plasmid are resolved by a similar process (30). In contrast, TLCΦ integration follows a XerD-first pathway like chromosome dimer resolution (9), but the process escapes the control of FtsK control because TLCΦ encodes for its own XerD activation factor, XafT (Figure 1C, (9,31)).
Eight bp of the XerD-arm of TLCΦ attP deviate from the canonical XerD-arm of dif sites (Figure 1B). In vitro experiments previously showed that XerD failed to bind TLCΦ attP under conditions in which it efficiently bound the dif1 site (9). In addition, XerC/XerD/TLCΦ attP complex formation was barely detectable (9). Those observations raised questions about how XerC and XerD could promote the previously observed highly efficient integration of TLCΦ in vivo (Figure 1D, (9,13,31)).
Here, we present a methodology based on deep sequencing that permits the in vivo assessment of the integration and excision frequencies of thousands of mobile elements with differing attachment sites. This methodology was applied to investigate the XerD-arm sequence-dependence of TLCΦ attP during Xer-mediated recombination reactions, in either FtsK- or XafT-driven conditions. By in vitro experiments we also compared the efficiency of XafT-driven and FtsKγ-driven Xer recombination reactions between two dif1 sites, and between dif1 and TLCΦ attP. Taken together, our results suggest that XafT is not only a XerD-activation factor, but also a synapse formation and/or stabilization factor. Our results also indicate that undesired excision events are prevented because the expression of XafT is limited after integration.
MATERIALS AND METHODS
Strains, plasmids and oligonucleotides
Strains, plasmids and oligonucleotides are listed in Supplementary Tables S1, S2 and S3, respectively. V. cholerae strains were built by natural transformation using appropriate selection markers and/or blue/white β-galactosidase screens (32). Plasmids pSM11 and pSM15 are derivatives of the pTLC8 and pSM12 non-replicative mini-TLCΦ respectively (31). They were constructed by replacing the TLCΦ attP locus with the BsaI-ccdB-BsaI cassette of pFB5 (33) using Gibson assembly (34). Pools of n8gtg, tagn8 and n5a2n3g TLCΦ attP-derived sites flanked by cognate BsaI sites were generated by primer extension of oligo 4069 annealed to degenerate 4066, 4067, 4068 oligonucleotides, respectively. The degenerate attP sites flanked by BsaI sites were inserted in place of the BsaI-ccdB-BsaI cassette of the XafT+ pSM11 and XafT− pSM15 mini-TLCΦ plasmids by restriction digestion and ligation. The plasmid libraries were transformed into an E. coli xerC− strain (FCV14). Transformation reactions were plated on 20 cm diameter petri dishes and repeated to obtain a total of about 106 colonies. Plasmid libraries were then extracted from ∼1 × 1010 FCV14 cells using a plasmid midi-prep kit (Qiagen), and re-transformed into the E. coli conjugation donor strain β2163. pSM11 and pSM15 derivatives harbouring dif1 in place of TLCΦ attP, pSM17 and pSM18, were constructed by the same technique, respectively. The use of a Type IIS restriction digest and ligation-based process ensured that the mini-TLCΦ plasmids from the XafT− and XafT+ libraries only differed from pTLC8 and pSM12 by the sequence of the XerD-arm of their attachment site.
Integration assays
To monitor integration, engineered V. cholerae strains with arabinose-inducible XerCD expression and integration-reporter dif1-LacZ constructs were used. CMV30 (lacZ-dif1 reporter at the dif1 locus) and EPV369 (lacZ-dif1 reporter at the lacZ locus) recipient cells were cultured to an optical density (OD 600 nm) of 0.3 in Lysogeny-broth (LB) supplemented with 0.2% of l-arabinose (l-ara) to induce the expression of XerD and XerC. The Escherichia coli β2163 conjugation donor cells containing the mini-TLCΦ libraries were grown to an OD600 of 0.6 in LB supplemented with 0.3 mM of diaminopimelic acid (dap). β2163 is a diaminopimelic acid auxotroph (dap−) which allows for simple counterselection after conjugation. E. coli donor and V. cholerae recipient cells were mixed in a 1:10 ratio and incubated for conjugation at 37°C for 3 h on LB-agar plates supplemented with dap and L-ara. The conjugation mixtures were subsequently resuspended in LB supplemented with 0.2% of L-ara and incubated for an additional 1 h of liquid culture. Serial dilutions of the conjugation mix were plated on LB-agar with X-gal to determine the total number of V. cholerae conjugants (blue dap+ clones), and on LB-agar supplemented with Chloramphenicol (Cm) and X-gal to determine the number of specific integration events into dif1 (white dap+ CmR clones). L-ara was omitted from the integration plates to limit the possibility of excision of the mini-TLCΦ plasmid during colony formation. The global frequency of integration of mini-TLCΦ plasmids carrying dif1, TLCΦ attP, and degenerate XerD-arm mini-TLCΦ plasmid libraries was calculated as the ratio between the number of specific integrants to the number of conjugants. Integrant libraries were harvested for NGS by scraping the plates after overnight growth and the recovered cells were subsequently stored at –80°C. Over 10 conjugations were performed for each library condition and were pooled together for sequencing or excision assays.
Excision assays
To analyse the excision of integrated attP library variants, the stored integration libraries were re-inoculated into liquid culture without antibiotic selection, thus the retention of the integrated elements was not actively encouraged. About 109 frozen cells from the integration libraries were precultured for 2 h in 100 ml of LB. One ml of the preculture was then used to seed 200 ml of LB supplemented with 0.2% of L-ara to induce the expression of XerD and XerC.
NGS analysis
The attP sites of the donor plasmid libraries were amplified by performing 17 PCR cycles on 50–100 ng of midiprep plasmid DNA with an equimolar mix of 4103–4105 P5 and 4174–4176/4243–4245 P7 primers (Table S4), which bind on the TLCΦ genome and the plasmid backbone, respectively. The genomic DNA from V. cholerae integration and excision libraries was recovered using a gDNA extraction kit (GenElute Bacterial Genomic DNA kit, Sigma). The attP sites specifically integrated into dif1 were then amplified by performing 17 PCR cycles on 50–100 ng of gDNA with an equimolar mix of 4103–4105 P5 and 4178–4180/4246–4248 P7 primers. The 4178–4180/4246–4248 P7 primers bind in E. coli lacZ, which ensures that we only analysed the sequence of non-replicative mini-TLCΦ plasmids specifically integrated inside the lacZ-dif1 reporter cassette. The products were purified from the P5 and P7 primers by double selection with AMPure beads (Beckman). NGS was performed on an Illumina NextSeq 500 machine. NextSeq reads were trimmed with Cutadapt (version 1.17). MatLab scripts were developed to analyse results, draw 2D-maps, butterfly plots, violin plots, bar plots, and sequence frequency logos, and are available upon request. The relative integration frequency of a mini-TLCΦ plasmid was calculated as the ratio of the frequency of its attP motif in the integration library,  , to the frequency of the attP motif in the donor library,
, to the frequency of the attP motif in the donor library, 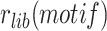 . The integration efficiency,
. The integration efficiency, 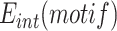 was then obtained by multiplying the relative integration frequencies by the global integration frequency of the corresponding library,
was then obtained by multiplying the relative integration frequencies by the global integration frequency of the corresponding library,  . The retention efficiency of a motif after growth in conditions permitting its excision,
. The retention efficiency of a motif after growth in conditions permitting its excision, 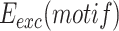 , was calculated as the ratio of the frequency of the motif after growth in the excision conditions,
, was calculated as the ratio of the frequency of the motif after growth in the excision conditions, 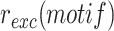 , to its frequency in the integration library,
, to its frequency in the integration library, 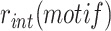 . It was used as an indicator of the stability of the corresponding plasmid.
. It was used as an indicator of the stability of the corresponding plasmid.
In vitro recombination assays
Short dif1 substrates were obtained by annealing oligonucleotides 3418 and 3419 (Table S4). Oligo 3418 possessed 5’-CY5 and 3’-CY3 fluorophores. The 152 bp dif1 and attP substrates were produced by PCR using 2206/3419 and 2470/1110 primer couples on pSM17 and pTLC8, respectively. Recombination reactions were performed in 25 mM Tris–HCl pH7.5, 1 mM EDTA, 10% glycerol, 100 mM NaCl and 100 ng/ml BSA. Reactions were incubated for 2 h at 37°C. To remove the reaction proteins, samples were treated with 0.5% SDS and 0.4 mg/mL proteinase K for 1 h at 37°C. Recombination reactions were separated by electrophoresis on native 5% 29:1 acrylamide gels containing TBE 0.5X buffer for a period of 80 min at 17 mA. Gels were fluorescently imaged with a Typhoon laser scanner (GE healthcare) using filters 670BP30 and 580BP30.
XafT immunoblotting
Expression profiles of XafT from either integrated or replicative forms of pTLC plasmids were performed in V. cholerae EPV369 (lacZ-dif1 reporter at the lacZ locus) by immunoblotting against XafT with C-terminal 3xFLAG epitopes. Clones harbouring integrated or replicative forms of the pTLC variants were selected on LB-agar supplemented with Cm and X-gal, with integrated forms giving rise to white colonies, and non-integrated replicative forms giving rise to blue colonies. Total cellular protein extracts were separated on precast 8–16% gradient acrylamide TGX ‘Stain-Free’ gels (Bio-Rad) in Tris-glycine–SDS electrophoresis buffer. Gels were electroblotted by wet-transfer onto nitrocellulose. The following antibodies were used for immunoprobing: 1:10 000 anti-FLAG M2 (F1804, Sigma), and 1:10 000 anti-mouse IgG-HRP (A9044, Sigma), both diluted in 5% fat-free milk TBS-T (20 mM Tris pH 7.6, 150 mM NaCl, 0.01% Tween-20). Chemiluminescence was performed using SuperSignal™ West Pico PLUS (ThermoFisher) ECL, and captured over a 60-min exposure with a ChemiDoc imager (Bio-Rad). Supplementary Figure S6 contains uncropped source images of the original SDS-PAGE, transfer, and immunoblot. Supplementary method contains an extended explanation of the plasmid construction and sample preparation used for this immunoblot.
Protein purification
MBP-XafT, MBP-XerD, MBP-XerD-FtsKγ and MBP-XerC fusions were produced in E. coli BL21(DE3) cells from plasmids pCM153, pCM162, pSM20 and pCM157, respectively. The AA linker between the C-terminus of MBP and the N-terminus of the proteins of interest contained a 6xHIS tag and a TEV protease cleavage site. Cultures were grown at 37°C under agitation until exponential phase, then induced with 1 mM IPTG. Induction was performed at 20°C overnight under agitation. Induced cultures were pelleted, washed in LB and lysed with a French press in presence of protease inhibitors (Merck, cOmplete EDTA free), 1 M NaCl (or 200 mM for XerD), 1 mM EDTA, 50 mM Tris–HCl pH 7. The following steps were all performed at 4°C. Lysates were centrifuged at 34 000 g, then the supernatants were removed and passed though 0.45 μm and 0.2 μm filters. FPLC purification was performed using an AKTA purifier (GE Healthcare). The MBP fusion proteins were bound on 5 ml MBPTrap columns (GE Healthcare) in a solution of 50 mM Tris pH 7, 0.2 M NaCl, 1 mM EDTA. MBP fusions were eluted within the same buffer supplemented with 10 mM maltose. Purified MBP-XerD, MBP-XerD-FtsKγ and MBP-XerC proteins were digested overnight with TEV protease to cleave their MBP-6xHis tags. Finally, MBP-XafT, XerD, XerD-FtsKγ and XerC were purified and concentrated using 5 ml HiTrap Heparin HP columns (GE Healthcare). The heparin affinity purification step removed cleaved MBP-6xHis moieties from the XerD, XerD-FtsKγ and XerC protein preparations. Protein aliquots were stored at –80°C.
RESULTS
NGS-based comparison of the efficiency of integration and excision of mobile elements
A classical methodology to analyse the mechanisms of integration of a mobile element is to monitor the frequency of integration, 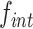 , of wild-type or genetically modified versions of the element in genetic mutants of its host. XafT was discovered by this approach (31). In brief, a non-replicative version of TLCΦ, lacking the coding sequence for the native TLCΦ phage replication protein Cri, was cloned into a plasmid backbone (pSW23T) that has a conditional R6K origin of replication and the cat chloramphenicol (Cm) resistance gene (Figure 2A, (35)). pSW23T derivatives will only replicate in bacterial strains possessing a chromosomally encoded R6K-replication protein pir (π). These conditionally-replicative ‘mini-TLCΦ’ plasmids were delivered to a π− Cm-sensitive N16961 V. cholerae strain by conjugation using a π+E. coli donor strain which requires diaminopimelic acid (dap) for growth. Conjugation mixtures were then plated onto LB-agar lacking dap (Figure 2A, (35)). As the V. cholerae strain lacks π, which is necessary for replication of the mini-TLCΦ plasmids, and as the E. coli donor strain is a dap auxotroph, any Cm-resistant colonies correspond to the integration of a mini-TLCΦ plasmid in the genome of a V. cholerae recipient cell. This permits the assessment of the fraction of Cm-resistant cells to the total number of recipient cells,
, of wild-type or genetically modified versions of the element in genetic mutants of its host. XafT was discovered by this approach (31). In brief, a non-replicative version of TLCΦ, lacking the coding sequence for the native TLCΦ phage replication protein Cri, was cloned into a plasmid backbone (pSW23T) that has a conditional R6K origin of replication and the cat chloramphenicol (Cm) resistance gene (Figure 2A, (35)). pSW23T derivatives will only replicate in bacterial strains possessing a chromosomally encoded R6K-replication protein pir (π). These conditionally-replicative ‘mini-TLCΦ’ plasmids were delivered to a π− Cm-sensitive N16961 V. cholerae strain by conjugation using a π+E. coli donor strain which requires diaminopimelic acid (dap) for growth. Conjugation mixtures were then plated onto LB-agar lacking dap (Figure 2A, (35)). As the V. cholerae strain lacks π, which is necessary for replication of the mini-TLCΦ plasmids, and as the E. coli donor strain is a dap auxotroph, any Cm-resistant colonies correspond to the integration of a mini-TLCΦ plasmid in the genome of a V. cholerae recipient cell. This permits the assessment of the fraction of Cm-resistant cells to the total number of recipient cells, 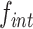 , by spreading serial dilutions on plates lacking dap and supplemented or not with Cm, respectively (Figure 2A).
, by spreading serial dilutions on plates lacking dap and supplemented or not with Cm, respectively (Figure 2A).
Figure 2.
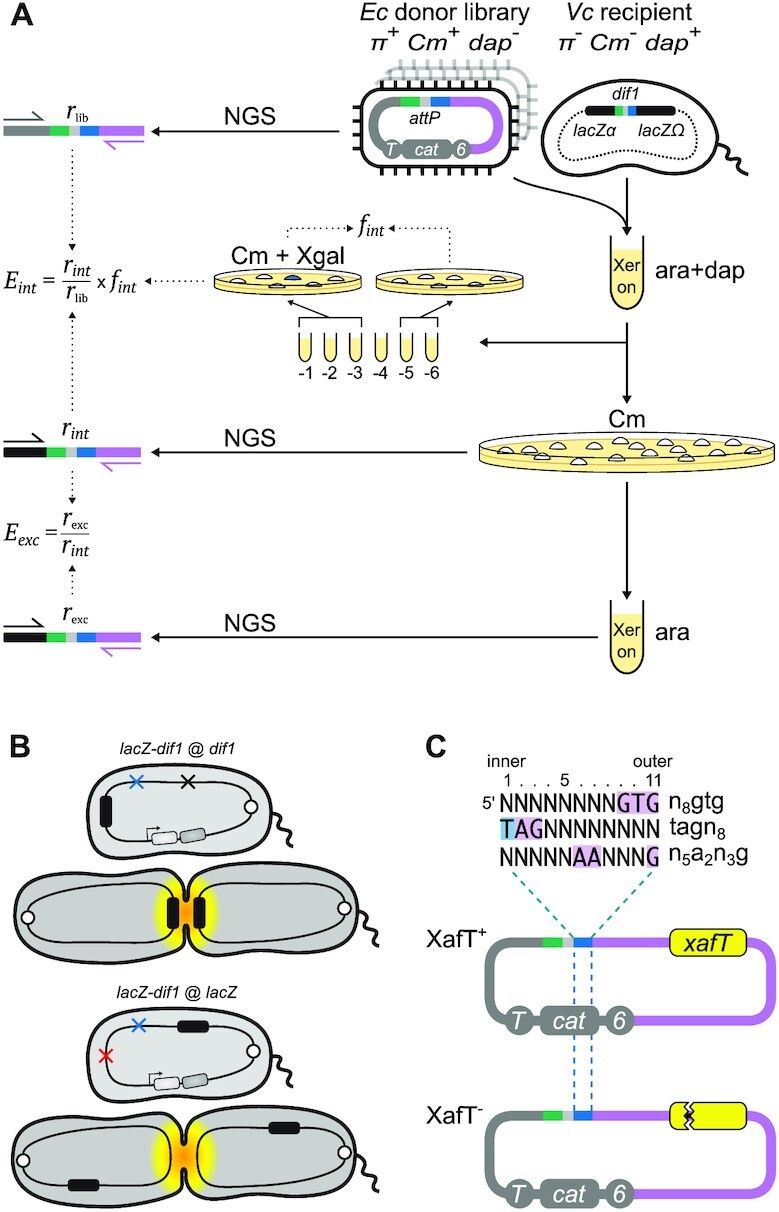
Parallel monitoring of the integration and excision of Mobile DNA elements. (A) Schematic of classical and NGS based strategies to monitor integration and excision efficiencies. ‘Xer on’: growth in the presence of arabinose. Pink: TLCΦ DNA; Grey: plasmid vector DNA; 6: R6Kγ replication origin; cat: chloramphenicol resistance gene; T: transfer origin; dap: diaminopimelic acid; Cm: chloramphenicol; ara: L-arabinose; Xgal: X-gal; lacZ-dif1 reporter: E. coli lacZ gene containing an internal dif1 site. Plasmid- and lacZ-specific P5, and TLCΦ-specific P7 adaptor primers used for next generation sequencing (NGS) are indicated by black, grey and pink arrows, respectively. (B) Scheme of the V. cholerae reporter strains. White circle: chromosome 1 replication origin; red, blue and black crosses: dif and adjoining IMEX deletion, xerD deletion and V. cholerae lacZ deletion, respectively; black rectangle: E. coli lacZ-dif1 reporter; light and dark grey rectangles: synthetic xerC and xerD operon under the control of the arabinose promoter (Para). Yellow shading depicts the subcellular region and timing of activity of FtsK during the cell cycle. (C) Non-replicative mini-TLCΦ plasmid libraries. Top panel: top strand of the XerD-arm of the degenerate attP motifs. Sequence legend as in Figure 1B. Bottom panel: scheme of XafT+ and XafT−mini-TLCΦ plasmids. Yellow rectangle: xafT gene; sawed lines: stop mutation.
In our previous work we have also established a robust colorimetric assay to distinguish specific integration events from random integration events, based on the classical Blue/White LacZ (β-galactosidase) phenotype of bacterial cells on LB-agar supplemented with X-gal. The colorimetric assay is based on the insertion of the dif1 recombination site within the coding sequence of the E. coli lacZ (eclacZ) gene in such a manner as not to perturb the β-galactosidase activity of the reporter gene product. The reporter is hereafter denoted as the ‘lacZ-dif1’ reporter. It has been utilised in several ΔlacZ V. cholerae strain backgrounds (ΔvclacZ). These include placing the lacZ-dif1 reporter at the normal chr1 dif1 locus (lacZ-dif1 at dif1) where recombination of the site will be subjected to regulation by FtsK, or at exogenous loci such as replacing vclacZ in a Δdif1 background (lacZ-dif1 at vclacZ) where the recombination site reporter will be outside the domain of action of FtsK (Figure 2B, (7)). The sensitivity of this technique allows us to monitor 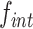 as low as 10−6.
as low as 10−6.
The classical methodology is simple and reliable, but cannot be reasonably applied to compare the efficiency of integration of thousands of different mobile elements. In addition, calculating integration rate by plating 10-fold serial dilutions has an intrinsic ten-fold precision limit. This leads to a 100-fold precision limit when estimating the fraction of integrants in the recipient population because the total number of cells is also measured using 10-fold dilutions. To solve those issues, we designed a high-throughput methodology based on Next Generation Sequencing (NGS), which we applied to libraries of conjugative non-replicative mini-TLCΦ plasmids carrying attP sites with different XerD-arm sequences. The relative frequency of each of the different XerD-arm sequence motifs in the plasmid libraries,  , was determined by NGS using a primer binding in the TLCΦ genome and a primer binding in the pSW23T vector backbone (Figure 2A). The plasmid libraries were then delivered to V. cholerae recipient cells in which the V. cholerae lacZ gene, the wild-type IMEX arrays, and the original dif1 site were deleted, and which were equipped with the lacZ-dif1 reporter. Cm-resistant V. cholerae colonies were collected on large selection plates and pooled. The relative frequency of each of the different XerD-arm sequence motifs in the Cm-resistant cell pools,
, was determined by NGS using a primer binding in the TLCΦ genome and a primer binding in the pSW23T vector backbone (Figure 2A). The plasmid libraries were then delivered to V. cholerae recipient cells in which the V. cholerae lacZ gene, the wild-type IMEX arrays, and the original dif1 site were deleted, and which were equipped with the lacZ-dif1 reporter. Cm-resistant V. cholerae colonies were collected on large selection plates and pooled. The relative frequency of each of the different XerD-arm sequence motifs in the Cm-resistant cell pools, 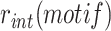 was determined by NGS (Figure 2A). PCR amplification using a primer binding in the TLCΦ genome in combination with a primer binding in lacZ ensured that only dif1-specific integration events were taken into account. The efficiency of integration of a plasmid with a given XerD-arm motif,
was determined by NGS (Figure 2A). PCR amplification using a primer binding in the TLCΦ genome in combination with a primer binding in lacZ ensured that only dif1-specific integration events were taken into account. The efficiency of integration of a plasmid with a given XerD-arm motif, 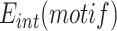 , was estimated as the ratio of
, was estimated as the ratio of  to
to 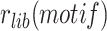 . It was multiplied with the global frequency of integration of the whole library,
. It was multiplied with the global frequency of integration of the whole library,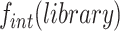 , to take into account the influence of the host genetic context (Figure 2A).
, to take into account the influence of the host genetic context (Figure 2A).
The NGS approach can also be used to monitor the retention efficiency of thousands of different integrated mobile elements, provided that the enzymes catalysing their excision are conditionally expressed. This is particularly interesting in the case of IMEX since they integrate into a locus controlled by the normal domain of action of FtsK, which could also lead to FtsK-driven excision events. To this end, we used V. cholerae reporter cells that were genetically modified to place the production of XerC and XerD under the control of the arabinose inducible promoter (Figure 2A, (28)). We then determined the relative frequency of each of the different XerD-arm sequences in the plasmids that specifically integrated at dif1 after growth in conditions that permitted their excision, 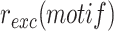 , i.e. in the absence of selection and in the presence of arabinose for XerC and XerD production (Figure 2A). The influence of each XerD-arm motif on the retention efficiency of the integrated mini-TLCΦ plasmids,
, i.e. in the absence of selection and in the presence of arabinose for XerC and XerD production (Figure 2A). The influence of each XerD-arm motif on the retention efficiency of the integrated mini-TLCΦ plasmids,  , was assessed by calculating the ratio of
, was assessed by calculating the ratio of 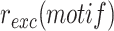 to
to 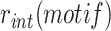 (Figure 2A). As the integration of TLCΦ in dif1 results in the formation of a prophage flanked by attP and dif1,
(Figure 2A). As the integration of TLCΦ in dif1 results in the formation of a prophage flanked by attP and dif1,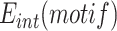 and
and  reflected the efficiency of recombination reactions between the same two sites (Figure 1A).
reflected the efficiency of recombination reactions between the same two sites (Figure 1A).
For the purpose of this study, we generated three pools of 65 536 (48) different TLCΦ attP sites using synthetic oligonucleotides carrying eight degenerate bases at different positions of the XerD-arm as templates (Figure 2C). Two of the pools were designed to explore the influence of the eight innermost and outermost positions of the XerD-arm. They are referred to as n8gtg and tagn8 on the basis of the sequence of their top strand. We added to the study a pool harbouring degenerate bases in both the inner and outer part of the XerD-arm, n5a2n3g. The three pools of sequences were cloned in place of the attP site of XafT+ and XafT− mini-TLCΦ plasmids (Figure 2C). NGS analysis showed that each resulting mini-TLCΦ library contained over 99.5% of the XerD-arm sequence motifs that was expected to be included by the pool from which it originated, with a median copy number in the order of 15 per million reads (Supplementary Figure S2). The XafT+ and XafT− mini-TLCΦ libraries contained over 99.9% and 99.8% of the 190 528 XerD-arm sequence motifs included in the three combined degenerate pools, respectively.
XafT expands the number of XerD-arm motifs that can be used for integration
We first explored the influence of the XerD-arm sequence of TLCΦ attP on the efficiency of integration in an FtsK-driven context. To this end, we conjugated the XafT− mini-TLCΦ libraries in a V. cholerae recipient strain in which the lacZ-dif1 reporter was inserted at the dif locus of chr1 (strain CMV30, Figure 2B, (9,28)). FtsK promoted the integration of the XafT− mini-TLCΦ plasmid harbouring dif1 at a very high frequency (Figure 3A, FtsK panel). However, the integration of the XafT− mini-TLCΦ plasmid harbouring the wild-type TLCΦ attP was below the detection level. The global integration frequency of the XafT− n8gtg, tagn8 and n5a2n3g mini-TLCΦ libraries was 1000-fold lower than that of the XafT− mini-TLCΦ plasmid harbouring dif1 (Figure 3A, FtsK panel). NGS revealed that FtsK/XerC/XerD had promoted the integration of only 2.5% of the total number of XerD-arm motifs present in the combined n8gtg, tagn8 and n5a2n3g libraries (Figure 3A, FtsK panel). We generated the sequence logos of the integrated sites from each library (Supplementary Figure S3) and a sequence logo combining the 4729 XerD-arm sequence of the integrated mini-TLCΦ plasmids of the three n8gtg, tagn8 and n5a2n3g libraries (Figure 3B, FtsK panel). The sequence logos correspond to the top strand of the XerD-arm. The 5’ end of the sequence logos is adjacent to the central region. We refer to it as the innermost side of the XerD-arm (Figure 3C). We refer to the 3’ end of the logos as the outermost side of the XerD-arm (Figure 3C). The logos revealed that the more similar the degenerate attP sequence was to dif1, the higher the integration frequency was in the Ftsk-driven context (Figure 3B and Supplementary Figure S3, FtsK panels). It was particularly evident for the innermost bp of the XerD-arm, but the outermost bp showed the same trend (Figure 3B and Supplementary Figure S3, FtsK panels).
Figure 3.
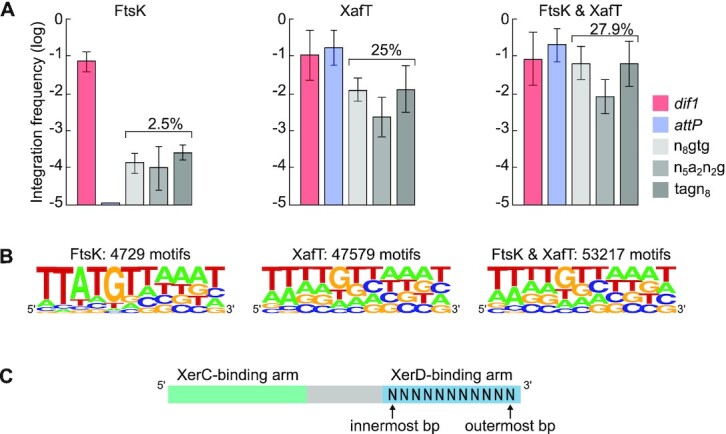
Non-replicative mini-TLCΦ plasmid integration. (A) Integration frequencies of mini-TLCΦ plasmids harbouring dif1 or TLCΦ attP, and global integration frequencies of n8gtg, n5a2n3g and tagn8 plasmid libraries (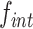 ). FtsK panel: XafT− plasmids conjugated in a strain harbouring dif1 at its natural locus; XafT panel: XafT+ plasmids conjugated in a strain harbouring dif1 at the lacZ locus; FtsK & XafT panel: XafT+ plasmids conjugated in a strain harbouring dif1 at its natural locus. Mean and standard deviations of at least 3 independent assays. (B) Combined sequence of the XerD-arm from the n8gtg, n5a2n3g and tagn8 integration libraries. The logos show the frequency of each base at the degenerate positions. The number of integrated motifs corresponding to each logo is indicated above. Individual logos are shown in Supplementary Figure S3. (C) Illustration of our naming scheme for bases of the XerD-binding arm.
). FtsK panel: XafT− plasmids conjugated in a strain harbouring dif1 at its natural locus; XafT panel: XafT+ plasmids conjugated in a strain harbouring dif1 at the lacZ locus; FtsK & XafT panel: XafT+ plasmids conjugated in a strain harbouring dif1 at its natural locus. Mean and standard deviations of at least 3 independent assays. (B) Combined sequence of the XerD-arm from the n8gtg, n5a2n3g and tagn8 integration libraries. The logos show the frequency of each base at the degenerate positions. The number of integrated motifs corresponding to each logo is indicated above. Individual logos are shown in Supplementary Figure S3. (C) Illustration of our naming scheme for bases of the XerD-binding arm.
We next explored the influence of the XerD-arm sequence of TLCΦ attP on the efficiency of integration in a XafT-driven context. To this end, we conjugated the XafT+ mini-TLCΦ libraries in the V. cholerae recipient strain in which lacZ-dif1 was inserted at the V. cholerae lacZ locus (strain EPV369, Figure 2B). Our previous work had shown that the choreography of chromosome segregation restricts the activity of FtsK to the terminus region of the two V. cholerae chromosomes (Figure 2B, (28,36–38)). Correspondingly the integration of a XafT− plasmid harbouring dif1 was found to be 100-fold less efficient when the chromosomal dif1 site was located at the lacZ locus compared to when it was located at its natural locus (9). Thus, the frequency of integration of a XafT+ mini-TLCΦ plasmid in a V. cholerae recipient strain harbouring lacZ-dif1 at the lacZ locus can be mainly attributed to the activation of Xer recombination by XafT. The integration of the XafT+dif1 and XafT+ TLCΦ attP plasmids at the lacZ locus was as efficient as the integration of the XafT−dif1 plasmid at the dif1 locus (Figure 3A, XafT panel). In addition, the global integration frequency of the XafT+ n8gtg, tagn8 and n5a2n3g mini-TLCΦ plasmid libraries was now only 10-fold lower than that of the XafT+dif1 plasmid (Figure 3A, XafT panel). NGS showed that 47579 different mini-TLCΦ plasmids had integrated, which corresponds to 25% of the total number of different plasmids present in the combined n8gtg, tagn8 and n5a2n3g mini-TLCΦ libraries (Figure 3B, XafT panel). The sequence logos of the first six positions of the sites integrated via XafT-driven recombination were statistically different from those of the sites integrated via FtsK-driven recombination (Supplementary Figure S3, XafT panels). The nature of the bases of the XerD-arm was less important for XafT-driven than for FtsK-driven integration (Figure 3B and Supplementary Figure S3, XafT panels).
Next, we explored the integration of the XafT+ mini-TLCΦ libraries in the V. cholerae recipient strain equipped with lacZ-dif1 at the natural dif1 locus, i.e. in a context in which both XafT and FtsK can act (strain CMV30, Figure 2B). The results were remarkably similar to those obtained for the integration of the XafT+ mini-TLCΦ plasmids in the strain carrying lacZ-dif1 at the lacZ locus (Figure 3 and Supplementary Figure S3). The only notable difference was a slightly higher global integration frequency (Figure 3A, FtsK & XafT panel) and greater number of different integrated XafT+ mini-TLCΦ plasmids (Figure 3B, FtsK & XafT panel). This could be explained by the growth advantage of the reporter strain carrying lacZ-dif1 at its natural locus over the reporter strain carrying it at the lacZ locus, since the latter is Δdif1 and therefore cannot resolve chr1 dimers (20). Neverthless, the XerD-arm sequence logos of the XafT+ mini-TLCΦ plasmids integrated at the dif1 locus were similar to those of the XafT+ mini-TLCΦ plasmids integrated at the lacZ locus (Figure 3B and Supplementary Figure S3).
Overall, FtsK-driven integration seemed restricted to the attP sites that most resembled dif1, with the inner positions of the XerD-arm playing a more important role than the outer positions, whereas XafT seemed to allow the integration of plasmids harbouring attP sites that significantly deviated from dif1 at any given position of the XerD-arm.
The first and fifth innermost bp of the XerD-arm are crucial for integration
Sequence logos allow a quick interpretation of pools of motifs, but the information that can be derived from them is limited. In particular, it is not possible to determine whether a logo results from the combination of motifs deviating from a single specific consensus sequence, or from the overlap of two or more distinct consensus sequences. For instance, the highly degenerate sequence logos of the XafT and FtsK & XafT integration libraries could be due to the addition of motifs slightly deviating from dif1 along with motifs slightly deviating from TLCΦ attP (Figure 3B). In addition, the relative influence of each particular bp on the efficiency of a process cannot be assessed with a sequence logo. The higher frequency of integration obtained with XafT+ mini-TLCΦ plasmids compared to XafT− mini-TLCΦ plasmids could not be simply explained by the higher number of different XerD-arm sequences that could be integrated (Figure 3B).
We developed a new two-dimensional map (2D-map) graphical representation to avoid such limitations. In these maps, each recovered XerD-arm motif is represented by a small coloured square at specific x and y coordinates based on its sequence (Figure 4A). Each square is coloured on the basis of the integration efficiency of the plasmids harbouring the corresponding motif. Colours range from black to bright yellow, with black, dark blue and bright yellow corresponding to non-recovered motifs, motifs with the poorer integration efficiency, and motifs with higher integration efficiency, respectively.
Figure 4.
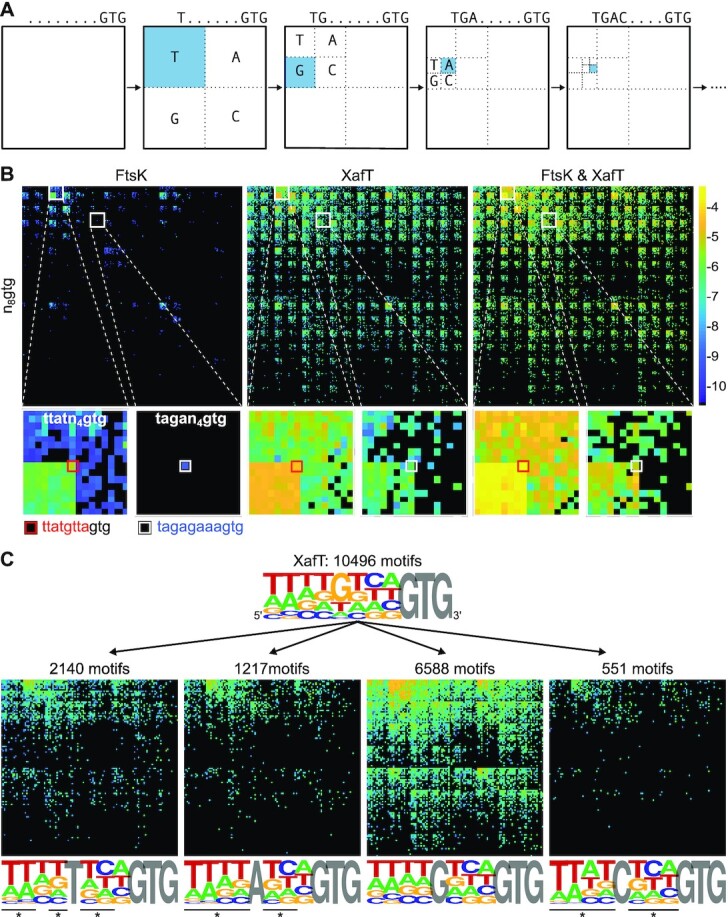
2D-maps representation of the integration results. (A) 2D-map representation methodology: Different [x, y] coordinates are assigned to each nucleotide for the 28 possible motifs as follows: the [x, y] coordinates are initially set to [1, 1]. Then, [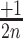 ,
, 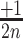 ], [
], [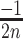 ,
, 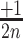 ], [
], [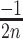 ,
,  ] or [
] or [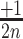 ,
, 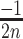 ] are added to [x, y] for each n position of the degenerate motif if the nth base is A, T, G or C, respectively. Relative integration efficiencies are represented with a dark-blue to bright yellow colour scale. Positions that were not found in the integration libraries are shown in black. (B) 2D-maps of the integrated n8gtg motifs. FtsK: results from the conjugation of the n8gtg XafT− mini-TLCΦ library in the strain harbouring lacZ-dif1 at the natural dif1 locus; XafT: results from the conjugation of the n8gtg XafT+ mini-TLCΦ library in the strain harbouring lacZ-dif1 at lacZ locus; FtsK & XafT: results from the conjugation of the n8gtg XafT+ mini-TLCΦ library in the strain harbouring lacZ-dif1 at the natural dif1 locus. Below each 2D-maps are shown zooms of the regions corresponding to the 24 motifs that carry the same first innermost bases as the XerD-arm of TLCΦ attP (tagan4gtg) and dif1 (ttatn4gtg). The position of the TLCΦ attP XerD motif and of the motif most similar to the XerD-arm of dif1 are highlighted by white and red squares, respectively. Their sequences are indicated below the panels. Bases identical to the XerD-arms of TLCΦ attP and dif1 are written in blue and red, respectively. 2D-maps of the integrated n5a2n3g and tagn8 motifs are shown in Supplementary Figure S4. (C) Sequence logo of the XerD-arm of the attP sites of the n8gtg XafT+ mini-TLCΦ plasmids that were found integrated in lacZ-dif1 at lacZ locus. The total number of n8gtg integrated motifs is indicated. 2D-maps of the integrated n4tn3gtg, n4an3gtg, n4gn3gtg and n4cn3gtg motifs, as indicated. The number of integrated motifs from each category is indicated. The sequence logo of the XerD-arm of the integrated sites from each category is shown below each 2D-map. Positions in which the bases of the integrated n4tn3gtg, n4an3gtg, and n4cn3gtg XerD-arm motifs are significantly different from the integrated n4gn3gtg XerD-arm motif are indicated by a star (chi2-test, P-value < 0.001).
] are added to [x, y] for each n position of the degenerate motif if the nth base is A, T, G or C, respectively. Relative integration efficiencies are represented with a dark-blue to bright yellow colour scale. Positions that were not found in the integration libraries are shown in black. (B) 2D-maps of the integrated n8gtg motifs. FtsK: results from the conjugation of the n8gtg XafT− mini-TLCΦ library in the strain harbouring lacZ-dif1 at the natural dif1 locus; XafT: results from the conjugation of the n8gtg XafT+ mini-TLCΦ library in the strain harbouring lacZ-dif1 at lacZ locus; FtsK & XafT: results from the conjugation of the n8gtg XafT+ mini-TLCΦ library in the strain harbouring lacZ-dif1 at the natural dif1 locus. Below each 2D-maps are shown zooms of the regions corresponding to the 24 motifs that carry the same first innermost bases as the XerD-arm of TLCΦ attP (tagan4gtg) and dif1 (ttatn4gtg). The position of the TLCΦ attP XerD motif and of the motif most similar to the XerD-arm of dif1 are highlighted by white and red squares, respectively. Their sequences are indicated below the panels. Bases identical to the XerD-arms of TLCΦ attP and dif1 are written in blue and red, respectively. 2D-maps of the integrated n5a2n3g and tagn8 motifs are shown in Supplementary Figure S4. (C) Sequence logo of the XerD-arm of the attP sites of the n8gtg XafT+ mini-TLCΦ plasmids that were found integrated in lacZ-dif1 at lacZ locus. The total number of n8gtg integrated motifs is indicated. 2D-maps of the integrated n4tn3gtg, n4an3gtg, n4gn3gtg and n4cn3gtg motifs, as indicated. The number of integrated motifs from each category is indicated. The sequence logo of the XerD-arm of the integrated sites from each category is shown below each 2D-map. Positions in which the bases of the integrated n4tn3gtg, n4an3gtg, and n4cn3gtg XerD-arm motifs are significantly different from the integrated n4gn3gtg XerD-arm motif are indicated by a star (chi2-test, P-value < 0.001).
The low density of coloured positions highlighted the limited number of XerD-arm sequences that could be integrated by FtsK (Figure 4B, Supplementary Figures S4A and S4B, FtsK 2D-maps). A single small green tile was visible on the n8gtg 2D-map, which corresponded to the 64 TTATGNNNNGTG top strand sequences. It demonstrated the importance of the first 5 inner positions of the XerD-arm for FtsK-driven integration (Figure 4A, zoom on the ttatn4gtg region). Results obtained with the n5a2n3g and tagn8 libraries confirmed this observation (Supplementary Figure S4A and S4B, FtsK panels). The n8gtg, n5a2n3g and tagn8 2D-maps also revealed that FtsK could promote the integration of a plasmid harbouring TLCΦ attP, albeit at a low integration frequency, which demonstrates the utility of combining the NGS approach with a 2D-map graphical representation (Figure 4B, Supplementary Figures S4A and B, zooms).
The 2D-maps of the n8gtg motifs recovered in the XafT and FtsK & XafT integration libraries presented a very striking checkered pattern (Figure 4B, Supplementary Figures S4A and B, XafT and FtsK & XafT panels). The black stripes (indicating sequences that were not recovered) of the n8gtg and n5a2n3g 2D-maps correspond to DNA motifs that lack a G at the fifth position of the top strand of the XerD-arm (Figure 4B and Supplementary Figure S4A, XafT and FtsK & XafT 2D-maps). The pattern of the tagn8 2D map fitted with the importance of the base at this position (Supplementary Figure S4B, XafT and FtsK & XafT panels). To confirm the importance of the fifth bp of the XerD-arm for XafT-driven integration, we grouped the n8gtg motifs of the XafT+ n8gtg integration library on the basis of their fifth base (Figure 4C). The 2D-maps showed that most of the different possible n4gn3gtg motifs were recovered, while the recovered n4tn3gtg, n8an3gtg and n4cn3gtg motifs were mainly limited to those that carried a T at the first and second innermost positions of the XerD-arm. It further suggested that C was the most detrimental nucleic acid at the fifth position, which needed to be compensated for by the presence of the same bases as dif1 at the four innermost positions. Correspondingly, base frequencies at each of the seven positions of the n4gn3gtg and n4cn3gtg motif lists were statistically different (Figure 4C). The presence of A or T at the fifth position was less detrimental. Yet, the n4an3gt and n4tn3gt motifs statistically differed from the n4gn3g motifs at all positions except the third and the eighth innermost positions, respectively (Figure 4C).
The 2D-maps of the XafT and FtsK & XafT integration libraries also showed the influence of the first innermost bp of the XerD-arm: yellow-coloured motifs were for the most part restricted to the top left quarter of the n8gtg and n5a2n3g 2D-maps, which indicates the presence of a T at the first position of the top strand of the XerD-arm (Figure 4B and Supplementary Figure S4A, XafT and FtsK & XafT panels). In addition, half of the plasmids harbouring an attP site with a tagn8 XerD-arm motif were recovered in the XafT and FtsK & XafT libraries, further highlighting the importance of a T at the first innermost bp of the XerD-arm for efficient integration (Supplementary Figure S4B).
Taken together, those results suggest that the highly degenerate XerD-arm sequence logo of the XafT and FtsK & XafT integration libraries derives from the superposition of two categories of motifs, those harbouring a G at the fifth position and those harbouring a T at the first position (Figure 3B).
XafT permits the highly efficient integration of sites deviating from dif1
XafT allows the integration of plasmids harbouring attP sites deviating from dif1 (Figure 4 and Supplementary Figure S4). Yet, the efficiency of integration of the sites that most resembled dif1 was higher than the efficiency of integration of the sites that most resembled TLCΦ attP (Figure 4B, Supplementary Figures S4A and 4B zooms). To further highlight differences of integration efficiency, we developed a second graphical representation of the data obtained by the NGS approach, which we refer to as butterfly plots. In this representation, recovered sites are separated in different bins as a function of the number of bases that differentiate them from TLCΦ attP (Figure 5A). For the n8gtg, n5a2n3g and tagn8 degenerate sites we studied, it corresponds to eight bins. Each bin was further divided into two (+) and (−) bins, corresponding to the sites that were more similar to dif1 than TLCΦ attP or not, respectively (Figure 5A). We then plotted the integration efficiency of each motif from the different bins.
Figure 5.
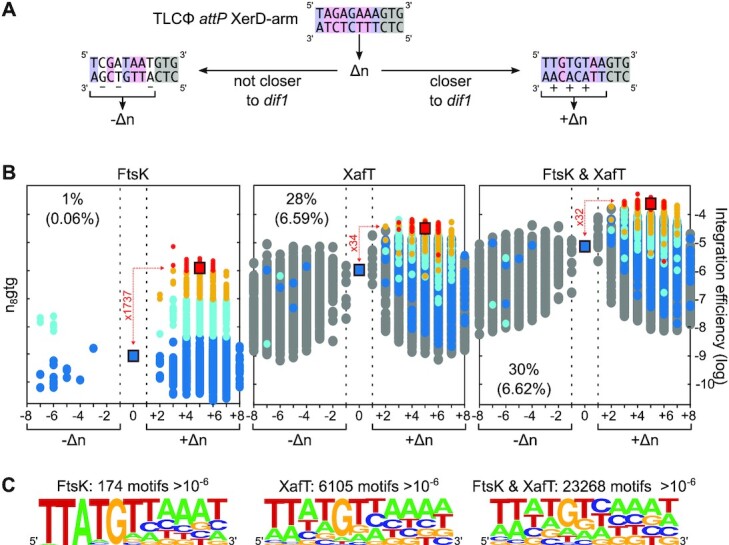
Butterfly plots showing the relative integration efficiency of attP sites harbouring the different possible n8gtg XerD-arm motifs. (A) Butterfly plot representation methodology. Δn: number of bases by which an XerD-arm motif deviates from TLCΦ attP. If the motif is more similar to dif1 than TLCΦ attP, its integration frequency is plotted at the +Δn position of the X-axis. If a motif is not closer to dif1 than TLCΦ attP, its integration frequency is plotted at the –Δn position of the X-axis. (B) Butterfly plots of the integrated n8gtg motifs. The proportion of integrated motifs falling in the (−) group is indicated. Their contribution to the global frequency of integration of the library is indicated between brackets. We coloured the various attP sites leading to significant integration driven by FtsK (FtsK panel) with respect to their integration frequency: from higher than 10−6, 10−7, 5 × 10−8 or lower than 5 × 10−8 in red, orange, cyan and blue, respectively. The corresponding sites in the XafT and FtsK & XafT panels are shown with the same colour code. Sites which were absent in the FtsK panel (motifs which were not capable of being integrated by FtsK) are shown in grey. The difference between the mean frequency of integration of red motifs and the integration frequency of TLCΦ attP (located in the middle of the graph) is indicated in red. The positions of TLCΦ attP and of the attP sites most similar to dif1 are indicated by squares. (C) Combined sequence logos of the n8gtg, n5a2n3g and tagn8 motifs with an integration frequency higher than 10−6. The logos show the frequency of each base at the degenerate positions. The number of motifs from which the logo was derived is indicated. Individual logos and butterfly plots of the integrated n5a2n3g and tagn8 motifs are shown in Supplementary Figure S5.
Only a third (21 070/ 65 536) of the possible n8gtg motifs are closer to dif1 than to TLCΦ attP. Nevertheless, over 99% of the plasmids whose integration could be driven by FtsK harboured an n8gtg XerD-arm that was closer to dif1 than TLCΦ attP (Figure 5B, FtsK panel). In addition, those sites accounted for over 99.4% of the integration events (Figure 5B, FtsK panel). The same observations could be made on the n5a2n3g and tagn8 butterfly plots (Supplementary Figure S5, FtsK panels). We attributed red, orange, cyan and blue colours to the XerD-arm motifs of the plasmids that integrated at dif1 at frequencies higher than 10−6, 10−7, 5 × 10−8 or lower than 5 × 10-8, respectively. The red sites comprised 53 n8gtg, 61 n5a2n3g and 61 tagn8 sequences (Supplementary Figure S5, FtsK panels). Together, those sites represented a total of 174 different sequences (Figure 5C, FtsK panel). The corresponding sequence logo was similar to dif1, highlighting that FtsK-driven integration was restricted to the attP sites that most resemble dif1 (Figure 5C, FtsK panel).
The butterfly representation further highlighted that the FtsK-driven integration frequency of a plasmid harbouring TLCΦ attP was over a 1000-fold lower than the mean integration frequency of the n8gtg and n5a2n3g red attP sites, and over a 100-fold lower than the mean integration frequency of the red tagn8attP sites (Figure 5 and Supplementary Figure S5, FtsK panels). In contrast, the efficiency of integration of TLCΦ attP jumped from about 10−9 to about 10−6 when it was driven by XafT (Figure 5 and Supplementary Figure S5, XafT and FtsK & XafT panels). Differences between the integration efficiency of the different attP sites were alleviated by the presence of XafT. In particular, the mean frequency of the red n8gtg attP sites was now only 30-fold higher than that of TLCΦ attP (Figure 5B XafT and FtsK & XafT panels). Furthermore, the proportion of sites with an integration efficiency higher than 10−6 dramatically increased. High integration efficiencies were not anymore limited to the (+) wing of the butterfly plots: XafT drove the integration of about 30% of the n8gtg mini-TLCΦ plasmids harbouring a site from the (−) bins (Figure 5B, XafT and FtsK & XafT panels). Results obtained with the n5a2n3g and tagn8 plasmid libraries confirmed those observations (Supplementary Figure S5, XafT and FtsK & XafT panels).
As a result, XafT promoted the integration of over 6000 different attP sites with a frequency higher than 10−6 in the recipient strain harbouring lacZ-dif1 at the lacZ locus and over 20000 different attP sites in the recipient strain harbouring lacZ-dif1 at the dif1 locus (Figure 5C, XafT and FtsK & XafT panels). The logo of those sites emphasized that efficient integration was no longer limited to sites that carried the same sequences as the XerD-arm of dif1 (Figure 5C and Supplementary Figure S5, XafT and FtsK & XafT panels).
Mini-TLCΦ plasmids with attP sites deviating from dif1 escape FtsK-driven excision
An unusually long IMEX, the gonococcal genomic island (GGI), is integrated at the dif site of the chromosome of most pathogenic Neisseria species (39,40). It is flanked by a dif-like site of varying sequence between the species (Supplementary Figure S1B). The FtsK DNA translocase was shown to strip XerC and XerD bound to one of those sites, GGI1 attP, thereby preventing excision events (41). This is in contrast to the activity of FtsK at canonical dif sites, where the translocase stalls upon interaction with dif-bound XerD, prior to the activation of recombination. As GGI1 attP only deviates from the canonical dif sequence by 4 bp within the 6 outermost bp of the XerD-arm, this suggests that sequence alterations in XerD-arm of IMEX attP sites could have evolved to prevent FtsK-driven excision events. To explore this hypothesis in the context of TLCΦ attP, we monitored the excision of XafT+ n8gtg, tagn8 and n5a2n3g mini-TLCΦ plasmids that had integrated at the dif1 locus. Inducing the expression of XerC and XerD did not reduce the total diversity of different attP sites in the cell population after 19 h of growth in the absence of selective pressure (Figure 6A and Supplementary Figure S6, Xer+ butterfly plots). However, the proportion of the attP sites from the red and orange categories decreased (Figure 6A and Supplementary Figure S6, Xer+ bar plots). No excision was observed when the expression of XerC and XerD was not induced, demonstrating that excision depended on Xer recombination (Figure 6A and Supplementary Figure S6, Xer− butterfly and bar plots). The efficiency of excision of the attP sites from the red and orange categories, i.e. those that most resembled dif1, was higher than the excision efficiency of TLCΦ attP (Figure 6A and Supplementary Figure S6, Xer+ butterfly plots). However, the efficiency of excision of all the sites, including those from the red and orange categories, was far lower than the 20% per generation excision frequency of a DNA cassette flanked by two dif1 sites (28).
Figure 6.

Retention efficiency of the different possible n8gtg XafT+ mini-TLCΦ libraries. (A) Retention efficiency from the natural dif1 locus after growth in LB in conditions of production of XerCD (Xer+) or not (Xer−). Bar plots show the proportion of n8gtg motifs belonging to the red, orange, cyan, blue and grey categories within the integrated motifs. Retention efficiencies of the n5a2n3g and tagn8 XafT+ mini-TLCΦ libraries are shown in Supplementary Figure S6. (B) Excision from the lacZ locus after growth in L-broth in conditions of production of XerCD (Xer+) or not (Xer−). (C) Anti-FLAG immunoblot of XafT-3xFLAG expression in integrated vs replicative pTLC conditions. The predicted molecular weight of XafT-3xFLAG C-terminal fusion protein (220 aa) is 25.5 kDa (EMBOSS Pepstats). Immunoblot contains normalized whole-cell samples of V. cholerae strain EPV369 over several plasmid conditions. Negative control is EPV369. Positive control is EPV369 containing an (induced) arabinose-inducible XafT-3xFLAG expression plasmid. Subsequent lanes contained cell extracts from either integrated or replicative versions of pTLC synthetic phage plasmids. White and blue illustrated colonies indicate samples derived from integrated (into the lacZ-dif1 reporter cassette) or replicative V. cholerae clones, respectively. Cri− and ΔattP denote pTLC variants which are unable to replicate or unable to integrate, respectively. Supplementary Figure S6C contains uncropped source images of the original SDS-PAGE, transfer, and immunoblot.
To determine the relative contribution of FtsK and XafT to the observed excision events, we analysed the excision of XafT+ n8gtg plasmids integrated at the lacZ locus, where the action of FtsK is very limited. Even after 42 h of growth, there was little change in the relative frequency of the different attP sites (Figure 6B, Xer+ butterfly plots) and in the relative proportion of the sites from the five colour code categories (Figure 6B, Xer+ bar plots). These results suggested that under laboratory growth conditions, FtsK drove most if not all of the excision events of mini-TLCΦ plasmids integrated at the dif locus of chr1.
As dif1 was used as the bacterial attachment site in the integration assays, excision was due to the reverse recombination reaction between the same initial sites (Figure 1). Therefore, we expected XafT to promote the excision of the plasmids for which it permitted integration. Two hypotheses could explain the inefficiency of XafT-driven excision: (i) FtsK could dismantle the formation of intramolecular synapses between dif1 and non-canonical Xer recombination sites as proposed for the GGI1 (41); (ii) XafT expression from an integrated copy of TLCΦ could be insufficient for its activity. The inefficiency of XafT-driven excision events at the lacZ locus ruled out the first hypothesis since this locus is outside of the normal FtsK activity zone. To test the second hypothesis, we directly compared the expression of XafT in a strain carrying an integrated copy of TLCΦ and a strain carrying a replicative non-integrated copy of TLCΦ. We utilised ‘pTLC’ plasmids, which contain the genome of TLCΦ harboured on a conjugatable plasmid with a conditional R6K origin of replication and a chloramphenicol resistance marker. To observe expression by immunoblotting, we inserted a triple FLAG epitope (3xDYKDDDDK) at the C-terminus of XafT (XafT-3FLAG), generating pTLC-3FLAG (Figure 6C). As controls, we constructed a non-integrative version of pTLC-3FLAG by precisely deleting its attP site (pTLC-3FLAG, ΔattP), and a non-replicative version of pTLC-3FLAG by mutating its HUH endonuclease (pTLC-3FLAG, cri−). As the R6K origin of replication of the plasmid vector is non-functional in V. cholerae, replicative forms of pTLC are only driven by the rolling-circle replication activity of Cri. The three constructs were conjugated in the V. cholerae recipient strain harbouring the lacZ-dif1 reporter at the lacZ locus. Conjugation of the non-integrative pTLC-3FLAG led to the formation of blue Cm-resistant colonies, indicating its efficient replication. Conjugation of the non-replicative pTLC-3FLAG led to the formation of white Cm-resistant colonies, indicating that it integrated at lacZ-dif1. Conjugation of the replicative and integrative XafT-3FLAG pTLC led to the formation of both white and blue Cm-resistant colonies. We could detect XafT-3FLAG in protein extracts from the blue Cm-resistant cells harbouring either the TLCΦ plasmid or its integration-defective version. In contrast, we could not detect XafT-3FLAG in protein extracts from white Cm-resistant cells harbouring an integrated copy of TLCΦ, whether it could replicate or not, which was in agreement with the idea that XafT was not sufficiently expressed to drive Xer recombination after integration (Figure 6C).
TLCΦ attP/dif1 synaptic complexes are rare and/or transient
FtsK only efficiently activates recombination between dif sites when it is bound on the XerD side of one or the other of the two DNA duplexes engaged in the recombination complex (42,43). It suggested the possibility that FtsK translocation could dismantle weakly assembled TLCΦ attP/dif1 synaptic complexes before the γ-domain of FtsK could activate XerD, thereby preventing FtsK-driven integration events (Figure 7A, (41)). C-terminal fusions of FtsKγ to XerD have been previously shown to maximize the efficiency of reactions mediated by the E. coli and N. gonorrhoea Xer recombinases. Therefore, if the XerD-FtsKγ interaction was the limiting factor for Xer recombination of dif1 vs TLCΦ attP, we expected to be able to promote the recombination of dif1 with TLCΦ attPin vitro using a V. cholerae XerD-FtsKγ fusion protein (22,41). As a point of comparison, we reconstituted XafT-mediated reactions using XerC and XerD proteins and an N-terminal fusion of XafT to the maltose binding protein, MBP-XafT (31). We used a short 34-bp synthetic double-stranded DNA (dsDNA) dif1 fragment and longer dsDNA fragments containing either dif1 or TLCΦ attP as substrates. To facilitate the differentiation of the HJ intermediate and the crossover products from the substrates, the 5’ and 3’ ends of the bottom strand of the short dif1 substrate were labelled with cy5 and cy3 fluorescent dyes, respectively (Figure 7B). Incubation of the two dif1 substrates with XerC and XerD-FtsKγ or with XerC, XerD and MBP-XafT yielded similar amounts of HJs and crossover products, suggesting that FtsKγ promoted recombination as efficiently as XafT when it was fused to XerD (Figure 7C, left panel). Yet, incubation of the short labelled dif1 substrate and the TLCΦ attP substrate with XerC and XerD-FtsKγ produced barely detectable amounts of HJs and crossover products, suggesting that FtsKγ does not greatly stimulate the reaction between attP and dif, and that this poor activation is not a direct result of the FtsK translocase activity (Figure 5C, right panel). In contrast, XerC, XerD and MBP-XafT mediated the formation of similarly high amounts of HJs and crossover products between the short labelled dif1 substrate and the long dif1 or TLCΦ attP substrates (Figure 7B). We conclude that TLCΦ attP/dif1 synaptic complexes are normally too rare and/or too transient to allow for FtsKγ-driven activation of XerD.
Figure 7.
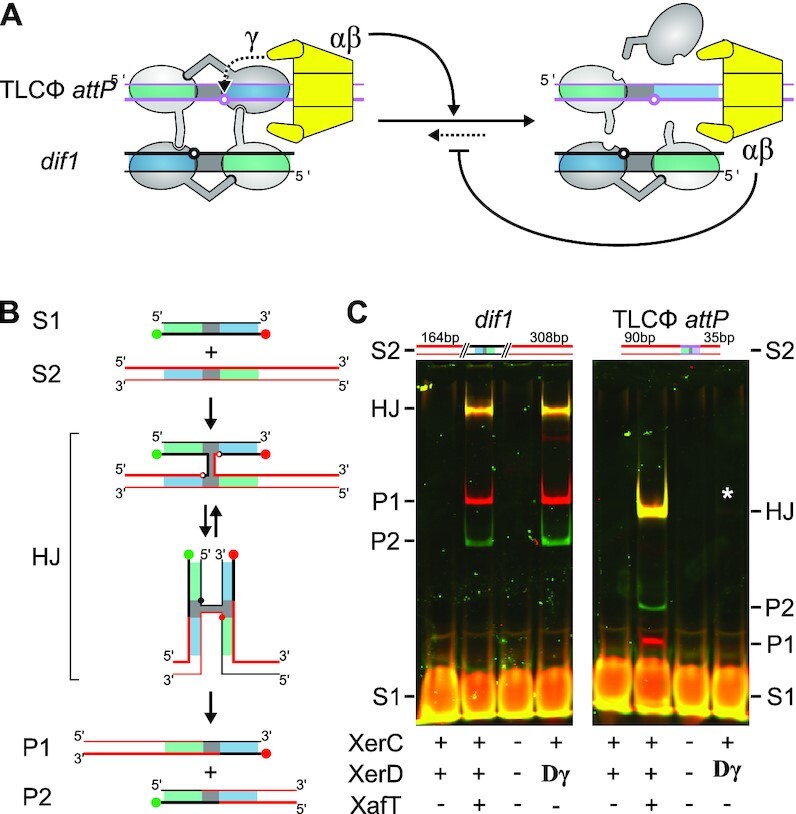
Relative efficiency of XafT- and FtsKγ-driven Xer recombination reactions. (A) Synapse formation between dif1 and TLCΦ attP. Scheme depicting that the non-canonical XerD-arm of TLCΦ attP should limit synapse formation and that FtsK translocation could dismantle and/or inhibit the formation of synapses, thereby preventing recombination. (B) Scheme of the in vitro recombination substrates, the HJ recombination intermediate and the crossover products. Green ball: 3’ Cy3 label; Red ball: 5’ Cy5 label; Grey, Green and Blue rectangles: Central region, XerC-binding and XerD-binding arm, respectively. The DNA strands exchanges by XerC and XerD are depicted as thin and fat lines, respectively. (C) XafT- and FtsKγ-promoted recombination reactions. Left panel: recombination of a short labelled dif1 substrate (S1) and a long non-labelled dif1 substrate (S2); Right panel: recombination of a short labelled dif1 substrate (S1) and a long non-labelled TLCΦ attP substrate (S2). The S2 substrates are depicted above each gel image. +: V. cholerae XerC, V. cholerae XerD or MBP-XafT, as indicated; Dγ: V. cholerae XerD-FtsKγ fusion; −: mock buffer of the corresponding purified proteins. A white star indicates the presence of a faint HJ band.
DISCUSSION
An NGS-based methodology to analyse the sequence requirements of a DNA recombination reaction
Studies on the sequence specificity of DNA transactions are generally limited to the consensus sequence targeted by DNA binding proteins. When there are multiple copies of a binding motif in the genome, it can be achieved with Chromatin Immuno-Precipitation (ChIP) or Whole-Genome Affinity Determination of protein-DNA interactions (WGADseq) methodologies (44). When the binding motif has low sequence variability, additional investigations may include electromobility gel assays, DNA footprinting, and/or systematic evolution of ligands by exponential enrichment (SELEX). The resulting consensus or sequence logo can be refined by replacing each base of the target motif one at a time and monitoring how it affects the binding of the protein. However, sequence logos can mask the ability of proteins to bind to several distinct consensus sequences. In addition, they do not permit the determination of the relative influence of each particular position of a sequence on the base requirements at the other positions. To bypass those limitations, we designed a methodology based on NGS to monitor the integration and excision efficiency of thousands of different mobile elements in vivo (Figure 2). We also developed two new graphical representations that facilitate the analysis of the results, 2D-maps (Figure 4) and butterfly plots (Figure 5). We applied the methodology to explore the influence of the XerD-arm of TLCΦ attP on the efficiency of Xer recombination reactions (Figures 1 and 2). The extreme sensitivity of the methodology permitted the evaluation of the integration efficiency of events that were too rare to be detected by the classical methodology, such as the integration of TLCΦ attP by a FtsK-driven Xer recombination reaction (Figures 3–5). 2D-map and butterfly plot graphical representations highlighted that efficient FtsK-driven Xer recombination reactions were limited to the attP sites whose XerD-arm only very slightly deviated from the consensus XerD-arm of dif sites (Figures 3–6). In contrast, they revealed that XafT promoted recombination reactions between dif1 and almost all of the different possible attP sites provided that the first or fifth innermost bases of their XerD-arm was identical to the corresponding dif bases (Figures 3–5). Our methodology and graphical representations could be easily adapted to explore the sequence specificity of any DNA binding protein using the relative retention of DNA motifs by the protein in vitro, or using the relative activity of an artificial promoter carrying the DNA motifs in vivo.
XafT allows Xer recombination between dif and attP sites with non-canonical XerD-arms
Xer recombination takes place in a nucleoprotein complex consisting of two recombining sites and a pair of each of the two XerC and XerD recombinases (Figure 1). The complex is held together by the affinity of the recombinases to their DNA binding sites and cyclic interactions between their C-terminal domains. However, binding of XerC to dif is relatively weaker than binding of XerD (2,20,45). Those observations suggested that contacts between XerD and dif played a primary role in the assembly and stability of the recombination synapses (2,20,45). In this regard, the efficiency with which TLCΦ was integrated into the primary chromosome of clinical and environmental clones of V. cholerae was surprising since the XerD-arm of TLCΦ attP was a poor substrate for XerD-binding whether its XerC protein partner was present or not in vitro (9,13). TLCΦ relies on its own XerD-activation factor for integration, XafT (9,31). No other V. cholerae or TLCΦ protein or sequence factors (in combination with XerCD) are required to promote a complete Xer recombination reaction between TLCΦ attP and dif1 in vitro (31).
Our analysis of the XerD-arm of TLCΦ attP on the integration efficiency and stability of mini-TLCΦ plasmids showed that FtsK only promotes the integration into dif1 of a very small subset of the different 190 528 TLCΦ attP derived sites we studied, restricted mostly to those that were the most similar to dif1 (Figures 3–5). In contrast, XafT promoted the recombination of dif1 with almost any of the 190 528 attP sites that carried the same residues as dif1 at the first or fifth position of the XerD-arm (Figures 3–5). In addition, it alleviated differences in the integration frequency of attP sites harbouring more or less canonical XerD-arms (Figure 5). The 2D map and butterfly plot representations of the integration of XafT+ mini-TLCΦ plasmid libraries in the V. cholerae recipient strain harbouring lacZ-dif1 at the native dif1 locus were similar to those obtained for the integration of XafT+ mini-TLCΦ plasmid libraries in the V. cholerae recipient strain harbouring lacZ-dif1 at the lacZ locus indicating that FtsK did not prevent XafT-driven integration of XafT+ mini-TLCΦ plasmids (Figures 4 and 5). The slightly higher integration efficiencies observed for the reporter strain carrying lacZ-dif1 at the native dif1 locus is probaly explained by the growth advantage of this strain over the reporter strain carrying dif1 at the lacZ locus (Figure 3). Finally, in vitro comparison of the efficiency with which XafT and FtsKγ drove recombination reactions between two dif1 sites or between dif1 and TLCΦ attP showed that TLCΦ attP/dif1 synaptic complexes are normally too rare and/or too transient to allow for FtsKγ-driven activation of XerD (Figure 7). Taken together, those results suggest that XafT can promote the efficient recombination of dif1 with TLCΦ attP-derived sites harbouring a non-canonical XerD-arm because XafT helps assemble and/or stabilize Xer synaptic complexes. As XafT contains a dimerization domain and directly interacts with XerD, it is tempting to propose that it recruits a second XerD recombinase at the dif1 locus by forming a proteinaceous bridge with an XerD recombinase bound to dif1 (31). The resulting dif1/XerC/XerD/XafT/XerD complex could then be engaged in a larger nucleoprotein complex with an attP site solely bound by XerC via the interactions of recombinases. Thus, a limited amount of homology between the XerD-arm of the attP site and the canonical XerD binding site would be required to ensure the formation of a productive recombination synapse. Future work will need to confirm this hypothesis by directly analysing synaptic complex assembly with or without XafT using biophysical methods.
This scenario is reminiscent of the control of Xer recombination on multicopy plasmids. The dimer resolution sites of multicopy plasmids exploiting Xer and the attachment sites of IMEX relying on the non-canonical XerC-first recombination pathway contain a few degenerate positions in the XerC and XerD-arms (Supplementary Figure S1B). In particular, there are two mutations in the inner region of the XerC-arm of the ColE1 dimer resolution site, cer, including one at the 5th position, and there is a mutation at the sixth position of the XerD-arm, which together would be expected to limit the formation of and/or the stability of synaptic complexes (Supplementary Figure S1B). The efficiency and directionality of Xer recombination then relies on accessory proteins that bind to accessory sequences flanking the core cer site, which bring together the core cer sites in a synaptic complex with a specific topological configuration (30,46). Future studies will also need to address the evolutionary pressures that seems to have set the non-canonical bases of each plasmid dimer resolution and IMEX attachment sites (Supplementary Figure S1B). The NGS methodology we present will be a great asset for future work on this subject.
Importance of the fifth innermost bp of the XerD-arm for recombination
The NGS data allowed us to revisit the importance of the different bases within the XerD-arm of dif sites. Whether integration was driven by FtsK or XafT, the mini-TLCΦ plasmids that integrated the most efficiently were those that harboured the most dif-like motifs (Figure 5). It further indicated that the five outermost positions of the XerD-arm were less constrained than the six innermost positions (Figure 5). This result could seem surprising since previous work showed that the specificity of binding of the E. coli Xer recombinases was ensured by the base composition of the sixth, seventh and ninth to eleventh non-palindromic positions of their binding arms (47,48). However, contacts between the E. coli recombinases and dif were shown to be mainly limited to the seven innermost bases of their binding sites, and in particular to the first five palindromic bases (Supplementary Figure S1, (45)). Correspondingly, the innermost bases of the XerC and XerD binding sites are more conserved than the outermost bases through evolution (49). Interestingly, it was recently reported that Bacillus subtilis and Staphylococcus aureus XerD serves to unload bacterial SMC complexes (50). This activity relies on the binding of XerD to additional chromosomal loci other than dif, independently from XerC. Those loci harbour a dif-like site composed of a bona fide XerD binding site, a 5–6 bp degenerate central region and the five innermost bp of the XerD binding site (50). Our results raise the possibility that those sites evolved to allow the cooperative binding of two XerD molecules.
The nature of the fifth bp of the XerD-arm of the attP sites seems the most important for XafT-driven recombination (Figures 3–5). Correspondingly, it was previously observed in E. coli that a dif site carrying a single mutation at the fifth bp of the XerD-arm was less proficient than wild-type dif for Xer recombination (45). In addition, methylation interference analysis showed that E. coli XerD interacted with the guanine at the fifth position of the top strand of its binding site (45). A model based on the crystal structure of E. coli XerD and the Catabolite Activator Protein-DNA complex further suggested that XerD contacts the guanine of the top strand of its binding site with a conserved arginine of its C-terminal domain (R220 in E. coli XerD and R224 in V. cholerae XerD, (51)). A cytosine base is present at the 5th innermost position of the top strand of the dif site of Helicobacter pylori and Lactococcus lactis, which harbour alternative Xer machineries composed of a single Xer recombinase (Supplementary Figure S1). This position was shown to be important for the binding and the activity of H. pylori Xer recombinase (52). In effect, this basepair is inverted with respect to the GC bp of canonical XerC and XerD-arms (Supplementary Figure S1B). X-ray structure analysis revealed that the H. pylori recombinase contacts the bottom-strand guanine of this bp with an arginine of its N-terminal domain, R65 (52). These observations highlight the evolutionary distance between conventional and alternative Xer machineries.
Preventing undesired Xer-mediated excision
IMEX such as CTXΦ and VGJΦ exploit the low basal ability of XerC to catalyse the formation of HJs, which are subsequently resolved by DNA replication (Figure 1C, (7,8,29)). This strategy of integration is advantageous because it escapes the requirement for FtsK and because the reverse reaction necessarily leaves an intact chromosomal copy of the phage (8). The stability of CTXΦ is further ensured because its attP site is formed by a hairpin of its single stranded DNA genome, which is masked in the double stranded integrated prophage (Supplementary Figure S1, (7,29)). In contrast, TLCΦ and the GGIs rely on an intrinsically fully reversible reaction (Figure 1D, (9,31)). It was proposed that FtsK translocation could prevent excision of the GGIs because it strips XerD from non-canonical XerD-binding sites (41). Our results lend further support to this hypothesis since all of the 190 528 mini-TLCΦ plasmids we studied, including those that harboured the attP sites that most resembled dif1, were far more stable than a DNA cassette flanked by two dif1 sites (Figure 6 and Supplementary Figure S6, (28,37)). However, this hypothesis does not explain the excision inefficiency of mini-TLCΦ plasmids that are integrated at the lacZ locus, which is outside the FtsK activity zone (Figure 6, (28,36–38)). It suggested that integrated mini-TLCΦ plasmids were stable because the expression of XafT was insufficient after integration. In support of this hypothesis, we could detect a C-terminal tagged 3xFLAG tag fusion of XafT, which is active in recombination, in cells harbouring a non-integrated replicative copy of TLCΦ but did not detect it in cells harbouring an integrated copy of TLCΦ (Figure 6). Future work will need to investigate the regulation of XafT expression and under which conditions it can be increased to promote the excision of integrated copies of TLCΦ.
DATA AVAILIBILITY
NGS plasmid library verification, integration and excision data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10951.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Raphaël Guérois and Virginia Lioy for helpful discussions, and Mhairi Davidson for critical reading.
Contributor Information
Solange Miele, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
James Iain Provan, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
Justine Vergne, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
Christophe Possoz, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
Françoise Ochsenbein, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
François-Xavier Barre, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [FP7/2007-2013/28159 to F.-X.B.]; Agence Nationale pour la Recherche [2016-CE12-0030-0 and 2021-CE35-0013-01 to F.-X.B., F.O., 2018-CE12-0012-03 to F.-X.B.]; Fondation pour la Recherche Médicale [EQU202003010328 to F.-X.B.]. Funding for open access charge: Agence Nationale pour la Recherche [2018-CE12-0012-03 to F.-X.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Midonet C., Barre F.-X.. Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol. Spectr. 2014; 2: 10.1128/microbiolspec.mdna3-0056-2014. [DOI] [PubMed] [Google Scholar]
- 2. Blakely G., May G., McCulloch R., Arciszewska L.K., Burke M., Lovett S.T., Sherratt D.J.. Two related recombinases are required for site-specific recombination at dif and cer in e. coli K12. Cell. 1993; 75:351–361. [DOI] [PubMed] [Google Scholar]
- 3. Colloms S.D., Sykora P., Szatmari G., Sherratt D.J.. Recombination at cole1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J. Bacteriol. 1990; 172:6973–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Das B., Martínez E., Midonet C., Barre F.-X.. Integrative mobile elements exploiting xer recombination. Trends Microbiol. 2013; 21:23–30. [DOI] [PubMed] [Google Scholar]
- 5. Balalovski P., Grainge I.. Mobilization of pdif modules in acinetobacter: a novel mechanism for antibiotic resistance gene shuffling. Mol. Microbiol. 2020; 114:699–709. [DOI] [PubMed] [Google Scholar]
- 6. Huber K.E., Waldor M.K.. Filamentous phage integration requires the host recombinases XerC and xerD. Nature. 2002; 417:656–659. [DOI] [PubMed] [Google Scholar]
- 7. Das B., Bischerour J., Val M.-E., Barre F.-X.. Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4377–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das B., Bischerour J., Barre F.-X.. VGJphi integration and excision mechanisms contribute to the genetic diversity of vibrio cholerae epidemic strains. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Midonet C., Das B., Paly E., Barre F.-X.. XerD-mediated ftsk-independent integration of TLCϕ into the vibrio cholerae genome. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16848–16853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramamurthy T., Mutreja A., Weill F.-X., Das B., Ghosh A., Nair G.B.. Revisiting the global epidemiology of cholera in conjunction with the genomics of vibrio cholerae. Front. Public Health. 2019; 7:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waldor M.K., Mekalanos J.J.. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996; 272:1910–1914. [DOI] [PubMed] [Google Scholar]
- 12. Rubin E.J., Lin W., Mekalanos J.J., Waldor M.K.. Replication and integration of a vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 1998; 28:1247–1254. [DOI] [PubMed] [Google Scholar]
- 13. Hassan F., Kamruzzaman M., Mekalanos J.J., Faruque S.M.. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010; 467:982–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chun J., Grim C.J., Hasan N.A., Lee J.H., Choi S.Y., Haley B.J., Taviani E., Jeon Y.-S., Kim D.W., Lee J.-H.et al.. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15442–15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mutreja A., Kim D.W., Thomson N.R., Connor T.R., Lee J.H., Kariuki S., Croucher N.J., Choi S.Y., Harris S.R., Lebens M.et al.. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011; 477:462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heidelberg J.F., Eisen J.A., Nelson W.C., Clayton R.A., Gwinn M.L., Dodson R.J., Haft D.H., Hickey E.K., Peterson J.D., Umayam L.et al.. DNA sequence of both chromosomes of the cholera pathogen vibriocholerae. Nature. 2000; 406:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weill F.-X., Domman D., Njamkepo E., Tarr C., Rauzier J., Fawal N., Keddy K.H., Salje H., Moore S., Mukhopadhyay A.K.et al.. Genomic history of the seventh pandemic of cholera in africa. Science. 2017; 358:785–789. [DOI] [PubMed] [Google Scholar]
- 18. Domman D., Quilici M.-L., Dorman M.J., Njamkepo E., Mutreja A., Mather A.E., Delgado G., Morales-Espinosa R., Grimont P.A.D., Lizárraga-Partida M.L.et al.. Integrated view of vibrio cholerae in the Americas. Science. 2017; 358:789–793. [DOI] [PubMed] [Google Scholar]
- 19. Aussel L., Barre F.X., Aroyo M., Stasiak A., Stasiak A.Z., Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002; 108:195–205. [DOI] [PubMed] [Google Scholar]
- 20. Val M.-E., Kennedy S.P., El Karoui M., Bonné L., Chevalier F., Barre F.-X.. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen vibrio cholerae. PLoS Genet. 2008; 4:e1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yates J., Aroyo M., Sherratt D.J., Barre F.-X.. Species specificity in the activation of xer recombination at dif by ftsK. Mol. Microbiol. 2003; 49:241–249. [DOI] [PubMed] [Google Scholar]
- 22. Grainge I., Lesterlin C., Sherratt D.J.. Activation of xercd-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011; 39:5140–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yates J., Zhekov I., Baker R., Eklund B., Sherratt D.J., Arciszewska L.K.. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol. Microbiol. 2006; 59:1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bigot S., Saleh O.A., Lesterlin C., Pages C., El Karoui M., Dennis C., Grigoriev M., Allemand J.-F., Barre F.-X., Cornet F.. KOPS: DNA motifs that control e. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005; 24:3770–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kennedy S.P., Chevalier F., Barre F.-X.. Delayed activation of xer recombination at dif by FtsK during septum assembly in escherichia coli. Mol. Microbiol. 2008; 68:1018–1028. [DOI] [PubMed] [Google Scholar]
- 26. Barre F.X., Aroyo M., Colloms S.D., Helfrich A., Cornet F., Sherratt D.J.. FtsK functions in the processing of a holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000; 14:2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cornet F., Louarn J., Patte J., Louarn J.M.. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 1996; 10:1152–1161. [DOI] [PubMed] [Google Scholar]
- 28. Galli E., Midonet C., Paly E., Barre F.-X.. Fast growth conditions uncouple the final stages of chromosome segregation and cell division in escherichia coli. PLoS Genet. 2017; 13:e1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Val M.-E., Bouvier M., Campos J., Sherratt D., Cornet F., Mazel D., Barre F.-X.. The single-stranded genome of phage CTX is the form used for integration into the genome of vibrio cholerae. Mol. Cell. 2005; 19:559–566. [DOI] [PubMed] [Google Scholar]
- 30. Colloms S.D., Bath J., Sherratt D.J.. Topological selectivity in xer site-specific recombination. Cell. 1997; 88:855–864. [DOI] [PubMed] [Google Scholar]
- 31. Midonet C., Miele S., Paly E., Guerois R., Barre F.-X.. The tlcφ satellite phage harbors a xer recombination activation factor. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:18391–18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meibom K.L., Blokesch M., Dolganov N.A., Wu C.Y., Schoolnik G.K.. Chitin induces natural competence in vibrio cholerae. Science. 2005; 310:1824–1827. [DOI] [PubMed] [Google Scholar]
- 33. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PloS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 35. Demarre G., Guérout A.-M., Matsumoto-Mashimo C., Rowe-Magnus D.A., Marlière P., Mazel D.. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate escherichia coli host strains. Res. Microbiol. 2005; 156:245–255. [DOI] [PubMed] [Google Scholar]
- 36. Espinosa E., Paly E., Barre F.-X.. High-resolution whole-genome analysis of sister-chromatid contacts. Mol. Cell. 2020; 79:857–869. [DOI] [PubMed] [Google Scholar]
- 37. Demarre G., Galli E., Muresan L., Paly E., David A., Possoz C., Barre F.-X.. Differential management of the replication terminus regions of the two vibrio cholerae chromosomes during cell division. PLoS Genet. 2014; 10:e1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. David A., Demarre G., Muresan L., Paly E., Barre F.-X., Possoz C.. The two Cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of vibrio cholerae chromosome I. PLoS Genet. 2014; 10:e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dillard J.P., Seifert H.S. A variable genetic island specific for neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 2001; 41:263–277. [DOI] [PubMed] [Google Scholar]
- 40. Domínguez N.M., Hackett K.T., Dillard J.P.. XerCD-mediated site-specific recombination leads to loss of the 57-kilobase gonococcal genetic island. J. Bacteriol. 2011; 193:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fournes F., Crozat E., Pages C., Tardin C., Salomé L., Cornet F., Rousseau P.. FtsK translocation permits discrimination between an endogenous and an imported Xer/dif recombination complex. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Massey T.H., Aussel L., Barre F.-X., Sherratt D.J.. Asymmetric activation of xer site-specific recombination by ftsK. EMBO Rep. 2004; 5:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bonné L., Bigot S., Chevalier F., Allemand J.-F., Barre F.-X.. Asymmetric DNA requirements in xer recombination activation by ftsK. Nucleic Acids Res. 2009; 37:2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poidevin M., Galli E., Yamaichi Y., Barre F.-X.. WGADseq: whole genome affinity determination of protein-dna binding sites. The Bacterial Nucleoid: Methods and Protocols, Methods in Molecular Biology. 2017; New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- 45. Blakely G.W., Sherratt D.J.. Interactions of the site-specific recombinases XerC and XerD with the recombination site dif. Nucleic Acids Res. 1994; 22:5613–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bregu M., Sherratt D.J., Colloms S.D.. Accessory factors determine the order of strand exchange in xer recombination at psi. EMBO J. 2002; 21:3888–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blakely G., Sherratt D.. Determinants of selectivity in xer site-specific recombination. Genes Dev. 1996; 10:762–773. [DOI] [PubMed] [Google Scholar]
- 48. Hayes F., Sherratt D.J.. Recombinase binding specificity at the chromosome dimer resolution site dif of escherichia coli. J. Mol. Biol. 1997; 266:525–537. [DOI] [PubMed] [Google Scholar]
- 49. Kono N., Arakawa K., Tomita M.. Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics. 2011; 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karaboja X., Ren Z., Brandão H.B., Paul P., Rudner D.Z., Wang X.. XerD unloads bacterial SMC complexes at the replication terminus. Mol. Cell. 2021; 81:756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Subramanya H.S., Arciszewska L.K., Baker R.A., Bird L.E., Sherratt D.J., Wigley D.B.. Crystal structure of the site-specific recombinase, xerD. EMBO J. 1997; 16:5178–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bebel A., Karaca E., Kumar B., Stark W.M., Barabas O.. Structural snapshots of xer recombination reveal activation by synaptic complex remodeling and DNA bending. Elife. 2016; 5:e19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


