Abstract
Human Positive Coactivator 4 (PC4) is a multifaceted chromatin protein involved in diverse cellular processes including genome organization, transcription regulation, replication, DNA repair and autophagy. PC4 exists as a phospho-protein in cells which impinges on its acetylation by p300 and thereby affects its transcriptional co-activator functions via double-stranded DNA binding. Despite the inhibitory effects, the abundance of phosphorylated PC4 in cells intrigued us to investigate its role in chromatin functions in a basal state of the cell. We found that casein kinase-II (CKII)-mediated phosphorylation of PC4 is critical for its interaction with linker histone H1. By employing analytical ultracentrifugation and electron microscopy imaging of in vitro reconstituted nucleosomal array, we observed that phospho-mimic (PM) PC4 displays a superior chromatin condensation potential in conjunction with linker histone H1. ATAC-sequencing further unveiled the role of PC4 phosphorylation to be critical in inducing chromatin compaction of a wide array of coding and non-coding genes in vivo. Concordantly, phospho-PC4 mediated changes in chromatin accessibility led to gene repression and affected global histone modifications. We propose that the abundance of PC4 in its phosphorylated state contributes to genome compaction contrary to its co-activator function in driving several cellular processes like gene transcription and autophagy.
INTRODUCTION
The integrity of the eukaryotic genome is maintained by multiple factors which are functionally interconnected by diverse mechanisms (1,2). Emerging evidences suggest that non-histone chromatin associated proteins play critical role in genome organization (3–8). Human positive coactivator 4 (PC4) is a highly abundant nuclear protein involved in multiple cellular processes (7,9–12). It is reported to activate gene transcription (9,10). Alternatively, it induces chromatin condensation and is critical for genome integrity (7,8). Its interaction with the core histones has been causally related to chromatin compaction in vitro (7). It is likely that chromatin compaction and transcriptional co-activation are mediated through different forms of the PC4 protein in a cell.
Post-translational modifications (PTMs) often contribute to multifunctionality of chromatin proteins by regulating their subcellular localization or their recruitment onto the genome via interaction with other cellular factors. Majority of PC4 is present in the phosphorylated state in cells and Casein Kinase-II (CKII) is predominantly responsible for the in vivo hyperphosphorylation of the N-terminal serine-rich acidic stretch (SEAC) (9). p300, on the other hand, acetylates PC4 and impacts its coactivator function through stimulation of DNA binding, DNA bending and binding to transcription factors like p53 (13–16). Till date, most of the studies that have focused on the roles of PC4 has been in the light of its coactivator function. Mechanistically, CKII-mediated phosphorylation of PC4 counters its co-activation function by gradual masking of the lysine-rich region (9,14,17). Notwithstanding these negative effects of phosphorylation on its coactivator roles, majority of the cellular PC4 still exist in the phosphorylated state and the implications are currently unclear.
Linker histone H1 is an essential component of the higher order chromatin organization (3,18,19). Single-base resolution *OH footprinting shows that H1 binds to the DNA entry and exit site of the nucleosome with the globular domain of histone H1 interacting with the DNA minor groove at the center of the nucleosome, symmetrically placed with respect to the nucleosomal dyad (20). Extensive crystal structure and cryo-electron microscopic studies have shown asymmetric binding and location of H1 with respect to the nucleosomal dyad which determines the twisting of the repeating tetra-nucleosomal structural units of 30 nm fibers (18). Such studies provide a picture of linker H1 changing the chromatin landscape. Linker H1 is also known to regulate chromatin dynamics by interacting with other non-histone proteins. H1 and the heterochromatin protein 1, HP1 interacts in a subtype specific manner upon methylation of lysine 26 which is inhibited upon ser27 phosphorylation (21–24). Acetylation of H1 at lysine, K,85 that occurs in response to DNA damage promotes HP1α recruitment to facilitate chromatin compaction (25). Thus, PTMs often regulate interactions between architectural proteins to confer the dynamic state of the chromatin.
It is observed that absence of PC4 leads to increased genomic instability because of its involvement in resolving G4DNA structures formed co-transcriptionally or its involvement in DNA damage response (26,27). A recent study has also shown PC4 depletion leading to drastic decompaction and altered nuclear architecture (8). However, these studies have only addressed the consequences of the absence of PC4 on the genome but could not address the exact molecular mechanism which regulates this function of PC4. We establish here that phosphorylation of PC4 by CKII is critical for the functional organisation of the genome by PC4.
Earlier observation stated that PC4 does not interact with linker histone H1, in vitro (7). However, here we report that PC4 interacts with linker histone H1, only when it is phosphorylated by CKII. This finding led us to explore the role of phosphorylated PC4 in chromatin compaction function. Intriguingly, we show that phospho-mimic PC4 efficiently compacts in vitro reconstituted nucleosomal array and substantiates the role of phosphorylated-PC4 in higher order chromatin organization in conjunction with linker histone H1. Cellular evidence also suggested that ectopic expression of phospho-mimic and wild type PC4, but not the phospho-deficient mutant, in PC4 knockdown cells could compact the chromatin as the control cells, restoring the global histone modification status. Encouraged by our biophysical and cellular assays, we carried out Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) to understand the genomic regions that are regulated by phospho-PC4. Presumably, this is the first study to show that PC4 indeed exhibits greater chromatin inaccessibility in a phosphorylation dependent manner and regulates a wide array of genes including both protein coding and non-coding RNAs. More importantly, expression of the wild type and phospho-mimic PC4 could not only induce greater depletion of ATAC-seq peaks and efficient chromatin binding but also led to the rescue of altered gene repression. Collectively, we present evidence to re-iterate that the chromatin function of PC4 is regulated by its phosphorylation and can impart implications on several cellular processes including autophagy.
MATERIALS AND METHODS
Expression and purification of recombinant proteins
Bacterially expressed recombinant N-terminal His6-tagged linker H1 variants (used for in vitro interaction studies) were subcloned in pET28b vector and purified using nickel-NTA (Ni-nitrilotriacetic acid) agarose (Millipore). Mutations in the N terminus region within the first serine rich tract of PC4 were introduced by using the site directed mutagenesis kit from Stratagene (QuikChange II XL Site-Directed Mutagenesis Kit, Agilent Technologies, Santa Clara, CA, USA) as per the manufacturer's instructions. A series of phospho-deficient mutants generated upon mutating serine 13, 15, 17 and 19 in different combinations to alanine were labelled as MTP3, MTP5, MTP6, MTP7, MTP8 and MTP9 (Supplementary Figure S1B). Bacterially expressed untagged recombinant PC4, phospho-mimic (PM-PC4) and phospho-deficient mutants of PC4 (MTPs) were expressed using pET11a vector and purified by heparin sepharose (GE Healthcare) and phosphocellulose P11 column chromatography (28). Recombinant core histones (Xenopus) H2A, H2B, H3 and H4, which come in inclusion bodies, were purified by denaturation in 8M urea followed by renaturation as described elsewhere (29). Purification of human linker histone H1was carried out as described elsewhere (30).
For cellular studies, Flag tagged Wild type PC4, PM-PC4 and MTP5 were subcloned in mammalian expression vector pFLAG-CMV-10.
Cell culture and stable transfection
Human cell lines (HEK293) were grown in the DMEM supplemented with sodium bicarbonate, 100U·ml−1 Penicillin 0.1mg·ml−1 Streptomycin, 0.25μg·ml−1 Amphotericin B and 10% heat inactivated FBS at 37°C in 80% humidifier air and 5% CO2. PC4 knockdown cell was generated using 10μg pGIPZ lentiviral shRNAs targeting PC4 (Open Biosystems, Dharmacon, Lafayette, CO, USA) and helper plasmids (5μg psPAX2, 1.5μg pRSV‐Rev, 3.5μg pCMV‐VSV‐G). 10ug of shRNA‐plasmid was mixed with helper plasmids (5μg psPAX2, 1.5μg pRSV‐Revs, 3.5μg pCMV‐VSV‐G) and were co transfected into HEK293T cells using lipofecatamine. Forty‐eight hours post transfection media containing assembled virus was collected and its titre was estimated. HEK293 cells were infected with the desired virus and the infected cells were subjected to selection pressure 72 h post transfection. Cells were grown in the presence of 3μg·ml−1 puromycin for three passages to establish the cell line. PC4 knockdown cells were then transfected with pFlag-CMV-10PC4, pFlag-CMV-10PM-PC4 and pFlag-CMV-10MTP5 plasmids using lipofectamine and media change was given within 24 hours of transfection and cultured under 1mg/ml dose of G418 selection.
Micrococcal nuclease (MNase) sensitivity assay
Cells were grown in DMEM medium supplemented with 10% fetal Bovine serum (FBS). The nuclei were prepared from packed cells suspended in hypotonic buffer (10mM Tris–HCl, 10mM KCl and 15mM MgCl2) followed by 10 min incubation at 4 degree C. The nuclei were digested with MNase (0.2U/μl) for different time points at room temperature in nuclei digestion buffer (10% glycerol, 10mM Tris–HCl pH 8, 3mM CaCl2, 150mM NaCl, 0.2mM PMSF). MNase digestion was stopped by the addition of 10mM EDTA and proteinase K at 37°C for 30 min followed by RNase A treatment for 1hr at 37°C. The DNA was then extracted by phenol chloroform extraction and ethanol precipitation. 1ug of each of the samples were loaded on 1% agarose gel and ran for 4–5 h in an Ethidium Bromide free agarose gel electrophoresis system at 50 V for better resolution of the MNase bands. The gels were then stained in Ethidium bromide staining solution for 15 min before visualization in Chemi-Doc system (Biorad).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X‐100. Cells were then blocked using 1% BSA in PBS and probed with the indicated primary antibodies and species‐specific secondary antibodies (Alexa 568-conjugated anti‐mouse). Cell nuclei were counterstained with 0.1μg·ml−1 Hoechst 33258 (Sigma). Samples were visualized using a confocal microscope (Zeiss LSM 880 Airyscan; Carl‐Zeiss, Feldbach, Switzerland). Images were captured at 63x magnification. Assay was performed in Flag expressing PC4 and PC4 mutant cell lines and probing was carried out using anti-Flag antibody.
Nucleosome and chromatin reconstitution
DNA template for nucleosome assembly:
DNA templates of 12 × 177 bp tandem repeats of the 601 sequence were cloned and purified as described (31).
The sequence for the 177 bp DNA repeat is listed below with 601 DNA sequence underlined:
GAGCATCCGGATCCCCTGGAGAATCCCGGT
GCCGAGGCC GCTCAATTGGTCGTAGACAGC
TCTAGCACCGCTTAAACGC ACGTACGCGCTG
TCCCCCGCGTTTTAACCGCCAAGGGGAT TAC
TCCCTAGTCTCCAGGCACGTGTCACATATATA
CATCCT GTTCCAGTGCCGGACCC
The respective histone octamers were reconstituted as previously described (4). Equimolar amounts of individual histones in unfolding buffer (7M guanidinium HCl, 20mM Tris–HCl, pH7.5, 10 mM DTT) were dialyzed into refolding buffer (2M NaCl, 10mM Tris–HCl, pH 7.5, 1mM EDTA, 5mM 2-mercaptoethanol), and purified through a Superdex S200 column. Chromatin arrays and 30-nm chromatin fibers were assembled using the salt dialysis method as previously described with a minor modification (29). The reconstitution reaction mixture with octamers and 601 based DNA templates were dialyzed over 16 h at 4°C in TEN buffer (10mM Tris–HCl, pH 8.0, 1mM EDTA, 2M NaCl), which was continuously diluted by slowly adding in TE buffer (10mM Tris–HCl, pH 8.0, 1mM EDTA) to lower the concentration of NaCl from 2M to 0.6M. For histone H1 incorporation, an different molar amount of histone H1 (relative to mononucleosomes) was added at this step and further dialyzed in TE buffer with 0.6M NaCl for 3h, followed by a final dialysis step in HE buffer (10mM HEPES, pH 8.0, 0.1mM EDTA) for 4h. For PC4 and PM-PC4 the proteins were added after dialysis in HE buffer. The reconstituted nucleosome was then subjected to AUC investigation and EM imaging.
Sedimentation velocity analytical ultracentrifugation
The chromatin samples with or without histone H1 or PC4 or PM–PC4 incorporation were prepared in HE buffer. Sedimentation experiments were performed on a Beckman Coulter ProteomeLab XL-I using a 4-hole An-60Ti rotor. Samples with an initial absorbance at 260 nm of ∼0.5–0.8 were equilibrated for 2 h at 20°C under a vacuum in a centrifuge prior to sedimentation. The absorbance at 260 nm was measured using a continuous scan mode during sedimentation at 32 000 × g in 12 mm double-sector cells. The data were analyzed using enhanced van Holde-Weischet analysis and Ultrascan II 9.9 revision 1504. The S20,w values (sedimentation coefficient corrected for water at 20°C) were calculated with a partial specific volume of 0.622ml/g for chromatin, and the buffer density and viscosity were adjusted. The average sedimentation coefficients were determined at the boundary midpoint, S50%(S). The data was then plotted in MICROSOFT EXCEL.
Electron microscopy
The reconstituted chromatin samples were negatively stained for which they are chemically fixed in 0.2% glutaraldehyde for 30min on ice. After fixation, the chromatin sample was absorbed onto the glow-discharged 300 mesh R2/1 Quantifoil (Quantifoil Micro Tools GmbH, Jena, Germany) holey grids for 1–1.5min, blotted using 4s blotting time at 100% humidity and stained with uranyl sulfate to stain the protein white against a black background for 45s, blotted and dried for 1 min before imaging. The nucleosomal array is prepared for metal shadowing for which the sample is fixed in 0.4% glutaraldehyde for 30min on ice and 1× spermidine is added before loading the sample on to Copper grids and incubated for 2min blotted and washed in water and alcohol gradient and dried. The copper grid is then placed on a glass slide for metal shadowing using a tungsten filament under a vacuum with 0.2 × 10−5 Torr pressure at low voltage and 15–18 A current for 15min in rotating condition. This allows the metal deposition to occur on the surface of the negatively charged DNA so it appears black against a white background during EM imaging. The grids were transferred into an FEI Titan Krios electron microscope (FEI, Eindhoven).
Isothermal calorimetry
Isothermal titration calorimetry(ITC) measurements were taken using a MicroCal iTC200 instrument (MicroCal, Inc.). Aliquots (2μl) of Linker H1.1 at a concentration of 21μM were injected from the syringe into the cell of 240μl filled with 2μM of phosphoPC4 or mock-phospho PC4. Titrations were conducted in 20mM Tris–HCl (pH 7.5), 0.2mM EDTA and 100 mM KCl buffer. To minimize the contribution of dilution to binding heat, the protein solutions were dialyzed against the same buffer prior to the ITC experiments. Injections weremade at intervals of 120s, and the duration of each injection was 0.4s. To ensure proper mixing after each injection, a constant stirring speed of 1000 rpm was maintained during the experiment. The control experiment was performed via injection of buffer in the ITC cell containing H1.1. All molarity calculations of PC4 were conducted according to its reported molecular mass 14.35kDa while H1 was taken as a monomer. The heat change versus the molar ratio of the titrated products was plotted and analyzed using the manufacturer's software, Origin 7.0, which yielded the stoichiometry (n, in terms of the number of molecules of H1 per phospho-PC4 or mock phospho-PC4) and the dissociation constant (Kd).
CD spectroscopy
The circular dichroism (CD) spectra of H1-stripped chromatin (0.6mg/ml) and complexes with individual different modified forms of PC4 were recorded after incubation at 25°C for 90 min or as indicated in the figures in 10mM Tris–HCl–25 mM NaCl (pH 7.4). The spectra were recorded at room temperature in a JASCO model J715 spectropolarimeter at settings from 250 to 300nm.
In vitro kinase assay
Bacterially expressed recombinant PC4/PC4 phospho-defective mutants(1ug) was incubated with 1ul of casein Kinase II (20 mU) from NEB in 1× Phospho buffer (50 mM HEPES–KOH, pH7.6, 125mM NaCl, 10mM MgCl2, 6% glycerol, 5mM DTT, 0.5mM PMSF) along with 10mM ATP in a 20ul reaction mixture at 30°C for 30min followed by three replenishments with the same amount of enzyme and ATP at a 1 h interval. The final incubation was prolonged for 12h after the last replenishment to ascertain the completion of the reaction. The phosphorylation was tracked by observing the mobility shift of phosphorylated PC4 in 15% SDS/PAGE by Coomassie staining.
In vitro interaction assays
The histone H1 interaction ability of PC4 was characterized by incubating 5ul of Ni-NTA beads with 1ug of His6-H1 and 250ng of recombinant untagged PC4 and PC4 mutants in a final volume of 300ul in biochemical buffer containing 200mM KCl supplemented with 30mM imidazole at 4°C for 3.0h. The beads were washed three times (1ml each) with the incubation buffers. The Ni-NTA agarose pulldown complexes were analyzed by western blotting using anti-PC4 polyclonal antibodies and anti-His antibody (sigma). Control experiments were performed with 5ul of Ni-NTA beads incubated with 250ng of individual recombinant untagged PC4 and PC4 mutants in the same buffer.
Chromatin fractionation
293 cells were collected from two 100-mm plates and washed with 2ml of cold 1× PBS (plus PI). The cell pellet was resuspended in 2.5 times the volume of buffer A (0.3M sucrose, 60mM KCl, 60mM Tris–HCl, pH 8.0, 2mM EDTA and 0.5% NP-40 plus PI) and incubated on ice for 30min, followed by a 5min spin at 2000 rcf at 4°C. The cytosolic fraction was collected. After one wash with buffer A, the nuclei was resuspended in 2 times the volume of pellet in 100mM NaCl and 0.2%NP-40 containing mRIPA (6 strokes in a 7-ml Dounce homogenizer) and spun at 6000rcf for 5 min. The supernatant was designated as nuclear extract (100mM). The pellet was similarly resuspended in 2× pellet volume of mRIPA with NaCl concentrations of 600mM and a 10min incubation on ice and this fraction was then kept for flag immuno pull-down using M2 agarose beads.
Immuno pull down
Flag-PC4, Flag-PM-PC4 and Flag-MTP5 stably transfected in PC4 knockdown HEK293 cells were harvested and washed once with 1× PBS and lysed by adding 1ml of chilled lysis buffer (50mM Tris–HCl, pH 7.4; 150mM NaCl; 5mM EDTA; 1% triton X-100; 1X-Complete protease inhibitor). Cells were incubated at 4°C for 1h with constant mixing. Lysate was clarified by centrifuging at 13000 rpm at 4°C for 10 min and incubated with 30μl of anti-Flag M2 agarose beads (pre-washed and equilibrated in lysis buffer; Sigma # F2426) for 3h at 4°C with uniform mixing. The beads were collected by centrifugation for 3 min at 2000 rpm at 4°C and the supernatants were removed by aspiration. The pellets were washed once with 1ml of cold lysis buffer, twice with tris buffered saline (TBS; 50mM Tris–HCl, pH 7.4; 150mM NaCl) and eluted in 30 μl of 500ng/μl of 3X-FLAG peptide (Sigma#F4799) in TBS buffer. The immunoprecipitated samples were then resolved in a 15% SDS-PAGE and immunoblot analysis was done for verifying the pulldown protein and their interacting partners.
Gene expression analysis using real time PCR
Total RNA from cells was isolated using the Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions. RNA was extracted from ∼2 × 104 cells. 0.5μg of RNA was reverse transcribed using MMLV reverse transcriptase (Sigma, M1302) using oligo‐dT primers. Resulting cDNAs were quantified by RT‐PCR using KAPA SYBR FAST Universal qPCR master mix (KAPA Biosystems, KK4601, Wilmington, MA, USA) on the Stepone Plus RT‐PCR platform (Life Technologies). Amplification with specific gene primers was performed. The housekeeping gene, β‐actin was used for normalization. The primers used for these analysis are listed in Supplementary Table S1. Relative expression was normalized to the endogenous control Actin using the 2−ΔΔCt method. Experiments were carried out in two technical triplicates, and three biological triplicates. qPCR was performed using SYBR Green (Sigma Aldrich, USA) and RT PCR primers (See Supplementary Table S1).
Chromatin immunoprecipitation (ChIP)
The cells were cross-linked in 1% formaldehyde. The crosslinking was quenched in 0.125M glycine, the cells were lysed in SDS buffer, and the chromatin was sheared using a Bioruptor® Diagenode (Belgium) instrument. Five to ten micrograms of antibody used in each ChIP. The antibody and BSA-blocked protein G sepharose beads (GE Healthcare, USA) were added to the cleared sheared cross-linked chromatin and incubated at 4°C overnight. The beads were then washed to remove non-specific binding with buffers containing different salt concentrations. The DNA–protein complexes were eluted from the washed beads using SDS-sodium bicarbonate elution buffer. The eluates and input were de-crosslinked at 65°C for 6h. Proteinase K (Thermo Fisher Scientific, USA) and RNase H (Sigma Aldrich, USA) were added to the de-crosslinked lysates. The DNA was extracted using the phenol:chloroform:isoamyl alcohol method, followed by ethanol precipitation (1/10th volume 3M sodium acetate, pH 5.2, 20mg glycogen, and 2.5 volumes of 100% ethanol) at –20°C overnight. The DNA pellet was washed in 70% ethanol and dissolved in nuclease-free water. ChIP-qPCR was performed using SYBR Green (Sigma Aldrich, USA) and loci-specific ChIP primers that were differentially accessible (see Supplementary Table S2).
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
Library for ATAC-seq was prepared according to a published protocol (32) using 50,000 intact cells as input. The cells were spun at 500 × g for 5 min followed by a wash using 50 μl of cold 1 × PBS and centrifugation at 500 × g for 5 min. Cells were lysed using cold lysis buffer (10mM Tris–HCl, pH 7.4, 10mM NaCl, 3mM MgCl2 and 0.1% IGEPAL CA-630) and spun at 500 × g for 10 min using a refrigerated centrifuge generating the nuclei prep. The nuclei prep was then resuspended in the transposase reaction mix (25μl 2 × TD buffer, 2.5μl transposase (Illumina) and 22.5μl nuclease-free water) 30min at 37°C. Tn5 transposase tagmentation simultaneously fragments the genome and tags the resulting DNA with Illumina sequencing adapters. DNA fragments were PCR amplified and subsequently purified using Qiagen MinElute PCR Purification Kit (Qiagen, Hilgen, Germany). Final library quality and quantity were analyzed and measured by Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara,CA, USA) and Life Technologies Qubit3.0 Fluorometer (Life Technologies), respectively. We conducted ATAC-seq from two biological replicates for each of the samples. Spearman's correlation of the ATAC-seq signal within consensus ATAC-seq peaks was compared between the replicate batches for each of the comparisons of PC4 versus PC4 KD, PM-PC4 versus PC4 KD and MTP5 versus PC4 KD respectively indicate reproducibility of the ATAC-seq analysis (Supplementary Figure S8D–F).
Quantification of western blots and MNase digested nucleosomal DNA
The western blots and agarose gel for MNAse assays were visualised using ChemiDoc Imaging system from Biorad. The intensities of the bands in the western blots were quantified using ImageJ NIH software and the intenisities of the MNase digested nucleosomal DNA bands for each lane were quantified using Image Lab software (Biorad). The band intensity values for both western blots and MNase digested nucleosomal DNA were plotted and analyzed using Prism 7 (GraphPad Software, La Jolla, CA, USA).
Statistical analysis
The statistical analysis for S50 values in AUC was done by paired t-test and ordinary one-way ANOVA, Sidak's multiple comparisons test. Two-way ANOVA, Dunnett's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001, ns, non-significant) was carried out for MNase sensitivity assay and for all the other assays ordinary one-way ANOVA, Sidak's multiple comparisons test was done considering P value *P< 0.05,**P< 0.01,***P< 0.001, ns-non-significant. All the tests were carried out by Prism 7 (GraphPad software, La Jolla, CA, USA).
RESULTS
Casein Kinase II mediated phosphorylation of PC4 is critical for its interaction with linker histone H1
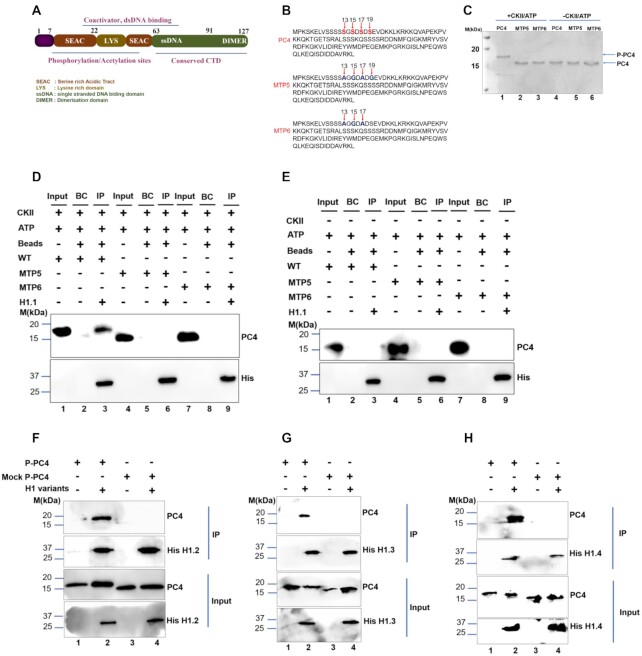
Although PC4 has been found to be predominantly phosphorylated in vivo, its physiological relevance has not been elucidated properly. The mass spectrometric analysis by Ge et al. showed that no significant phosphorylation is observed between residues 29 and 127 (9) and the potential CKII phosphorylation sites are located within the residues 2–28 or the first serine rich acid tract (9) highlighted in brown and labelled as SEAC domain of the protein (Figure 1A). Several phospho-defective mutant clones were generated mutating different serine residues to alanine either singly or in combinations, labelled as MTP (Supplementary Figure S1A and B, also see Materials and Methods). Two such phospho-defective mutants, MTP5 and MTP6 (Figure 1B), were phosphorylated in an in vitro kinase assay using commercially available Casein Kinase II (CKII) and their phosphorylation status was confirmed by shift in the mobility of PC4 in 15% SDS-PAGE (Figure 1C). The results indicate that the serine residues of PC4 indicated in Figure 1B; 13, 15, 17 are critical for CKII mediated phosphorylation (Figure 1C and Supplementary Figure S1A, B). The association of PC4 with chromatin is suggested to be through its preferential interaction with different core histones except the centromeric variant of histone H3, CENPA (7). Interestingly, unmodified bacterially expressed wild type recombinant PC4 does not interact with linker histone H1 in vitro (7). This might be because posttranslational modification of these proteins is critical in mediating such association. The linker histone H1 being an important chromatin component has been reported to regulate functions of other non-histone chromatin proteins like HP1α and HMG through direct interactions with them (22,25,33). Since phosphorylation of PC4 is one of the most predominant posttranslational modifications which inhibits transcription, we speculated that it could have an important role in chromatin condensation. Further, since linker histone H1 is also involved in chromatin compaction, we first intended to visualize whether there is any direct association between linker histone H1 and phospho-PC4. We expressed His-tagged linker H1.1 and unmodified wild type recombinant PC4 which is untagged (mentioned as PC4 in Figure 1D) in bacterial expression system and observed that PC4 interacts with linker histone H1 only when it is phosphorylated (Figure 1D, lane 3). As expected, the phospho-defective mutants (MTP5 and MTP6) failed to do so (Figure 1D, lane 3 versus lanes 6 and 9). The reduced migration of PC4 upon phosphorylation (Figure 1C, lane 1) were also seen in IP upon pulling down His-tagged H1.1 (Figure 1D, lane 3). The mock phosphorylated PC4 without CKII in a kinase reaction did not show any change in the migration pattern (Figure 1C, lane 4) nor did it interact with linker histone H1 (Figure 1E, lane 3). Similarly, in vitro interaction studies were carried out with other somatic linker H1 variants besides H1.1; H1.2, H1.3 and H1.4, that are majorly expressed during S phase in a replication dependent manner. The somatic H1 variants despite having some specific functions do share a considerable amount of functional redundancy. PC4 interacted with the H1 variants; H1.2, H1.3 and H1.4 (Figure 1F–H), only upon phosphorylation by CKII since only P-PC4 (phosphorylated PC4) was present in the IP. Mock P-PC4 (mock phosphorylated PC4), where PC4 had been subjected to kinase assay without the CKII enzyme, was not present in the IP upon pulling down His-tagged H1 variants (Figure 1F–H, lane 2 versus lane 4 for IP panel). Further, the strength of PC4 interaction with linker histone was quantified by isothermal titration calorimetry (Supplementary Figure S1C, D). Unlike, unmodified recombinant PC4 which did not show any significant enthalpy change in the presence of linker histone H1.1 (Supplementary Figure S1D), phosphorylated PC4 showed a robust association leading to significant enthalpy change, with a dissociation constant of 5 nM (Supplementary Figure S1C) exhibiting a one-site binding model of the isotherm. Collectively, these data suggested that phosphorylation of PC4 is indeed critical for mediating its interaction with linker histone H1 in vitro.
Figure 1.
Casein Kinase II mediated phosphorylation of PC4 is critical for its interaction with linker histone H1. (A) Domain organisation of PC4. (B) Amino acid sequence of PC4 highlighting serine residues at 13,15,17 and 19 position, critical for CKII phosphorylation and Amino acid sequence of phospho-defective mutants (MTP) indicating the critical serine residues mutated in different combinations in MTP5 and MTP6 mutants respectively. (C) In vitro kinase assay performed using 600ng of wild type and phospho-defective mutants (MTP5 and MTP6) using ATP and CKII enzyme (lanes 1–3) and only ATP for mock reactions (lanes 4–6). (D) An in vitro interaction assay was conducted following the kinase assay shown in Figure 1C of phosphorylated PC4, MTP5 and MTP6 with His-tagged linker histone H1.1 was carried out by Ni-NTA pulldown and probed with anti-His antibody for the His tagged H1.1 along with PC4 antibody. (E) In vitro interaction with his-tagged linker H1.1 using the mock controls (kinase assay without CKII enzyme) of PC4 and phospho-defective serine mutants (MTP5 and MTP6). BC refers to bead control which indicates the in vitro reaction set up without linker H1.1 for both panels D and E. (F–H) In vitro interaction assay with P-PC4 (PC4 phosphorylated by CKII) and Mock P-PC4 (mock phosphorylated PC4 without CKII) with different His-tagged linker histone H1 variants H1.2, H1.3, H1.4 respectively was carried out by Ni-NTA pulldown following an in vitro kinase assay by CKII and probed with anti-His antibody for the His-tagged H1 variants along with PC4 antibody (compare lane 2 versus lane 4 for IP panel in Figure 1F–H).
Phosphomimic PC4 (PM-PC4) interacts with linker histone H1
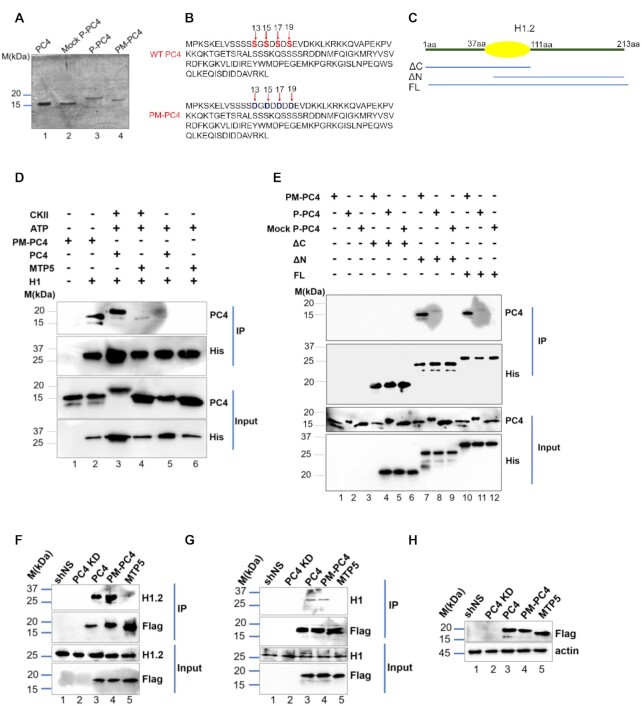
As the in vitro biochemical data suggested that phosphorylation of PC4 is essential to interact with linker histone H1, we further investigated the significance of this interaction in the physiological context using a phosphomimic clone. This was generated by two sequential site directed mutagenesis, where the four critical serine residues 13, 15, 17 and 19 was replaced by aspartic acid (PC4S13DS15DS17DS19D) denoted as PM-PC4 (Figure 2B). Concurring with previous studies (9) these four serine residues which were predicted to be potential CKII sites by mass spectrometric analysis lies within the first serine rich tract and is highly conserved as shown by sequence alignment of PC4/Sub1 across different species (Supplementary Figure S2A). The mobility of the phosphomimic PC4 (PM-PC4) was found to be comparable to P-PC4 (phosphorylated PC4) as shown in 15% SDS-PAGE (Figure 2A, compare lane 3 versus lane 4). In contrast, the mobility of wild type unmodified recombinant PC4 (PC4) and Mock P-PC4 (mock phosphorylated PC4) was found to be faster as compared to P-PC4 or PM-PC4 (Figure 2A, compare lanes 1 and 2 with lanes 3 and 4). Interestingly, we observed that indeed both PM-PC4 and P-PC4 interacts with linker histone H1.2 (Figure 2D, lane 2 versus lane 3). To further understand which domain of H1 could be critical for this interaction, an in vitro interaction assay was carried out using N-terminal and globular domain (ΔC) of linker H1.2, C-terminal and globular domain (ΔN) constructs of linker histone H1.2 (Figure 2C) with PM-PC4, P-PC and MockP-PC4 (Figure 2E). The results indicate that PM-PC4 exhibits more potent interaction with linker histone H1 harbouring globular domain along with the C-terminal domain (ΔN) instead of the N-terminal (ΔC) (Figure 2E, lane 7 versus lane 4). The strength of interaction between PM-PC4 with full length linker histone H1 (FL) was found to be lesser when compared to the C-terminal globular domain ΔN (Figure 2E, lane 10 compared to lane 7) alone. P-PC4 also exhibits interaction with ΔN and FL linker H1 (Figure 2E, compare lane 7 and lane 11 respectively). PM-PC4 exhibited greater interaction with ΔN (Figure 2E, compare lane 7 versus lane 8) and FL (Figure 2D, compare lane 10 versus lane 11) compared to P-PC4. No interaction of linker histone H1 full length or its deletion constructs (ΔN and ΔC) with the mock P-PC4 was observed (Figure 2D, lane 5 and Figure 2E, lanes 6, 9 and 12). These observations indicate that C-terminal domain of linker H1 could be mediating its interaction with PM-PC4 and P-PC4. Based on these observations, it was important to understand the status of this interaction in a in vivo scenario. To study whether phosphorylation of PC4 mediates its interaction with linker histone H1 in cells, 3X-flag tagged mammalian constructs of wild type PC4 (PC4), phosphomimic PC4 (PM-PC4) and phospho-defective PC4 (MTP5) were ectopically expressed in the background of PC4 knockdown (Supplementary Figure S2B). The stable knockdown of PC4 confirmed by the GFP expression (Supplementary Figure S2B) was achieved by shRNA mediated silencing where the shRNA was targeted to the 3’UTR of the PC4 mRNA enabling us to overexpress the clones in the knockdown background (PC4 KD). We confirmed that the PC4 expression upon transfecting Flag-conjugated constructs of wild type (PC4), phosphomimic (PM-PC4) and phospho-mutant (MTP5) PC4 in PC4 knockdown cells, is at an equivalent level to shNS and higher than PC4 KD cells (Supplementary Figure S2D). Further, the nuclear localization of all these constructs (Supplementary Figure S2C) indicates that the results obtained from the transient transfection closely resemble the in vivo situation. Subsequently, through flag immuno-pull down of different PC4 constructs, it was observed that both wild type PC4 and PM-PC4 could efficiently interact with linker histone H1 (as shown by probing with either H1.2 variant specific or with pan H1 antibody) (Figure 2F and G respectively). The phospho-defective mutant MTP5 however failed to interact with linker histone H1 (Figure 2F and G). However, the expression of the flag tagged PC4, PM-PC4 and MTP5 proteins were found to be comparable (Figure 2H) which emphasizes that the alteration in the association of PC4 with linker histone H1 is phosphorylation driven.
Figure 2.
Phosphomimic PC4 (PM-PC4) interacts with linker histone H1. (A) The mobility pattern of PM-PC4, P-PC4, Mock P-PC4 and PC4 (wild type unmodified recombinant) in 15% SDS-PAGE stained with Coomassie. (B) Amino acid sequence of PC4 highlighting serine residues at 13, 15, 17 and 19 position mutated to aspartic acid, D, in PM-PC4 sequence (C) A diagrammatic representation of the different domains of linker histone H1.2 used for the interaction studies. (D) In vitro interactions were performed by Ni-NTA pull down using 250 ng of phosphorylated wild type PC4, PM-PC4 and phospho-defective mutant (MTP5) with His tagged linker histone H1.2 (E) In vitro interactions were carried out with different domains of His-tagged H1.2: N-terminal globular domain (ΔC), C-terminal globular domain (ΔN) and full length (FL) with PM-PC4, P-PC4 and MockP-PC4 by Ni-NTA pull down and probed with anti-His and anti-PC4 antibodies.(F and G) Immuno-pulldown using Flag antibody carried out in PC4, PM-PC4 and MTP5 expressing 293cells in PC4 knockdown background and probed with both H1.2 variant specific (F) as well as with pan H1 (G) antibody to detect H1 in the IP. (H) The expression of different Flag-tagged PC4 constructs in PC4 knockdown HEK293 cells were checked by anti-flag antibodies.
Phosphomimic PC4 promotes condensation of an in vitro reconstituted nucleosomal array
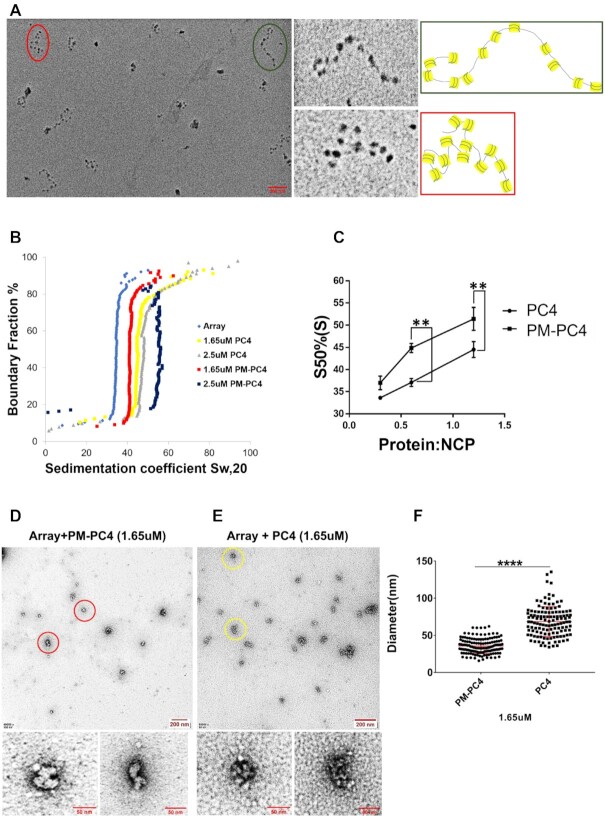
Upon establishing that phosphorylation of PC4 is essential for its interactions with the linker histone H1, both in vitro and in vivo, we wanted to address the role of PC4 phosphorylation in its chromatin compaction functions. At the onset we reconstituted a nucleosomal array using 12 × 177bp of 601 DNA and recombinant Xenopus core histones which was subsequently characterized by metal shadowing followed by Electron Microscopy (EM) imaging (Figure 3A). The EM images clearly showed presence of 12-nucleosome containing arrays with varying degrees of compaction. The close-up view of two such arrays encircled in green and red with the representation of their nucleosome positions alongside indicate the different degrees of compaction (Figure 3A). In order to study the effect of wild type unmodified recombinant PC4 or PM-PC4 protein-mediated nucleosomal compaction, the recombinant proteins were individually incubated with the nucleosomal array and subjected to sedimentation velocity analysis by analytical ultra-centrifugation or imaging by EM. The compaction of the nucleosomal array analysed by sedimentation velocity analytical ultracentrifugation showed that the PM-PC4 incubated nucleosomal array reaches higher sedimentation coefficient value as compared to the array incubated with wild type unmodified recombinant PC4 (by ∼7S) (Figure 3B). Further, concentration-dependent increase in compaction of the nucleosomal array mediated by PM-PC4 was observed (Supplementary Figure S3A and B). The sedimentation coefficient values at a 50% boundary fraction were plotted against the stoichiometric ratio of PC4 or PM-PC4 to the nucleosome core particle (NCP) is represented in Figure 3C. The data clearly indicates that PM-PC4 significantly increases the level of compaction of the nucleosome array in vitro than PC4 (wild type unmodified recombinant) (Figure 3B and C), despite the relative levels of both the proteins binding to the array being similar as seen in the SDS-PAGE (Supplementary Figure S3I). Upon visualizing the PM-PC4 or PC4 (wild type unmodified recombinant) incubated nucleosome array by EM imaging, we observed that in agreement with the sedimentation velocity analysis, the EM images also showed highly condensed nucleosome array formation in vitro by PM-PC4 as compared to PC4 (wild type unmodified recombinant) (Figure 3D, E). One of the condensed nucleosome arrays has been zoomed in to represent the highly compacted ordered arrays formed by PM-PC4 as compared to the unmodified recombinant PC4 (Figure 3D and E). To quantify the comparative effects of PM-PC4 and PC4 (wild type unmodified recombinant) on chromatin compaction, we measured the diameter of the condensed nucleosome arrays using ImageJ software, counting 20–30 particles from five different fields. The diameter of each condensed array was measured from its one end to the other using ImageJ software. The scale of the distance measured in ImageJ was set in reference to the scale bar of the image. The diameter distribution of the PM-PC4 bound array ranged between 30 and 60 nm, which was significantly lower than the unmodified recombinant PC4 indicating that more compacted arrays are formed by PM-PC4 (Figure 3F). The EM images of PM-PC4 versus PC4 (wild type unmodified recombinant) bound array indicates initiation of compaction by PM-PC4 at 1uM concentration (Supplementary Figure S3C versus S3D, Supplementary Figure S3K), which reaches maximal compaction at 1.65uM concentration. However, at these concentrations, PC4 (wild type unmodified recombinant) showed minimal effect on the nucleosome array (Figure 3D-F). EM images of nucleosome array bound with increasing concentration of PM-PC4 versus PC4 suggests that PM-PC4 forms ordered structures of smaller diameter till 2.5uM, unlike PC4 (Supplementary Figure S3E, F, J). However, upon increasing the molar concentration to 3.3uM PM-PC4 begins to form aggregated structures (Supplementary Figure S3G, H). Thus, these data collectively showed that PM-PC4 has a better compaction ability, forming highly compacted arrays, as compared to the wild type unmodified recombinant PC4.
Figure 3.
Phosphomimic PC4 promotes condensation of an in vitro reconstituted nucleosomal array. (A) EM image of nucleosomal arrays after metal shadowing. Two dodecameric arrays, marked in green and red circles within the overview EM image, shown in a close up view alongside. The models of representative 12 × 177 oligonucleosomes of the two indicated arrays with green and red borders shown alongside. (B) Sedimentation velocity analysis by analytical ultracentrifugation of PM-PC4 (phosphomimc PC4) and PC4 (wild type unmodified recombinant) incubated chromatin arrays. A nucleosome array without any ectopic addition of protein was used as a control. (C) Sedimentation coefficient values from panel C represented for the nucleosome core particle (NCP) containing increasing concentrations of PM-PC4 and PC4 at a 50% boundary fraction (n = 3). Data represent the means ± SEM. The S50%(S) were statistically analysed by Student's paired t-test (*P< 0.05,**P< 0.01,***P< 0.001, ns-non-significant). (D, E) EM images of PM-PC4 (phosphomimc PC4) and PC4 (wild type unmodified recombinant) incubated nucleosome array respectively. Two condensed arrays from each of the respective overview EM images have been encircled in red (for PM-PC4 bound array) and yellow (for PC4 bound array) circles and their close up view has been shown below. (F) Distribution of diameters (in nm) of PM-PC4 and PC4 bound compacted array particles. No. of particles measured = 142. Data represent the means ± SD. The S50%(S) were statistically analysed by Student's unpaired t-test (*P< 0.05,**P< 0.01,***P< 0.001, ns-non-significant).
Phosphorylation enhances PC4 mediated condensation of H1 bound array forming higher order structures
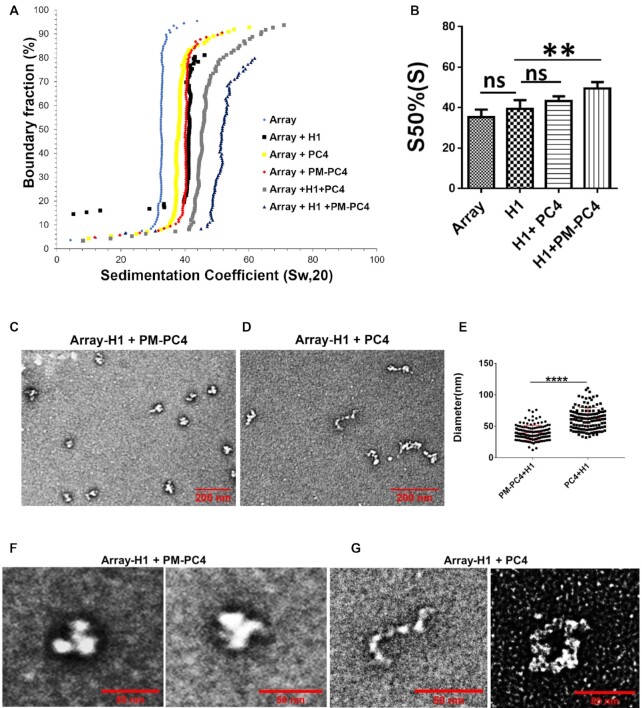
Linker H1 is an important component of the chromatin that not only stabilises the nucleosomal structure and spacing (3,34) but it also functions through interactions with other proteins which in turn brings about chromatin modification and regulates heterochromatin formation. Since we observed that linker histone H1 interacts with PC4 only when it is phosphorylated, we wanted to study whether phosphomimic PC4 (PM-PC4) and wild type unmodified recombinant PC4 (PC4) could complement linker H1 in mediating compaction of array into highly compacted fibers. Previous studies have already shown that H1 could condense nucleosome array into compact fibers when ratio of H1 to nucleosomal core particle (H1:NCP) is 1 (18). Therefore we selected H1-bound nucleosome array at a sub-optimal ratio, 0.5, of H1 to Nucleosome core particle (NCP), where it does not exhibit complete compaction (Supplementary Figure S4A and B) followed by complementation with either PM-PC4 or wild type unmodified recombinant PC4 at an optimal concentration of 1uM (Supplementary Figure S4C or D) that did not augment precipitation of the nulceosomal DNA. The sedimentation velocity analysis showed that PM-PC4 complements H1 acquiring higher sedimentation coefficient value as compared to PC4 (wild type unmodified recombinant) (by ∼6S) (Figure 4A, B). Thus, PM-PC4 could condense H1 containing array to form highly compacted chromatin fibers unlike PC4 (wild type unmodified recombinant) at the concentration used. EM imaging suggested that at the indicated concentrations, the proteins H1 (Supplementary Figure S4A-B), PM-PC4 and PC4 (Supplementary Figure S4C and D, respectively) could not form compacted fibers alone; but in presence of PM-PC4, H1 was able to form highly compacted fibers (Figure 4C, F and E) unlike with unmodified recombinant PC4 (Figure 4D, G and E). PM-PC4 further compacted the H1-bound array into condensed higher order structures (Figure 4C and F) resembling chromatin fibers, indicating that PM-PC4 is involved in higher order chromatin organisation. On the other hand, PC4 (wild type unmodified recombinant) was unable to further compact H1-bound array which still exhibited unfolded nucleosome array formation (Figure 4D and G, Supplementary Figure S4E). The corresponding SDS-PAGE of the arrays also indicate that PM-PC4 is bound to the array in the presence of linker H1 but not PC4 (wild type unmodified recombinant) (Supplementary Figure S4G). The diameter of PM-PC4 bound H1-array was significantly smaller than the unmodified recombinant PC4 bound H1-array (Figure 4E). PM-PC4 started aggregating H1-bound chromatin at a concentration (1.65 uM) where unmodified PC4 began to compact H1-bound array (Supplementary Figure S4F). These results indicate the ability of PM-PC4 to further compact and assist linker histone H1 to form higher order structures. Thus, phosphorylation assists PC4-mediated compaction function and thereby complementing another chromatin protein such as linker H1 to form higher order nucleosomal structures.
Figure 4.
Phosphorylation enhances PC4 mediated condensation of H1 bound array forming higher order structures. (A) Sedimentation velocity analysis by analytical ultracentrifugation of H1 bound to nucleosome array at 0.5 ratio (H1:NCP) with 1 uM of PM-PC4 (phosphomimic PC4) and PC4 (wild type unmodified recombinant). A nucleosome array without any any ectopic addition of protein was used as a control. (B) Sedimentation coefficient values from the panel A,at a boundary fraction of 50% (n = 3). Data represent the means ± SD.The S50%(S) were statistically analysed by ordinary one-way ANOVA, Sidak's multiple comparisons test as well as Student's paired t-test (*P< 0.05,**P< 0.01,***P< 0.001, ns, non-significant (C, D) EM images showing the overview of the structures formed upon PM-PC4 (phosphomimic PC4) (1 uM) and PC4 (wild type unmodified recombinant) (1 uM) mediated compaction of H1-array respectively. (E) Distribution of diameters (in um) of PM-PC4 (phosphomimic PC4) and PC4 (wild type unmodified recombinant) compacted H1 bound array particles. No. of particles measured = 132. Data represent the means ± SD. The S50%(S) were statistically analysed by Student's unpaired t-test (*P< 0.05,**P< 0.01,***P< 0.001,ns-non-significant). (F, G) The close up view showing the EM image of two H1 bound array compacted by 1uM of PM-PC4 and PC4 respectively.
Phosphorylation of PC4 regulates chromatin compaction and consequent histone modification status
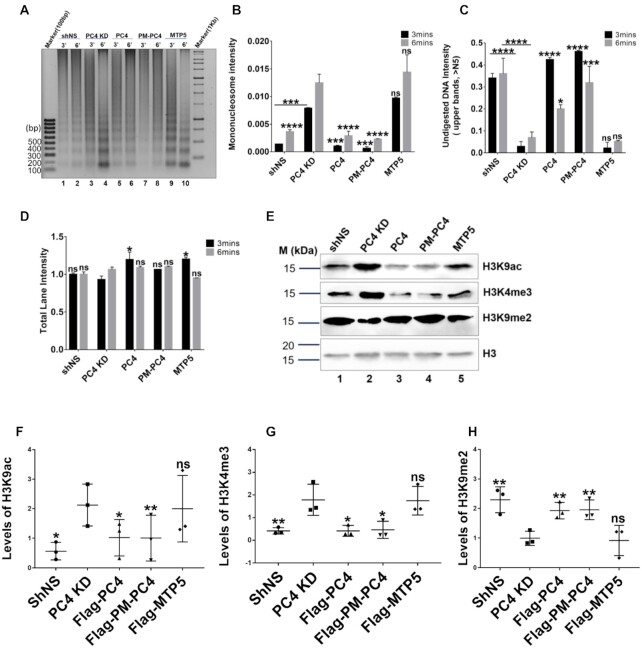
In order to understand the role of phosphorylation of PC4 in mediating chromatin condensation in vivo, we performed Micrococcal nuclease (MNase) accessibility assays of the cellular chromatin following stable transfection of 3X-flag tagged mammalian constructs of PC4, PM-PC4 and MTP5 in PC4 knockdown HEK293 cells. In agreement to our previous report, we found that knocking down of PC4 indeed results in chromatin decompaction, as revealed by more efficient digestion by MNase and generation of higher amount of mononucleosomes compared to the vector control cell line (shNS) (Figure 5A, lane 3 versus lane 1 and lane 4 versus lane 2 and Figure 5B, C). PC4 and PM-PC4 transfection in PC4 KD cells significantly induced chromatin compaction as revealed by higher intensity of undigested chromatin DNA (Figure 5A, lane 3 versus lanes 5 and 7 respectively, lane 4 versus lanes 6 and 8 respectively and Figure 5C) and lower mono-nucleosome intensity compared to PC4 KD cells (Figure 5A, lane 3 versus lanes 5 and 7 respectively, lane 4 versus lanes 6 and 8 respectively and Figure 5B). However, transfection of MTP5 into PC4 KD cells did not alter the MNase accessibility of PC4 KD cells (Figure 5A, lane 3 versus lane 9, lane 4 versus lane 10). This is evident from the intensity of mono-nucleosome (Figure 5B) and undigested chromatin DNA (Figure 5C) in MTP5 expressing cells compared to PC4 KD. The band intensity profile for each of the lanes shown in Supplementary Figure S5 clearly indicates higher intensity of mononucleosome for PC4 KD and MTP5 transfected cells, whereas the intensity of mononucleosome in shNS, PC4 and PM-PC4 transfected cells is lower (the band intensity profile pattern is from the top of the lane till the bottom of the lane, therefore the peak at the end corresponds to the mononucleosome) despite the total amount of digested chromatin DNA loaded onto the gel being similar across all the samples (Figure 5D).
Figure 5.
Phosphorylation of PC4 regulates chromatin compaction and consequent histone modification status. (A) PC4 knockdown 293 cells were transfected with the indicated plasmids and chromatin fractions were extracted and analyzed by micrococcal nuclease (MNase) sensitivity assay. (B) Quantification of band signal intensities corresponding to mono-nucleosome for every lane shown in (A) normalised to its total lane intensity (n = 2). (C) Quantifications of lane signal intensities corresponding to undigested genomic DNA (above the fifth band, >N5, starting from the bottom) for every lane shown in (A) normalised to its total lane intensity (n = 2). (D) Quantifications of total lane signal intensities in (A) (n = 2). Data represent the means ± SD. The data is statistically analysed by two-way ANOVA, Dunnett's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001,ns-non-significant). (E) Levels of different histone modifications checked by western blotting after transfecting PC4, PM-PC4 and MTP5 plasmids in PC4 knockdown cells. (F) Volcano plot of the differential ATAC–seq peaks between PC4 KD and PC4. (F–H) Quantification of levels of different histone modifications (n = 3). Data represent the means ± SD.The data is statistically analysed by ordinary one-way ANOVA, Sidak's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001,ns-non-significant).
Similar to our MNase sensitivity results, circular dichroism spectroscopy analysis showed significant decrease in the mean residue ellipticity value for linker H1 stripped HeLa cell chromatin upon incubation with P-PC4 compared to unmodified PC4 for 30 mins (Supplementary Figure S6A and B) implying greater condensation. These results indicate the probable role of PC4 phosphorylation in inducing chromatin compaction thereby maintaining higher order chromatin structure in vivo.
Since, analytical ultracentrifugation, electron microscopy and MNase sensitivity assays showed that phosphorylation of PC4 is critical for maintaining the condensed state of chromatin both in vitro and in cells, we were interested to find out whether phospho-PC4, is also required for maintaining the histone modification status in cells. Knocking down (KD) of PC4 exhibited alteration in the epigenetic state of the cell showing enhanced levels of activation marks like histone H3 lysine9 acetylation (H3K9ac) (Figure 5E, F), histone H3 lysine 4 trimethylation (H3K4me3) (Figure 5E and G). Concurring with greater chromatin decompaction observed in PC4 KD cells (Figure 5A–C), they also exhibited significant reduction in heterochromatin mark, histone H3 lysine di-methylation (H3K9me2) (Figure 5E and H). The H3K9ac and H3K4me3 are the signatures of transcriptional active marks present at the gene promoters. The activation marks, H3K9ac and H3K4me3, remarkably reduced upon transfection of PC4 and PM-PC4 (Figure 5E–G) as compared to PC4 KD cells but not in cells transfected with MTP5 (Figure 5E-G). The heterochromatin mark H3K9me2, which is indicative of more condensed chromatin state, significantly increased in PC4 and PM-PC4 transfected PC4 KD cells but not in case of MTP5 transfection (Figure 5E and H). Thus, transfecting PC4 and PM-PC4 in PC4 KD cells, significantly restored the histone modification state of the PC4 KD cells (Figure 5E–H). In contrast, MTP5, the phospho-deficient mutant PC4 could not alter the histone modification marks significantly (Figure 5E–H). These results thus clearly indicate the significant role of PC4 phosphorylation in maintaining chromatin compaction and thereby mediating an epigenetically repressive chromatin state inside the cells.
Earlier studies suggested that PC4 interacts preferentially with histone H3 and H2B which might contribute to PC4 mediated chromatin condensation (7). Therefore, we studied the effect of PC4 phosphorylation on its interaction with core histones. Flag immuno-pull down from chromatin fraction enriched in histones, showed that PC4 and PM-PC4 exhibited greater interaction with histone H3 and H2B (Supplementary Figure S6C, lane 1 and lane 2) whereas MTP5 showed no interaction with H2B upon respective pull-down assays (Supplementary Figure S6C, lane 3). Perhaps MTP5 does not compact the chromatin efficiently like PC4 and PM-PC4 (Figure 7A–C, F) as a result of its differential interaction with both core histones and linker H1. These results collectively suggest that in the cellular context phosphorylation of PC4 might have greater impact than just interaction with linker histone H1, resulting in the compaction of the chromatin.
Figure 7.
Phosphorylation of PC4 affects gene transcription by inhibiting chromatin accessibility. (A, B) Integrative Genomics Viewer (IGV) images showing ATAC-seq intensities in shNS, PC4 KD, PC4, PM-PC4 and MTP5 cells, on representative loci of two autophagy genes UVRAG and MAP3K7 respectively along with the published histone modification ChIP seq tracts in HEK293 cells (81–83). (C) Integrative Genomics Viewer (IGV) images showing ATAC-seq intensities in shNS, PC4 KD, PC4, PM-PC4 and MTP5 cells, on representative loci of the acetylated histone reader and regulator, YEATS2, along with the published histone modification ChIP seq tracts in HEK293 cells (81–83). (D) Relative expression of UVRAG, IPMK, MAP3K7 and YEATS2 in PC4 KD and PC4 knockdown cells stably-transfected with flag tagged PC4, PM-PC4 or MTP5 along with shNS (vector control cells) determined by real-time PCR. All data represent the means ± SD for n = 3. The data is statistically analysed by ordinary one-way ANOVA, Sidak's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001, ns, non-significant). (E) ChIP-qPCR to analyze the fold enrichment for Flag tagged PC4, PM-PC4 and MTP5 over the differentially depleted ATAC-seq peaks of the corresponding genes; UVRAG, IPMK and MAP3K7. Flag antibody was used to pull down sonicated chromatin. All the values of fold enrichment were normalized to IgG control and HNF4A gene was taken as a negative control chromatin region. All data represent the means ± SD for n = 3. The data is statistically analysed by ordinary one-way ANOVA, Sidak's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001, ns, non-significant). (F) Integrative Genomics Viewer (IGV) images showing ATAC-seq intensities in shNS, PC4 KD, PC4, PM-PC4 and MTP5 cells, on representative loci of IPMK, a PI3 Kinase and transcription co-activator regulating nuclear signaling events, along with the published histone modification ChIP seq tracts in HEK293 cells (81–83). (G) Relative expression of pro-autophagy genes ULK1, ULK2, AMPKα1, AMPKα2 and DRAM1 that are regulated by MAPK signalling and IPMK regulated PI3kinase-Akt signalling and (H) Relative expression of neural genes SCN2, M4 and GAD1 along with PC4 gene expression in PC4 knockdown cells stably-transfected with flag tagged PC4, PM-PC4 or MTP5 along with PC4 KD and shNS (vector control cells) determined by real-time PCR. All data represent the means ± SD for n = 3. The data is statistically analysed by ordinary one-way ANOVA, Sidak's multiple comparisons test (*P< 0.05,**P< 0.01,***P< 0.001, ns, non-significant).
Phosphorylation of PC4 affects chromatin accessibility of various genomic regions harboring diverse histone marks
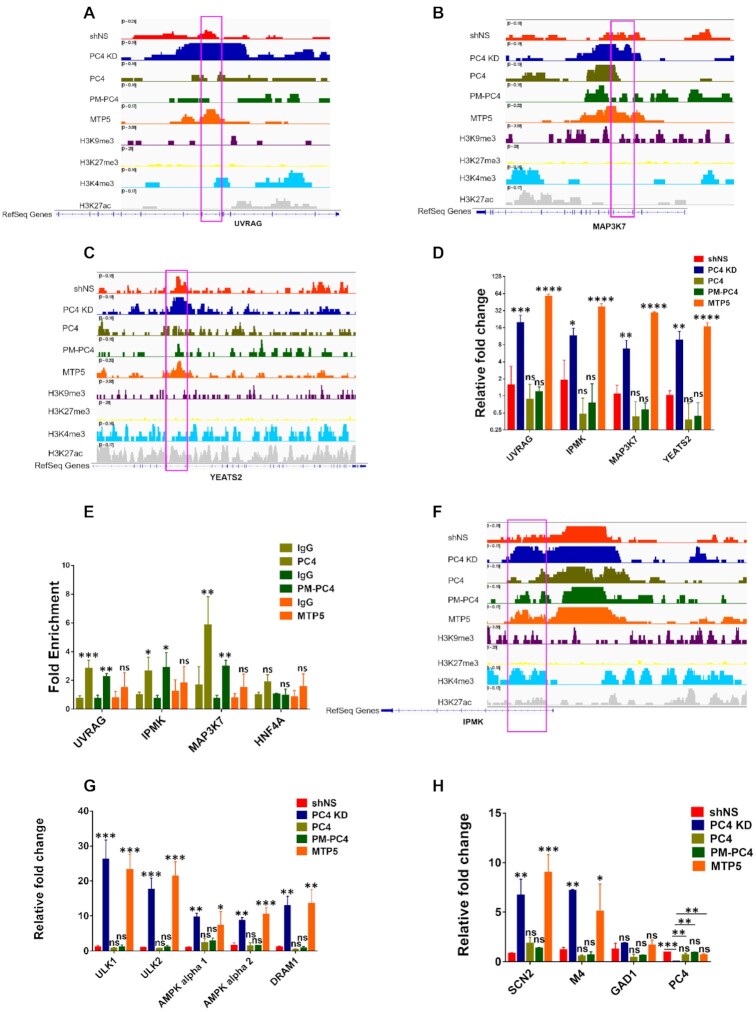
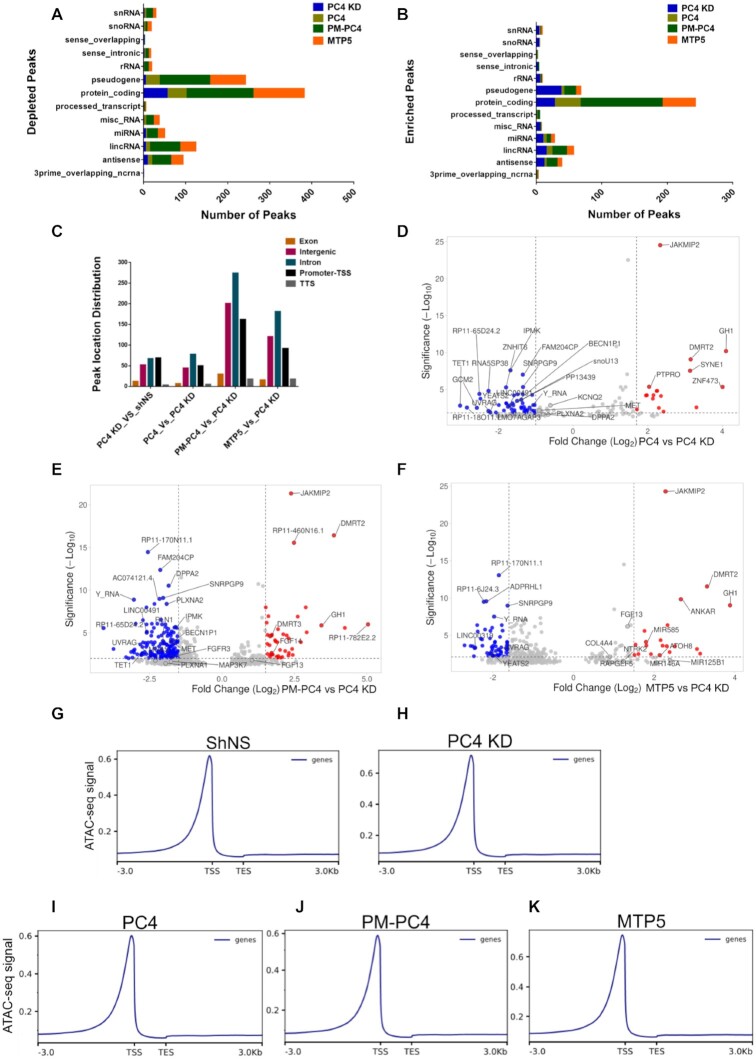
Based on the compaction studies of both nucleosome template as well as on chromatin DNA assessed by MNase sensitivity and CD spectroscopy, it was evident that phosphorylation regulates the chromatin compaction function of PC4. After exploring the role of PC4 phosphorylation through biochemical approach, we wanted to address its impact on the genomic organisation and its physiological implications. Therefore, we carried out ATAC-seq of the PC4, PM-PC4 and MTP5 expressing PC4 KD cells along with PC4 KD and shNS cells. The intensity of the ATAC-seq peaks obtained for PC4 KD cells were compared to shNS cells to assign the differential ATAC-seq peaks. The intensity of the ATAC-seq peaks obtained for PC4, PM-PC4 and MTP5 expressing in PC4 KD cells were compared to PC4 KD cells to assign the differential ATAC-seq peaks. Gene-type annotation of the differentially accessible peaks in PC4, PM-PC4 and MTP5 expressing cells indicates that besides protein coding genes, the chromatin of a substantial number of non-coding genes including long intergenic non-coding RNAs (lincRNAs), microRNA (miRNAs), anti-sense RNAs along with a smaller proportion of small nucleolar RNAs (snoRNAs) and small nuclear RNA (snRNA) are regulated by phosphorylation of PC4 (Figure 6A-B). The genomic distribution of all the differentially accessible ATAC-seq peaks in PC4 KD, PC4, PM-PC4 and MTP5 expressing cells is shown in Figure 6C. Interestingly, PC4 regulates chromatin opening majorly in the intronic regions, followed by promoter-TSS and intergenic chromatin (Figure 6C). The open chromatin profile of PC4, PM-PC4 and MTP5 transfected PC4 KD cells shows higher number of ATAC-seq peaks that are significantly depleted than enriched with respect to PC4 KD (adjusted P < 0.05) log2(Fold change) < –1.5) (Figure 6D–F). However, the ATAC-seq read density profiles imply lesser accessibility of the chromatin upon PC4 and PM-PC4 expression in PC4 KD cells (Figure 6I, J) unlike upon MTP5 expression (Figure 6K). The ATAC-seq read density profiles of PC4 and PM-PC4 transfected PC4 KD cells show comparable ATAC-seq signal to shNS cells (Figure 6I, J and G) but lower than PC4 KD cells (Figure 6H). The ATAC-seq read density profile of MTP5 and PC4 KD cells depict comparable ATAC-seq signal (Figure 6H and K), which is lower than shNS cells (Figure 6G, H and K). Taken together, these results show that phosphorylation of PC4 reduces chromatin accessibility of the genome. The differentially accessible ATAC-seq peaks in PC4, PM-PC4 transfected cells revealed autophagy associated genes (UVRAG, IPMK, MAP3K7, BECN1P1) to be more compacted relative to PC4 KD (Figure 6D, E). A list of the top 15 significantly altered ATAC-seq peaks in PC4, PM-PC4 and MTP5 expressing PC4 KD cells show that chromatin regions of various non-coding RNA genes (Y_RNA, LINC30019, MIR585) are affected in PC4 phosphorylation dependent manner (Supplementary Figure S7A–C). There are, however, several overlapping chromatin regions compacted by all the three proteins (PC4, PM-PC4 and MTP5) as well as some unique chromatin regions that are exclusive to each cell type (Figure 6D–F and Supplementary Figure S7A–C).
Figure 6.
Phosphorylation of PC4 affects chromatin accessibility of various genomic regions harbouring diverse histone marks. (A) Gene type annotation of the differentially depleted ATAC-seq peaks assessed in PC4 KD with respect to shNS and assessed in PC4, PM-PC4 and MTP5 expressing knockdown cells with respect to PC4 KD. (B) Gene type annotation of the differentially enriched ATAC-seq assessed in PC4 KD with respect to shNS and assessed in PC4, PM-PC4 and MTP5 expressing knockdown cells with respect to PC4 KD. (C) Annotation of the differential ATAC-seq peaks in PC4 KD, PC4, PM-PC4 and MTP5 expressing PC4 KD cells across the different functional regions of the genome. (D) Volcano plot of the differential ATAC–seq peaks obtained in PC4 expressing knockdown cells in comparison to PC4 KD. (E) Volcano plot of the differential ATAC–seq peaks obtained in PM-PC4 expressing knockdown cells in comparison to PC4 KD. (H) Volcano plot of the differential ATAC–seq peaks obtained in MTP5 expressing knockdown cells in comparison to PC4 KD. The blue and red dots in (D–F) represent the differential ATAC-seq peaks whose read density decreased and increased respectively by 1.5-fold or more (adjusted P < 0.05 and log2(Fold change) > 1.5 or log2(Fold change) < –1.5). The top 10 significantly altered ATAC-seq peaks for each of the comparisons in (D–F) has been marked. Additionally, ATAC-seq peaks that are associated with autophagy and are unique to each comparison have been marked in (D-F). These genes are referred later in the subsequent sections. (G–K) Average ATAC-Seq read density profiles of shNS, PC4 KD, PC4, PM-PC4 and MTP5 respectively showing normalized RPKM ± 3Kb of TSS/average gene peak centers.
We have observed that absence of PC4 and also expression of PM-PC4 (in PC4 knockdown cells) alter the histone modification status in cells (Figure 5E–H), therefore we further characterised the genomic regions compacted by PC4, PM-PC4 and MTP5 upon overlapping with chromatin bound peaks of a transcriptional activation mark, H3K4me3; an enhancer mark, H3K27ac and a heterochromatin mark, H3K9me3 from the available ChIP-seq data of HEK293 cells. Remarkably, around 40 percent of the PC4, PM-PC4 and MTP5 regulated genomic regions overlapped with merely 5% of the global histone modification marks: H3K4me3, H3K9me3 and H3K27ac (Supplementary Figure S7D–F). Around 25–30% of the genomic regions where the chromatin opening is reduced by PC4, PM-PC4 and MTP5 accounts for half of the overlap or around 2.5% of the global histone marks (H3K4me3, H3K9me3 and H3K27ac) as represented in the Venn diagram representation (Supplementary Figure S7G–I). These findings are suggestive of two important implications: firstly, PC4 phosphorylation regulated chromatin regions harbor both heterochromatin (H3K9me3) as well as active (H3K4me3) chromatin marks (Supplementary Figure S7D–I) and secondly, PC4 phosphorylation may affect the histone modification state predominantly in the intronic regions, followed by intergenic and promoter–TSS regions. Intronic chromatin and more specifically the first introns which constitutes majority of the phospho-PC4 depleted peaks (Figures 6C, 7B–F and Supplementary Figure S8B), is known to harbor higher density of chromatin marks affecting gene expression (35) as also observed in this study Figure 7D.
These results collectively suggest that PC4 phosphorylation impacts genome organisation through compaction of both protein coding and non-coding genes.
Phosphorylation of PC4 affects gene transcription by inhibiting chromatin accessibility
The open chromatin profile assayed by ATAC-sequencing revealed that phosphorylation of PC4 plays an important role in affecting accessibility of diverse chromatin regions and were supported by MNase sensitivity studies and histone modification levels. We were therefore, interested to study the implications of phospho-PC4 mediated chromatin compaction on gene expression. Gene Set Enrichment Analysis (GSEA)-defined KEGG pathway analysis obtained from mapping the differential ATAC-seq peaks present in PC4 and PM-PC4, but absent in MTP5, indicated that several signaling pathways like autophagy, MAPK signaling, metabolic pathways as well as microRNAs contributing to tumorigenesis are significantly less accessible as compared to PC4 KD (Supplementary Figure S8A). This is also observed in the IGV images showing reduced ATAC-seq intensities of two important regulators of autophagy; UVRAG and MAP3K7 (36,37) in shNS, PC4 and PM-PC4 cells compared to PC4 KD and MTP5 (Figure 7A-B). We also observed similar accessibility differences in the chromatin region of IPMK gene (Figure 7F) and a core autophagy gene, ATG5 (Supplementary Figure S8B). IPMK encodes for inositol polyphosphate multi-kinase which is a critical metabolic sensor that directly integrates glucose signaling to the activation of the AMPK pathway (38). IPMK has been reported to enhance autophagy-related transcription by stimulating AMPK-dependent Sirt-1 activation (39,40). We observed that intronic region of IPMK gene is indeed more closed in PC4 and PM-PC4 transfected knockdown cells as compared to PC4 KD and MTP5 (Figure 7F) cells. Besides autophagy and signaling pathway associated genes, we observed phosphorylation dependent reduction in the chromatin accessibility of an epigenetic regulator, YEATS2 gene (Figure 7C). YEATS2 binds to acetylated histone H3 via its YEATS domain is part of the ATAC complex and is required for the maintenance of the global H3K9ac levels in cells and knockdown of YEATS2 revealed marked reduction in global H3K9ac levels (41). Both PC4 and PM-PC4 expression not only exhibited greater compaction of YEATS2 gene (Figure 7C) but also resulted in YEATS2 gene repression (Figure 7D) which is induced in PC4 KD and MTP5 expressing KD cells (Figure 7D).
Gene expression of the representative chromatin regions which are compacted in PC4 and PM-PC4 but not in MTP5 expressing PC4 KD cells as compared to PC4 KD indicated that phosphorylation dependent chromatin compaction resulted in gene repression (Figure 7D). On the other hand, MTP5 showed minimal effect on the chromatin accessibility as compared to PC4 KD, thus inducing the expression of these genes (Figure 7D). The compaction efficiency of PC4 and PM-PC4 cells was correlative to their significant enrichment on to these chromatin regions unlike MTP5, as indicated by ChIP-PCR conducted upon immunoprecipitating Flag tagged-PC4/PM-PC4/MTP5 bound chromatin using Flag antibody (Figure 7E). However, no significant enrichment was observed for PC4, PM-PC4 and MTP5 for a gene, HNF4A, that is not differentially compacted by either of these three constructs expressing cells (Figure 7E).
We argued that the implications of PC4 phosphorylation-mediated chromatin compaction was not only imposed on the expression of the genes whose chromatin regions were inaccessible upon PC4 and PM-PC4 expression, but the effect could be indirectly translated to downstream event(s). To this end, we studied the expression of pro-autophagy genes ULK1, ULK2, AMPKα1, AMPKα2 and DRAM1 which are directly regulated by IPMK as well as MAPK signalling pathways (40,42,43). IPMK also act as a transcriptional co-activator of p53-dependent transcription in inducing cell death (44) and p53 has been shown to upregulate ULK1 and ULK2 gene expression in response to DNA-damage induced autophagy (45). We found that the expression of the pro-autophagy genes is indeed induced upon PC4 KD or MTP5 expression but repressed significantly by PC4 and PM-PC4 which were comparable to the levels of vector control cells (shNS) (Figure 7G).
Earlier studies have shown the involvement of PC4 in REST-CoREST-mediated repression of neuronal gene expression in non-neuronal cells (46). Silencing of PC4 has been shown to induce histone acetylation at the promoter of neural specific gene, sodium channel 2 (SCN2) and showed reduced REST-CoREST occupancy at the SCN2 promoter (46). Here as we observe that phosphorylation of PC4 is involved in chromatin compaction and knocking down of this protein induces histone H3 acetylation at lysine residue 9 (H3K9ac) (Figure 5E-F). We observed that SCN2 and M4 genes’ expression were significantly upregulated in PC4 KD and MTP5 expressing PC4 KD cells as compared to shNS but showed no significant difference with PC4 and PM-PC4 expressing PC4 KD cells. However, upon transfection of MTP5 in PC4 KD cells SCN2 and M4 expression remained significantly high as compared to shNS cells, suggesting that the repression of these neural genes occurred in a PC4-phosphorylation dependent manner (Figure 7H). However, such significant differences upon complementing PC4 and PM-PC4 was not observed for another REST target gene GAD1, whose expression remains unchanged upon PC4 KD (Figure 7H). Besides expression of the autophagy genes that are regulated as a result of the differential compaction by phospho-PC4 versus phospho-defective mutant PC4, there are also these neural genes that are repressed due to differential interaction of PC4 with another heterochromatin protein HP1α (data not shown).
To establish the functional link between the differential chromatin opening of the core autophagy genes UVRAG (Figure 7A), ATG5 (Supplementary Figure S8B), the expression results of the downstream pro-autophagy genes (Figure 7D) and the cellular autophagy status, we looked into the Microtubule-associated proteins 1A/1B light chain 3B (LC3) and Sequestosome-1 (p62) levels (Supplementary Figure S8C). The conversion of LC3I to lipidated LC3II form reflects the induction of the autophagy process, which was upregulated in PC4 KD cells but decreased upon expressing Flag tagged PC4, PM-PC4 (Supplementary Figure S8C, lane 2 versus lanes 3 and 4) but not by MTP5 (Supplementary Figure S8C, lane 2 versus lane 5). The degradation of the autophagosomal adaptor protein, p62, which is indicative of the autophagic flux, was found to be significantly higher in PC4 and PM-PC4 expressing knockdown cells compared to PC4 KD but comparable to shNS cells. However, this did not happen in MTP5 expressing knockdown cells. MTP5 expression in PC4 knockdown background resulted in similar levels of p62 expression as compared to PC4 KD cells (Supplementary Figure S8C, lane 2 versus lane 5). Thus, these results indicate that both induction of autophagy (LC3II) as well as autophagic flux (p62) is regulated by phosphorylated PC4 (Supplementary Figure S8C) through chromatin compaction, wherein its interactions with other chromatin proteins may play a critical role (Figure 8).
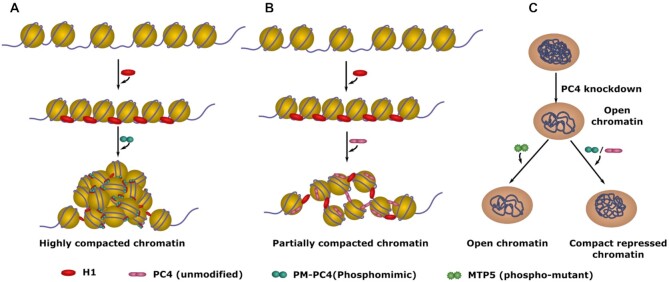
Figure 8.
Model depicting PC4 mediated chromatin organisation upon phosphorylation. (A) Nucleosome array bound by linker H1 folds into highly compacted chromatin in the presence of Phosphomimic-PC4 (PM-PC4) (B) Nucleosome array bound by H1 forms partially folded array in the presence of unmodified PC4. (C) Silencing of PC4 leads to opening of the chromatin and subsequent gene activation in cells. This phenomenon is rescued upon expressing PC4, that is phosphorylated inside cells, and PM-PC4, as they mediate chromatin compaction and reinstate the repressed chromatin state. On the other hand, expression of Phospho-mutant PC4 (MTP5) fails to compact the chromatin and repress the genome, activated upon PC4 knockdown.
DISCUSSION
Post-translational modifications of proteins often act as the fine tuners of their functions which result in greater implications on the cellular processes. Phosphorylation of histones is one such example that has impacted chromatin structure in response to DNA damage (47) or during meiosis and mitosis (48–51). Phosphorylation of linker H1 has been shown to assists RNAP I and RNAP II-dependent transcription (52–54). Here, the implications of phosphorylation of a non-histone chromatin protein, human positive coactivator 4, PC4, has been addressed. We were intrigued by the observation that despite phosphorylation of PC4 negatively impacting its coactivation function (9,16,17), it is majorly present in the phosphorylated state inside the cells. The present study shows that phosphorylation of PC4 could be critically linked to genome compaction and organization. One of the significant observations made in this study is that phosphorylation is crucial for its interaction with chromatin protein, linker histone H1 (Figure 1D and Supplementary Figure S1A, B), and its different variants (Figure 1F–H) (with a Kd value of 5nM as shown by ITC) (Supplementary Figure S1C). The phosphorylation dependent interaction of PC4 with both H1 and core histones (Supplementary Figure S6C) is an attributing factor in defining PC4 mediated chromatin compaction as observed in in vitro nucleosomal array compaction assays as well as in cellular system.
Our ATAC-seq data suggest that PC4 regulates chromatin opening majorly in the intronic regions, followed by promoter-TSS and intergenic chromatin (Figure 6C). A recent study shows that dynamic intron retention (IR) regulates gene expression and increased chromatin accessibility contributes substantially to this in a cell-specific manner (55). The importance of genomic introns is slowly emerging as they serve as repositories for cis elements, participating in transcriptional regulation and genome organization (56,57). Several specific intron-hosted DNA elements such as enhancers (58–61), or silencers (58,62,63) have been identified that modulate the function of the main upstream promoter (64,65). Such regulatory elements are found within the first intron (58–66) which also exhibits the highest density of regulatory chromatin marks (35). Interestingly majority of the phospho-PC4 mediated chromatin compaction occurred within the first intron. We observe diverse histone modification marks to be present on the intronic chromatin compacted by PC4 phosphorylation (Figure 7B, C, Supplementary Figures S7F and S8B). Introns carry cis-acting elements; such as intronic splicing silencers and intronic splicing enhancers which regulates alternative splicing (67,68). Introns are also associated with nonsense-mediated decay (NMD) of mRNAs (69). Genome compaction by a non-histone protein like PC4 may have serious implications on alternative splicing and mRNA processing which needs to be explored further.
The differential ATAC-seq peaks indicate several miRNA genes, causally linked to tumorigenesis, are specifically enriched in PC4 knockdown MTP5 expressing cells. These miRNA genes include; MIR585, MIR146A and MIR125B1 (70–72) (Figure 6F and Supplementary Figure S7C). There are non-coding RNA genes (LINC00491) that are more inaccessible in wild type-PC4 and PM-PC4 expressing knockdown cells (Figure 6D, E, Supplementary Figure S8A). Our ATAC-seq data suggests that phosphorylation of PC4 could indeed influence chromatin compaction of wide array of genes (Figure 6A, B), which paves way for exploring non-coding RNA mediated regulations of several biological processes in specific cellular context. Collectively these observations suggest that phospho-PC4 mediated chromatin compaction (organization) may not be random. There could be phospho-PC4 interactome involved in the preferential compaction of the genome.
The multifunctional chromatin protein, PC4, presumably play critical roles in differentiation and pluripotency (73–75). Phospho-PC4 mediated chromatin compaction underscores the necessity of an overwhelming majority of the phosphorylated form of PC4 in vivo, with the possibilities of its involvement in determining the cellular fate by regulating the chromatin state. PC4 regulates genes that are associated with stabilization phase of reprogramming like Dppa2, Dppa4 and Gdf3 (74). ATAC-sequencing has revealed the chromatin regions of several such pluripotency and reprogramming factors like DPPA2, FGF13, FGF14 and SOX11 (76–78) to be differentially regulated by phospho-PC4 (Figure 6D–F and Supplementary Figure S7A–C).
Our study further showed that several of the differentially regulated chromatin regions in wild type PC4 and PM-PC4 expressing cells involve signaling genes like MAP3K7 and IPMK (Figure 7B and F). We observed that phosphorylation not only induces better compaction of these chromatin regions but also relays repression of these genes (Figure 7D). On the other hand, expression of phospho-defective mutant of PC4, MTP5, in knockdown background still shows sufficient chromatin opening in these genomic regions (Figure 7B and F), thus, favoring a transcriptionally active chromatin state (Figure 7D). It may be reasonable to argue that the absence of phosphorylation could result in inefficient chromatin binding of MTP5 (Figure 7E), possibly due to weaker or no interaction with histones or H1.
The neural genes SCN2 and M4 which were overexpressed upon PC4 knockdown was observed to be repressed upon complementing PM-PC4 and PC4, but not by MTP5 (Figure 7H). The repression of these genes could be a result of phospho-PC4 mediated compaction of the chromatin loci of several neuronal pathway associated genes (Supplementary Figure S8A). However, the role of other repressive factors interacting with phosphorylated PC4 cannot be ruled out. Similarly, some of the pro-autophagy genes which were upregulated upon PC4 silencing, were repressed in a PC4 phosphorylation dependent manner (Figure 7G). These are downstream effectors of genes like IPMK, MAP3K7, or UVRAG. Phospho-PC4 reduces the chromatin opening of the intronic region of an epigenetic reader and regulator, YEATS2 (Figure 7C). Downregulation of YEATS2 expression by PC4 phosphorylation (Figure 7D) could attribute to the reduction in the global H3K9ac levels observed upon knockdown of PC4 (41) (Figure 5E, F). Our findings show that wild type PC4 and PM-PC4 expression in the PC4 knockdown background not only restores genomic compaction but also the global histone modification status (Figure 5E-H), thereby creating a repressive chromatin state.
We establish a functional link between phospho-PC4 mediated compaction of upstream regulatory genes (Figure 7A, B, F and Supplementary Figure S8B) and a downstream cellular process, autophagy (Supplementary Figure S8A). PC4 phosphorylation reduces the autophagy levels in the basal state of the cell which is upregulated upon PC4 knockdown (Supplementary Figure S8C). Thus, phosphorylation assists in the negative regulation of autophagy by PC4. A recent study has revealed that PC4 accumulation occurs during ageing and accelerates the ageing process via inhibition of autophagy (79). The implications of PC4 phosphorylation on cellular autophagy could affect the process of natural ageing and would be an interesting axis to the regulation of this newly discovered function of PC4 (79).
PC4 undergoes multiple post-translational modifications (PTMs), these include phosphorylation by CKII (9), Aurora Kinase B (80) and acetylation by lysine acetyl transferase p300 (13–16) and possibly by other KATs too. In this study we address the CKII mediated phosphorylation which promotes its interaction with both linker histone H1 and core histones. On the other hand, p300 mediated acetylation induces its double stranded DNA binding, its interactions with tumor suppressor, p53 and thereby its co-activation function (14–16). Interestingly, phosphorylation of PC4 by CKII but not Aurora kinase B inhibits its acetylation mediated by p300 (14) and counters the activities of PC4 that are induced upon acetylation (9,14–17). Emerging evidences of cross-talks of PC4 post-translational modifications (14,16) suggest that the dynamic nature of chromatin organisation in signal dependent genome function, could be critically regulated by the highly abundant nuclear protein, PC4. The different players (factors): such as phosphatases and deacetylases are yet to be identified. The cross-talks of these modifications in the physiological context could be highly significant.
DATA AVAILABILITY
The ATAC-seq data for this study are available in the GEO database under accession number GSE191333. The ChIP seq datasets used in this study are available from the GEO database with the following accession numbers: GSE66530 for H3K4me3 and H3K9me3 ChIP, GSM2711409 for H3K27ac ChIP. UCSC genome browser links for the genomic tracts are as follows: UVRAG https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq1 MAP3K7 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq2 YEATS2 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq3 ATG5 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq4 IPMK https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq5 IPMK https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq6.
Supplementary Material
Contributor Information
Pallabi Mustafi, Transcription and Disease Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, India.
Mingli Hu, National laboratory of Bio-macromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Science, Beijing 100101, China.
Sujata Kumari, Transcription and Disease Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, India.
Chandrima Das, Transcription and Disease Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, India; Biophysics and Structural Genomics Division, Saha Institute of Nuclear Physics, 1/AF Bidhannagar, Kolkata 700064, India.
Guohong Li, National laboratory of Bio-macromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Science, Beijing 100101, China.
Tapas K Kundu, Transcription and Disease Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, India; Division of Neuroscience and Ageing Biology, CSIR-Central Drug Research Institute, Sitapur Road, Sector 10, Jankipuram Extension, Lucknow 226031, India.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JNCASR, Department of Biotechnology, Government of India; Kundu T.K. is a recipient of Sir J.C. Bose National Fellowship; Mustafi P. is supported by JNCASR integrated PhD fellowship; JNCASR intramural funds to Kundu T.K.; Li G. supported by HHMI fellowship. Funding for open access charge: Sir J.C. Bose National Fellowship (to T.K.K.).
Conflict of interest statement. None declared.
REFERENCES
- 1. van Driel R., Fransz P.F., Verschure P.J.. The eukaryotic genome: a system regulated at different hierarchical levels. J. Cell Sci. 2003; 116:4067–4075. [DOI] [PubMed] [Google Scholar]
- 2. Berretta J., Morillon A.. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009; 10:973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson P.J.J., Rhodes D.. Structure of the “30 nm” chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006; 16:336–343. [DOI] [PubMed] [Google Scholar]
- 4. Lu X., Wontakal S.N., Emelyanov A.v, Morcillo P., Konev A.Y., Fyodorov D.v, Skoultchi A.I.. Linker histone H1 is essential for drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009; 23:452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A.. Transcriptional repression by the methyl-CpG-binding protein mecp2 involves a histone deacetylase complex. Nature. 1998; 393:386–389. [DOI] [PubMed] [Google Scholar]
- 6. Reeves R. HMG nuclear proteins: linking chromatin structure to cellular phenotype. Biochim. Biophys. Acta - Gene Regul. Mech. 2010; 1799:3. [Google Scholar]
- 7. Das C., Hizume K., Batta K., Kumar B.R.P., Gadad S.S., Ganguly S., Lorain S., Verreault A., Sadhale P.P., Takeyasu K.et al.. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 2006; 26:8303–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sikder S., Kumari S., Mustafi P., Ramdas N., Padhi S., Saha A., Bhaduri U., Banerjee B., Manjithaya R., Kundu T.K.. Nonhistone human chromatin protein PC4 is critical for genomic integrity and negatively regulates autophagy. FEBS J. 2019; 286:4422–4442. [DOI] [PubMed] [Google Scholar]
- 9. Ge H., Zhao Y., Chait B.T., Roeder R.G.. Phosphorylation negatively regulates the function of coactivator PC4. Proc. Nat. Acad. Sci. U.S.A. 1994; 91:12691–12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garavís M., Calvo O.. Sub1/PC4, a multifaceted factor: from transcription to genome stability. Curr. Genet. 2017; 63:1023–1035. [DOI] [PubMed] [Google Scholar]
- 11. Pan Z.Q., Ge H., Amin A.A., Hurwitz J.. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J. Biol. Chem. 1996; 271:22111–22116. [DOI] [PubMed] [Google Scholar]
- 12. Wang J.-Y., Sarker A.H., Cooper P.K., Volkert M.R.. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 2004; 24:6084–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M.. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994; 78:525–534. [DOI] [PubMed] [Google Scholar]
- 14. Kumar B.R.P., Swaminathan V., Banerjee S., Kundu T.K.. p300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphorylation. J. Biol. Chem. 2001; 276:16804–16809. [DOI] [PubMed] [Google Scholar]
- 15. Banerjee S., Kumar B.R.P., Kundu T.K.. General transcriptional coactivator PC4 activates p53 function. Mol. Cell. Biol. 2004; 24:2052–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Batta K., Kundu T.K.. Activation of p53 function by human transcriptional coactivator PC4: role of protein-protein interaction, DNA bending, and posttranslational modifications. Mol. Cell. Biol. 2007; 27:7603–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jonker H.R.A., Wechselberger R.W., Pinkse M., Kaptein R., Folkers G.E.. Gradual phosphorylation regulates PC4 coactivator function. FEBS J. 2006; 273:1430–1444. [DOI] [PubMed] [Google Scholar]
- 18. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.M., Zhu P., Li G.. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014; 344:376–380. [DOI] [PubMed] [Google Scholar]
- 19. Bednar J., Garcia-Saez I., Boopathi R., Cutter A.R., Papai G., Reymer A., Syed S.H., Lone I.N., Tonchev O., Crucifix C.et al.. Structure and dynamics of a 197 bp nucleosome in complex with linker histone h1. Mol. Cell. 2017; 66:384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syed S.H., Goutte-Gattat D., Becker N., Meyer S., Shukla M.S., Hayes J.J., Everaers R., Angelov D., Bednar J., Dimitrov S.. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Nat. Acad. Sci. U.S.A. 2010; 107:9620–9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R.. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001; 7:729–739. [DOI] [PubMed] [Google Scholar]
- 22. Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R.. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 2005; 280:38090–38095. [DOI] [PubMed] [Google Scholar]
- 23. Hale T.K., Contreras A., Morrison A.J., Herrera R.E.. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol. Cell. 2006; 22:693–699. [DOI] [PubMed] [Google Scholar]
- 24. Trojer P., Zhang J., Yonezawa M., Schmidt A., Zheng H., Jenuwein T., Reinberg D.. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the jumonji domain-containing JMJD2/KDM4 proteins. J. Biol. Chem. 2009; 284:8395–8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., Li Z., Dong L., Tang M., Zhang P., Zhang C., Cao Z., Zhu Q., Chen Y., Wang H.et al.. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018; 46:7716–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez C.R., Singh S., Hambarde S., Griffin W.C., Gao J., Chib S., Yu Y., Ira G., Raney K.D., Kim N.. Yeast sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017; 45:5850–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mortusewicz O., Evers B., Helleday T.. PC4 promotes genome stability and DNA repair through binding of ssDNA at DNA damage sites. Oncogene. 2016; 35:761–770. [DOI] [PubMed] [Google Scholar]
- 28. Ge H., Martinez E., Chiang C.M., Roeder R.G.. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996; 274:57–71. [DOI] [PubMed] [Google Scholar]
- 29. Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K.. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2003; 375:23–44. [DOI] [PubMed] [Google Scholar]
- 30. Hizume K., Yoshimura S.H., Takeyasu K.. Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry. 2005; 44:12978–12989. [DOI] [PubMed] [Google Scholar]
- 31. Luger K., Rechsteiner T.J., Richmond T.J.. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999; 304:3–19. [DOI] [PubMed] [Google Scholar]
- 32. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zlatanova J., van Holde K.. Linker histones versus HMG 1/2: a struggle for dominance?. Bioessays. 1998; 20:584–588. [DOI] [PubMed] [Google Scholar]
- 34. Woodcock C.L., Skoultchi A.I., Fan Y.. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006; 14:17–25. [DOI] [PubMed] [Google Scholar]
- 35. Jo S.S., Choi S.S., Hurst L.. Analysis of the functional relevance of epigenetic chromatin marks in the first intron associated with specific gene expression patterns. Genome Biol. Evol. 2019; 11:786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song Y., Quach C., Liang C.. UVRAG in autophagy, inflammation, and cancer. Autophagy. 2020; 16:387–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia J., Bissa B., Brecht L., Allers L., Choi S.W., Gu Y., Zbinden M., Burge M.R., Timmins G., Hallows K.et al.. AMPK is activated during lysosomal damage via a galectin-ubiquitin signal transduction system. Autophagy. 2020; 16:1550–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bang S., Kim S., Dailey M.J., Chen Y., Moran T.H., Snyder S.H., Kim S.F.. AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc. Nat. Acad. Sci. U.S.A. 2012; 109:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guha P., Snyder S.H.. Noncatalytic functions of IPMK are essential for activation of autophagy and liver regeneration. Autophagy. 2019; 15:1473–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guha P., Tyagi R., Chowdhury S., Reilly L., Fu C., Xu R., Resnick A.C., Snyder S.H.. IPMK mediates activation of ULK signaling and transcriptional regulation of autophagy linked to liver inflammation and regeneration. Cell Rep. 2019; 26:2692–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi W., Guan H., Lyu J., Zhao D., Xi Y., Jiang S., Andrews F.H., Wang X., Gagea M., Wen H.et al.. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat. Commun. 2017; 8:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maag D., Maxwell M.J., Hardesty D.A., Boucher K.L., Choudhari N., Hanno A.G., Ma J.F., Snowman A.S., Pietropaoli J.W., Xu R.et al.. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Nat. Acad. Sci. U.S.A. 2011; 108:1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goodall M.L., Fitzwalter B.E., Zahedi S., Wu M., Rodriguez D., Mulcahy-Levy J.M., Green D.R., Morgan M., Cramer S.D., Thorburn A.. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell. 2016; 37:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu R., Sen N., Paul B.D., Snowman A.M., Rao F., Vandiver M.S., Xu J., Snyder S.H.. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci. Signal. 2013; 6:ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao W., Shen Z., Shang L., Wang X.. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 2011; 18:1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Das C., Gadad S.S., Kundu T.K.. Human positive coactivator 4 controls heterochromatinization and silencing of neural gene expression by interacting with REST/NRSF and CoREST. J. Mol. Biol. 2010; 397:1–12. [DOI] [PubMed] [Google Scholar]
- 47. Rossetto D., Avvakumov N., Côté J.. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012; 7:1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei Y., Mizzen C.A., Cook R.G., Gorovsky M.A., Allis C.D.. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in tetrahymena. Proc. Nat. Acad. Sci. U.S.A. 1998; 95:7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei Y., Yu L., Bowen J., Gorovsky M.A., David Allis C.. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999; 97:99–109. [DOI] [PubMed] [Google Scholar]
- 50. Sauvé D.M., Anderson H.J., Ray J.M., James W.M., Roberge M.. Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J. Cell Biol. 2000; 145:225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de La Barre A.E., Gerson V., Gout S., Creaven M., Allis C.D., Dimitrov S.. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 2000; 19:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng Y., John S., Pesavento J.J., Schultz-Norton J.R., Schiltz R.L., Baek S., Nardulli A.M., Hager G.L., Kelleher N.L., Mizzen C.A.. Histone H1 phosphorylation is associated with transcription by RNA polymerases i and iI. J. Cell Biol. 2010; 189:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee H.L., Archer T.K.. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998; 17:1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koop R., di Croce L., Beato M.. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 2003; 22:588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petrova V., Song R., Nordström K.J.V., Walter J., Wong J.J.-L., Armstrong N.J., Rasko J.E.J., Schmitz U.. Chromatin accessibility determines intron retention in a cell type-specific manner. 2021; bioRxiv doi:30 March 2021, preprint: not peer reviewed 10.1101/2021.02.17.431609. [DOI] [PMC free article] [PubMed]
- 56. Chorev M., Carmel L.. The function of introns. Front. Genet. 2012; 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rose A.B. Introns as gene regulators: a brick on the accelerator. Front. Genet. 2019; 10:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tourmente S., Chapel S., Dreau D., Drake M.E., Bruhat A., Couderc .J.L., Dastugue B. Enhancer and silencer elements within the first intron mediate the transcriptional regulation of the beta 3 tubulin gene by 20-hydroxyecdysone in drosophila kc cells. Insect. Biochem. Mol. Biol. 1993; 23:137–143. [DOI] [PubMed] [Google Scholar]
- 59. Scohy S., Gabant P., Szpirer C., Szpirer J.. Identification of an enhancer and an alternative promoter in the first intron of the α-fetoprotein gene. Nucleic Acids Res. 2000; 28:3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bianchi M., Crinelli R., Giacomini E., Carloni E., Magnani M.. A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin c gene. Gene. 2009; 448:88–101. [DOI] [PubMed] [Google Scholar]
- 61. Beaulieu E., Green L., Elsby L., Alourfi Z., Morand E.F., Ray D.W., Donn R.. Identification of a novel cell type-specific intronic enhancer of macrophage migration inhibitory factor (MIF) and its regulation by mithramycin. Clin. Exp. Immunol. 2011; 163:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaunitz F., Heise K., Gebhardt R.. A silencer element in the first intron of the glutamine synthetase gene represses induction by glucocorticoids. Mol. Endocrinol. 2004; 18:63–69. [DOI] [PubMed] [Google Scholar]
- 63. Gaunitz F., Deichsel D., Heise K., Werth M., Anderegg U., Gebhardt R.. An intronic silencer element is responsible for specific zonal expression of glutamine synthetase in the rat liver. Hepatology. 2005; 41:1225–1232. [DOI] [PubMed] [Google Scholar]
- 64. Bornstein P., McKay J., Liska D.J., Apone S., Devarayalu S.. Interactions between the promoter and first intron are involved in transcriptional control of alpha 1(I) collagen gene expression. Mol. Cell. Biol. 1988; 8:4851–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang G.R., Li X., Cao H., Zhao H., Geller A.I.. The vesicular glutamate transporter-1 upstream promoter and first intron each support glutamatergic-specific expression in rat postrhinal cortex. Brain Res. 2011; 1377:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vasil V., Clancy M., Ferl R.J., Vasil I.K., Hannah L.C.. Increased gene expression by the first intron of maize shrunken-1 locus in grass species. Plant Physiol. 1989; 91:1575–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sorek R., Ast G.. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003; 13:1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roy M., Kim N., Xing Y., Lee C.. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. RNA. 2008; 14:2261–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kalyna M., Simpson C.G., Syed N.H., Lewandowska D., Marquez Y., Kusenda B., Marshall J., Fuller J., Cardle L., McNicol J.et al.. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in arabidopsis. Nucleic Acids Res. 2012; 40:2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu C., Yang J., Wu H., Li J.. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. OncoTargets Ther. 2019; 12:6525–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen J., Jiang Q., Jiang X.-Q., Li D.-Q., Jiang X.-C., Wu X.-B., Cao Y.-L.. 2020) miR-146a promoted breast cancer proliferation and invasion by regulating NM23-H1. J. Biochem. 167:41–48. [DOI] [PubMed] [Google Scholar]
- 72. Wang Y., Zeng G., Jiang Y.. The emerging roles of miR-125b in cancers. Cancer Manage. Res. 2020; 12:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jo J., Hwang S., Kim H.J., Hong S., Lee J.E., Lee S.G., Baek A., Han H., Lee J.il, Lee I.et al.. An integrated systems biology approach identifies positive cofactor 4 as a factor that increases reprogramming efficiency. Nucleic Acids Res. 2016; 44:1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tai C.I., Ying Q.L.. Gbx2, a LIF/Stat3 target, promotes reprogramming to and retention of the pluripotent ground state. J. Cell Sci. 2013; 126:1093–1098. [DOI] [PubMed] [Google Scholar]
- 75. Ochiai K., Yamaoka M., Swaminathan A., Shima H., Hiura H., Matsumoto M., Kurotaki D., Nakabayashi J., Funayama R., Nakayama K.et al.. Chromatin protein PC4 orchestrates b cell differentiation by collaborating with IKAROS and IRF4. Cell Rep. 2020; 33:108517. [DOI] [PubMed] [Google Scholar]
- 76. Lim P.S.L., Meshorer E.. 2020) Dppa2 and dppa4 safeguard bivalent chromatin in order to establish a pluripotent epigenome. Nat. Struct. Mol. Biol. 27:685–686. [DOI] [PubMed] [Google Scholar]
- 77. Mossahebi-Mohammadi M., Quan M., Zhang J.-S., Li X.. FGF signaling pathway: a key regulator of stem cell pluripotency. Front. Cell Dev. Biol. 2020; 8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tsang S.M., Oliemuller E., Howard B.A.. Regulatory roles for SOX11 in development, stem cells and cancer. Semin. Cancer Biol. 2020; 67:3–11. [DOI] [PubMed] [Google Scholar]
- 79. Chen L., Liao F., Wu J., Wang Z., Jiang Z., Zhang C., Luo P., Ma L., Gong Q., Wang Y.et al.. Acceleration of ageing via disturbing mTOR-regulated proteostasis by a new ageing-associated gene PC4. Aging Cell. 2021; 20:e13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dhanasekaran K., Kumari S., Boopathi R., Shima H., Swaminathan A., Bachu M., Ranga U., Igarashi K., Kundu T.K.. Multifunctional human transcriptional coactivator protein PC4 is a substrate of aurora kinases and activates the aurora enzymes. FEBS J. 2016; 283:968–985. [DOI] [PubMed] [Google Scholar]
- 81. Hattori T., Lai D., Dementieva I.S., Montaño S.P., Kurosawa K., Zheng Y., Akin L.R., Świst-Rosowska K.M., Grzybowski A.T., Koide A.et al.. Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation. Proc. Nat. Acad. Sci. U.S.A. 2016; 113:2092–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lamb K.N., Bsteh D., Dishman S.N., Moussa H.F., Fan H., Stuckey J.I., Norris J.L., Cholensky S.H., Li D., Wang J.et al.. Discovery and characterization of a cellular potent positive allosteric modulator of the polycomb repressive complex 1 chromodomain, CBX7. Cell Chem. Biol. 2019; 26:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morgan M.A.J., Rickels R.A., Collings C.K., He X., Cao K., Herz H.-M., Cozzolino K.A., Abshiru N.A., Marshall S.A., Rendleman E.J.et al.. A cryptic tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev. 2017; 31:2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ATAC-seq data for this study are available in the GEO database under accession number GSE191333. The ChIP seq datasets used in this study are available from the GEO database with the following accession numbers: GSE66530 for H3K4me3 and H3K9me3 ChIP, GSM2711409 for H3K27ac ChIP. UCSC genome browser links for the genomic tracts are as follows: UVRAG https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq1 MAP3K7 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq2 YEATS2 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq3 ATG5 https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq4 IPMK https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq5 IPMK https://genome.ucsc.edu/s/pallabidgp123/ATAC-seq6.