Abstract
Magnetotactic bacteria (MTB) have been found in a wide variety of marine habitats, ranging from intertidal sediments to deep-sea seamounts. Deep-sea hydrothermal fields are rich in metal sulfides, which are suitable areas for the growth of MTB. However, MTB in hydrothermal fields have never been reported. Here, the presence of MTB in sediments from the Tangyin hydrothermal field was analyzed by 16S rRNA gene amplicon analysis, metagenomics, and transmission electron microscopy. Sequencing 16S rRNA gene yielded a total of 709 MTB sequences belonging to 20 OTUs, affiliated with Desulfobacterota, Alphaproteobacteria, and Nitrospirae. Three shapes of magnetofossil were identified by transmission electron microscopy: elongated-prismatic, bullet-shaped, and cuboctahedron. All of these structures were composed of Fe3O4. A total of 121 sequences were found to be homologous to the published MTB magnetosome-function-related genes, and relevant domains were identified. Further analysis revealed that diverse MTB are present in the Tangyin hydrothermal field, and that multicellular magnetotactic prokaryote (MMPs) might be the dominant MTB.
Keywords: magnetotactic bacteria, 16S rRNA gene, magnetosome genes, magnetofossil, hydrothermal field
Introduction
Magnetotactic bacteria (MTB) represent a group of prokaryotes that can migrate along geomagnetic field lines (Blakemore, 1975; Bellini, 2009). MTB are diverse in their morphology, phylogeny, and physiology. There are single-cell forms (cocci, ovoid, rod, curved rod, and spirillum) and multicellular forms that are known as the multicellular magnetotactic prokaryotes (MMPs). The MMPs can be divided into two types based on their shape: spherical MMPs (sMMPs) and ellipsoidal MMPs (eMMPs) (Bazylinski et al., 2013; Amor et al., 2020). MTB exhibits great taxonomic diversity. Most are phylogenetically affiliated with phyla of Proteobacteria, Desulfobacterota (previously known as Deltaproteobacteria), Nitrospirae, Planctomycetes, the candidate phylum Omnitrophica, and the candidate phylum Latescibacteria (Bazylinski et al., 2013; Lin et al., 2018). Recent studies of reconstructed metagenome-assembled MTB genomes have expanded our knowledge of the taxonomy of MTB, several of which belonged to the phyla Nitrospinota, UBA10199, Bdellovibrionota, Bdellovibrionata_B, Fibrobacterota, Riflebacteria phyla, Elusimicrobia, and Candidatus Hydrogenedentes (Lin et al., 2020; Uzun et al., 2020).
The MTB can use intracellular biomineralization to synthesize special organelles called magnetosomes, which help them find and remain near suitably chemically stratified water columns or sediments. Magnetosomes are composed of magnetite or/and greigite (Zhang et al., 2014) and exhibit different shapes (e.g., elongated-prismatic, bullet/tooth-shaped, and cuboctahedron) (Bazylinski et al., 2013). There is a strong correlation between the morphology of biogenetic magnetosomes and MTB phylogeny (Li et al., 2020b). Alphaproteobacteria MTB always synthesizes cuboctahedral or elongated-prismatic magnetite magnetosomes, while MTB with bullet-shaped magnetite crystals is associated with the Desulfobacterota, Nitrospirae, and the candidate phylum Omnitrophica (Lin et al., 2014; Amor et al., 2020). Desulfobacterota MTB can also biomineralize greigite crystals of diverse morphologies (Lefèvre et al., 2011; Zhang et al., 2014). Inside the cell, magnetosomes are arranged as single chains, double chains, or multiple chains, and disorderly arrangements have also been reported (Amor et al., 2020). Magnetosome formation in MTB is a biomineralization process that is strictly controlled by conserved genes found in magnetosome gene clusters (MGCs) (Lin et al., 2020). Generally, there are four steps in the formation of magnetosomes, which arrange in chains: cytoplasmic membrane invagination forms vesicles, proteins are targeted to the vesicle (magnetosome) membrane, iron is transported to vesicles (membranous invagination of magnetosomes) and mineralized into magnetite crystals, and the crystals are assembled into chains of magnetosomes (Uebe and Schüler, 2016). Studies on pure cultures of the MTB, like Magnetospirillum gryphiswaldense MSR-1, revealed that the MGC genes are found in five operons: the mms6 operon, the mamGFDC operon, the mamAB operon, the mamXY operon, and the feoAB1 operon (Uebe and Schüler, 2016). Magnetosome genes that compose MGCs of different MTB species are not identical, as reflected in species-level differences in magnetosome type. However, almost all MGCs contain the mamAB operon (Lefèvre and Wu, 2013). This operon contains the core gene, mamABEKMOPQI, which is thought to play an important role in magnetosome formation (Lefèvre and Wu, 2013; Lin et al., 2018). After the death of a magnetotactic bacterium, the magnetosomes are released; over time, they accumulate in sediments, forming fossil magnetosomes (magnetofossils) (Lin et al., 2014). The magnetic mineral ultrastructure, morphology, composition, size, and other characteristics of magnetofossils can be observed and effectively distinguished using a transmission electron microscope (TEM) (Li et al., 2020a). The study of magnetofossils can provide paleoecological and paleoenvironmental information (Hesse, 1994).
The MTB is widespread in sediments and water at the oxic-anoxic interface (OAI, previously called the OATZ) of freshwater, brackish, marine, and hypersaline environments (Bazylinski et al., 2013). The abundance of marine MTB is usually higher in the intertidal zone, where most reports indicate that magnetotactic cocci are relatively common and the dominant species of MTB (Lin et al., 2012; Abreu et al., 2016). Most of these studied environments have near-neutral pH, moderate temperatures, and are concentrated in the Northern hemisphere. However, there are some reports of MTB in special habitats, including mangrove swamps, coral reefs, seamounts, deep seas, etc. (Torres de Araujo et al., 1986; Dong et al., 2016; Liu et al., 2017; Teng et al., 2018). Vibrio and rod-shaped MTB, but not cocci, have been found in hemipelagic sediments of the Santa Barbara Basin (Stolz et al., 1986). Other studies indicated that the MTB found in extreme environments often belonged to Desulfobacterota and Nitrospirae (Abreu et al., 2007; Nash, 2008; Lefèvre et al., 2010). In addition, biogenic magnetite, with elongated octahedral and prismatic morphologies in ferromanganese nodules, has also been reported (Hassan et al., 2020).
In deep-sea hydrothermal fields, which are rich in metal sulfides, microorganisms mainly obtain energy from reductive sulfur oxidation (McCollom and Shock, 1997; Amend et al., 2011; Meier et al., 2019), and chemolithoautotrophic microorganisms represent the only primary producers (Jannasch and Mottl, 1985). Simultaneously, most cultured MTB can grow chemolithoautotrophically using reductive sulfides. Desulfobacterota MTB are sulfate-reducing anaerobes that grow only chemoorganoheterotrophically (Bazylinski et al., 2013). MTB might dwell in and adapt to hydrothermal environments. To assess this possibility, we combined 16S rRNA gene amplicon and metagenomic data and TEM observations to study the presence, diversity, and characteristics of MTB in the Tangyin hydrothermal field of Okinawa Trough.
Materials and Methods
Sample Collection
The Tangyin hydrothermal field is located atop an upland found 38 km northeast of Yonaguni Knoll IV field, at the southern end of the Okinawa Trough. Surface sediment samples were collected by a box sampler during a cruise conducted by the R/V Kexue in May 2014. All sediment samples were quick-frozen in liquid nitrogen and stored at −80°C until laboratory analysis.
16S rRNA Gene Sequencing and Analysis
Total genomic DNA was extracted from sediment samples as described by Zhou et al. (1996). The V3–V4 regions of the 16S rRNA gene were amplified using universal primer sets 338F and 806R. PCR products were purified, quantified, and paired-end sequencing was performed on the Illumina MiSeq PE300 platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Detailed protocols were previously published by Wang et al. (2018). The paired-end reads of fastq files were merged and quality-filtered using Usearch (version 8.1) (Edgar, 2016). The filtered reads were clustered into operational taxonomic units (OTUs), using UPARSE with the threshold set to 97% (Edgar, 2013). Representative reads for OTUs were aligned with the GenBank nucleic acid database (NT) using BLASTn, and MTB-related OTUs were screened using an identity threshold of 90% (Altschul et al., 1990). The CLUSTALW multiple alignment software was used for sequence alignment (Larkin et al., 2007). The phylogenetic tree was constructed based on the neighbor-joining method using MEGA6 (Tamura et al., 2013) with the bootstrap p value of 1,000.
Rock Magnetic Measurements
We measured the isothermal remanent magnetization (IRM) acquisition curve and first-order reversal curves (FORC) on the bulk sample using a vibrating sample magnetometer (VSM, Princeton Measurements Corporation's MicroMag™ 3900) at the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS) (Roberts et al., 2000; Kruiver et al., 2001; Egli et al., 2010). For the IRM acquisition curve, the field was added from 10 μT to 1 T with 120 data points in a log distribution of the field steps and the averaging time is 1 s for each data point. Coercivity unmixing analyses were conducted using the Max Unmix model (Maxbauer et al., 2016). For the FORC diagram, the saturation field is set to 1 T and 200 partial hysteresis curves were measured with 350 ms averaging time. We used FORCinel software (v 3.06) to create a FORC diagram (Harrison and Feinberg, 2008). The FORC were smoothed using VARIFORC parameters: Sc0 = 6, Sb0 = 5, Sc1 = Sb1 = 8.
Magnetofossils Observation
The so-called magnetic fingers were used to extract magnetofossils, as previously described (Von Dobeneck et al., 1987; He and Pan, 2020). We made a minor modification based on the method mentioned above. Briefly, 2 ml of sediment and 0.1 g sodium hexametaphosphate were mixed with 30 ml Milli-Q water in a 50-ml centrifuge tube, and a magnetic finger was put into the mixture for at least 12 h. Magnetic minerals were washed from the surface of the magnetic finger into a 15-ml centrifuge tube using Milli-Q water. Two circular magnets were attached to the outside of the 15-ml centrifuge tube, and the enriched particles were shaken until they were evenly distributed throughout the tube and then allowed to settle for at least 12 h. A Pasteur tube was used to recover the magnetic particles that were adsorbed on the inner wall and transfer them to a 1.5-ml centrifuge tube. After rinsing the tube three times using Milli-Q water, 30 μl of anhydrous ethanol was used to re-suspend the magnetic particles. Then, 8 μl of this suspension was dropped onto a copper double grid for TEM observation.
The morphological characteristics of the magnetofossils were observed using a Hitachi H8100 microscope operating at 100 kV at the Institute of Oceanology, Chinese Academy of Sciences (IOCAS). High-resolution transmission electron microscopy (HRTEM), selected area electron diffraction (SAED), and X-ray energy-dispersive spectroscopy (XEDS) were obtained using a JEOL JEM-2100 TEM operated at 200 kV at the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS).
Metagenome Sequencing and MGCs Analysis
The genomic DNA was sequenced in BGI Co., Ltd. (Wuhan, China) via shotgun sequencing on the Illumina HiSeq platform (150 bp paired-end strategy). The raw sequencing data were filtered and trimmed to generate high-quality clean data. Clean reads were assembled using the IDBA-UD software (Peng et al., 2012). Genes were predicted using Prodigal (Hyatt et al., 2010) and aligned with known MGC genes using BLASTp, with thresholds of 35% for similarity and 50% for gene coverage. MGC genes (mam, mad, feo, mms, man) of MTB strains were downloaded from the MAGE website (Vallenet et al., 2019). All genes were manually checked, and putative magnetosome genes with higher homology to known MGC genes were selected for further analysis. The abundances of these putative magnetosome genes were calculated, and the Pfam database was used to predict the domains of the selected genes and known magnetosome genes (Mistry et al., 2021). At the same time, some selected putative magnetosome gene sequences have higher similarities with known homologous genes. These sequences were used as representative sequences to construct phylogenetic trees. The construction method is the same as above. The sequences homologous to the two known MTB magnetome genes with the most abundant number and species were selected, and the sequences with the highest similarity to the known sequences in each putative magnetome gene were selected for gene organization comparison. Jalview was used to analyze the conserved regions of predictive protein sequences of magnetosome genes (Waterhouse et al., 2009).
Results
MTB Community Analysis
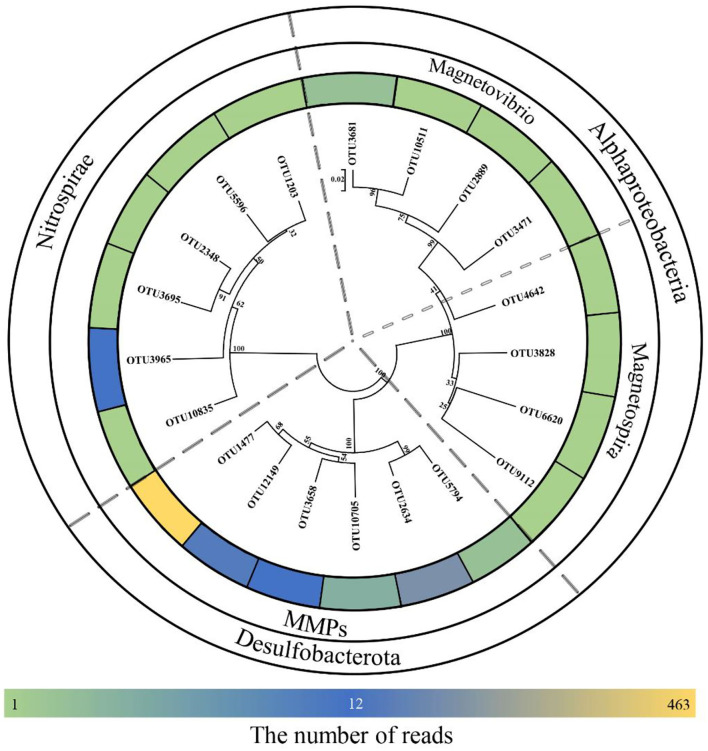
Using high-throughput sequencing, we obtained a total of 153,056 tags from the Tangyin hydrothermal field sediment samples, corresponding to a total of 3,115 OTUs using the threshold of 97% sequence identity. After alignment to published MTB 16S rRNA gene sequences, a total of 709 sequences were screened out (0.46% of all tags). These sequences belonged to 20 OTUs (0.64% of all OTUs) (Supplementary Table S1). Of them, eight (10 reads) belonged to genera Magnetovibrio and Magnetospira in the class of Alphaproteobacteria, six (17 reads) belonged to Nitrospirae MTB, and six (682 reads) belonged to MMPs affiliated with Desulfobacterota. Notably, two dominant MMP OTUs accounted for 463 reads and 146 reads and were highly similar to sMMP Ca. Magnetomorum litorale. Of the remaining four MMP-related OTUs, three (60 reads) were most similar to sMMP Ca. Magnetoglobus multicellularis Araruama and one (13 reads) was similar to eMMP Ca. Magnetananas rongchenensis (Table 1). Results of the phylogenetic tree based on representative sequences of the OTUs were consistent with our sequence alignment results (Figure 1).
Table 1.
Numbers of OTUs and reads corresponding to different species of magnetotactic bacteria.
| OTUs | Reads | |||
|---|---|---|---|---|
| Desulfobacterota | sMMPs | Ca. Magnetomorum litorale | 2 | 609 |
| Ca. Magnetoglobus multicellularis Araruama | 3 | 60 | ||
| eMMPs | Ca. Magnetananas rongchenensis | 1 | 13 | |
| Nitrospirae | Magnetobacterium sp. P1B_23 | 2 | 13 | |
| Ca. Magnetobacterium bavaricum | 3 | 3 | ||
| Uncultured magnetotactic rod MHB-1 | 1 | 1 | ||
| Alphaproteobacteria | Magnetovibrio | Magnetovibrio blakemorei MV-1 | 4 | 6 |
| Magnetospira | Magnetospira sp. QH-2 | 3 | 3 | |
| Magnetospira thiophila MMS-1 | 1 | 1 | ||
| Total | 20 | 709 | ||
| Bacteria | 3,115 | 153,056 | ||
| MTB percentage of bacteria | 0.64% | 0.46% |
Figure 1.
Neighbor-joining tree for environmental magnetotactic bacteria based on 16S rRNA gene sequences. Scale bar: 0.02.
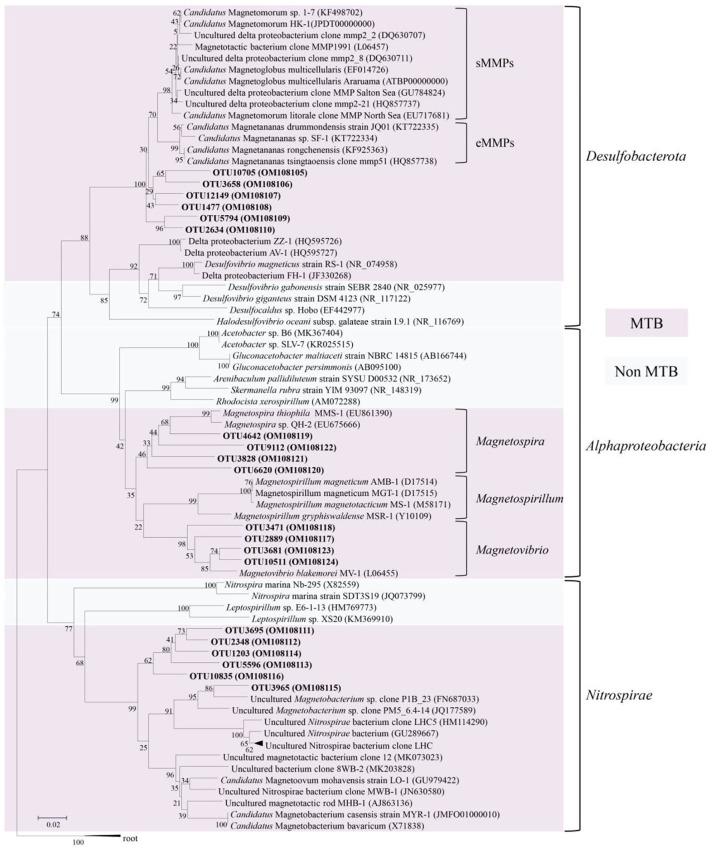
The 16S rRNA gene sequences clustered six OTUs into a branch within Desulfobacterota, which was branched with known MMPs but formed a different group (Figure 2). The five MTB-related OTUs belonging to the phylum Nitrospirae were also found in a different clade from the known MTB in the phylogenetic tree, while OTU3965 was clustered in the same clade with the known thermophilic MTB (Figure 2). Besides, the MTB-related OTUs affiliated with Magnetovibrio and Magnetospira were also found in the same branch as the known MTB (Figure 2).
Figure 2.
Phylogenetic tree constructed based on 16S rRNA sequence gene reads related to magnetotactic bacteria. The sequences determined in this study are shown in bold text. GenBank accession numbers of the sequences used are indicated in parentheses. Scale bar: 0.02.
Detection of Magnetofossils
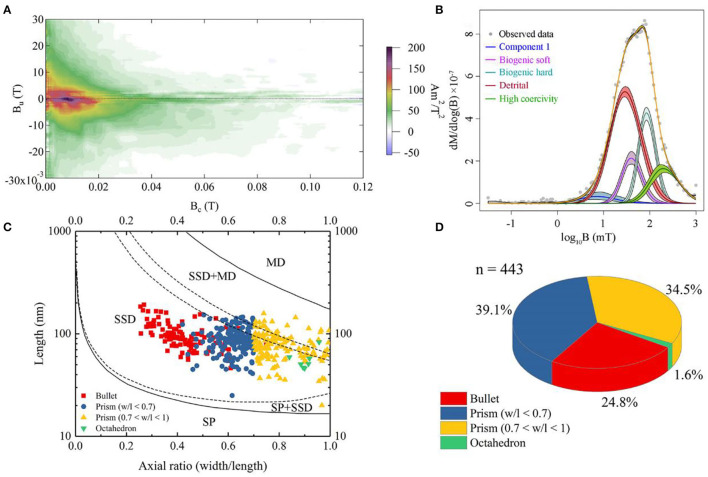
The FORC diagram does not show a clear central ridge which indicates non-interacting uniaxial single domain grains (Figure 3A). It suggests that the bulk sample probably contains multi-domain (MD) and/or vortex magnetic minerals and magnetofossils with broken chains. We decomposed the sample into five components using IRM unmixing analysis, including component 1, biogenic soft (BS, with a median coercivity of 41.1 ± 1 mT), biogenic hard (BH, with a median coercivity of 88.4 ± 1 mT), detrital (with a median coercivity of 30.1 ± 1 mT), and high coercivity component (with a median coercivity of 217.3 ± 1 mT). The dispersion parameter (DP) of BS and BH components is 0.21 and 0.18, respectively (Figure 3B). Component 1 might indicate coarse magnetic minerals with lower coercivity (7.4 ± 1.1 mT). The high coercivity components probably indicate that the sample contains hematite or goethite. Detrital magnetic minerals are the dominant component which contribute 48.8% to the remanent magnetization of the bulk sample (Figure 3B). The proportions of BS and BH, which usually represents isotropic and elongated magnetofossils, are 12.4% and 21.1%, respectively.
Figure 3.
Rock magnetic and statistical data on magnetofossil morphologies for the sediment. (A) First-order reversal curve (FORC) diagram. The Bc and Bu axis indicates coercivity and magnetostatic interaction, respectively. (B) Isothermal remanent magnetization (IRM)-unmixing result. The horizontal axis stands for coercivity (expressed on a base 10 logarithmic scale), and the gradient of the IRM acquisition curve (yellow) is fitted by lognormal distributions. The different color stands for different components; Purple and turquoise curves are biogenic soft and hard components, respectively. Red and green curves are detrital and high coercivity components, respectively. The blue curve may indicate coarse-grained minerals. (C) Magnetofossil size distributions. Red square, navy blue dot, and yellow triangle represent bullet-shaped, elongated prism, and short prism, respectively. The green triangle denotes octahedral magnetofossil. The domain-state phase diagram is modified after Muxworthy and Williams (2009). SP, SD, SSD, and MD represent superparamagnetic, single domain, stable single domain, and multidomain, respectively. (D) Statistics based on panel (C). The colors representing different morphotypes of magnetofossils are consistent with those in panel (C).
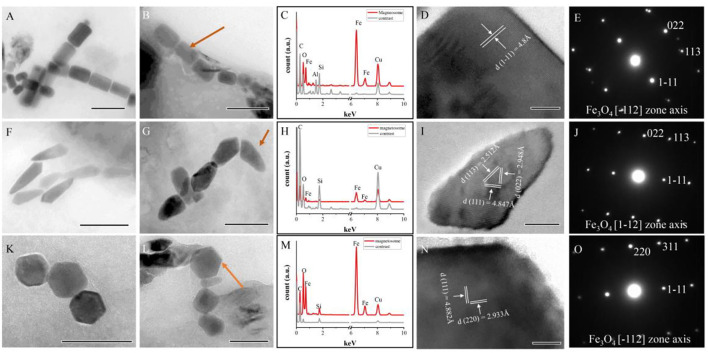
Subsequently, a total of 443 magnetofossils were observed by TEM. Three different magnetofossil morphologies were recognized: elongated-prismatic, bullet-shaped, and cuboctahedral (Figures 4A,B,F,G,K,L). Most magnetofossils fall within a stable single domain (SSD) size range (Figure 3C). Prismatic magnetofossils (84 ± 25 × 59 ± 18 nm, n = 326) were the dominant type, accounting for 73.6% of the total magnetofossils (Figure 3D). Here, we define prisms with an axial ratio smaller than 0.7 to be elongated prismatic magnetofossils while those with an axial ratio larger than 0.7 to be short prisms. Bullet-shaped (98 ± 29 × 40 ± 10 nm, n = 110) magnetofossils accounted for 24.8% of the total, while the cuboctahedron type accounted for only 1.6% (56 ± 12 × 51 ± 12 nm, n = 7). Energy-dispersive X-ray analysis indicated that the variously shaped magnetofossils were all composed of iron and oxygen (Figures 4C,H,M). Measurement of the crystal lattice and analysis of the electron diffraction pattern showed that the particles were magnetite (Figures 4D,E,I,J,N,O).
Figure 4.
Characteristics of magnetofossils with three main shapes. (A,B) Elongated-prismatic magnetofossils. (C–E) Energy dispersive X-ray spectra (C), HRTEM image (D), and electron diffraction patterns (E) of the magnetofossil indicated by an arrow in panel (B). (F,G) Bullet-shaped magnetofossils. (H–J) Energy dispersive X-ray spectra (H), HRTEM image (I), and electron diffraction patterns (J) of the magnetofossil indicated by an arrow in panel (G). (K,L) Cuboctahedron magnetofossils. (M–O) Energy dispersive X-ray spectra (M), HRTEM image (N), and electron diffraction patterns (O) of the magnetofossil are indicated by an arrow in panel (L) (cuboctahedron magnetofossil). Scale bars (A,B,F) 200 nm; (G,K,L) 100 nm; (I) 20 nm; and (D,N) 10 nm.
Analysis of Homologous Sequences of Magnetosome Genes
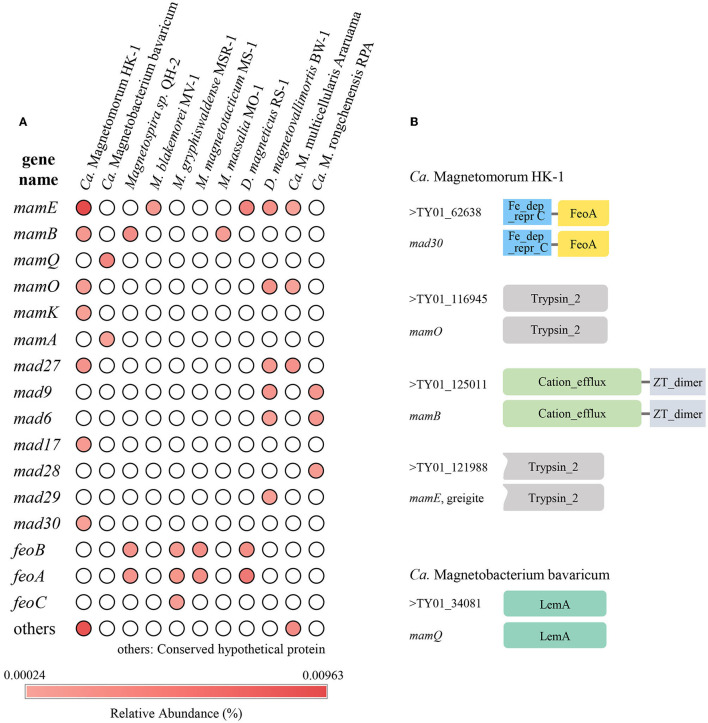
Through functional annotation and comparison with known MGC genes, a total of 121 homologous sequences of magnetosome genes were found (Supplementary Table S2), yielding a relative abundance of 0.078%. These homologous sequences of magnetosome genes were assigned to different genera or species based on similarity and showed relatively high similarity to 11 species of MTB. The homologous magnetosome genes with high similarity to MMPs (Ca. Magnetomorum HK-1, Ca. Magnetoglobus multicellular Araruama, Ca. Magnetananas Rongchenensis) accounted for 44.2% with 0.035% relative abundance. Also, the sequences that were homologous to Ca. Magnetomorum HK-1 had the highest relative abundance (0.025%). Four types of magnetosome genes were identified: mam, mad, feo, and others (i.e., conserved hypothetical protein and magnetosome-associated genes). The homologous sequence of mam gene clusters had the highest (0.037%) relative abundance among the four types; the homologous sequences of mamE accounted for the majority (0.024%), and the homologous sequences of mamABOKQ were also found. The various homologous sequences of mad genes (mad6, mad9, mad17, mad28, mad29, and mad30) were found, as were three feo genes (feoA, feoB, and feoC-like) (Figure 5A).
Figure 5.
Homologous magnetosome genes analyses. (A) Relative abundance of homologous sequences of magnetosome genes in the sampled sediment. (B) Structural domain prediction for sequences that were homologous to Ca. Magnetomorum HK-1 and Ca. Magnetobacterium bavaricum. Fe_dep_repr C, Iron dependent repressor, metal binding, and dimerization domain; FeoA, FeoA domain; Trypsin_2, trypsin-like peptidase domain; Cation_efflux, Cation efflux family; ZT_dimer, Dimerization domain of Zinc Transporter; LemA, LemA protein family domain.
Domain prediction was carried out on the homologous protein sequences. Most of the homologous protein sequences had the same domains as known magnetosome proteins. For example, TY01_125011, which was homologous to mamB of Ca. Magnetomorum HK-1, the homologous MamB protein, was predicted to have ZT-dimer and cation efflux domains, while TY01_34081, which was homologous to mamQ of Ca. Magnetobacterium Bavaricum, the homologous MamQ protein had a LemA domain (Figure 5B).
Discussion
Since MTB was first discovered independently by Bellini and Blakemore (Blakemore, 1975; Bellini, 2009), the phylogenetic information reported for them has been based primarily on 16S rRNA gene sequence analysis. As it has proven to be difficult to cultivate MTB, the 16S rRNA gene analysis has been used to infer the diversity and phylogenetic affiliations of MTB in various environments (Lin et al., 2018; Teng et al., 2018; Amor et al., 2020). Tan et al. (2021) analyzed MTB communities in tropical marine environments using the 16S rRNA genes from the metagenomic analysis. MTB at seamounts was also identified by 16S rRNA gene analysis (Liu et al., 2017). Dong et al. (2016) used the 16S rRNA gene and magnetofossil analyses to infer the existence of MTB in deep-sea surface sediments. Most mature biogenic magnetosomes are single-domain and arranged in chains in most MTB cells. According to these two basic characteristics, combined with the size, crystallographic structure, and composition of the magnetosomes, TEM can be used to identify magnetofossils effectively (Kopp and Kirschvink, 2008; Li et al., 2020a). Previous studies found that biogenic magnetite was distributed widely in deep-sea sediments (Petersen et al., 1986; Yamazaki et al., 2019; He and Pan, 2020; Usui and Yamazaki, 2021), and the proportions of bullet-shaped magnetofossils increased in relatively reductive and less oxic environments, while isotropic magnetofossils dominated in relatively oxic environments (Hesse, 1994; Yamazaki and Kawahata, 1998; Yamazaki and Shimono, 2013; He and Pan, 2020; Lu et al., 2021). Recently, the known extent of MTB diversity has undergone a significant expansion due to the identification of magnetosome gene cluster (MGC)-containing genomes and studies screening for sequences homologous to known MGC genes (Lin et al., 2020; Uzun et al., 2020). Lin et al. (2020) performed a large-scale reconstruction of metagenome-assembled MTB genomes from diverse ecosystems, and 13 bacterial phyla were detected, six of which were not previously known, including MTB. Thus, the existing literature indicates that analyses of 16S rRNA gene sequences, magnetofossil diversity, and magnetosome gene-homologous sequences obtained from an environment can be used to predict the existence of MTB diversity.
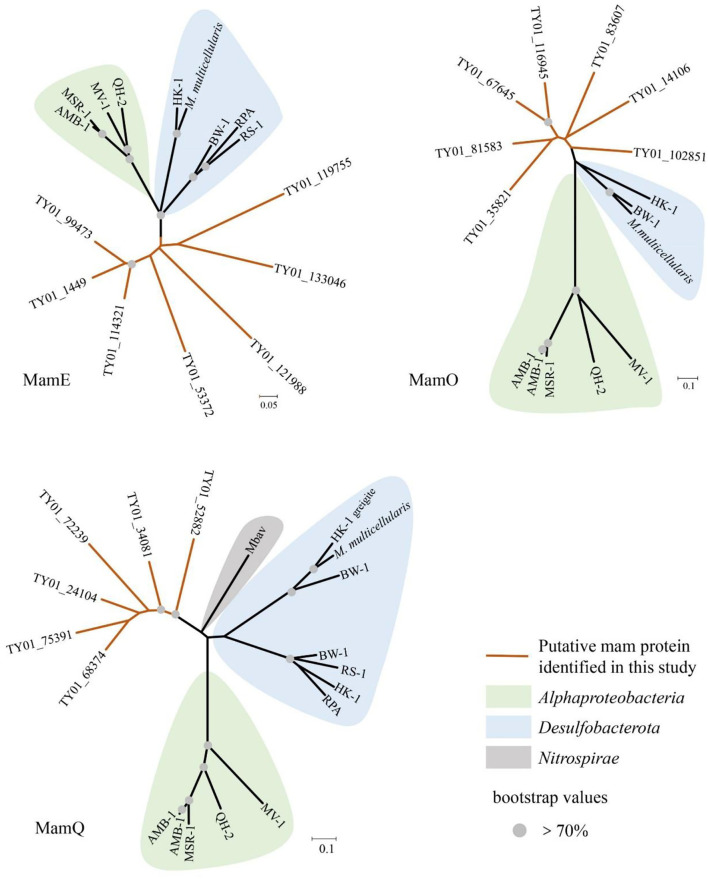
In our study, 16S rRNA gene analysis showed that MTB affiliated with Desulfobacterota, Alphaproteobacteria, and Nitrospirae had been found. FORC diagrams have no central ridge and the coercivity obtained from FORC is smaller than 20 mT, which is different from common FORCs of magnetofossils (Jovane et al., 2012). Vali and Kirschvink (1989) found partial magnetofossil dissolved in deep-sea sediment. Particle corrosion and clumping probably decreased the coercivity and increased the level of interparticle interaction, which is shown in our FORC diagram (Figure 3A). We also observed that partial magnetofossils experienced moderate corrosion in our sample (Supplementary Figure S1). Moreover, the diversity of magnetofossil was also affected by early diagenesis (Rodelli et al., 2019; Yamazaki, 2020). Yamazaki (2020) concluded that bullet-shaped magnetofossils were dissolved easier than prismatic ones, which may explain that the proportions of bullet-shaped magnetofossils are only 24.8% in our sample (Figure 3D). According to the diversity of magnetofossils observed from TEM images, the ancient MTB living in the Tangyin hydrothermal field of the Okinawa Trough might belong to Alphaproteobacteria, Etaproteobacteria, Gammaproteobacteria, Desulfobacterota, Nitrospirae, and the candidate phylum Omnitrophica (Figure 4) (Amor et al., 2020; Liu et al., 2021a,b). Moreover, we had detected some homologous sequences of magnetosome genes, and these homologous magnetosome genes did not form a complete magnetosome gene cluster. However, these homologous sequences have the same domain as the known magnetosome genes. This indicates they may have the same functions, and these homologous sequences could be magnetosome genes. Then, the phylogenetic trees of MamE, O, and Q protein sequences showed that these homologous sequences are evolutionarily distinct from known MTB (Figure 6). The homologous sequences of MamE and MamO representative sequences were homologous to Desulfobacterota. These branches were far from known MTB in the phylogenetic tree. Whereas, compared with the known MTB, most of these homologous Mam protein sequences have conserved regions (Supplementary Figure S2). Therefore, the MamE, O, and Q in hydrothermal have different evolutionary statuses and show high diversity. Of course, in consideration of these homologous sequences having big differences evolutionarily from known MTB, the homologous sequences of MamE and mamO are not necessarily Desulfobacterota either; they might be originated from other MTB identified by 16S rRNA gene sequences shown in Figure 2 (Alphaproteobacteria or Nitrospirae). Meanwhile, the possibility that they are paralogous genes or pseudogenes from non-MTB cannot be ruled out. Nevertheless, combining the 16S rRNA gene results with the magnetofossils, we are more inclined that these are magnetosome genes from MTB. Here, our analysis of MTB-related 16S rRNA gene sequences, magnetofossils produced by MTB, and homologous sequences of magnetosome genes indicate that MTB could inhabit the deep-sea hydrothermal sediments.
Figure 6.
Neighbor-joining tree of homologous sequences of MamE, MamO, and MamQ protein from sediment of Tangyin hydrothermal field. HK-1, Candidatus Magnetomorum HK-1; RPA, Candidatus Magnetananas rongchenensis; BW-1, Desulfamplus magnetovallimortis BW-1; M. multicellularis, Candidatus Magnetoglobus multicellularis Araruama; RS-1, Desulfovibrio magneticus RS-1; QH-2, Magnetospira sp. QH-2; MV-1, Magnetovibrio blakemorei MV-1; AMB-1, Magnetospirillum magneticum AMB-1; MSR-1, Magnetospirillum gryphiswaldense MSR-1; Mbav, Candidatus Magnetobacterium bavaricum.
Our analysis revealed that 65.3% of the 16S rRNA gene sequence reads associated with MTB belonged to OTU1477, which is affiliated with the MMP branch. All MMP-related 16S rRNA gene sequence reads accounted for 96.2% of all MTB-related reads. Moreover, the homologous sequences of magnetosome genes are most similar to known MTB belonging to Desulfobacterota, and MMPs accounted for 44.2% of all magnetosome gene homology sequences. The proportions of bullet-shaped magnetic particles are higher than those of MTB magnetosomes from the intertidal zone of Huiquan Bay in Qingdao (unpublished data) (24.8% vs. 4.0%). Bullet-shaped magnetosomes have only been found in the MTB belonging to Desulfobacterota, Nitrospirae, and the candidate phylum Omnitrophica (Kolinko et al., 2012; Chen et al., 2015; Li et al., 2015; Qian et al., 2019). Interestingly, both rock magnetic results and TEM analysis indicate that the abundance of BH components (bullet and elongated prisms) is ~1.7 times higher than that of BS components (cuboctahedron and short prisms) (Figures 3B,D). All the homologous sequences of mam genes identified herein correspond to the core genes of magnetosome synthesis. Given this, 16S rRNA gene sequences, magnetofossils, and homologous sequences of magnetosome genes consistently indicate that MMPs might be the dominant species among the MTB in this region. The hydrothermal field is an anoxic environment and is rich in reducing sulfides and sulfates (Orcutt et al., 2011). Previous research found that sulfate-reducing bacteria were the dominant species of the microbial community in the Tangyin hydrothermal field (Wang et al., 2018). Among the MTB, MMPs are anaerobic sulfate-reducing bacteria (Kolinko et al., 2014). Therefore, MMPs may represent the dominant group of MTBs at the sampled location, likely reflecting their adaptation to the hydrothermal environment.
As we all know, the hydrothermal zone is a candidate place for the origin of life (Trolard et al., 2022). Meanwhile, although the origins of MTB remain unclear, previous research has shown that the MTB of Nitrospirae and Proteobacteria differentiated near the Archean, and this suggests that MTB has existed at least in the Archean Eon (Lin et al., 2017). The geochemical conditions in the hydrothermal zone are thought to be similar to those on the early Earth (Baross and Hoffman, 1985; Trolard et al., 2022). The evolution of MTB here may be different from known MTB, which has been implied by the phylogenetic tree of 16S rRNA and homolog sequences of magnetosome genes. The dominant MTB in the hydrothermal field may represent a particular MTB species associated with the early Earth-like environment. There are microorganisms of ancient origin in hydrothermal vents (Takai and Nakamura, 2011), and MMPs are also an important model for studying evolution in prokaryotes (Keim et al., 2007). If MMPs are present in the hydrothermal field, the MMPs may be a potential model microorganism for understanding the early evolution of life on Earth.
Conclusion
In this study, a total of 709 MTB-related 16S rRNA gene sequence reads were found in the Tangyin hydrothermal field. The 20 related OTUs represent Desulfobacterota, Alphaproteobacteria, and Nitrospirae. MMPs represented the largest number total of 121 homologous magnetosome gene sequences were annotated. The results collectively suggest that MTB exists in the Tangyin hydrothermal field and MMPs might be the dominant MTB in this region.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM108105-OM108124 and https://www.ncbi.nlm.nih.gov/genbank/, PRJNA514953.
Author Contributions
TX, WYZ, and WCZ designed the research. MY and X-HZ collected the sample and performed the metagenomic sequencing. WYZ and MY carried out a metagenomic analysis. SC carried out rock magnetic measurement and TEM experiments. SC and KH carried out the magnetism and TEM data analysis. SC, WYZ, KH, TX, and WCZ carried out data and statistical analysis. SC, TX, WYZ, WCZ, KH, and L-FW prepared the manuscript. All authors participated in the discussion of the results. All authors contributed to the article and approved the submitted version.
Funding
This study was supported financially by the National Natural Science Foundation of China (U1706208 and 41976137).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jinhua Li at the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS) for his assistance in rock magnetic measurements and Xu Tang at the IGGCAS for the efforts to maintain operation in TEM experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.887136/full#supplementary-material
References
- Abreu F., Carolina A., Araujo V., Leão P., Silva K. T., de Carvalho F. M., et al. (2016). Culture-independent characterization of novel psychrophilic magnetotactic cocci from Antarctic marine sediments. Environ. Microbiol. 18, 4426–4441. 10.1111/1462-2920.13388 [DOI] [PubMed] [Google Scholar]
- Abreu F., Martins J. L., Silveira T. S., Keim C. N., Lins de Barros H. G. P., Gueiros Filho F. J., et al. (2007). ’Candidatus Magnetoglobus multicellularis', a multicellular, magnetotactic prokaryote from a hypersaline environment. Int. J. Syst. Evol. Microbiol. 57, 1318–1322. 10.1099/ijs.0.64857-0 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amend J. P., McCollom T. M., Hentscher M., Bach W. (2011). Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim. Cosmochim. Acta 75, 5736–5748. 10.1016/j.gca.2011.07.041 [DOI] [Google Scholar]
- Amor M., Mathon F. P., Monteil C. L., Busigny V., Lefevre C. T. (2020). Iron-biomineralizing organelle in magnetotactic bacteria: function, synthesis and preservation in ancient rock samples. Environ. Microbiol. 22, 3611–3632. 10.1111/1462-2920.15098 [DOI] [PubMed] [Google Scholar]
- Baross J. A., Hoffman S. E. (1985). Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life Evol. Biosphere 15, 327–345. 10.1007/BF01808177 [DOI] [Google Scholar]
- Bazylinski D. A., Lefèvre C. T., Schüler D. (E). (2013). “Magnetotactic Bacteria,” in The Prokaryotes: Prokaryotic Physiology and Biochemistry, eds Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. L. (Berlin, Heidelberg: Springer Berlin Heidelberg; Springer e-books; Imprint: Springer; ), 453–494. [Google Scholar]
- Bellini S. (2009). On a unique behavior of freshwater bacteria. Chin. J. Ocean. Limnol. 27, 3–5. 10.1007/s00343-009-0003-5 [DOI] [Google Scholar]
- Blakemore R. (1975). Magnetotactic bacteria. Science 190, 377–379. 10.1126/science.170679 [DOI] [PubMed] [Google Scholar]
- Chen Y.-R., Zhang R., Du H.-J., Pan H.-M., Zhang W.-Y., Zhou K., et al. (2015). A novel species of ellipsoidal multicellular magnetotactic prokaryotes from Lake Yuehu in China. Environ. Microbiol. 17, 637–647. 10.1111/1462-2920.12480 [DOI] [PubMed] [Google Scholar]
- Dong Y., Li J., Zhang W., Zhang W., Zhao Y., Xiao T., et al. (2016). The detection of magnetotactic bacteria in deep sea sediments from the east Pacific Manganese Nodule Province. Environ. Microbiol. Rep. 8, 239–249. 10.1111/1758-2229.12374 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2016). UCHIME2: Improved chimera prediction for amplicon sequencing. bioRxiv [Preprint]. 10.1101/074252 [DOI] [Google Scholar]
- Egli R., Chen A. P., Winklhofer M., Kodama K. P., Horng C.-S. (2010). Detection of noninteracting single domain particles using first-order reversal curve diagrams. Geochem. Geophys. Geosyst. 11, Q01Z11. 10.1029/2009GC002916 [DOI] [Google Scholar]
- Harrison R. J., Feinberg J. M. (2008). FORCinel: An improved algorithm for calculating first-order reversal curve distributions using locally weighted regression smoothing. Geochem. Geophys. Geosyst. 9, Q05016. 10.1029/2008GC001987 [DOI] [Google Scholar]
- Hassan M. B., Rodelli D., Benites M., Abreu F., Murton B., Jovane L. (2020). Presence of biogenic magnetite in ferromanganese nodules. Environ. Microbiol. Rep. 12, 288–295. 10.1111/1758-2229.12831 [DOI] [PubMed] [Google Scholar]
- He K., Pan Y. (2020). Magnetofossil abundance and diversity as paleoenvironmental proxies: a case study from southwest Iberian margin sediments. Geophys. Res. Lett. 47, e2020GL087165. 10.1029/2020GL087165 [DOI] [Google Scholar]
- Hesse P. P. (1994). Evidence for bacterial palaeoecological origin of mineral magnetic cycles in oxic and sub-oxic Tasman Sea sediments. Mar. Geol. 117, 1–17. 10.1016/0025-3227(94)90003-5 [DOI] [Google Scholar]
- Hyatt D., Chen G.-L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannasch H. W., Mottl M. J. (1985). Geomicrobiology of deep-sea hydrothermal vents. Science 229, 717–725. 10.1126/science.229.4715.717 [DOI] [PubMed] [Google Scholar]
- Jovane L., Florindo F., Bazylinski D. A., Lins U. (2012). Prismatic magnetite magnetosomes from cultivated Magnetovibrio blakemorei strain MV-1: a magnetic fingerprint in marine sediments? Environ. Microbiol. Rep. 4, 664–668. 10.1111/1758-2229.12000 [DOI] [PubMed] [Google Scholar]
- Keim C. N., Lopes Martins J., Lins de Barros H., Lins U., Farina M. (2007). “Structure, Behavior, Ecology and Diversity of Multicellular Magnetotactic Prokaryotes,” in Magnetoreception and magnetosomes in bacteria, ed Schüler D. (Berlin; New York, NY: Springer; ), 103–132. [Google Scholar]
- Kolinko S., Jogler C., Katzmann E., Wanner G., Peplies J., Schüler D. (2012). Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 14, 1709–1721. 10.1111/j.1462-2920.2011.02609.x [DOI] [PubMed] [Google Scholar]
- Kolinko S., Richter M., Glöckner F.-O., Brachmann A., Schüler D. (2014). Single-cell genomics reveals potential for magnetite and greigite biomineralization in an uncultivated multicellular magnetotactic prokaryote. Environ. Microbiol. Rep. 6, 524–531. 10.1111/1758-2229.12198 [DOI] [PubMed] [Google Scholar]
- Kopp R. E., Kirschvink J. L. (2008). The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth-Sci. Rev. 86, 42–61. 10.1016/j.earscirev.2007.08.001 [DOI] [Google Scholar]
- Kruiver P. P., Dekkers M. J., Heslop D. (2001). Quantification of magnetic coercivity components by the analysis of acquisition curves of isothermal remanent magnetisation. Earth Planet. Sci. Lett. 189, 269–276. 10.1016/S0012-821X(01)00367-3 [DOI] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lefèvre C. T., Abreu F., Schmidt M. L., Lins U., Frankel R. B., Hedlund B. P., et al. (2010). Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl. Environ. Microbiol. 76, 3740–3743. 10.1128/AEM.03018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre C. T., Menguy N., Abreu F., Lins U., Pósfai M., Prozorov T., et al. (2011). A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334, 1720–1723. 10.1126/science.1212596 [DOI] [PubMed] [Google Scholar]
- Lefèvre C. T., Wu L.-F. (2013). Evolution of the bacterial organelle responsible for magnetotaxis. Trends Microbiol. 21, 534–543. 10.1016/j.tim.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Li J., Liu Y., Liu S., Roberts A. P., Pan H., Xiao T., et al. (2020a). Classification of a complexly mixed magnetic mineral assemblage in Pacific Ocean surface sediment by electron microscopy and supervised magnetic unmixing. Front. Earth Sci. 8:110. 10.3389/feart.2020.609058 [DOI] [Google Scholar]
- Li J., Menguy N., Gatel C., Boureau V., Snoeck E., Patriarche G., et al. (2015). Crystal growth of bullet-shaped magnetite in magnetotactic bacteria of the Nitrospirae phylum. J. R. Soc. Interface 12, 20141288. 10.1098/rsif.2014.1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Menguy N., Roberts A. P., Gu L., Leroy E., Bourgon J., et al. (2020b). Bullet-shaped magnetite biomineralization within a magnetotactic deltaproteobacterium: implications for magnetofossil identification. J. Geophys. Res. Biogeosci. 125, 257. 10.1029/2020JG005680 [DOI] [Google Scholar]
- Lin W., Bazylinski D. A., Xiao T., Wu L.-F., Pan Y. (2014). Life with compass: diversity and biogeography of magnetotactic bacteria. Environ. Microbiol. 16, 2646–2658. 10.1111/1462-2920.12313 [DOI] [PubMed] [Google Scholar]
- Lin W., Paterson G. A., Zhu Q., Wang Y., Kopylova E., Li Y., et al. (2017). Origin of microbial biomineralization and magnetotaxis during the Archean. Proc. Natl. Acad. Sci. U. S. A. 114, 2171–2176. 10.1073/pnas.1614654114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Wang Y., Li B., Pan Y. (2012). A biogeographic distribution of magnetotactic bacteria influenced by salinity. ISME J. 6, 475–479. 10.1038/ismej.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Zhang W., Paterson G. A., Zhu Q., Zhao X., Knight R., et al. (2020). Expanding magnetic organelle biogenesis in the domain bacteria. Microbiome 8, 152. 10.1186/s40168-020-00931-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Zhang W., Zhao X., Roberts A. P., Paterson G. A., Bazylinski D. A., et al. (2018). Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J. 12, 1508–1519. 10.1038/s41396-018-0098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang W., Li X., Li X., Chen X., Li J.-H., et al. (2017). Bacterial community structure and novel species of magnetotactic bacteria in sediments from a seamount in the Mariana volcanic arc. Sci. Rep. 7, 17964. 10.1038/s41598-017-17445-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Liu Y., Zhao X., Roberts A. P., Zhang H., Zheng Y., et al. (2021a). Diverse phylogeny and morphology of magnetite biomineralized by magnetotactic cocci. Environ. Microbiol. 23, 1115–1129. 10.1111/1462-2920.15254 [DOI] [PubMed] [Google Scholar]
- Liu P., Tamaxia A., Liu Y., Qiu H., Pan J., Jin Z., et al. (2021b). Identification and characterization of magnetotactic Gammaproteobacteria from a salt evaporation pool, Bohai Bay, China. Environ. Microbiol. 24, 938–950. 10.1111/1462-2920.15516 [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang D., Jiang X., Lin Z., Yang Y., Liu Q. (2021). Paleoenvironmental significance of magnetofossils in pelagic sediments in the equatorial Pacific Ocean before and after the eocene/oligocene boundary. J. Geophys. Res. Solid Earth 126, e2021JB022221. 10.1029/2021JB022221 [DOI] [Google Scholar]
- Maxbauer D. P., Feinberg J. M., Fox D. L. (2016). MAX UnMix: a web application for unmixing magnetic coercivity distributions. Comput. Geosci. 95, 140–145. 10.1016/j.cageo.2016.07.009 [DOI] [Google Scholar]
- McCollom T. M., Shock E. L. (1997). Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim. Cosmochim. Acta 61, 4375–4391. 10.1016/S0016-7037(97)00241-X [DOI] [PubMed] [Google Scholar]
- Meier D. V., Pjevac P., Bach W., Markert S., Schweder T., Jamieson J., et al. (2019). Microbial metal-sulfide oxidation in inactive hydrothermal vent chimneys suggested by metagenomic and metaproteomic analyses. Environ. Microbiol. 21, 682–701. 10.1111/1462-2920.14514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muxworthy A. R., Williams W. (2009). Critical superparamagnetic/single-domain grain sizes in interacting magnetite particles: implications for magnetosome crystals. J. R. Soc. Interface 6, 1207–1212. 10.1098/rsif.2008.0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash C. (2008). Mechanisms and evolution of magnetotactic bacteria (Ph.D. degree). California Institute of Technology, Pasadena, CA, USA. [Google Scholar]
- Orcutt B. N., Sylvan J. B., Knab N. J., Edwards K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. 10.1128/MMBR.00039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- Petersen N., von Dobeneck T., Vali H. (1986). Fossil bacterial magnetite in deep-sea sediments from the South Atlantic Ocean. Nature 320, 611–615. 10.1038/320611a0 [DOI] [Google Scholar]
- Qian X.-X., Liu J., Menguy N., Li J., Alberto F., Teng Z., et al. (2019). Identification of novel species of marine magnetotactic bacteria affiliated with Nitrospirae phylum. Environ. Microbiol. Rep. 11, 330–337. 10.1111/1758-2229.12755 [DOI] [PubMed] [Google Scholar]
- Roberts A. P., Pike C. R., Verosub K. L. (2000). First-order reversal curve diagrams: A new tool for characterizing the magnetic properties of natural samples. J. Geophys. Res. Solid Earth 105, 28461–28475. 10.1029/2000JB900326 [DOI] [Google Scholar]
- Rodelli D., Jovane L., Giorgioni M., Rego E. S., Cornaggia F., Benites M., et al. (2019). Diagenetic fate of biogenic soft and hard magnetite in chemically stratified sedimentary environments of Mamanguá Ría, Brazil. J. Geophys. Res. Solid Earth 124, 2313–2330. 10.1029/2018JB016576 [DOI] [Google Scholar]
- Stolz J. F., Chang S.-B. R., Kirschvink J. L. (1986). Magnetotactic bacteria and single-domain magnetite in hemipelagic sediments. Nature 321, 849–851. 10.1038/321849a0 [DOI] [Google Scholar]
- Takai K., Nakamura K. (2011). Archaeal diversity and community development in deep-sea hydrothermal vents. Curr. Opin. Microbiol. 14, 282–291. 10.1016/j.mib.2011.04.013 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. M., Ismail M. H., Cao B. (2021). Biodiversity of magnetotactic bacteria in the tropical marine environment of Singapore revealed by metagenomic analysis. Environ. Res. 194, 110714. 10.1016/j.envres.2021.110714 [DOI] [PubMed] [Google Scholar]
- Teng Z., Zhang Y., Zhang W., Pan H., Xu J., Huang H., et al. (2018). Diversity and characterization of multicellular magnetotactic prokaryotes from coral reef habitats of the Paracel Islands, South China Sea. Front. Microbiol. 9:2135. 10.3389/fmicb.2018.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres de Araujo F. F., Pires M. A., Frankel R. B., Bicudo C. E. M. (1986). Magnetite and magnetotaxis in algae. Brief Commun. 50, 375–378. 10.1016/S0006-3495(86)83471-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolard F., Duval S., Nitschke W., Ménez B., Pisapia C., Ben Nacib J., et al. (2022). Mineralogy, geochemistry and occurrences of fougerite in a modern hydrothermal system and its implications for the origin of life. Earth-Sci. Rev. 225, 103910. 10.1016/j.earscirev.2021.103910 [DOI] [Google Scholar]
- Uebe R., Schüler D. (2016). Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 14, 621–637. 10.1038/nrmicro.2016.99 [DOI] [PubMed] [Google Scholar]
- Usui Y., Yamazaki T. (2021). Non-chained, non-interacting, stable single-domain magnetite octahedra in deep-sea red clay: a new type of magnetofossil? Geochem. Geophys. Geosyst. 22, e2021GC009770. 10.1029/2021GC009770 [DOI] [Google Scholar]
- Uzun M., Alekseeva L., Krutkina M., Koziaeva V., Grouzdev D. (2020). Unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. Sci Data 7, 252. 10.1038/s41597-020-00593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vali H., Kirschvink J. L. (1989). Magnetofossil dissolution in a palaeomagnetically unstable deep-sea sediment. Nature 339, 203–206. 10.1038/339203a0 [DOI] [Google Scholar]
- Vallenet D., Calteau A., Dubois M., Amours P., Bazin A., Beuvin M., et al. (2019). MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 48, D579–D589. 10.1093/nar/gkz926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dobeneck T., Petersen N., Vali H. (1987). Bakterielle magnetofossilien. Geowiss. Unserer Zeit 1, 27–35. 10.2312/geowissenschaften.1987.5.27 [DOI] [Google Scholar]
- Wang L., Yu M., Liu Y., Liu J., Wu Y., Li L., et al. (2018). Comparative analyses of the bacterial community of hydrothermal deposits and seafloor sediments across Okinawa Trough. J. Mar. Syst. 180, 162–172. 10.1016/j.jmarsys.2016.11.012 [DOI] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009): Jalview Version 2—a multiple sequence alignment editor analysis workbench. Bioinformatics 25, 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T. (2020). Reductive dissolution of biogenic magnetite. Earth Plan. Space 72, 150. 10.1186/s40623-020-01290-3 [DOI] [Google Scholar]
- Yamazaki T., Kawahata H. (1998). Organic carbon flux controls the morphology of magnetofossils in marine sediments. Geology 26, 1064. [DOI] [Google Scholar]
- Yamazaki T., Shimono T. (2013). Abundant bacterial magnetite occurrence in oxic red clay. Geology 41, 1191–1194. 10.1130/G34782.1 [DOI] [Google Scholar]
- Yamazaki T., Suzuki Y., Kouduka M., Kawamura N. (2019). Dependence of bacterial magnetosome morphology on chemical conditions in deep-sea sediments. Earth Planet. Sci. Lett. 513, 135–143. 10.1016/j.epsl.2019.02.015 [DOI] [Google Scholar]
- Zhang R., Chen Y.-R., Du H.-J., Zhang W.-Y., Pan H.-M., Xiao T., et al. (2014). Characterization and phylogenetic identification of a species of spherical multicellular magnetotactic prokaryotes that produces both magnetite and greigite crystals. Res. Microbiol. 165, 481–489. 10.1016/j.resmic.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Zhou J., Bruns M. A., Tiedje J. M. (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322. 10.1128/aem.62.2.316-322.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM108105-OM108124 and https://www.ncbi.nlm.nih.gov/genbank/, PRJNA514953.