Abstract
The rapid detection of food-borne bacterial pathogens as part of a quality control program is necessary for the maintenance of a safe food supply. In this report, we present our findings for an immunocapture PCR method for the detection of Campylobacter jejuni in foods. The method permits direct detection of the pathogen without an enrichment step and can be performed in approximately 8 h. Assay results are quantitative, and one cell in a milliliter sample can be detected. Application of the method to spiked milk samples and chicken skin washes did not affect the sensitivity of the assay.
Campylobacter enteritis is a significant cause of morbidity throughout the world, with an estimated annual incidence of 400 million cases (15). In the United States, it has been estimated that 2.4 million cases occur annually, resulting in 120 to 360 deaths (17). Campylobacter jejuni is the species most commonly associated with the disease, although the prevalence of Campylobacter coli infections rivals that of C. jejuni in some countries (7). Despite the low mortality rate associated with Campylobacter enteritis, the disease has a significant economic impact, with an estimated annual cost of nearly $1 billion in the United States alone (17).
Campylobacters inhabit the intestinal tracts of a variety of mammals and birds (14), and they are transmitted from host to host via the fecal-oral route. In humans, infection results from the ingestion of contaminated foods, with as few as 500 CFU having been reported to cause disease (20). Campylobacters have been isolated from foods of both animal and nonanimal origin (13). Contaminated raw milk and untreated water are commonly associated with outbreaks of the disease (7); however, poultry, in particular chicken, is the most commonly implicated food source (18). Widespread contamination of poultry carcasses occurs during processing due to the release of intestinal contents during evisceration. Contamination rates of chicken carcasses as high as 93 to 98% (2, 18) have been reported, with C. jejuni counts on retail chicken often exceeding 103 cells per 100 g (1). Both the ubiquitousness and the potentially low infectious dose of these pathogens make their presence in the food supply a significant health hazard.
Due to the prevalence of Campylobacter species in the food supply, routine and reliable monitoring for these pathogens is necessary in order to reduce their impact upon human health. Cultivation methods involving enrichment, isolation, and biochemical characterization require 4 to 5 days to complete (4, 5). Due to the perishable nature of many food items, a more rapid detection method is necessary to feasibly monitor the potential sources of these pathogens. For this reason, we have developed an immunocapture PCR method for the detection of Campylobacter in foods. The assay utilizes an immunomagnetic capture technique to isolate the bacteria from food samples, followed by PCR amplification of the genomic DNA encoding rRNA. The PCR products are detected in a chromogenic assay, thus permitting quantitative analysis. This rapid and highly sensitive method requires approximately 8 h to perform and can detect a single cell in a milliliter of sample.
C. jejuni strain 81-176 was used for immunological reagent production and assay development. The organism was originally isolated from a 9-year-old girl suffering from Campylobacter enteritis during an outbreak in Minnesota (3), and it was provided to us by researchers at the Navy Medical Research Institute in Bethesda, Md. The organism was grown on Mueller-Hinton (M-H) agar (Difco Laboratories, Detroit, Mich.) under microaerophilic conditions (5% CO2, 10% O2, 85% N2) in Campy pouches (BBL, Cockeysville, Md.) at 37°C overnight. For easier handling, cells were killed by suspension in sterile 13 mM phosphate-buffered saline (PBS; 0.85% NaCl, pH 7.2) followed by the addition of formalin (37% formaldehyde) to a final concentration of 0.5% formaldehyde. The mixture was incubated at room temperature for 1 h. The killed cells were collected by centrifugation (3,200 × g), and the pellet was washed once with PBS containing 0.02% sodium azide (PBS-azide). Cell suspensions were tested for kill efficiency by plating on M-H agar. The killed cell preparations were stored at 4°C. Prior to injection into animals, the cells were removed from the PBS-azide by centrifugation and then resuspended in sterile PBS. Cell suspensions were quantified by optical density measurements at 600 nm and visual counts with a phase-contrast hemocytometer.
For studies on assay specificity, Escherichia coli strain 11775, Salmonella enterica serovar Enteritidis strain 13076, and Vibrio parahaemolyticus strain 17802 were purchased from the American Type Culture Collection (Manassas, Va.). The E. coli and Salmonella serovar Enteritidis strains were grown aerobically on M-H agar at 37°C, while the V. parahaemolyticus strain was grown on nutrient agar containing 3% NaCl at 37°C under aerobic conditions. Formalin-killed cell preparations were prepared as described for C. jejuni.
Polyclonal rabbit anti-Campylobacter antibodies were developed for the immunomagnetic capture of C. jejuni. New Zealand White rabbits (8 to 10 lb) were immunized subcutaneously and intradermally with 2 × 109 formalin-killed cells at intervals of 3 weeks. The primary inoculum was administered in complete Freund's adjuvant, while subsequent boosts were given in incomplete Freund's adjuvant. Antibody production was monitored using a whole-cell enzyme immunoassay (19). Following the third boost, the rabbits were bled intracardially under anesthesia. The immunoglobulin G (IgG) antibodies in the antisera were purified by affinity chromatography on protein A-Sepharose.
To facilitate the isolation of the target bacteria, the polyclonal anti-Campylobacter IgG was added directly to the sample to a final concentration of approximately 0.5 μg/ml. The mixture was then incubated for 30 min at room temperature with constant rotation to form antigen-antibody complexes. One hundred to two hundred micrograms of sheep anti-rabbit IgG-coated Dynabeads M-280 (Dynal, Lake Success, N.Y.) was then added to the suspension. The sample was mixed for 30 min, and then the beads and bound antibody-bacterium complexes were isolated from the mixture using a magnet. The bead pellet was washed once with TBST (10 mM Tris [pH 8], 150 mM NaCl, 0.05% Tween 20).
To release the C. jejuni genomic DNA, the captured cells were lysed using a modification of the method of Goldenberger et al. (6). The magnetic beads and captured cells were placed in 20 μl of digestion buffer (50 mM Tris-HCl [pH 8.5], 1 mM EDTA, 0.1% sodium dodecyl sulfate, and a 1-mg/ml concentration of proteinase K), vortexed, and then incubated at 55°C for 1 h. During incubation, the samples were vortexed at intervals of 15 min. After the lysis was completed, the proteinase K was heat inactivated by incubating the mixture at 95°C for 1 min. The lysate was cooled to 4°C and then centrifuged briefly, and 15 μl of the supernatant was used in PCR.
For amplification of C. jejuni genomic DNA by PCR, the primers reported by Giesendorf et al. (5) were used. Primers C442 (5′-GGAGGATGACACTTTTCGGAGC-3′) and C490 (5′-ATTACTGAGATGACTAGCACCCC-3′) were synthesized by a commercial vendor (Midland Certified Reagent Company, Midland, Tex.). Each primer was modified at the 5′-terminal nucleotide with the addition of a biotin group. These primers have been demonstrated to be specific for C. jejuni, C. coli, and Campylobacter lari and to recognize genomic regions that encode 16S rRNA.
For each amplification reaction, 1.25 U of Amplitaq Gold (Perkin-Elmer Corp., Norwalk, Conn.) and 0.1 U of HK-uracil N-glycosylase (Epicentre Technologies, Madison, Wis.) were utilized. In addition, the concentration of each primer was 70 nM; dATP, dCTP, dGTP, and dUTP were included at 200 μM each; fluoresceinated dUTP (FL-11-dUTP; Amersham Life Science, Arlington Heights, Ill.) was used at 1 μM; the MgCl2 concentration was 2.5 mM; and Tween 20 was used at 5% (vol/vol). The reaction mixtures (total volume = 50 μl) were heated at 50°C for 15 min to permit destruction of carryover PCR product by the uracil glycosylase. Each mixture was then incubated at 95°C for 10 min to inactivate the glycosylase and to activate the Amplitaq Gold. After this, the sample was heated at 52°C for 1 min to facilitate primer annealing, followed by 1 min at 72°C for chain extension. Subsequently, 34 cycles of the same annealing and extension steps, along with a denaturation step of 30 s at 95°C, were performed.
The PCR products were detected using an avidin capture assay. To achieve this, wells of an Immulon 2 microtiter plate (Dynatech Laboratories, Chantilly, Va.) were coated with 1 μg of avidin in 100 μl of 100 mM NaHCO3, pH 9.5, by incubation at room temperature for 2 h. The wells were washed with TBST and then blocked with 200 μl of 3% bovine serum albumin (BSA)–TBS–azide (10 mM Tris, pH 8, containing 150 mM NaCl, 0.02% NaN3, and 3% BSA). The block was performed at room temperature for 1 h, and then the wells were washed with TBST. Ten to twenty microliters of test sample was added to each well, along with sufficient 3% BSA–TBS–azide to produce a final volume of 100 μl. The sample was incubated in the well for 30 min at room temperature to facilitate immobilization of the PCR products via the biotin moieties that were coupled to the 5′-terminal nucleotides of primers C442 and C490. After the avidin capture was completed, the wells were washed with TBST. The immobilized PCR products were then labeled with an alkaline phosphatase (AP)-labeled antibody that recognized the fluorescein groups bound to the DNA as a result of FL-11-dUTP incorporation during chain elongation (10). Following a 30-min incubation at room temperature with the anti-fluorescein conjugate, the wells were washed, and 200 μl of 1-mg/ml para-nitrophenyl phosphate in AP development buffer (25 mM Tris [pH 9.5], 150 mM NaCl, 5 mM MgCl2, 0.02% NaN3) was added to each well. Color development was determined by measuring the absorbance at 405 nm using a microtiter plate reader.
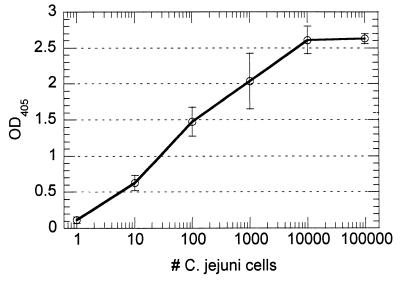
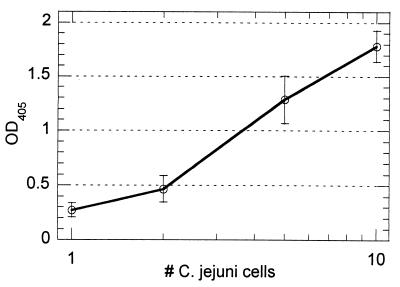
The combination of PCR amplification and the avidin capture assay produced a highly sensitive quantitative assay. A typical standard curve utilizing crude C. jejuni cell lysate is presented in Fig. 1. The linear range of the curve spans 3 to 4 logarithms, thus permitting analysis over a large range of cell concentrations. Reproducibility was good, with the coefficients of variation for duplicate samples falling in the range of 10 to 20%. For the standard curve in Fig. 1, statistical analysis of the data by Student's t test (one-tailed hypothesis) demonstrated a statistically significant (0.005 < P < 0.01) signal above background from a single C. jejuni cell, thus demonstrating the sensitivity of the approach.
FIG. 1.
Representative standard curve for detection of formalin-killed C. jejuni using PCR combined with avidin capture assay. PCR products were labeled by incorporation of FL-11-dUTP. Cell numbers were based upon visual counts prior to serial dilution. The average absorbance reading for the negative control (OD at 405 nm [OD405] = 0.014) was eightfold less than the reading for the 1-cell/ml sample.
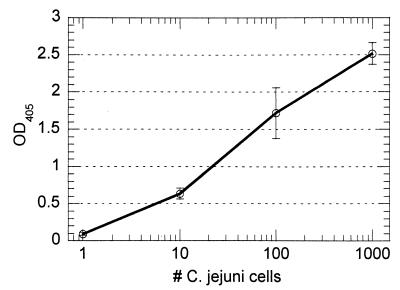
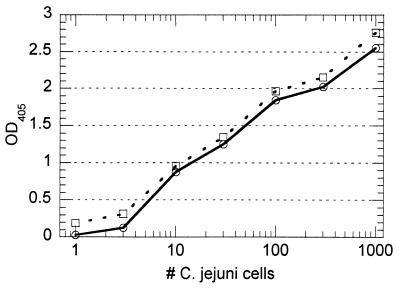
With pure suspensions of formalin-killed C. jejuni, the performance of the immunocapture PCR method was similar to that of direct PCR amplification of the crude cell lysate. A definite signal (P < 0.0005) was produced from one cell in a 1-ml sample volume (Fig. 2). The reproducibility of the method was good; the coefficients of variation for replicate samples ranged from 6 to 23%, with the greatest variability occurring at lower cell counts. The approach produced good target specificity, since the inclusion of 106 cells of E. coli (ATCC 11775) per ml did not have a significant effect upon the detection of C. jejuni (Fig. 3). Additional experimentation in the presence of 106 cells of either Salmonella serovar Enteritidis (ATCC 13076) or V. parahaemolyticus (ATCC 17802) per ml produced similar results (data not shown).
FIG. 2.
Performance of immunocapture PCR assay on suspensions of C. jejuni in PBS containing 3% BSA. Sample volumes were 1 ml, and samples had cell counts ranging from 0 to 1,000. A signal could be obtained from a single cell following 35 cycles of PCR. The background reading for the 0-cell control was 0.003. OD405, optical density at 405 nm.
FIG. 3.
Effect of background flora upon performance of immunocapture PCR. Standard curves developed in the absence (○) and presence (□) of 106 formalin-killed E. coli cells per ml are presented. The presence of a high concentration of formalin-killed E. coli did not affect the capacity of the assay to detect the target pathogen. OD405, optical density at 405 nm.
Since milk and chicken are commonly associated with Campylobacter infections, we evaluated the performance of the immunocapture PCR with these foods. Samples of both foods were purchased at local markets. For testing with milk, pasteurized whole milk was used, since raw milk was unavailable. Milk samples were diluted in sterile PBS and then spiked with formalin-killed cells. For testing with chicken samples, washes were prepared by removing the skin from chicken thighs and then placing it in sterile PBS (0.5 g of skin/ml). The sample was mixed for a few minutes at room temperature, after which the liquid was collected by decanting and spiked with formalin-killed cells.
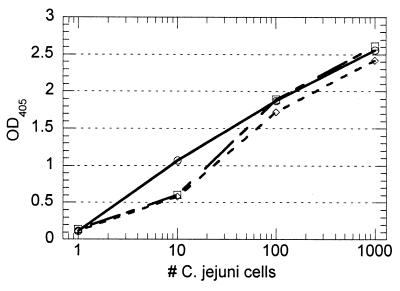
The presence of pasteurized whole milk (diluted 1/2 or 1/10 in buffer) had no adverse effect upon the performance of the assay, as demonstrated by the similar standard curves obtained in the presence and absence of milk (Fig. 4). The performance of the assay with chicken skin washes was also good, providing a detectable signal from one cell/ml (Fig. 5). These results were an indication that the immunocapture effectively removed the bacteria from the sample without transferring significant amounts of inhibitory components to the PCR.
FIG. 4.
Performance of immunocapture PCR with spiked pasteurized whole milk. Standard curves were developed with spiked milk samples diluted in buffer 1/2 (○) or 1/10 (□). Cell concentrations ranging from 0 to 1,000/ml were used. No apparent differences in the test curves were determined relative to the buffer control (◊). OD405, optical density at 405 nm.
FIG. 5.
Performance of immunocapture PCR with spiked chicken skin wash (0.5 g of skin/ml of PBS). Based upon analysis of readings by Student's t test (one-tailed hypothesis), a signal could be achieved from a single cell per ml of sample (0.025 < P < 0.010). The coefficients of variation for replicates ranged from 8 to 24%. The average reading for the 0-cell control was 0.109. OD405, optical density at 405 nm.
Subsequent experimentation utilized chicken skin samples that were spiked with formalin-killed C. jejuni and then washed. This work was intended to evaluate the effectiveness of sample processing with regard to the recovery of cells from the starting material. Using the wash procedure, cells spiked onto the chicken skin could be recovered, resulting in the quantitation of four cells per g of skin during an in-house single blind study. Although they are preliminary, the results from two such studies suggest that the efficiency of recovery achieved by the wash method is at best 50%, relative to standards prepared by direct inoculation of skin washes with known cell counts.
The complexity of food samples complicates the reliable performance of direct PCR, as is the case for clinical and environmental specimens (9, 11, 12, 16, 22). To avoid inhibition of PCR by food components, the reported method involves isolation of the target pathogen by immunocapture prior to the PCR amplification step. This approach permits the detection of low concentrations of C. jejuni without the need for selective enrichment of the bacteria (5, 8) or purification of the target DNA (21), which are required by other reported direct PCR methods. As an added advantage, utilization of immunocapture permits cultivation of the isolated bacteria by simply inoculating the immunomagnetic beads onto culture medium. The isolation method also provides concentration of the target bacteria, thus permitting analysis of samples that would be too dilute for use in other direct PCR protocols due to sample volume restrictions.
The majority of reported PCR methods, including those for Campylobacter, require gel electrophoresis for the detection of PCR products. Although these methods are sensitive and relatively rapid, they do not permit easy analysis of large numbers of samples unless expensive automated machinery is utilized. In addition, quantitative analysis of electrophoresis results requires densitometric analysis and thus additional equipment. For the reported method, the PCR products are labeled during the amplification process through the incorporation of FL-11-dUTP as reported by Luk in 1994 (10). The procedure capitalizes upon the ability of Taq DNA polymerase to incorporate modified dUTP in place of dTTP into the PCR product and the ability of the modified PCR product to be used as a template for subsequent rounds of amplification. Following their capture in avidin-coated wells, the PCR products containing the FL-11-dUTP are subsequently detected using an AP-labeled, antifluorescein antibody, followed by incubation with a chromogenic AP substrate.
One limitation of the fluorescein incorporation method is that it is susceptible to background signal resulting from primer-dimer formation. When conditions are optimized, however, the assay system can be reliably performed, providing highly sensitive quantitative results. In situations where primer-dimer formation cannot be controlled without significantly reducing the performance of the assay, we have adapted the system to utilize a hybridization probe to detect the PCR products in the avidin capture assay. This modified approach has provided good performance with a slight reduction in sensitivity (results not presented). Both this system and real-time PCR provide quantitative results. However, the reported assay system is much more affordable, as it does not require specialized equipment but rather utilizes laboratory devices that are commonly available (i.e., a thermal cycler and a plate reader).
The dependence of the system upon immunocapture for isolation of the target organism mandates the utilization of antibody reagents with the appropriate affinity and specificity for the target pathogen. The work presented in this paper was performed utilizing polyclonal antibodies that were produced by immunizing rabbits with a single strain (81-176) of C. jejuni. These antibodies provided efficient capture, as evidenced by the production of a signal from a target concentration of one cell/ml. In limited testing with pure cultures, we have scaled up the approach to test a larger sample size (i.e., 10 ml) and have found that detection of cell concentrations of less than one cell/ml can be achieved. Also, we have seen no differences in assay performance between live and formalin-killed cells or cells with different morphologies (i.e., spiral rods versus coccobacilli) in pure culture, thus indicating comparable capture efficiencies among different C. jejuni 81-176 cell types. The strain specificity of the antibodies, however, has proven to be too narrow, as we have encountered significant variance among capture efficiencies of different C. jejuni strains. Therefore, future work will involve the development of antibody reagents with broader strain specificity.
Detection of pathogenic Campylobacter by cultivation and biochemical characterization is a time-consuming process; thus, more rapid methods of detection are highly desirable. In this report, we have presented our results from an immunocapture PCR method for the detection of C. jejuni in foods. The method permits direct detection of this bacterial pathogen without the need for enrichment and can be performed in approximately 8 h, thus making it more rapid than other direct PCR methods or cultivation. It is also extremely sensitive, producing a signal from a single cell in 1 ml of sample. The assay is highly specific, being unaffected by the presence of 106 cells of E. coli, Salmonella serovar Enteritidis, or V. parahaemolyticus per ml. Finally, the method produces quantitative results, thus providing information beyond simply indicating the presence or absence of the pathogen. Based upon these characteristics, we believe that the assay has potential as a surveillance method for the maintenance of food safety.
Acknowledgments
This work was supported by Small Business Innovation Research grant R44 FD-01536 from the Food and Drug Administration.
We thank Julia Leung for reviewing the manuscript.
REFERENCES
- 1.Altekruse S F, Stern N J, Fields P I, Swerdlow D L. Campylobacter jejuni—an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berndtson E, Tivemo M, Engvall A. Distribution and numbers of Campylobacter in newly slaughtered broiler chickens and hens. Int J Food Microbiol. 1992;15:45–50. doi: 10.1016/0168-1605(92)90134-o. [DOI] [PubMed] [Google Scholar]
- 3.Black R E, Perlman D, Clements M L, Levine M M, Blaser M J. Human volunteer studies with Campylobacter jejuni. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 207–215. [Google Scholar]
- 4.Docherty L, Adams M R, Patel P, McFadden J. The magnetic immuno-polymerase chain reaction assay for the detection of Campylobacter in milk and poultry. Lett Appl Microbiol. 1996;22:288–292. doi: 10.1111/j.1472-765x.1996.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 5.Giesendorf B A J, Quint W G V, Henkens M H C, Stegeman H, Huf F A, Niesters H G M. Rapid and sensitive detection of Campylobacter spp. in chicken products by using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3804–3808. doi: 10.1128/aem.58.12.3804-3808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenberger D, Perschil I, Ritzler M, Altwegg M. A simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths P L, Park R W A. Campylobacters associated with human diarrhoeal disease. J Appl Bacteriol. 1990;69:281–301. doi: 10.1111/j.1365-2672.1990.tb01519.x. [DOI] [PubMed] [Google Scholar]
- 8.Hazeleger W, Arkesteijn C, Toorop-Bouma A, Beumer R. Detection of the coccoid form of Campylobacter jejuni in chicken products with the use of the polymerase chain reaction. Int J Food Microbiol. 1994;24:273–281. doi: 10.1016/0168-1605(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 9.Islam D, Lindberg A A. Detection of Shigella dysenteriae type 1 and Shigella flexneri in feces by immunomagnetic isolation and polymerase chain reaction. J Clin Microbiol. 1992;30:2801–2806. doi: 10.1128/jcm.30.11.2801-2806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luk J M C. A PCR enzyme immunoassay for detection of Salmonella typhi. BioTechniques. 1994;17:1038–1042. [PubMed] [Google Scholar]
- 11.Oyofo B A, Rollins D M. Efficacy of filter types for detecting Campylobacter jejuni and Campylobacter coli in environmental water samples by polymerase chain reaction. Appl Environ Microbiol. 1993;59:4090–4095. doi: 10.1128/aem.59.12.4090-4095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panaccio M, Georgesz M, Lew A M. FoLT PCR: a simple PCR protocol for amplifying DNA directly from whole blood. BioTechniques. 1993;14:238–243. [PubMed] [Google Scholar]
- 13.Park C E, Sanders G W. Occurrence of thermotolerant campylobacters in fresh vegetables sold at farmers' outdoor markets and supermarkets. Can J Microbiol. 1992;38:313–316. doi: 10.1139/m92-052. [DOI] [PubMed] [Google Scholar]
- 14.Park R W A, Griffiths P L, Moreno G S. Sources and survival of campylobacters: relevance to enteritis and the food industry. Soc Appl Bacteriol Symp Suppl. 1991;70:97S–106S. [PubMed] [Google Scholar]
- 15.Rollwagen F M, Pacheco N D, Clements J D, Pavlovskis O, Rollins D M, Walker R I. Killed Campylobacter elicits immune response and protection when administered with an oral adjuvant. Vaccine. 1993;11:1316–1320. doi: 10.1016/0264-410x(93)90101-3. [DOI] [PubMed] [Google Scholar]
- 16.Scholl D R, Kaufmann C, Jollick J D, York C K, Goodrum G R, Charache P. Clinical application of novel sample processing technology for the identification of salmonellae by using DNA probes. J Clin Microbiol. 1990;28:237–241. doi: 10.1128/jcm.28.2.237-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skirrow M B, Blaser M J. Clinical and epidemiologic considerations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 3–8. [Google Scholar]
- 18.Stern N J. Reservoirs for Campylobacter jejuni and approaches for intervention in poultry. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni. Current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 49–60. [Google Scholar]
- 19.van den Bosch J F, Hendriks J H I M, Gladigau I, Willems H M C, Storm P K, de Graaf F K. Identification of F11 fimbriae on chicken Escherichia coli strains. Infect Immun. 1993;61:800–806. doi: 10.1128/iai.61.3.800-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker R I, Caldwell M B, Lee E C, Guerry P, Trust T J, Ruiz-Palacios G M. Pathophysiology of Campylobacter enteritis. Microbiol Rev. 1986;50:81–94. doi: 10.1128/mr.50.1.81-94.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegmüller B, Lüthy J, Candrian U. Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl Environ Microbiol. 1993;59:2161–2165. doi: 10.1128/aem.59.7.2161-2165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]