Abstract
We have developed a rapid procedure for the detection of virulent Yersinia enterocolitica in ground pork by combining a previously described PCR with fluorescent dye technologies. The detection method, known as the fluorogenic 5′ nuclease assay (TaqMan), produces results by measuring the fluorescence produced during PCR amplification, requiring no post-PCR processing. The specificity of the chromosomal yst gene-based assay was tested with 28 bacterial isolates that included 7 pathogenic and 7 nonpathogenic serotypes of Y. enterocolitica, other species of Yersinia (Y. aldovae, Y. pseudotuberculosis, Y. mollaretti, Y. intermedia, Y. bercovieri, Y. ruckeri, Y. frederiksenii, and Y. kristensenii), and other enteric bacteria (Escherichia, Salmonella, Citrobacter, and Flavobacterium). The assay was 100% specific in identifying the pathogenic strains of Y. enterocolitica. The sensitivity of the assay was found to be ≥102 CFU/ml in pure cultures and ≥103 CFU/g in spiked ground pork samples. Results of the assay with food enrichments prespiked with Y. enterocolitica serotypes O:3 and O:9 were comparable to standard culture results. Of the 100 field samples (ground pork) tested, 35 were positive for virulent Y. enterocolitica with both 5′ nuclease assay and conventional virulence tests. After overnight enrichment the entire assay, including DNA extraction, amplification, and detection, could be completed within 5 h.
Yersinia enterocolitica is a foodborne pathogen which causes gastrointestinal disorders with a wide range of clinical manifestations, from mild diarrhea to mesenteric lymphadenitis (5). The association of human illness with consumption of Y. enterocolitica-contaminated food, animal wastes, and unchlorinated water is well documented. This organism is known to contaminate refrigerated foods due to its psychotrophic nature and is frequently associated with ground pork and other ground meats. The species comprises a heterogeneous group of organisms with more than 50 serotypes and several biotypes (27). However, the pathogenic serotypes commonly associated with human yersiniosis are limited to European strains (O:1,3; O:3; O:9; and O:5) (26) and American strains (O:8; O:13a,13b; O:20; O:21; O:18; and O:4) (31). Virulence in Y. enterocolitica results from a complex interplay between a series of plasmid-borne and chromosomal genes (7, 20, 32). The latter include yst, the chromosomal gene encoding a low-molecular-weight, heat-stable enterotoxin, characteristic only of the virulent strains of Y. enterocolitica (10, 27). Pathogenic Y. enterocolitica strains are characterized by their ability to adhere to and invade epithelial cells (21, 26). This function is encoded by a genetic locus on the chromosome known as the ail gene (20). Once localized within the target cells, Y. enterocolitica is able to resist the primary immune response of the host. This resistance depends on the presence of a 70-kb virulence plasmid, pYV, which directs secretion of two major groups of proteins, called Yops and YadA, that are known to interfere with the functioning of phagocytic cells. Most human isolates of Y. enterocolitica produce a heat-stable enterotoxin with properties similar to those of the heat-stable enterotoxin of enterotoxigenic Escherichia coli (1).
Phenotypic differentiation between virulent and avirulent strains of Y. enterocolitica requires a series of biochemical and serological tests that are time consuming and that produce inconsistent results (23). Several investigators have developed PCR and DNA probe techniques for the detection of pathogenic Yersinia (9, 10, 14, 19, 22, and 28). The PCR technique has shown great promise as a highly sensitive and specific method but has several limitations. The amplified product must be detected in order to prove its presence, and a variety of methods, like gel electrophoresis and Southern blotting and dot blot hybridizations with labeled (chemical or radioactive) probes, have been used. However, these methods are time consuming and laborious, requiring skill and multistep processing that add to the cost and complexity of the test. The number of samples that can be analyzed at any one time is also limited. Furthermore, ethidium bromide, which is used to stain agarose gels, is a mutagen and is not appropriate for routine use in food-monitoring laboratories (24).
Recently, 5′ nuclease assays have been described that allowed the automated PCR amplification, detection, and analysis of Salmonella spp. (4, 13, and 18), Listeria monocytogenes (2, 3), E. coli O157:H7 (24), and Shiga-like toxin genes (30; M. S. Y. Ho, S. J. A. Flood, and C. Paszko-Kolva, Abstr. 97th Gen. Meet. Am. Soc. Microbiol. 1997, abstr. P-17, p. 439, 1997) in various foods. The 5′ assay exploits the 5′→3′ activity of Thermus aquaticus DNA polymerase (8, 17) to hydrolyze an internal TaqMan probe labeled with a fluorescent reporter dye and a quencher dye (16). The probe is designed to hybridize to an internal region of the targeted sequence. For the intact probe, the fluorescence from the reporter is suppressed by the quencher dye due to its spatial proximity to the reporter. As the PCR amplification proceeds, the annealed probe is hydrolyzed by the Taq DNA polymerase, separating the two dyes and increasing the reporter fluorescence signal that can be detected on a fluorometer (ABI Prism 7200 sequence detection system; PE Applied Biosystems). Because the increase in fluorogenic reporter signals is a direct consequence of a successful PCR, this procedure can be used in the detection of specific DNA sequences. The fluorometric data can be automatically read and interpreted using a 96-sample format and presented as positive or negative conclusions as to the presence or absence of the DNA within 15 min of completion of the PCR.
In this report, we describe the development and validation of a 5′ nuclease assay for the rapid (within 24 h) detection and analysis of virulent Y. enterocolitica in ground pork. The assay targets the heat-stable enterotoxin gene (yst), characteristic only of the virulent strains of Y. enterocolitica. The sequences for primers Pr2a and Pr2c and the probe previously described by Ibrahim et al. (10) were modified to suit the 5′ nuclease assay.
Bacterial strains and culture conditions.
Bacterial strains that were evaluated are listed in Table 1. The Y. enterocolitica serotypes O:3 and O:9 were used as the reference strains in all optimization and sensitivity experiments. The Y. enterocolitica strains were enriched in peptone-sorbitol-bile broth (PSBB), incubated (35°C, 8 to 16 h), plated on cefsulodin-Irgasan-novobiocin (CIN) agar (Oxoid, Basingstoke, Hampshire, England), and incubated again (35°C, 18 to 24 h).
TABLE 1.
Bacterial strains evaluated and the ΔRQ values generated in PCR with the Pr2a and Pr2c primers and the Yer-prb fluorogenic probe
| Bacterium evaluated | ΔRQ valuea | Interpretationb |
|---|---|---|
| Y. enterocolitica (virulent serotypes) | ||
| O:3c | 29.26 | Positive |
| O:9c | 29.95 | Positive |
| O:8c | 29.65 | Positive |
| O:18c | 29.56 | Positive |
| O:20c | 29.13 | Positive |
| O:21c | 29.38 | Positive |
| O:13c | 29.39 | Positive |
| Y. enterocolitica (avirulent serotypes) | ||
| O:22c | 3.36 | Negative |
| O:22d | 3.19 | Negative |
| O:7c | 2.89 | Negative |
| O:7e | 3.26 | Negative |
| O:10d | 3.05 | Negative |
| O:10e | 3.20 | Negative |
| O:34e | 3.33 | Negative |
| Other species of Yersinia | ||
| Y. aldovaec | 2.24 | Negative |
| Y. bercovieric | 2.05 | Negative |
| Y. frederikseniic | 2.67 | Negative |
| Y. intermediaic | 2.71 | Negative |
| Y. kristenseniic | 1.93 | Negative |
| Y. mollarettic | 2.29 | Negative |
| Y. pseudotuberculosisf | 1.85 | Negative |
| Y. ruckeric | 2.82 | Negative |
| Other bacteria | ||
| Citrobacter freudiniif | 2.24 | Negative |
| E. colig | 1.26 | Negative |
| E. coli O157:H7g | 1.49 | Negative |
| Flavobacterium spp.g | 1.24 | Negative |
| Salmonella enterica serotype chloleraesiusg | 0.81 | Negative |
| S. enterica serotype Typhimuriumg | 1.54 | Negative |
Determined with ∼107 CFU/ml for each strain.
A positive score is assigned when the ΔRQ is greater than the ΔRQ threshold, based on a fourfold average value of the no-template controls.
From the National Animal Disease Center, United States Department of Agriculture. Ames, Iowa.
From the Centers for Disease Control and Prevention, Atlanta, Ga.
From the Food Microbiology Culture Collection, Kansas State University, Manhattan.
From M. M. Chengappa, Veterinary Diagnostic Laboratory, Kansas State University.
From R. D. Oberst, Food Animal Health and Management Center, Kansas State University.
Development of 5′ nuclease assay probe for detection of virulent Y. enterocolitica.
The primers (Prla, 5′ AATGCTGTCTTCATTTGGAGC 3′, and Prlb, 5′ ATCCCAATCACTACTGACTTC 3′) and probe sequences reported by Ibrahim et al. (10) were modified to suit the requirements of the 5′ nuclease assay. The fluorogenic probe (Yer-Prb FAM-CAAGCAAGCTTGTGATCCTCCG-TAMRA) specific to the yst gene (EMBL database, accession number X69218) was synthesized as previously described (2). The probe was an internal fluorogenic probe labeled with the reporter dye (FAM-6-carboxy-fluorescein) at the 5′ end and a quencher (TAMRA-6-carboxytetramethylrhodamine) at the 3′ end (PE Applied Biosystems). Fluorescence was detected using a luminescence spectrometer (ABI Prism 7200 sequence detection system; PE Applied Biosystems) with a 96-well plate reader using the equation ΔRQ = RQ+ − RQ− (2). A positive interpretation for pathogenic Y. enterocolitica was based on a threshold of four times the average ΔRQ value of no-template controls (25) from individual 96-well optical reaction plates (three no-template controls per plate). This enabled us to eliminate a lot of avirulent strains that fell in the positive interpretation range.
DNA extraction procedure.
The DNA extraction procedure utilized the chelating properties of Chelex resin. Tenfold serial dilutions were made of PSBB cultures. Aliquots of 1 ml from each dilution were centrifuged (14,000 × g, 3 min), the supernatant was decanted carefully, and the pellet was resuspended in 200 μl of thoroughly mixed PrepMan sample preparation reagent (PE Applied Biosystems). The tubes were vortexed for 5 to 10 s or as long as required to resuspend the pellet, floated in boiling water (10 min), and chilled on ice (5 min). Then the tubes were centrifuged (14,000 × g, 3 min), and the supernatants were carefully transferred to new microcentrifuge tubes. A 2.5-μl aliquot of the supernatant served as the template for each PCR amplification in the 5′ nuclease assay.
PCR conditions.
The PCR amplification conditions were different from those described by Ibrahim et al. (10). Briefly, 2.5 μl of sample containing the DNA template to be evaluated was added to 22.5 μl of PCR master mix (2.5 μl of 1× PCR buffer II [Perkin-Elmer], 2.5 to 6.0 mM MgCl2, 300 nM concentrations of each primer [Pr2a and Pr2c], 200 μM deoxynucleoside triphosphate, 0.025 U of AmpliTaq DNA polymerase [Perkin-Elmer], 40 nM fluorogenic probe, and 22.5 μl of water) in disposable 96-well optical reaction plates (PE Applied Biosystems). Each set of reaction mixtures included a single row of wells of Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA) for the autozero control and triplicate wells that were no-template controls (containing no Y. enterocolitica DNA templates). Each assay also included DNA from the avirulent strains of Y. enterocolitica, other species of Yersinia, and other bacteria listed in Table 1. All other bacteria were cultivated in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) at 37°C for 15 h. The PCR had an initial denaturing step (95°C, 5 min) followed by 35 amplification cycles of a two-step PCR (94°C, 30 s; 44°C, 30 s, and 72°C, 30 s), with a final extension (72°C, 10 min) on a thermocycler (GeneAmp PCR system 9600; Perkin-Elmer).
Specificity studies with pure cultures.
Specificity studies were performed utilizing DNA extracted from the Yersinia species and serotypes and other bacteria (Table 1). Pure cultures of virulent and avirulent Y. enterocolitica were grown in PSBB (35°C, 8 to 16 h) and the other Yersinia spp. and bacteria were grown in brain heart infusion broth (37°C, 8 to 16 h). Sensitivity studies utilizing pure cultures of Y. enterocolitica serotypes O:3 and O:9 also were performed to identify the lower detection limit of the 5′ nuclease assay. The cultures were grown overnight, serially diluted (10-fold), and enumerated on CIN plates. Then DNA was extracted using PrepMan sample preparation reagent and was run through the 5′ nuclease assay. The experiment was replicated three times. All the virulent Y. enterocolitica strains gave a positive reaction with the ΔRQ values above the threshold of 4.44 (four times the average ΔRQ of the no-template controls), whereas the avirulent Y. enterocolitica, Yersinia spp., and other bacterial species gave a “no” interpretation with ΔRQ values below the threshold. Similar to the PCR results described by Ibrahim et al. (10), the assay was found to be 100% specific in identifying the pathogenic strains of Y. enterocolitica.
Sensitivity studies with pure cultures and spiked ground pork samples.
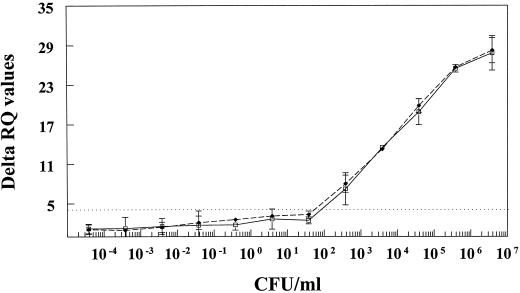
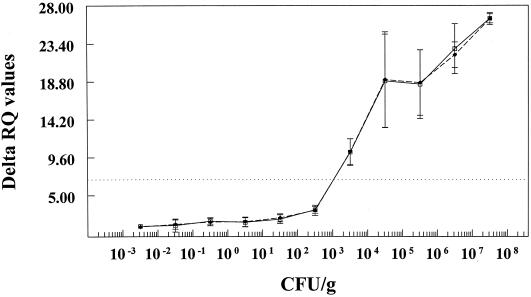
Sensitivity studies were performed on pure cultures of virulent Y. enterocolitica serotypes (O:3 and O:9) to test the lower detection limit of the fluorogenic 5′ nuclease assay. When pure cultures of O:3 and O:9 grown in PSBB for 8 to 16 h were enumerated and run through the DNA extraction and fluorogenic 5′ nuclease assay, the ΔRQ values were greater than the detection threshold of 4.44 when ≥102 CFU/ml were present (Fig. 1). Dividing the lowest dilution of the culture that gave a positive reaction by the approximate final volume of the DNA extraction (CFU per microliter) gave a lower detection limit of ≥10 CFU per PCR. In our assay, the lowest detection limit was 9.4 CFU/PCR. This was followed by DNA extraction, PCR, and fluorescence detection. This experiment was repeated three times.
FIG. 1.
Sensitivity of the fluorogenic 5′ nuclease assay for detecting pathogenic Y. enterocolitica in disposable 96-well optical reaction plates (PE Applied Biosystems). Tenfold dilutions of Y. enterocolitica serotypes O:3 (□) and O:9 (⧫) were made in PSBB in triplicate. One-milliliter aliquots from each dilution were subjected to DNA extraction using the PrepMan method, and 2.5 μl of the recovered DNA solution was PCR-amplified using the Pr2a and Pr2c primers in the presence of the Yer-prb fluorogenic probe. Detection and analysis were completed with the ABI Prism sequence detection system. All amplification and detection reactions were completed in 96-well optical reaction plates. The average ΔRQ values for each dilution were plotted against the average number of CFU per milliliter as determined by plating each dilution on CIN agar. The adjusted ΔRQ threshold value was calculated to be 4.44 (dashed horizontal line). Error bars indicate the standard deviations of the means.
Ground pork obtained from local grocery stores in Manhattan, Kans., was confirmed to be culture negative for Y. enterocolitica. Then 25-g samples were spiked with 103 CFU of Y. enterocolitica serotypes O:3 and O:9, enriched in 225 ml of PSBB, and incubated at 35°C for 12 h. The sensitivity of the 5′ nuclease assay in identifying virulent Y. enterocolitica in spiked ground pork samples was found to be approximately 103 CFU/g with a threshold ΔRQ value of 6.68 and above (Fig. 2). The sensitivity for spiked samples was calculated to be ≥102 CFU/PCR. In our assay, the lowest detection limit per PCR was 36 CFU for spiked samples.
FIG. 2.
Sensitivity of the fluorogenic 5′ nuclease assay for detecting pathogenic Y. enterocolitica in spiked ground pork samples. Tenfold dilutions of Y. enterocolitica serotypes O:3 (□) and O:9 (⧫) in PSBB in triplicate after enrichment at 35°C for 12 h. Aliquots (1 ml) were collected for DNA recovery using the PrepMan extraction method, and 2.5 μl of the recovered DNA solution was amplified with the Pr2a and Pr2c primers in the presence of the Yer-prb fluorogenic probe. Detection and analysis were completed with the ABI Prism sequence detection system. All amplification and detection reactions were completed in 96-well optical reaction plates. The average ΔRQ values determined from DNA recovered from both serotypes were plotted against the average number of CFU per milliliter determined by plating each ground pork dilution on CIN agar. The ΔRQ threshold value at four times the average of no-template controls was calculated to be 6.68 (dashed horizontal line). Error bars indicate the standard deviations of the means.
Unknown sample study.
A hundred samples of ground pork purchased at three different grocery stores in Manhattan, Kans., were enriched in PSBB (25 g of sample in 225 ml of PSBB) for 18 h and then tested for the presence of virulent Y. enterocolitica by conventional culture methods, two virulence tests, and the 5′ nuclease assay. One loopful of the preenriched sample was streaked on CIN agar plates which were incubated for 24 to 48 h at 35°C. After 48 h, suspected Y. enterocolitica colonies were tested using the API 20E system (bioMerieux, Hazelwood, Mo.). All the colonies that tested positive for Y. enterocolitica by the API 20E System were further tested for virulence by autoagglutination and crystal violet binding tests (29). The conventional methods identified Y. enterocolitica in 45 of the 100 samples of ground pork. Out of these 45 samples, only 35 were identified as being virulent by 5′ nuclease assay. These 35 isolates were also the only samples that were positive for virulence by the crystal violet binding and autoagglutination tests.
The described fluorogenic 5′ nuclease assay was successful in detecting virulent strains of Y. enterocolitica within 5 h after an 18 h enrichment. Positive interpretations were obtained for all reference strains of virulent Y. enterocolitica that were evaluated for pure cultures of serotypes O:3 and O:9 and for ground pork samples spiked with serotypes O:3 and O:9. This automated amplification and detection procedure could be a reliable rapid-screening method for detecting virulent Y. enterocolitica DNA.
Sensitivity studies involving pure cultures and spiked ground pork enrichments demonstrated that the assay was reliably sensitive, with lower detection limits of 102 to 103 CFU/ml or CFU/g under both conditions. Based on these data, a ΔRQ value of 4.44 and above was considered positive for the presence of virulent Y. enterocolitica in pure cultures, and a ΔRQ value of 6.68 and above was considered positive for spiked ground pork samples.
The virulent Y. enterocolitica strains among the 28 bacterial cultures tested for specificity gave positive interpretations, with ΔRQ values above 6.68 in all three replications. Therefore, this assay was found to be 100% specific for detecting pathogenic strains of Y. enterocolitica.
The unknown sample study was carried out to evaluate the use of the 5′ nuclease assay in actual food testing. The results of this assay were consistent with those of the genotypic virulence tests and those of cultural methods. Because not all strains of Y. enterocolitica are virulent, virulence tests are needed in order to determine if the food sample harbors a virulent strain. Most of these virulence tests are time consuming and laborious and require 24 to 30 h of incubation after selective enrichment and isolation, which require 2 to 3 days. Identifying virulent Y. enterocolitica in food or clinical samples by cultural methods requires 4 to 5 days, which adds even more time and expense to the detection procedure.
There are considerable difficulties associated with the isolation of Y. enterocolitica from foods (11, 15). Most methods require time-consuming enrichments to achieve optimal isolation. Moreover, no currently available method allows optimal recovery of all virulent serotypes. However, the 5′ nuclease assay developed by us requires only 5 h of processing (DNA extraction, PCR amplification, and detection) after overnight enrichment. The problem with all selective media described so far, including CIN, is that they provide inadequate differentiation between virulent and avirulent Y. enterocolitica (11). The avirulent variants are common in many foods, and their colony morphology makes them difficult to distinguish from the virulent strains (15).
Other molecular systems for detecting pathogenic Y. enterocolitica, such as multiplex riboprobes (7) that used a pool of RNA probes specific for various chromosomal and plasmid-borne virulence genes, oligonucleotide probes (11) for the detection of virulent Y. enterocolitica and PCR (31) require additional processing steps that increase the analysis time. The main advantage of this 5′ nuclease assay is the rapidity with which it screens for the presence of virulent Y. enterocolitica. The detection system uses a 96-well fluorescence plate reader with optical tubes and caps, so 96 samples can be analyzed at a time. Plate readings do not require tubes to be opened after amplification, so the potential for carryover is reduced. Therefore, this 5′ nuclease assay is more applicable than other systems for the detection of pathogenic Y. enterocolitica in food production and processing facilities.
Acknowledgments
This research was funded by the State Research Extension Educational Service, USDA, under agreement no. 34211-8362, Food Safety Consortium.
We thank Irene Wesley, National Animal Disease Center, USDA (Ames, Iowa), for providing the cultures for the study.
Footnotes
Contribution no. 00-171-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Amirmozafari N, Robertson D C. Nutritional requirements for the synthesis of heat-stable enterotoxin by Yersinia enterocolitica. Appl Environ Microbiol. 1993;59:3314–3320. doi: 10.1128/aem.59.10.3314-3320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batt C A. Molecular diagnostics for dairy-borne pathogens. J Dairy Sci. 1997;80:220–229. doi: 10.3168/jds.S0022-0302(97)75931-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Yee A, Griffiths M, Larkin C, Yamashiro C T, Behari R, Paszko-Kolva C, Rahn K, De Grandis S A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol. 1997;35:239–250. doi: 10.1016/s0168-1605(97)01241-5. [DOI] [PubMed] [Google Scholar]
- 5.Cover T L, Aber R C. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 6.Fliss I, Blais B W, Holley R, Simard R E. Multiplex riboprobes for the detection of virulent Yersinia enterocolitica and simple methods for their preparation. J Appl Bacteriol. 1995;79:195–202. doi: 10.1111/j.1365-2672.1995.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 7.Gemski P, Lazere J R, Casey T. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect Immun. 1980;27:682–685. doi: 10.1128/iai.27.2.682-685.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim A, Liesack W, Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992;30:1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim A, Liesack W, Griffiths M W, Robins-Browne R M. Development of a highly specific assay for rapid identification of pathogenic strains of Yersinia enterocolitica based on PCR amplification of the Yersinia heat-stable enterotoxin gene (yst) J Clin Microbiol. 1997;35:1636–1638. doi: 10.1128/jcm.35.6.1636-1638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapperud G, Dommarsnes K, Skurnik M, Hornes E. A synthetic oligonucleotide probe and a cloned polynucleotide probe based on the yopA gene for detection and enumeration of virulent Yersinia enterocolitica. Appl Environ Microbiol. 1990;56:17–23. doi: 10.1128/aem.56.1.17-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapperud G. Yersinia enterocolitica in food hygiene. Int J Food Microbiol. 1991;12:1242–1248. doi: 10.1016/0168-1605(91)90047-s. [DOI] [PubMed] [Google Scholar]
- 13.Kimura B, Kawasaki S, Fujii T, Kusunoki J, Itoh T, Flood S J A. Evaluation of TaqMan PCR assay for detecting Salmonella in raw meat and shrimp. J Food Prot. 1999;62:329–335. doi: 10.4315/0362-028x-62.4.329. [DOI] [PubMed] [Google Scholar]
- 14.Kwaga J, Iversen J O, Misra V. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J Clin Microbiol. 1992;30:2668–2673. doi: 10.1128/jcm.30.10.2668-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee L A, Gerber A R, Lonsway D R, Smith J D. Yersinia enterocolitica O:3 infections in infants and children, associated with the household preparation chitterlings. N Engl J Med. 1990;322:984–987. doi: 10.1056/NEJM199004053221407. [DOI] [PubMed] [Google Scholar]
- 16.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyamichev V, Brow M A D, Dahlberg J E. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 18.Matsuura M, Yamashiro C, Flood S, Paszko-Kolva C. Detection of Salmonella in food using a fluorogenic 5′ nuclease assay. Am Environ Lab. 1997;March:24–25. [Google Scholar]
- 19.Michiels T, Vanooteghem J-C, Lambert de Rouroit C L, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of the epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller V L, Farmer III J J, Hill W E, Falkow S. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect Immun. 1989;57:121–131. doi: 10.1128/iai.57.1.121-131.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima H, Inoue M, Mori T, Itoh K I, Arakawa E, Watanabe H. Detection and identification of Yersinia pseudotuberculosis and pathogenic Yersinia enterocolitica by an improved polymerase chain reaction method. J Clin Microbiol. 1992;30:2484–2486. doi: 10.1128/jcm.30.9.2484-2486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble M A, Barteluk R L, Freeman H J, Subramaniam R, Hudson J B. Clinical significance of virulence-related assay of Yersinia species. J Clin Microbiol. 1987;25:802–807. doi: 10.1128/jcm.25.5.802-807.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberst R D, Hays M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J A, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Vizcaino J M, Cambro-Alvarez M. Enzyme immunoassay techniques, ELISA, in animal and plant disease. 2nd ed. Paris, France: Office International des Epizooties; 1987. Execution of the ELISA technique; pp. 23–24. [Google Scholar]
- 26.Schiemann D A, Devenish J A. Relationship of HeLa cell infectivity to biochemical, serological, and virulence characteristics of Yersinia enterocolitica. Infect Immun. 1982;35:497–506. doi: 10.1128/iai.35.2.497-506.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiemann D A. Yersinia enterocolitica and Yersinia pseudotuberculosis. In: Doyle M P, editor. Food-borne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 601–672. [Google Scholar]
- 28.Skurnik M, Wolf-Watz H. Analysis of the yop gene encoding the yop1 virulence determinants of Yersinia spp. Mol Microbiol. 1989;3:517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 29.Weagant S D, Feng P, Stanfield J J. Bacteriological analytical manual. Gaithersburg, Md: AOAC International; 1995. Yersinia enterocolitica; pp. 8.01–8.13. [Google Scholar]
- 30.Witham P K, Yamashiro C T, Livak K J, Batt C A. A PCR-based assay for the detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl Environ Microbiol. 1996;62:1347–1353. doi: 10.1128/aem.62.4.1347-1353.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wren B W, Tabaqchali S. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction. Lancet. 1990;336:693. doi: 10.1016/0140-6736(90)92191-j. [DOI] [PubMed] [Google Scholar]
- 32.Zink D L, Feelay J C, Wells J G. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature (London) 1980;288:224–226. doi: 10.1038/283224a0. [DOI] [PubMed] [Google Scholar]