Abstract
Although the cyanobacterium Anabaena circinalis occurs worldwide, Australian isolates are believed to exclusively possess the saxitoxin group neurotoxins (paralytic shellfish poisons). Identification of A. circinalis in a mixed population is complicated due to limited morphological differences between Anabaena species. Sequence analysis of the DNA-dependent RNA polymerase (rpoC1) gene from 24 Anabaena isolates, including 12 designated A. circinalis, permitted a phylogenetic analysis to be performed. In addition, an A. circinalis-specific PCR was developed and tested successfully on environmental samples.
Anabaena circinalis produces neurotoxins, anatoxin-a and paralytic shellfish poisons. While A. circinalis is distributed worldwide, the production of PSPs is believed to be exclusive to Australian strains (5). Paralytic shellfish poisons, including saxitoxin, neosaxitoxin, and gonyautoxin, are alkaloids that are potent sodium channel blockers in nerve axons which cause progressive paralysis and death from respiratory failure (3, 12). In a study of the cyanobacterial component of the surface waters of the Murray-Darling Basin in Australia, Baker and Humpage (1) identified the cyanobacterial genus Anabaena as most abundant in natural-bloom samples, with up to seven coiled Anabaena morphotypes coexisting at any one time. A. circinalis was found in 41% of samples, with 55% of A. circinalis blooms shown to produce neurotoxins. Differentiation of A. circinalis from other coiled Anabaena species using microscopic methods is difficult and time consuming. In view of the potential toxicity of A. circinalis, it is paramount that this species be correctly identified in environmental samples. A combination of genotypic and microscopic identification techniques is of potential benefit.
The 16S rRNA gene has been used extensively for cyanobacterial identification (7, 8, 10); however, the DNA-dependent RNA polymerase (rpoC1) gene has been described as a more discriminatory marker (9). A phylogenetic examination of cyanobacterial rpoC1 gene sequence data is presented, focusing on the genus Anabaena and in particular the species A. circinalis. We report here for the first time an A. circinalis-specific PCR assay targeting the rpoC1 gene to detect this organism directly in environmental water samples.
Molecular techniques.
The cyanobacterial strains used in this study (Table 1) were grown under constant light intensity for up to 14 days at 25°C in ASM-1 medium (4). Genomic DNA was extracted from reference strains as previously described (15) and from environmental samples using the InstaGene matrix (Bio-Rad) and phenol-chloroform treatment. Briefly, 10-ml environmental samples were pelleted by centrifugation and resuspended in 90% InstaGene matrix and 10% Triton X-100 to 200 μl. Following incubation at 55°C for 30 min, the cells were vortexed for 1 min, heated to 95°C for 10 min, and then centrifuged. DNA was extracted once with an equal volume of phenol-chloroform and then once with chloroform. The DNA was precipitated, resuspended in 50 μl of water, and used directly in PCRs.
TABLE 1.
Characterization of cyanobacterial strains used in the present study
| Strain | Locationa | rpoC1 sequence | A. circinalis PCR | Saxitoxin productiond | A. circinalis type |
|---|---|---|---|---|---|
| Anabaena circinalis | |||||
| ANA019A | Chaffey Dam, NSW | Yb | + | + | I |
| ANA059C | Renmark, SA | Y | + | NA | I |
| ANA118A | Booligal, NSW | Y | + | + | I |
| ANA118C | Booligal, NSW | Y | + | + | I |
| ANA125A | Yarrawonga, VIC | NDc | + | NA | ND |
| ANA150A | Burrinjuck Reservoir, NSW | Y | + | + | I |
| ANA175A | Collarenebri, NSW | Y | + | NA | I |
| ANA209F | Myponga Reservoir, SA | Y | + | NA | I |
| ANA301A | Riverslea, QLD | Y | + | NA | I |
| ANA306A | Palm Island, QLD | ND | + | − | ND |
| ANA311E | Geelong, VIC | Y | + | NA | II |
| ANA323B | Perth, WA | ND | + | + | ND |
| ANA332H | Yarraman, QLD | Y | + | − | II |
| ANA349H | Hope Valley Reservoir, SA | Y | + | NA | II |
| ANA350C | Craigbourne Dam, TAS | Y | + | NA | II |
| Anabaena flos-aquae | |||||
| ANA051A | Murtho Reserve, SA | ND | − | NA | |
| ANA076C | Iraak Creek, VIC | ND | − | NA | |
| ANA264A | Darling Anabranch, NSW | Y | − | − | |
| ANA354A | River Murray, SA | Y | − | NA | |
| Anabaena spiroides f. spiroides | |||||
| ANA139A | Torrumbarry Weir, VIC | Y | + | NA | II |
| ANA281A | Windamere Dam, NSW | ND | − | NA | |
| ANA292C | Lake Ballyrogan, NSW | Y | − | NA | |
| Anabaena spiroides f. minima | |||||
| ANA044A | Lake Albert, SA | ND | − | NA | |
| ANA084E | Lake Alexandrina, SA | ND | − | NA | |
| ANA234B | Menindee Lake, NSW | ND | − | NA | |
| Anabaena sp. | |||||
| ANA062C | Moolooroo, SA | ND | − | NA | |
| ANA098C | Lagoon, NSW | ND | − | NA | |
| ANA193E | Binghi, NSW | ND | − | NA | |
| ANA238A | Darling Anabranch, NSW | ND | − | NA | |
| ANA238C | Darling Anabranch, NSW | Y | − | NA | |
| ANA255C | Darling Anabranch, NSW | Y | − | NA | |
| Anabaena aphanizomenoides | |||||
| ANA023C | Murtho Park, SA | ND | − | NA | |
| ANA214B | Nildottie Lagoon, SA | ND | − | NA | |
| ANA217A | Nildottie Lagoon, SA | ND | − | NA | |
| ANA259C | Darling Anabranch, NSW | ND | − | NA | |
| ANA280A | Chaffey Dam, NSW | Y | − | − | |
| ANA303C | Riverslea, QLD | ND | − | NA | |
| Anabaena pertubarta f. tumida | |||||
| ANA112D | Cobram, VIC | ND | − | NA | |
| ANA146C | Fish Creek Farm Dam, VIC | ND | − | NA | |
| ANA187A | Mungindi, NSW | ND | − | NA | |
| ANA221E | Yarramundi, SA | Y | − | NA | |
| ANA297B | Deniliquin, NSW | Y | − | NA | |
| ANA313C | Bundaleer Reservoir, SA | ND | − | NA | |
| ANA313D | Bundaleer Reservoir, SA | ND | − | NA | |
| Anabaena solitaria | |||||
| ANA060B | Renmark, SA | ND | − | NA | |
| ANA075C | Iraak Creek, VIC | ND | − | NA | |
| ANA207A | Bridgewater Farm Dam, SA | Y | − | NA | |
| ANA282B | Windamere Dam, NSW | Y | − | NA | |
| ANA337A | Goulbourn, NSW | Y | − | NA | |
| Aphanizomenon issatschenkoi | |||||
| APH016B | Torrumbarry Weir, VIC | ND | − | NA | |
| APH025A | Wongulla Lagoon, SA | ND | − | − | |
| APH027A | Rockhampton, QLD | ND | − | − | |
| Aphanizomenon gracile | |||||
| APH015B | Bundaleer Reservoir, SA | ND | − | NA | |
| APH026E | Joan Powling, VIC | Y | − | NA | |
| Aphanizomenon ovalisporum | |||||
| APH028A | Palm Lakes, QLD | Y | − | NA |
NSW, New South Wales; SA, South Australia; VIC, Victoria; QLD, Queensland; WA, Western Australia; TAS, Tasmania.
Y, yes.
ND, not determined.
According to reference 14. NA, not analyzed.
PCR amplification using cyanobacterium-specific primers targeted to the rpoC1 gene (rpoC1-1 [5′-GAGCTCYAWNACCATCCAYTCNGG] and rpoC1-T [5′-GGTACCNAAYGGNSARRTNGTTGG]) has been previously described (9). Primers used for the A. circinalis-specific PCR assay detailed in this study were Ana2 (5′-GATAGCATCCTCAATTTCTAGCCATTGG), Ana4 (5′-CTCTGAAGCCAGAAATGGACGGC), and Ana-ICF (5′-TAGCCATTGGCATATCCAAGAGAATAGC) and were constructed from the sequence determined in this study. Each 50-μl PCR mixture contained 1 to 10 ng of genomic DNA, 20 pmol of Ana2, 20 pmol of Ana4, 200 μM deoxynucleoside triphosphates, 2.5 mM magnesium chloride, 1× PCR buffer II, 2.5 U of Ampli Taq Gold, and 2 pg of an internal control fragment (ICF). The following protocol was used: 94°C for 10 min for 1 cycle; 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 35 cycles; 72°C for 15 min for 1 cycle; and holding at 4°C. Nucleotide data was analyzed with GeneJockeyII (Biosoft, Cambridge, United Kingdom), sequence alignments were done with ClustalX (13), and phylogenetic trees were constructed with the MEGA analysis platform (6). The Jukes-Cantor method was used to calculate pairwise distances, and a tree was constructed with the neighbor-joining algorithm. Bootstrap analyses were performed with 500 replicates.
rpoC1 sequence and phylogeny.
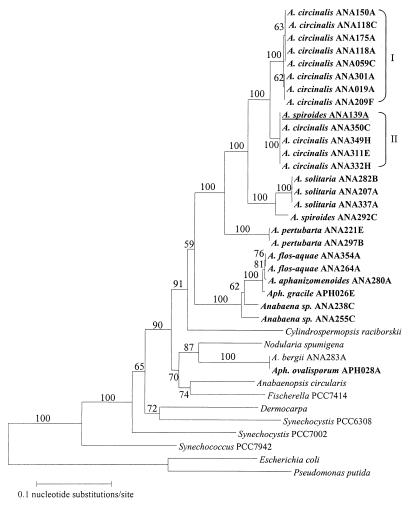
A 612-bp fragment of the rpoC1 gene from 24 Anabaena strains and 2 Aphanizomenon strains was amplified and sequenced (Table 1). An alignment of the sequences showed that in 4 out of 12 A. circinalis strains examined, identical nucleotide changes were observed at 18 of 540 positions. This suggests that genotypic groups of A. circinalis exist (termed types I and II). It is interesting that at position 57, three out of eight type I strains (ANA019A, ANA209F, and ANA301A) had the same nucleotide change, which coincided with the same change in type II isolates. Subsequent analysis failed to associate the observed differences in rpoC1 sequence to strain origin or toxin production. Importantly, the rpoC1 gene provided sufficient sequence variation to differentiate the genus Anabaena to the species level and A. circinalis to the strain level.
A phylogenetic analysis of a range of cyanobacteria based on partial rpoC1 sequences has previously been presented (15) and is combined here with the additional rpoC1 gene sequences obtained for a range of Anabaena and Aphanizomenon species (Fig. 1). The two types of A. circinalis are evident, with the Anabaena genus fitting into the previously identified heterocyst-forming cluster (15). Strain ANA139A was originally identified as A. spiroides on the basis of morphological criteria (Table 1). However, analysis of its rpoC1 sequence showed 100% identity to A. circinalis type II and as shown in Fig. 1, it is within the A. circinalis grouping. This result prompted its identification to be changed and highlighted the importance of rpoC1 typing for Anabaena, as clearly species assignment based on morphological criteria alone can lead to misidentification. Due to the association of A. circinalis with toxicity, misidentification of this species is of considerable concern.
FIG. 1.
Phylogenetic tree constructed from rpoC1 nucleotide sequence alignments. Strains sequenced in this study are in bold type (GenBank accession no. AF199423 to AF199433). The A. spiroides ANA139A isolate reclassified as A. circinalis ANA139A type II is underlined. Bootstrap values derived from 500 replicates of the sequence data are shown.
Interestingly, Anabaena bergii ANA283A clusters with Aphanizomenon ovalisporum APH028A (11), separately from the Anabaena genus. In addition, this cluster is distant from the position of Aphanizomenon gracile APH026E (Fig. 1). Both Aphanizomenon ovalisporum and A. bergii are also known to produce the cyanobacterial toxin cylindrospermopsin. rpoC1 sequence analysis revealed 100% identity between the two isolates, and this is supported by 99% 16S rRNA gene sequence identity (M. A. Schembri, unpublished data). Despite morphological data initially indicating that strain ANA283A belongs to the Anabaena genus, sequence analysis demonstrated that this is not the case and that this isolate and Aphanizomenon ovalisporum are morphological variants of the same cyanobacterium. A more detailed genetic and morphological analysis of the taxonomy of these two organisms must now be performed.
Morphological differences observed between strains of Anabaena flos-aquae II led Cronberg and Komárek (2) to define a new cyanobacterial species, Anabaena pertubarta f. tumida. A number of strains from the culture collection of the Australian Water Quality Centre were reclassified accordingly. While previously based on morphological observations, definition of the species A. pertubarta f. tumida is now also supported by genetic data, with the rpoC1 sequence analysis presented here clearly indicating distinct phylogenetic separation between A. flos-aquae and A. pertubarta strains (Fig. 1).
A. circinalis PCR assay.
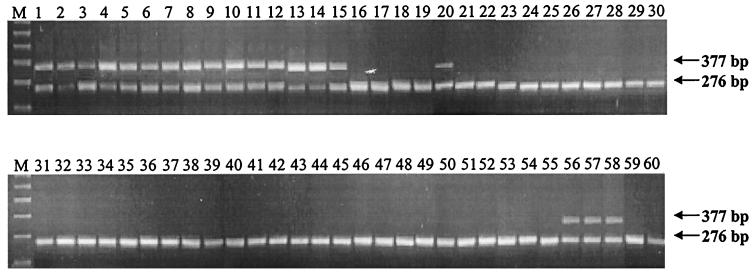
A PCR assay to specifically detect A. circinalis in reference and environmental samples was developed using primers designed on the basis of conserved regions of the A. circinalis rpoC1 gene. An ICF was constructed using a previously described method (15) employing primers Ana2, Ana4, and Ana-ICF. Ana-ICF recognized a sequence internal to the region bounded by Ana2 and Ana4. A PCR with Ana2 and Ana-ICF yielded a fragment which was used in a subsequent PCR with Ana2 and Ana4, which recognizes a terminal decameric sequence incorporated using Ana-ICF. The final product was 276 bp in length and contained 3′ and 5′ sequences which exactly matched Ana2 and Ana4. Two picograms of this internal control was spiked into each PCR mixture. Amplification of the ICF yielded a 276-bp fragment, with the presence of A. circinalis chromosomal DNA resulting in an additional 377-bp product. The ICF indicates if failure to amplify the diagnostic product is due to genuine absence of the target sequence or because the PCR failed due to sample inhibition. In this case, the detection limit is approximately 2,000 cells but lower levels should be detectable with careful optimization of PCR conditions.
Reference cultures of Anabaena and Aphanizomenon (Table 1) and four environmental samples were screened with the assay, including an environmental sample from Bahia Blanca, Argentina (Fig. 2). This sample was known to contain A. circinalis and Microcystis and Ceratium spp. The 377-bp diagnostic band was amplified in all A. circinalis reference cultures (lanes 1 to 15) and in three of the environmental samples known to contain A. circinalis following microscopic analysis (lanes 56 to 58). Sequence analysis of the three diagnostic PCR products confirmed the identification of A. circinalis in the environmental samples. In addition, the specific PCR also supported the definition of A. spiroides f. spiroides ANA139A as A. circinalis (lane 20). This is the first report of a PCR assay that is specific for A. circinalis and able to identify this species in an environmental water sample. A PCR test which targets toxin-encoding genes in A. circinalis would be ideal. However, in the absence of DNA sequence information regarding these genes, but the strong association of this species with toxicity, the rapid and specific identification of A. circinalis is a useful tool in the management of toxic algal blooms.
FIG. 2.
A. circinalis-specific PCR assay. The diagnostic product (377 bp) and the ICF (276 bp) are indicated. Lanes 1 to 15 are A. circinalis isolates (019A, 059C, 118A, 118C, 125A, 150A, 175A, 209F, 301A, 306A, 311E, 323B, 332H, 349H, and 350C), lanes 16 to 19 are A. flos-aquae isolates (051A, 076C, 264A, and 354A), lanes 20 to 22 are A. spiroides f. spiroides isolates (139A, 281A, and 292C), lanes 23 to 25 are A. spiroides f. minima isolates (044A, 084E, and 234E), lanes 26 to 31 are Anabaena sp. isolates (062C, 098C, 193E, 238A, 238C, and 255C), lanes 32 to 37 are A. aphanizomenoides isolates (023C, 214B, 217A, 259C, 280A, and 303C), lanes 38 to 44 are A. pertubarta f. tumida isolates (112D, 146C, 187A, 221E, 297B, 313C, and 313D), lanes 45 to 49 are A. solitaria isolates (060B, 075C, 207A, 282B, and 337A), lanes 50 to 52 are Aphanizomenon issatschenkoi isolates (016B, 025A, and 027A), lanes 53 and 54 are Aphanizomenon gracile isolates (015B and 026E), lane 55 is an Aphanizomenon ovalisporum isolate (028A), lane 56 is an environmental sample containing A. circinalis (Geelong, Victoria, Australia), lane 57 is an environmental sample containing A. circinalis (Gawler, South Australia, Australia), lane 58 is an environmental sample containing A. circinalis (Bahia Blanca, Argentina), lane 59 is an environmental sample containing C. raciborskii (Broken Hill, New South Wales, Australia), lane 60 is the ICF only, and lane M contains molecular size markers (700, 600, 500, 400, 300, and 200 bp).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this work have been deposited with the GenBank database under the following accession numbers: A. circinalis type I rpoC1, AF199423; A. circinalis type IA (representing strains ANA019A, ANA209F, and ANA301A), AF199424; A. circinalis type II rpoC1, AF199425; Anabaena solitaria rpoC1, AF199426; A. spiroides ANA292C rpoC1, AF199427; A. pertubarta rpoC1, AF199428; A. flos-aquae rpoC1, AF199429; Anabaena aphanizomenoides rpoC1, AF199430; Aphanizomenon gracile rpoC1, AF199431; Anabaena sp. strain ANA238C rpoC1, AF199432; Anabaena sp. strain ANA255C rpoC1, AF199433.
Acknowledgments
This work was supported by the Cooperative Research Centre for Water Quality and Treatment.
We thank Peter Baker for expert advice and provision of strains, Mark Schembri for unpublished data on A. bergii, Glen Shaw for provision of Aphanizomenon ovalisporum, Dennis Steffensen for the sample from Argentina, and Paul Monis for assistance with phylogenetic tree analysis.
REFERENCES
- 1.Baker P D, Humpage A R. Toxicity associated with commonly occurring cyanobacteria in surface waters of the Murray-Darling basin, Australia. Aust J Mar Freshwater Res. 1994;45:773–786. [Google Scholar]
- 2.Cronberg G, Komárek J. Planktic cyanoprokaryotes found in South Swedish lakes during the XIIth International Symposium on Cyanophyte Research, 1992. Algol Stud. 1994;75:323–352. [Google Scholar]
- 3.Falconer I R. An overview of problems caused by toxic blue-green algae (cyanobacteria) in drinking and recreational water. Environ Toxicol. 1999;14:5–12. [Google Scholar]
- 4.Gorham P R, McLachlan J, Hammer U T, Kim W K. Isolation and culture of toxic strains of Anabaena flos-aquae (Lyngb.) deBréb. Verh Int Ver Limnol. 1964;15:796–804. [Google Scholar]
- 5.Humpage A R, Rositano J, Bretag A H, Brown R, Baker P D, Nicholson B C, Steffensen D A. Paralytic shellfish poisons from Australian cyanobacterial blooms. Aust J Mar Freshwater Res. 1994;45:761–771. [Google Scholar]
- 6.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.0. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 7.Lyra C, Hantula J, Vainio E, Rapala J, Rouhiainen L, Sivonen K. Characterization of cyanobacteria by SDS-PAGE of whole-cell proteins and PCR/RFLP of the 16S rRNA gene. Arch Microbiol. 1997;168:176–184. doi: 10.1007/s002030050485. [DOI] [PubMed] [Google Scholar]
- 8.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palenik B, Haselkorn R. Multiple evolutionary origins of prochlorophytes, the chlorophyll-containing prokaryotes. Nature (London) 1992;355:265–267. doi: 10.1038/355265a0. [DOI] [PubMed] [Google Scholar]
- 10.Rudi K, Larsen F, Jakobsen K S. Detection of toxin-producing cyanobacteria by use of paramagnetic beads for cell concentration and DNA purification. Appl Environ Microbiol. 1998;64:34–37. doi: 10.1128/aem.64.1.34-37.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw G R, Sukenik A, Livne A, Chiswell R K, Smith M J, Seawright A A, Norris R L, Eaglesham G K, Moore M R. Blooms of the cylindrospermopsin containing cyanobacterium, Aphanizomenon ovalisporum (Forti), in newly constructed lakes, Queensland, Australia. Environ Toxicol. 1999;14:167–177. [Google Scholar]
- 12.Steffensen D, Burch M, Nicholson B, Drikas M, Baker P. Management of toxic blue-green algae (cyanobacteria) in Australia. Environ Toxicol. 1999;14:183–195. [Google Scholar]
- 13.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velzeboer, R. M. A., P. D. Baker, J. Rositano, T. Heresztyn, G. A. Codd, and S. L. Raggett. Geographic patterns of occurrence and composition of saxitoxins of the cyanobacterial genus Anabaena in Australia. Phycologia, in press.
- 15.Wilson K M, Schembri M A, Baker P D, Saint C P. Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl Environ Microbiol. 2000;66:332–338. doi: 10.1128/aem.66.1.332-338.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]