Abstract
We describe two prolonged bacteriophage blooms within sugar beet rhizospheres ensuing from an artificial increase in numbers of an indigenous soil bacterium. Further, we provide evidence of in situ competition between these phages. This is the first in situ demonstration of such microbial interactions in soil. To achieve this, sugar beet seeds were inoculated with Serratia liquefaciens CP6RS or its lysogen, CP6RS-ly-Φ1. These were sown, along with uninoculated seeds, in 36 field plots arranged in a randomized Latin square. The plots were then sampled regularly over 194 days, and the plants were assayed for the released bacteria and any infectious phages. Both the lysogen and nonlysogen forms of CP6RS survived equally well in situ, contradicting earlier work suggesting lysogens have a competitive disadvantage in nature. A Podoviridae phage, identified as ΦCP6-4, flourished on the nonlysogen-inoculated plants in contrast to those plants inoculated with the lysogen. Conversely, the Siphoviridae phage ΦCP6-1 (used to construct the released lysogen) was isolated abundantly from the lysogen-treated plants but almost never on the nonlysogen-inoculated plants. The uninoculated plants also harbored some ΦCP6-1 phage up to day 137, yet hardly any ΦCP6-4 phages were found, and this was consistent with previous years. We show that the different temporal and spatial distributions of these two physiologically distinct phages can be explained by application of optimal foraging theory to phage ecology. This is the first time that such in situ evidence has been provided in support of this theoretical model.
Bacteria are ubiquitous in the environment, with a global estimate of 4 × 1030 to 6 × 1030 cells (26). With this ubiquity comes an importance to the biosphere that is well recognized; thus, any process that substantially affects natural bacterial communities will also be significant. One such process may be predation by bacteriophages (phages). It is thought that predatory phages could control the numbers of bacteria and facilitate gene transfer between bacteria by transduction (5, 6, 14). Certainly phages are as common as bacteria. In addition, estimates of phage abundance in aquatic habitats suggest their numbers are 10 times greater than those of bacteria (5). Extrapolating this estimate to the biosphere at large would make phages the most abundant organisms on earth.
Clearly then, phages have a potentially significant global impact. But is this potential realized? This is a difficult question to answer, as the natural population ecology of phages has been little studied. Most knowledge derives from investigations using chemostats, mainly because phage-bacterium interactions serve as a useful paradigm of predator-prey interactions generally, and chemostats afford the opportunity to test the validity of mathematical models (see reference 16 for a review). However, chemostat conditions are far removed from the complexity of nature. A few studies have attempted to follow long-term phage population changes in situ, but these concentrated on aquatic habitats, considering only gross overall changes in bacterial and phage populations (6, 24). In addition, some microcosm studies have considered specific bacterium-phage systems over short time scales (7, 9, 10, 20–22).
Our present study is the first to describe interactions between competing phages within a natural habitat over a prolonged (i.e., 6-month) time scale. The data we present provide compelling evidence of competition between two indigenous predatory phages for the same prey bacterium within a natural environment that is consistent with established interspecific-competition theory (19).
MATERIALS AND METHODS
Bacteria used in this study.
The bacteria used in this study (Table 1) were derived from Serratia liquefaciens CP6, previously isolated from a sugar beet grown at our field site (3). For the present study, a spontaneous spectinomycin- and nalidixic acid-resistant mutant of S. liquefaciens CP6 was isolated and called CP6SpN. In addition, a lysogen of S. liquefaciens CP6RS was isolated (4) from a CP6RS culture inoculated with the temperate Siphoviridae phage ΦCP6-1, and this lysogen was named CP6RS-ly-Φ1. These isogenic forms of the wild-type CP6 grew equally well in soil in the laboratory. All bacteria were maintained on nutrient agar (CM3; Oxoid) at 4°C, with stocks kept at −80°C in 50% glycerol.
TABLE 1.
S. liquefaciens strains used in this study
| Strain | Phenotypea | Comments | Reference |
|---|---|---|---|
| CP6 | Wild type | Sugar beet rhizosphere isolate from farm | 3 |
| CP6-ly-Φ1 | ΦCP6-1r | Phage ΦCP6-1 lysogen of CP6 | 4 |
| CP6-ly-Φ2 | ΦCP6-2r | Phage ΦCP6-2 lysogen of CP6 | 4 |
| CP6-ly-Φ3 | ΦCP6-3r | Phage ΦCP6-3 lysogen of CP6 | 4 |
| CP6-ly-Φ5 | ΦCP6-5r | Phage ΦCP6-5 lysogen of CP6 | 4 |
| CP6RS | Rifr Strepr | Spontaneous antibiotic-resistant mutant of CP6 | 4 |
| CP6RS-ly-Φ1 | Rifr Strepr ΦCP6-1r | Phage ΦCP6-1 lysogen of CP6RS | This study |
| CP6SpN | Spr Nalr | Spontaneous antibiotic-resistant mutant of CP6 | This study |
Phages ΦCP6-1 to ΦCP6-5 were isolated from sugar beet rhizosphere samples collected from our field site at Oxford University Farm (3). Rif, rifampin; Strep, streptomycin; Sp, spectinomycin; Nal, nalidixic acid.
Phages ΦCP6-1 and ΦCP6-4 are double-stranded-DNA-tailed phages belonging to the families Siphoviridae and Podoviridae, respectively (4). They both infect S. liquefaciens CP6 and were previously isolated from our field site (3).
The field site.
Fieldwork took place during 1997 and 1998 at a site on Oxford University Farm, Wytham, Oxford, United Kingdom. The site was originally pastureland, and sugar beets had been grown there since 1990. The soil is an Evesham series heavy clay soil with 53% clay, 25% sand, 22% silt (pH 7.7), and 8.5% organic content. As in previous years, the soil was fertilized with an NPK (13:13:20) fertilizer (dosage, 0.15 kg m−2) and treated with herbicide (Roundup; 2 kg ha−1) and insecticide (Gamma-col; ICI; 1.4 liter ha−1) prior to sowing (3).
First field experiment, 1997.
On 16 May 1997 (day zero), five soil samples were randomly collected from a 2.25- by 5.1-m plot within the field site. Next, around 500 sugar beet seeds (EB3 pellets; Germains UK Ltd., Kings Lynn, United Kingdom) were soaked in sterile quarter-strength Ringer's solution (QSR) (BR52; Oxoid) for 5 min and sown at 15-cm intervals within the plot.
Homogenates were prepared from the soil samples by mixing 1 g of soil with 20 ml of QSR and thoroughly homogenizing the resulting suspension by adding sterile 5-mm-diameter glass beads, vortex mixing the suspension for 1 min, and then shaking it on an orbital shaker for 10 min. The homogenates were screened for phages antagonistic towards CP6 by the overlay agar technique (2). The base medium was nutrient agar, while the overlay agar was made from nutrient broth (CM1; Oxoid) (13 g liter−1) and bacteriological agar (L11; Oxoid) (6.5 g liter−1). The homogenates (1 ml) were centrifuged for 5 min at 14,000 × g, and 100 μl of the resulting supernatant was used to inoculate the overlay agar (2.5 ml) with an equal volume of CP6 culture.
The plates were incubated overnight at a plaque size-optimizing temperature of 15°C, and any resulting plaques were counted. Phages from these plaques were identified using the methodologies detailed in our previous studies (3, 4). That is, the plaques were initially classified according to appearance (the two most dominant phages at the field site have very distinct plaque morphologies [4]). The classifications were then tested by producing phage lysates from the plaques and assaying them for the ability to lyse the CP6 lysogens listed in Table 1 (i.e., superimmunity testing [4]). As a final confirmatory step, DNA was extracted from representative lysates (3) and cut with EcoRI (Promega) as described by the manufacturer. Digests were run on 0.7% agarose gels at 0.32 V cm−2, along with HindIII-cut lambda DNA (D-9780; Sigma), and the resulting restriction profiles were compared with the expected banding patterns (4). When no phages were detected, the homogenates were enriched with nutrient broth, spiked with an overnight culture of CP6, and reassayed after overnight incubation at 20°C to optimize phage proliferation.
Soil bacteria were counted by plating the homogenates on tryptone soy broth agar (TSBA) (17), which, when enumerated, gave an estimate of total viable heterotrophic bacterial numbers, and on Pseudomonas selection isolation agar (PSIA) (15) to determine total pseudomonad counts. TSBA plates were incubated at 15°C for 7 days before enumeration, while PSIA plates were incubated at 30°C for 48 h.
On days 25, 52, 82, 123, 160, 209, and 286 after being sown, 10 sugar beet plants were collected at random from the field site and weighed. After as much loose soil as possible was dislodged, the rhizosphere from each plant was sampled by scraping the surface of each root with a sterile scalpel and collecting the resulting thin layer of soil in a sterile 50-ml centrifuge tube. Homogenates were then prepared and assayed for phages and bacteria, as described for the soil samples. Additionally, the homogenates were enumerated on a Serratia selective medium (SSM) that we had designed for estimating total CP6-like bacterial counts (i.e., D3 Erwinia medium [13] modified by eliminating arabinose and increasing the sucrose to 40 g liter−1). Prior tests with CP6-spiked soil samples had shown that, when incubated for 48 h at 30°C, this medium selected against most non-Serratia bacteria while CP6 appeared as small (∼1-mm-diameter) orange colonies with yellow borders that were clearly discernible against the dark-blue background of the surrounding agar. The identities of randomly selected CP6-like isolates were confirmed by checking their sensitivities to our CP6 phage collection (3) and their API 20E strip (bioMérieux sa) profiles.
Second field experiment, 1998.
On 22 May 1998 (day zero), in a separate region of the field site, 3.9 by 4.8 m was partitioned off and divided into 36 plots in a six-by-six matrix, with a 30-cm-wide border separating each plot from its neighbor. Soil was collected from each plot. The plots were then randomly assigned to one of five sugar beet seed treatments (Fig. 1). The seeds were treated as follows: for treatment 1, the seeds were uninoculated as in 1997; for treatment 2, the seeds were inoculated with CP6SpN; for treatment 3, the seeds were inoculated with CP6SpN and CP6RS; for treatment 4, the seeds were inoculated with CP6RS-ly-Φ1; for treatment 5, the seeds were inoculated with CP6SpN and CP6RS-ly-Φ1. The purpose of these inoculations was to release differently marked lysogen (CP6RS-ly-Φ1) and nonlysogen (CP6SpN) forms of S. liquefaciens to investigate cross infection of the temperate phage ΦCP6-1 in situ and to see whether transduction might occur. CP6RS was released as a nonlysogen control of CP6RS-ly-Φ1. The inoculations were achieved by soaking the seeds for 5 min in the appropriate bacterial suspensions prepared with QSR (3). Estimates of inoculum density were obtained by drop plate counting (12) on nutrient agar with the appropriate selective agents. Inocula were also checked for phages by similarly drop plating serial dilutions of samples filtered through 0.2-μm-pore-size membranes onto CP6-inoculated overlay plates.
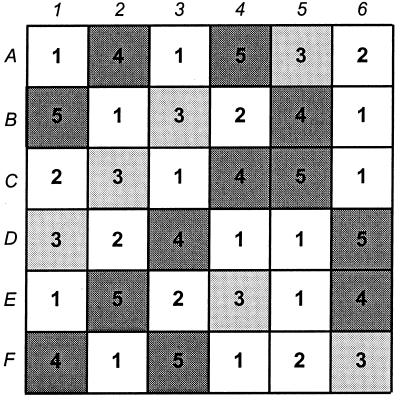
FIG. 1.
Arrangement of plots during 1998 field experiment. Each square represents a separate plot. Plots were assigned to one of five treatments by following a randomized Latin square arrangement. The squares shaded light gray indicate the plots containing plants inoculated with CP6RS, squares shaded dark gray indicate the plots containing plants inoculated with CP6RS-ly-Φ1, and unshaded squares represent plots containing uninoculated plants, either by design (i.e., treatment 1), or through inoculation failure (i.e., treatment 2); see the text for descriptions of treatments.
Directly after inoculation, the seeds were sown within their designated plots in ascending order of treatment. Latex gloves and sterile forceps were used throughout for handling the seeds. Within each plot, the seeds were sown at 15-cm intervals in a 4-by-3 matrix. Three seeds were planted in each hole to maximize germination success, with multiple germinations being thinned to single seedlings as they emerged.
As in 1997, homogenates were prepared from the collected soil samples, except that this time 1 g of soil was suspended in 10 ml of QSR. The homogenates were screened for phages antagonistic towards CP6 as described for 1997. Bacterial numbers were estimated by plating the homogenates onto TSBA, SSM, SSM supplemented with streptomycin at 1,000 μg ml−1 (to select for bacteria with the CP6RS phenotype), and SSM supplemented with spectinomycin at 100 μg ml−1 (to select for the CP6SpN phenotype). Plates were incubated as described above.
On days 19, 47, 68, 96, 137, and 194 after being sown, one plant from each of the 36 plots was randomly selected and weighed. The rhizosphere of each plant collected was then sampled and assayed for phages and bacteria as for the soil samples. Putative S. liquefaciens CP6RS or S. liquefaciens CP6SpN colonies were confirmed by growing them on nutrient agar supplemented with rifampin (100 μg ml−1) or nalidixic acid (200 μg ml−1), respectively, which selected for their second phenotypic markers. Additionally, selected isolates were assayed for sensitivity towards phages ΦCP6-1 to ΦCP6-6 (4). This confirmed whether CP6 bacterial strains (i.e., CP6RS and CP6SpN) had been reisolated and identified any ΦCP6-1 lysogens (i.e., CP6RS-ly-Φ1) by their sensitivities to all phages except ΦCP6-1. Lysogeny was confirmed by stabbing colonies onto CP6-inoculated overlay lawns to detect zones of lysis after incubation.
Statistics.
Calculations were done using the MINITAB version 11 computer package (Minitab Inc., University Park, Pa.). Sugar beet weights and bacterial counts were compared statistically using analysis of variance, after log10 (x + 1) transformation, with group means compared by calculating the minimum significant difference at a P value of 0.05, according to the Tukey-Kramer method (8). Phage counts were compared using the mood-median test, and linear regression lines were compared by analysis of covariance (8). Contour plots showing in situ phage distributions were generated using MINITAB.
RESULTS
The 1997 field experiment.
Prior to sowing, the mean total-viable bacteria count per gram of soil was 2.5 × 107 CFU, as estimated on TSBA medium. A mean of 2.6 × 106 CFU of pseudomonads g−1 was also calculated from PSIA plates. No phages were detected from any of the five soil homogenates directly after preparation (limit of detection, 200 PFU per g of soil). Only after the homogenates had been enriched with host bacteria were phage detected in one of the soil samples, and these were all identified as ΦCP6-1.
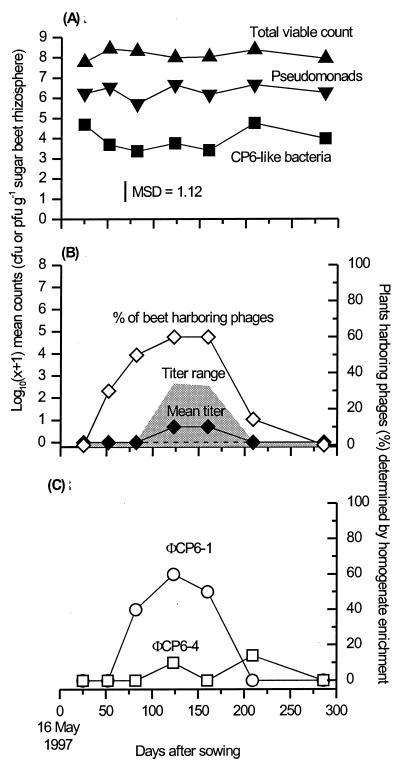
The sugar beets grew, reaching their maximum weight by around day 123 (mean, 1,107.8 g). We monitored the densities of indigenous Pseudomonas and CP6-like bacteria (Fig. 2A) and those phages antagonistic towards S. liquefaciens CP6 (Fig. 2B). Phage abundances determined through enrichment (Fig. 2B) illustrated the fact that phages were present throughout the experiment, but often at densities below our preenrichment limit of detection (mean, 2.3 × 102 PFU g−1). Small significant (i.e., P < 0.05) variations in numbers of both CP6-like bacteria and pseudomonads were seen (Fig. 2A), but these did not coincide with significant changes in the populations of their phages.
FIG. 2.
Temporal variation in abundance of bacteria and phages within the sugar beet rhizosphere recorded during 1997. (A) Bacteria on sugar beets. Bacterial counts were determined on TSBA plates to give total viable counts (apex-up triangles), on PSIA plates to give total pseudomonad counts (apex-down triangles), and on SSM plates to give total S. liquefaciens CP6 counts (squares). (B) Total CP6 phages on sugar beets. (C) Breakdown of CP6 phage population into its two major components. Phage titers (panel B, solid diamonds) were estimated from freshly prepared rhizosphere homogenates by plaque counting. The percentages of sugar beets harboring CP6 phages generally (panel B, open diamonds) and phages ΦCP6-1 and ΦCP6-4 in particular (panel C, circles and squares, respectively) were determined after homogenates had been nutrient enriched and incubated overnight. Each plotted point represents 10 replicate sugar beets, except for day 209, when 14 plants were sampled. MSD, minimum significant difference at a P value of 0.05 (8). The gray shading (panel B) indicates the range of phage titers recorded, while the dashed line shows the position of both the upper and lower quartiles for this data (i.e., both Q1 and Q3 = 0 for all means shown).
As in 1996 (3), we also analyzed the isolated phages antagonistic to bacterium CP6 in depth. Plaque morphology, superimmunity tests, and restriction fragment length polymorphism analysis showed that this phage population consisted predominately of the Siphoviridae phage ΦCP6-1 (Fig. 2C) and, to a lesser extent, ΦCP6-4, a Podoviridae phage frequently isolated from our site in 1996 (3). A few ΦCP6-3 (Myoviridae) phage (3) and several previously unisolated phages were also present.
The 1998 field experiment.
Prior to sowing, there was no significant difference among the treatment plots in terms of total viable bacterial counts (P = 0.897) or total CP6-like bacteria (P = 0.879). The mean total viable count was 3.3 × 107 CFU g−1 of soil, while for CP6-like bacteria it was 8.6 × 103 CFU g−1. As in 1997, no CP6-antagonistic phages were isolated from freshly prepared soil homogenates (limit of detection, 100 PFU g−1). Only after the samples had been enriched were 3 of the 36 samples shown to harbor phages for this bacterium. Specifically, soil from plot A6 was shown to contain an unidentified CP6-antagonistic phage and plot B4 soil carried ΦCP6-4, while soil from plot E4 harbored phage ΦCP6-3.
All inocula had bacterial counts of around 109 CFU ml−1 prior to seed inoculation. Cultures of the lysogen CP6RS-ly-Φ1 contained 103 PFU ml−1 of phage ΦCP6-1 due to spontaneous lysis. No free phages were detected from either the CP6SpN or CP6RS cultures.
The sugar beets reached maximum weight by around day 137. The resulting mean weight, 275.4 g, was much less than in the previous year, probably due to the combined effects of later sowing and a particularly dry summer. In addition, analysis of variance showed that the overall mean total viable bacterial count recorded from the rhizosphere samples of these 1998 plants (9.3 × 107 CFU g−1) was significantly lower (P < 0.05) than that determined from the 1997 plants (1.3 × 108 CFU g−1).
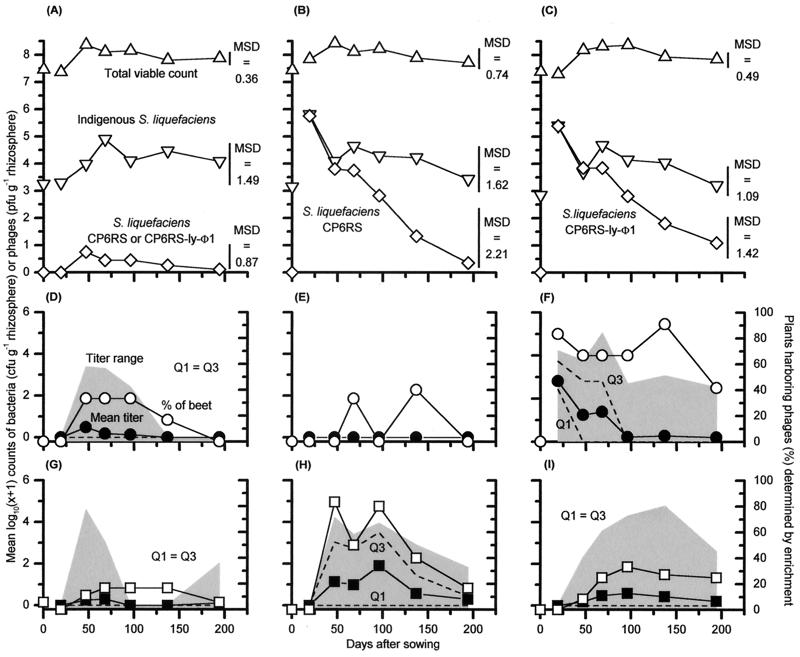
Both S. liquefaciens CP6RS and S. liquefaciens CP6RS-ly-Φ1 inocula survived well after release (Fig. 3B and C). S. liquefaciens CP6SpN, however, was never detected on any plants sampled, showing that this bacterium had not survived in situ. Moreover, a comparison of sugar beet weights and bacterium and phage counts between treatments revealed that the S. liquefaciens CP6SpN inoculation had no detectable effect on the experiment (all P > 0.05). Consequently, for the purposes of subsequent analysis, treatments 1 and 2 were judged to be the same (i.e., “uninoculated” controls), and their results were combined (Fig. 3A, D, and G). Treatments 4 and 5 were also determined to be equivalent (i.e., a CP6RS-ly-Φ1 release), and their data sets were merged (Fig. 3C, F, and I). Treatment 3 was effectively a release of CP6RS alone (Fig. 3B, E, and H).
FIG. 3.
1998 field experiment, showing the fate of the S. liquefaciens CP6RS and CP6RS-ly-Φ1 releases within the sugar beet rhizospheres and the consequent phage blooms they triggered. Bacterial counts (A to C) are compared with those of phage ΦCP6-1 (D to F) and phage ΦCP6-4 (G to I). Due to the nonsurvival of S. liquefaciens CP6SpN in situ, a comparison is made between uninoculated beets (a pooling of results from treatments 1 and 2) (A, D, and G), nonlysogen-treated beets (treatment 3) (B, E, and H), and lysogen-treated beets (pooled results from treatments 4 and 5) (C, F, and I). Because of this pooling of results, each plotted point represents either 18 plants (uninoculated beets), 6 plants (nonlysogen-treated beets), or 12 plants (lysogen-treated beets). See the text for an explanation of why the treatments were combined. Means of the phage titers are indicated by solid symbols. The gray shading indicates the range of the phage titers, whereas the dashed lines show the positions of both the upper (Q3) and lower (Q1) quartiles of this data. Also shown (D to I) are phage abundances detected after nutrient enrichment (open symbols).
The nonlysogen and lysogen forms of S. liquefaciens CP6RS survived equally well in the sugar beet rhizosphere and established large populations within the rhizospheres that were not different (P = 0.747) from each other. These populations declined at the same rate, with no significant difference in regression line gradients (P = 0.179) (Fig. 3B and C). Furthermore, neither release noticeably changed the population dynamics of the indigenous bacteria from that seen on the uninoculated sugar beets (total viable bacterial count, P = 0.882; CP6-like bacterial count, P = 0.390).
On the first sampling occasion after sowing (day 19), no released bacteria were detected on any of the uninoculated control plants. However, by the next sampling occasion (day 47), 5 of the 18 untreated plants sampled harbored released bacteria (geometric mean, 5.1 × 102 CFU g−1). Subsequently, fewer control plants carried released bacteria, until by day 194, only one plant harbored these organisms. When bacteria were detected, the abundances were around 102 CFU g−1. In total, 14 out of the 108 control plants collected over the experiment had detectable quantities of released bacteria.
Overall, five different types of S. liquefaciens-infecting phages were identified from sugar beet samples, and these corresponded to phages ΦCP6-1 to ΦCP6-5 (3, 4). The vast majority isolated (84.4%) were either ΦCP6-1 or ΦCP6-4.
(i) Phage ΦCP6-1.
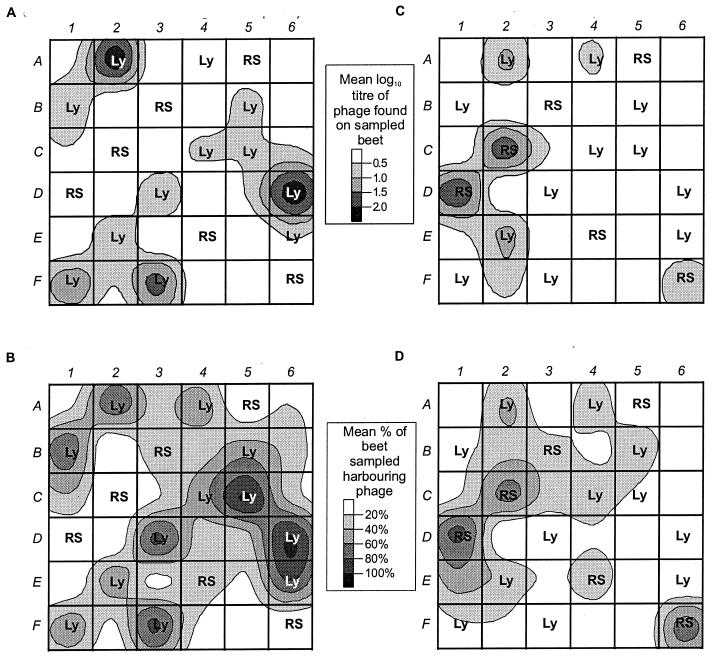
Relatively small numbers of ΦCP6-1 phage were isolated from uninoculated plants (Fig. 3D and 4A), and they were only very apparent when the homogenates were enriched (Fig. 3D and 4B). No ΦCP6-1 phage were isolated from plants inoculated with bacterium CP6RS (Fig. 3E and 4A) unless their homogenates were also enriched, whereupon plants from two plots (B3 and E4) were shown to harbor small amounts of ΦCP6-1 on days 68 and 137 (Fig. 3E and 4B).
FIG. 4.
Distribution of phages within the field site over the entire 1998 experiment, shown as contour plots of average phage abundance. (A and B) Abundance of phage ΦCP6-1 as determined by plaque counts from fresh homogenates (A) and as a percentage of sugar beets sampled after enrichment (B). (C and D) Distribution of ΦCP6-4 as determined by plaque counts from fresh homogenates (C) and after enrichment (D). Means were determined from the sum of all counts over the entire season. RS, plots containing CP6RS-inoculated plants (i.e., treatment 3); Ly, plots containing lysogen-inoculated plants (i.e., treatments 4 and 5). The unlabeled plots are uninoculated controls. Plaque counts are given as PFU per gram of rhizosphere.
In contrast, all but one of the plots inoculated with lysogen CP6RS-ly-Φ1 repeatedly produced plants harboring high densities of ΦCP6-1 from fresh homogenates (Fig. 3F and 4A), and high titers (up to 1.6 × 104 PFU g−1) were regularly recorded. The only exception was plot A4, and even then, when enriched, homogenates from this plot occasionally elicited phages (Fig. 4B). The ΦCP6-1 titers decreased with time, and this decline mirrored the observed drop in CP6RS-ly-Φ1 numbers on the same plants (Fig. 3C). From days 19 to 96 inclusive, the mean phage and lysogen counts appeared to decrease at the same rate, and a comparison of the slopes confirmed this, with no significant difference in gradient detected (P = 0.972).
In all, ΦCP6-1 counts were significantly greater (P < 0.001) in lysogen-inoculated plots (mean, 2.7 × 103 PFU g−1) than in both uninoculated (mean, 5.8 × 101 PFU g−1) and nonlysogen-inoculated plots, where ΦCP6-1 were below the limit of detection unless enriched.
(ii) Phage ΦCP6-4.
Phage ΦCP6-4 was rarely isolated from untreated plants (Fig. 3G and 4C) even after nutrient enrichment (Fig. 3G and 4D). However, for inoculated plants the patterns of significant difference between treatments were different for ΦCP6-4 and ΦCP6-1. That is, phage ΦCP6-4 titers were significantly higher in nonlysogen-treated plots (mean, 9.1 × 102 PFU g−1) than in either the uninoculated (mean, 3.6 × 102 PFU g−1; P < 0.001) or the lysogen-inoculated (mean, 3.5 × 102 PFU g−1; P = 0.009) plots. There was no difference between uninoculated and lysogen-inoculated plots (P = 0.110). Phage ΦCP6-4 was not inoculated into the site, and so it was not as abundant or as widely distributed as ΦCP6-1 (Fig. 4). More of the nonlysogen-inoculated plants (44.1%) carried ΦCP6-4 than either the lysogen-inoculated plants (19.7%) or uninoculated plants (12.0%). Furthermore, 83.3% of all nonlysogen-inoculated plots, 66.7% of all lysogen-inoculated plots, and 55.5% of uninoculated plots harbored plants with detectable ΦCP6-4 at some point during the experiment (Fig. 4D).
DISCUSSION
This is the first in situ study to unambiguously show that an increase in numbers of an indigenous soil bacterium can lead to an equally substantial rise in a naturally occurring bacteriophage (ΦCP6-4). This work is unique, because it took place in a completely natural environment with native bacteria and phages and occurred over a long, ecologically relevant time scale. In no other natural habitat has this been achieved. Previous equivalent terrestrial studies have all employed microcosms, over far shorter time scales, and often with very simplified microbial communities (7, 9, 21, 22). A few aquatic studies have been undertaken in situ. However, these followed gross changes in total bacterial and virus populations (6, 24). Interactions between individual bacterial and phage species in water have only been reported from microcosms (10, 20).
What makes our results so remarkable is that another S. liquefaciens phage, ΦCP6-1, failed to benefit from the release of S. liquefaciens CP6RS. Contrast the almost complete absence of this phage within CP6RS-inoculated plots (Fig. 3E) with its repeated occurrence elsewhere (Fig. 3D and F). This statistically significant difference leads us to conclude that phages ΦCP6-1 and ΦCP6-4 competed with each other in situ and that the different state of health of the released CP6RS, relative to wild-type CP6 indigenous within the soil, predisposed it to successful predation by ΦCP6-4 in preference to ΦCP6-1.
We assert these two conclusions for the following reasons. We already have strong evidence of temporal succession, and hence competition, between ΦCP6-1 and ΦCP6-4 occurring in situ in 1996 (3). During that field experiment, we observed an explosion in ΦCP6-1 numbers between days 48 and 99. This situation continued until day 156, when a dramatic decline in abundance occurred; thereafter numbers remained low until the end of the experiment. Concurrent with this decline was an even more substantial increase in the numbers of phage ΦCP6-4, which until that point had been almost completely absent.
Our subsequent research (4) confirmed ΦCP6-1 and ΦCP6-4 to be very different. (i) We found no DNA homology between the two phages. (ii) ΦCP6-1 was shown to be a Siphoviridae phage, while ΦCP6-4 was a member of the family Podoviridae. (iii) ΦCP6-1 was temperate for CP6, while ΦCP6-4 was entirely virulent. (iv) The latent period for ΦCP6-1 was almost three times that of ΦCP6-4, while its burst size was over five times greater. The last attributes are particularly pertinent to this discussion, as they have been identified as possible phage survival strategies (1, 23, 25).
For example, Stewart and Levin (23) theorized that virulence would be favored as a survival mechanism over lysogeny in those environments where there are high numbers of a physiologically “suitable” host available. According to their theory, ΦCP6-4 would predominate over ΦCP6-1 at our field site when such host cells became abundant. CP6RS may have been this physiologically suitable host. Besides being abundant as a consequence of our release, CP6RS would have been physiologically different from contemporaneous indigenous CP6.
The scenario we outline is also consistent with the work of Abedon (1) and Wang et al. (25), who applied optimal foraging theory to phage ecology. From their theoretical models, they concluded that phages with short latent periods and small burst sizes (like ΦCP6-4) would outcompete phages with longer latent periods and larger burst sizes (like ΦCP6-1) when the numbers of physiologically suitable host bacteria are high (as for CP6RS). Taken together, all these factors provide strong evidence of competition occurring between phages in situ.
We did not add ΦCP6-4 phage to our site, so the bloom we triggered derived entirely from naturally present virions. Phage ΦCP6-1 was also native; however, in this experiment its numbers were only increased substantially by a lysogen release. This inoculation, coincident with the CP6RS release, generated large numbers of ΦCP6-1 phage in all but one of the lysogen-inoculated plots. In these plots, ΦCP6-1 was up to 1,000-fold more numerous than in uninoculated plots. This and the pattern of significant differences in observed phage titers showed that these large titers came from the inoculated lysogen.
Several points arise from this concurrent release of lysogen and nonlysogen. First, it is clear that the proximity of all the plots to one another led to some movement of phage and released bacteria between plots, with small numbers of released bacteria repeatedly occurring in untreated controls (Fig. 3A) and the apparent spread of ΦCP6-1 from lysogen-inoculated plots to neighboring plots (Fig. 4B). Yet, in spite of these factors favoring ΦCP6-1, it was ΦCP6-4 that entirely dominated the nonlysogen plots, emphasizing its competitive advantage over ΦCP6-1 for CP6RS.
Second, ΦCP6-4 did not do so well in the lysogen-inoculated plots. It is unclear why this should be, as in the laboratory the lysogen was ΦCP6-4 sensitive. Perhaps the lysogen had a level of resistance to ΦCP6-4 that was only discernible under the nonoptimal growth conditions experienced in situ.
Third, this is the first study to simultaneously release lysogen and nonlysogen forms of the same bacterium into a natural environment and, in doing so, to demonstrate that a bacterium “burdened” with prophage DNA can survive as well as its wild type. This contradicts earlier microcosm studies (11, 18) that found lysogens surviving less well than nonlysogens.
Fourth, our study also illustrates what can happen to an environment into which a lysogen is released artificially. Not only is the microbial community altered by the new bacterium, but phages released from that lysogen also have the potential to affect indigenous bacteria and facilitate gene transfer through transduction. ΦCP6-1, for example, is a transducing phage (4).
A final important point to be drawn from our study is the unique observation that the temporal dynamics of specific phage populations in soil are repeated over successive years. Specifically, our results show that, over three consecutive years, ΦCP6-1 predominated at the beginning of the growing season (3) (Fig. 2C and 3D). In contrast, ΦCP6-4 was never abundant at that time. If it did bloom, it did so some time after the sugar beets had fully matured and ΦCP6-1 numbers had begun to fall (3). Thus, we conclude that the seasonality described in this paper highlights the potential predictability of bacterium-phage interactions in soil.
ACKNOWLEDGMENT
This work was supported by the Ministry of Agriculture Fisheries and Food (UK) grant RG0112.
REFERENCES
- 1.Abedon S T. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb Ecol. 1989;18:79–88. doi: 10.1007/BF02030117. [DOI] [PubMed] [Google Scholar]
- 2.Adams M H. Bacteriophages. New York, N.Y: Interscience Publishers; 1959. [Google Scholar]
- 3.Ashelford K E, Day M J, Bailey M J, Lilley A K, Fry J C. In situ population dynamics of bacterial viruses in a terrestrial environment. Appl Environ Microbiol. 1999;65:169–174. doi: 10.1128/aem.65.1.169-174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford K E, Fry J C, Bailey M J, Jeffries A R, Day M J. Characterization of six bacteriophages of Serratia liquefaciens CP6 isolated from the sugar beet phytosphere. Appl Environ Microbiol. 1999;65:1959–1965. doi: 10.1128/aem.65.5.1959-1965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergh O, Borcheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 6.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cresswell N, Herron P R, Saunders V A, Wellington E M H. The fate of introduced streptomycetes, plasmid and phage populations in a dynamic soil system. J Gen Microbiol. 1992;138:659–666. [Google Scholar]
- 8.Fry J C, editor. Biological data analysis: a practical approach. Oxford, United Kingdom: IRL Press; 1993. [Google Scholar]
- 9.Germida J J. Population dynamics of Azospirillum brasilense and its bacteriophage in soil. Plant Soil. 1986;90:117–128. [Google Scholar]
- 10.Hennes K P, Suttle C A, Chan A M. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol. 1995;61:3623–3627. doi: 10.1128/aem.61.10.3623-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herron P R, Wellington E M H. Population dynamics of phage-host interactions and phage conversion of streptomycetes in soil. FEMS Microbiol Ecol. 1994;14:25–32. [Google Scholar]
- 12.Jones J G. A guide to methods for estimating microbial numbers and biomass in fresh water. United Kingdom: Freshwater Biological Association, Windermere; 1979. [Google Scholar]
- 13.Kado C I, Heskett M G. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology. 1970;60:969–976. doi: 10.1094/phyto-60-969. [DOI] [PubMed] [Google Scholar]
- 14.Kokjohn T A, Miller R V. Gene transfer in the environment: transduction. In: Fry J C, Day M J, editors. Release of genetically engineered and other microorganisms. Cambridge, United Kingdom: Cambridge University Press; 1992. pp. 54–81. [Google Scholar]
- 15.Krueger C L, Sheikh W. A new selective medium for isolating Pseudomonas spp. from water. Appl Environ Microbiol. 1987;53:895–897. doi: 10.1128/aem.53.4.895-897.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenski R E. Dynamics of interactions between bacteria and virulent bacteriophage. In: Marshall K C, editor. Advances in microbial ecology. Vol. 10. New York, N.Y: Plenum Press; 1988. pp. 1–44. [Google Scholar]
- 17.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh P, Toth I, Meijer M, Schilhabel M B, Wellington E M H. Survival of the temperate actinophage φC31 and Streptomyces lividans in soil and the effects of competition and selection on lysogens. FEMS Microbiol Ecol. 1993;13:13–22. [Google Scholar]
- 19.May R M. Models for two interacting populations. In: May R M, editor. Theoretical ecology, principles and applications. 2nd ed. Oxford, United Kingdom: Blackwell Scientific Publications; 1981. pp. 78–104. [Google Scholar]
- 20.Ogunseitan O A, Sayler G S, Miller R V. Dynamic interactions of Pseudomonas aeruginosa and bacteriophage in lake water. Microb Ecol. 1990;19:171–185. doi: 10.1007/BF02012098. [DOI] [PubMed] [Google Scholar]
- 21.Pantastico-Caldas M, Duncan K E, Istock C A, Bell J A. Population dynamics of bacteriophage and Bacillus subtilis in soil. Ecology. 1992;73:1888–1902. [Google Scholar]
- 22.Stephens P M, O'Sullivan M, O'Gara F. Effect of bacteriophage on colonization of sugar beet roots by fluorescent Pseudomonas spp. Appl Environ Microbiol. 1987;53:1164–1167. doi: 10.1128/aem.53.5.1164-1167.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart F M, Levin B R. The population biology of bacterial viruses: why be temperate. Theor Popul Biol. 1984;26:93–117. doi: 10.1016/0040-5809(84)90026-1. [DOI] [PubMed] [Google Scholar]
- 24.Tuomi P, Torsvik T, Heldal M, Bratbak G. Bacterial population dynamics in a meromictic lake. Appl Environ Microbiol. 1997;63:2181–2188. doi: 10.1128/aem.63.6.2181-2188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang I-N, Dykhuizen D E, Slobodkin L B. The evolution of phage lysis timing. Evol Ecol. 1996;10:545–558. [Google Scholar]
- 26.Whitman W B, Coleman D C, Wiebe W J. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]