Abstract
The three pan-genotypic HCV NS3/4A protease inhibitors (PIs) currently in clinical use—grazoprevir, glecaprevir and voxilaprevir—are quinoxaline-based P2–P4 macrocycles and thus exhibit similar drug resistance profiles. Using our quinoxaline-based P1–P3 macrocyclic lead compounds as an alternative chemical scaffold, we explored structure-activity relationships (SARs) at the P2 and P4 positions to develop pan-genotypic PIs that avoid drug resistance. A structure-guided strategy was used to design and synthesize two series of compounds with different P2 quinoxalines in combination with diverse P4 groups of varying size and shape, with and without fluorine substitutions. Our SAR data and cocrystal structures revealed that interplay between the P2 and P4 groups influenced inhibitor binding and overall resistance profile. Optimizing inhibitor interactions in the S4 pocket led to PIs with excellent antiviral activity against clinically relevant PI-resistant HCV variants and genotype 3, providing potential pan-genotypic inhibitors with improved resistance profile.
Graphical Abstract

INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, cirrhosis, liver failure, and hepatocellular carcinoma.1 HCV is highly diverse with seven genotypes and numerous subtypes distributed across different regions; genotypes 1 and 3 are the most prevalent and account for 75% of all infections globally.2, 3 The genetic diversity of HCV has presented a challenge in developing effective pan-genotypic therapies. The recent approval of combination therapies with direct-acting antivirals (DAAs) has remarkably improved HCV treatment outcomes with ~95% cure rates for treatment-naive patients.4, 5 However, the occurrence of baseline polymorphisms among diverse genotypes and preexisting resistance-associated substitutions (RASs) can still lead to treatment failure in some patients.6–10 Nevertheless, the development of pan-genotypic DAAs targeting essential viral proteins NS3/4A, NS5A, and NS5B represents a major medicinal chemistry triumph.11
The HCV NS3/4A protease is a major target for developing pan-genotypic DAAs for the treatment of HCV infections.12 The earlier generation NS3/4A protease inhibitors (PIs) were not effective against all HCV genotypes and variants with common NS3 RASs, limiting their use in the clinic.6, 13 Generally, all NS3/4A PIs lose potency due to polymorphisms across genotypes and/or RASs in NS3 at positions R155, A156, and D168; the D168Q polymorphism is responsible for the reduced inhibitor potency against genotype (GT)-3.14–18 Grazoprevir (GZR) (Figure 1) was the first NS3/4A inhibitor to show activity against most HCV genotypes and viral variants containing mutations at two of the most common NS3 RAS sites R155 and D168; however, GZR is highly susceptible to RASs at A156.19 The two recently approved PIs glecaprevir (GLE) and voxilaprevir (VOX), which are structurally similar to GZR, exhibit pan-genotypic activity.14, 15 However, similar to GZR, GLE and VOX show significantly reduced potency against HCV variants with RASs at position A156 and double-mutant variants containing substitutions at position D168.14, 20–23 The structural similarity of the three clinically used PIs, which are quinoxaline-based P2–P4 macrocycles, poses a major risk of cross-resistance.20 Thus, there is a need to develop potent and robust NS3/4A PIs with alternative chemical scaffolds to effectively target all HCV genotypes while simultaneously avoiding resistance.
Figure 1.
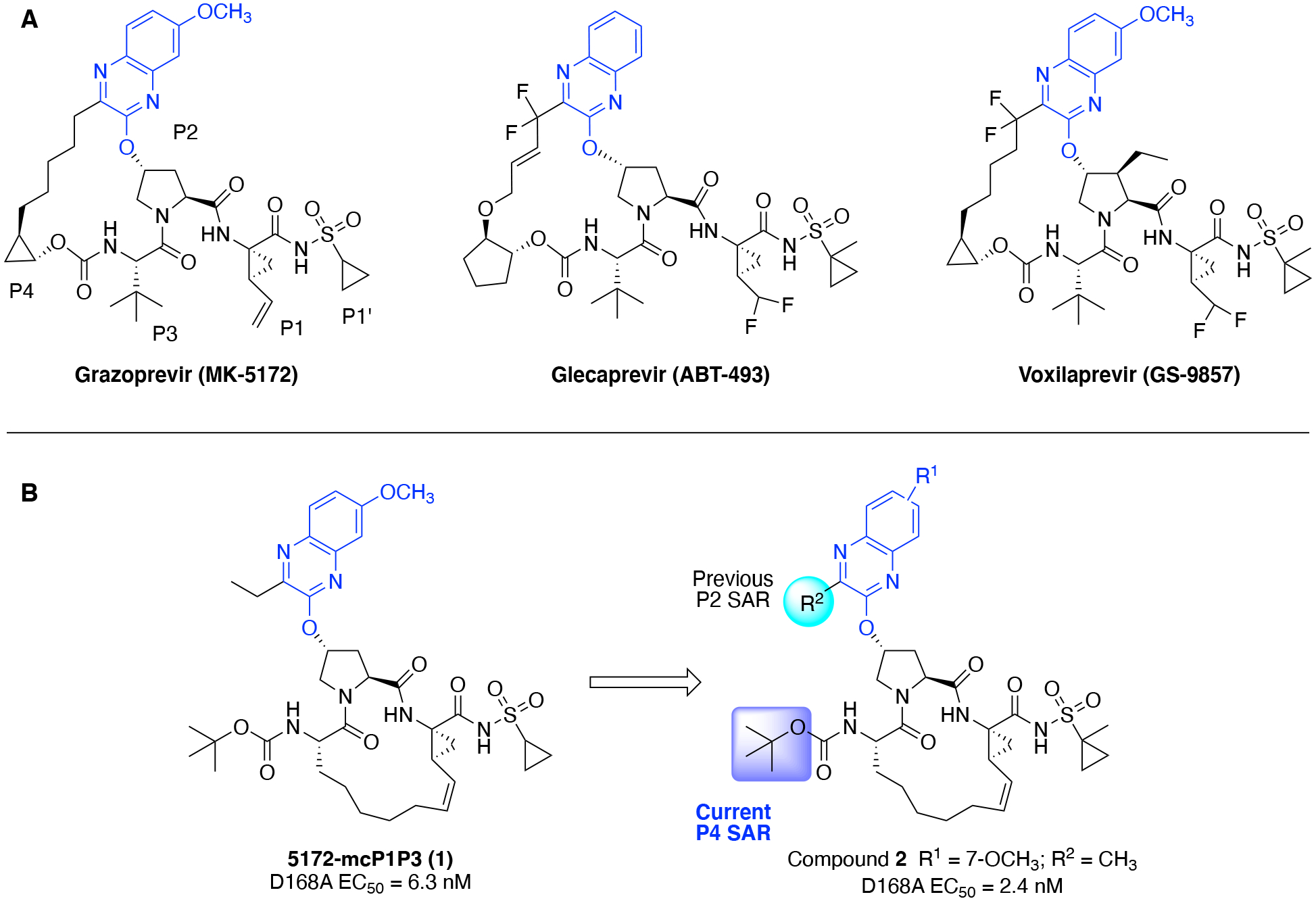
(A) Structures of FDA approved P2–P4 macrocyclic HCV NS3/4A protease inhibitors grazoprevir, glecaprevir, and voxilaprevir; (B) structures of the P1–P3 macrocyclic analogue 5172-mcP1P3 (1) and lead compound 2. The common P2 quinoxaline moiety is in blue and the positions modified are highlighted in cyan and purple, respectively.
We elucidated the molecular basis of drug resistance against HCV NS3/4A PIs showing that disruption of the electrostatic network involving residues R155, D168, and R123, which is critical for efficient inhibitor binding, underlies drug resistance due to RASs and/or polymorphisms.16–18 The improved potency of GZR against common RASs and HCV genotypes was revealed to be due to packing of the P2 quinoxaline moiety on the catalytic triad (H57, D81, S139).16 However, the P2–P4 macrocycle, which binds outside the substrate binding region (termed the substrate envelope)24–26 sterically clashes with RASs A156T/V, resulting in drastically reduced potency.16 To overcome this vulnerability, we designed a P1–P3 macrocyclic analogue of GZR, 5172-mcP1P3 (1) (Figure 1), which maintains better potency across HCV variants with common RASs including at position A156.27 The cocrystal structures showed that the P2 quinoxaline moiety in 1 still packs on the catalytic triad and largely avoids direct contacts with residues R155 and A156.28 Also, the conformational flexibility gained with the P1–P3 macrocyclic scaffold allows 1 to better accommodate RASs in the S2 and S4 subsites of the protease, resulting in an overall improved resistance profile in comparison to PIs with the P2–P4 macrocyclic scaffold.27, 28
In the HCV NS3/4A protease, the common RAS sites R155, A156, and D168 are located around the S2 and S4 subsites.16 The naturally occurring polymorphisms in GT-3a, R123T and D168Q, which cause reduced susceptibility to most PIs, are also located around the S4 subsite.17 Therefore, it is no surprise that modifications at the P2 heterocyclic moiety and P4 capping group, which bind in close proximity to these RAS sites, have the greatest impact on the resistance profile of PIs.27 While the role of the P2 moiety in determining inhibitor potency and resistance profile has been characterized, the effect of P4 modifications on the resistance profile has not been thoroughly investigated.27 Previous structure-activity relationship (SAR) studies in the linear and P1–P3 macrocyclic PIs have explored a variety of P4 capping groups;29–33 however, there is limited SAR data on the antiviral activity of these PIs against resistant variants.33, 34 In addition, the P4 capping group and part of the P2 heterocyclic moiety bind close to each other, but how the interplay between these groups impacts potency against resistant variants is not clear.
The P1–P3 macrocyclic analogue 1 is an ideal scaffold for investigating the impact of modifications at the P2 quinoxaline moiety and P4 capping group on antiviral activity against drug resistant variants. Our previous SAR studies focusing on the P2 moiety elucidated that PIs incorporating small hydrophobic groups at the 3-position of quinoxaline maintained excellent potency against drug resistant variants R155K, A156T, and D168A/V.35 The 3-methylquinoxaline derivative 2 was identified as a promising lead compound with exceptional antiviral activity and resistance profile. Recently, we carried out an extensive structural study to characterize the impact of cyclic P4 ring size on activity against the D168A protease.36 However, in the quinoxaline-based P1–P3 macrocyclic scaffold, the impact of P4 modifications on antiviral activity against drug resistant HCV variants and GT-3a remain unexplored. Investigation of SARs at the P4 position in combination with different P2 quinoxaline moieties can provide insights into how the interplay between the P2 and P4 moieties impact antiviral activity and resistance profile.
Here, we used a structure-guided design strategy to improve the antiviral activity and resistance profile of quinoxaline-based P1–P3 macrocyclic inhibitors against diverse HCV genotypes and resistant variants. Two series of inhibitors were explored with different P2 quinoxaline moieties in combination with diverse P4 capping groups, including acyclic and cyclic moieties, with and without fluorine substitutions. Fluorine has unique electronic properties and a larger van der Waals (vdW) radius.37–39 Since RASs around the NS3/4A protease active site change the shape, contour, and electrostatic properties of the S4 pocket, we reasoned that fluorinated motifs could maintain favorable interactions, thereby leading to robust pan-genotype PIs that are less susceptible to resistance. Our structure-guided SAR approach led to the design of PIs with excellent antiviral activity against drug resistant HCV variants D168A and A156T, and GT-3a. The cocrystal structures of four PIs bound to the D168A protease variant revealed a strong influence of substituent at the 3-position of P2 quinoxaline on the binding of P4 capping groups. The results indicate that incorporating fluorine substitutions proximal to sites of RASs provides a general strategy to improve antiviral activity across HCV genotypes and resistant variants.
RESULTS AND DISCUSSION
In the NS3/4A protease, the S4 subsite is variable across HCV variants, as polymorphisms (GT-3) and RASs at positions R155, D/Q168, and A156 both directly and indirectly influence the size and shape of the pocket (Figure 2). As a result, minor modifications at the P4 capping group significantly impact potency against HCV variants with RASs. Therefore, our SAR efforts focused on exploring various approaches to circumvent the effects of structural changes in the S4 pocket due to RASs including: (1) enhancing hydrophobic packing in the S4 pocket using P4 groups of optimal shape and size, and (2) optimizing binding interactions using fluorine substitutions. The selection of P4 groups was informed by the prior SAR work on different NS3/4A inhibitor scaffolds, detailed structural analysis of analogues of compound 2 bound to WT NS3/4A protease and the D168A variant, and molecular modeling.29, 36, 40 We hypothesized that enhanced binding interactions in the S4 pocket, in combination with optimal P2 quinoxaline moieties, would improve potency against HCV variants with RASs as well as GT-3a.
Figure 2.
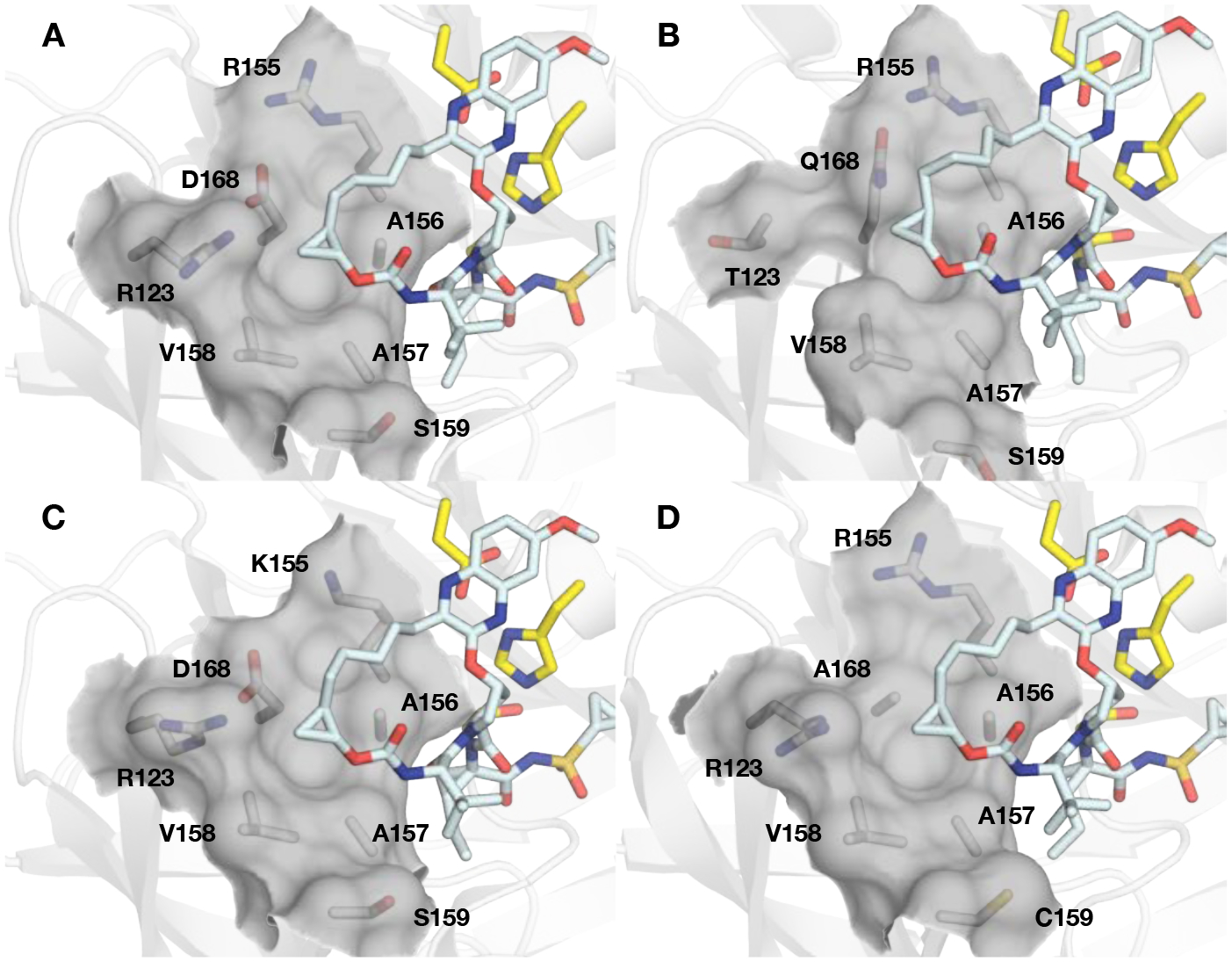
The variability of the HCV NS3/4A protease S4 subsite. The cocrystal structures of grazoprevir with the (A) GT-1a wild-type NS3/4A protease (PDB 3SUD), (B) GT-1a3a chimera (PDB 6P6Q), (C) R155K (PDB 3SUE), and (D) D168A (PDB 3SUF) protease variants. The catalytic triad (yellow) and residues comprising the S4 pocket (R/T123, R/K155, A156, A157, V158, S/C159, and D/A168 are shown as sticks. The surface of the S4 pocket is outlined in grey.
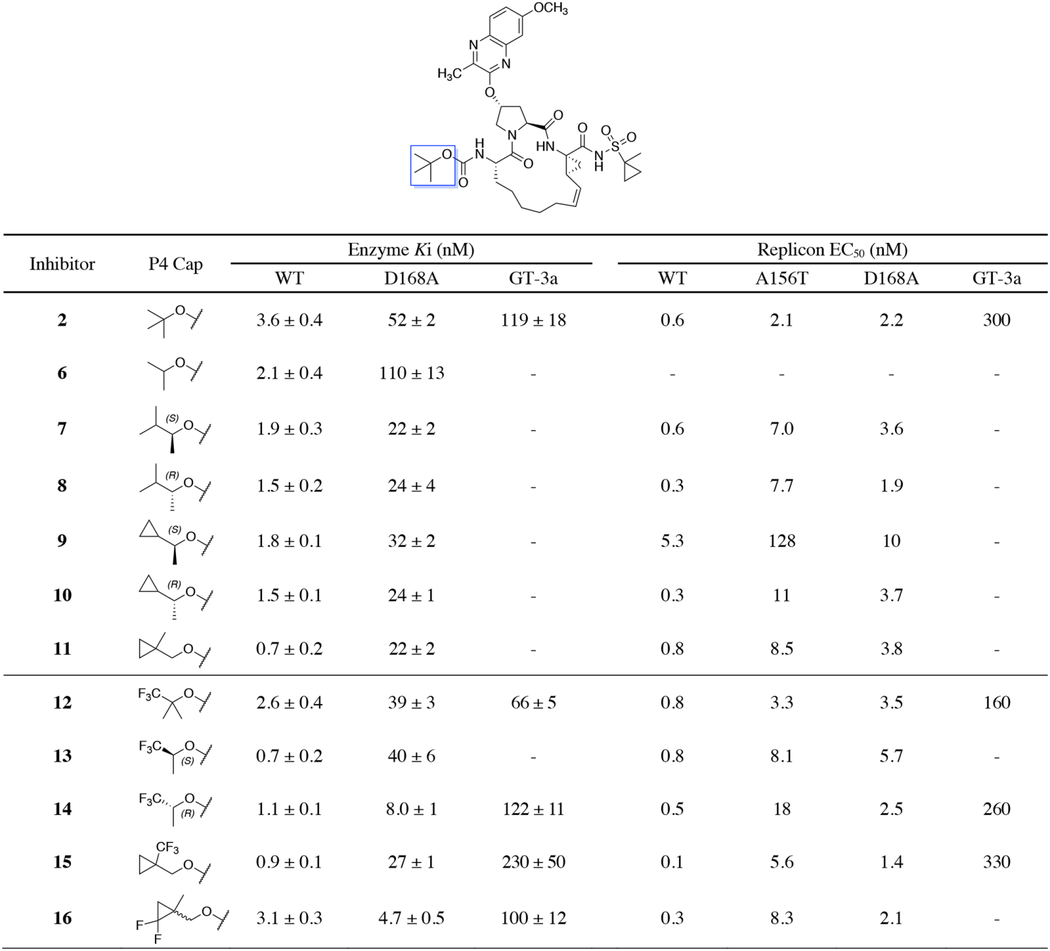
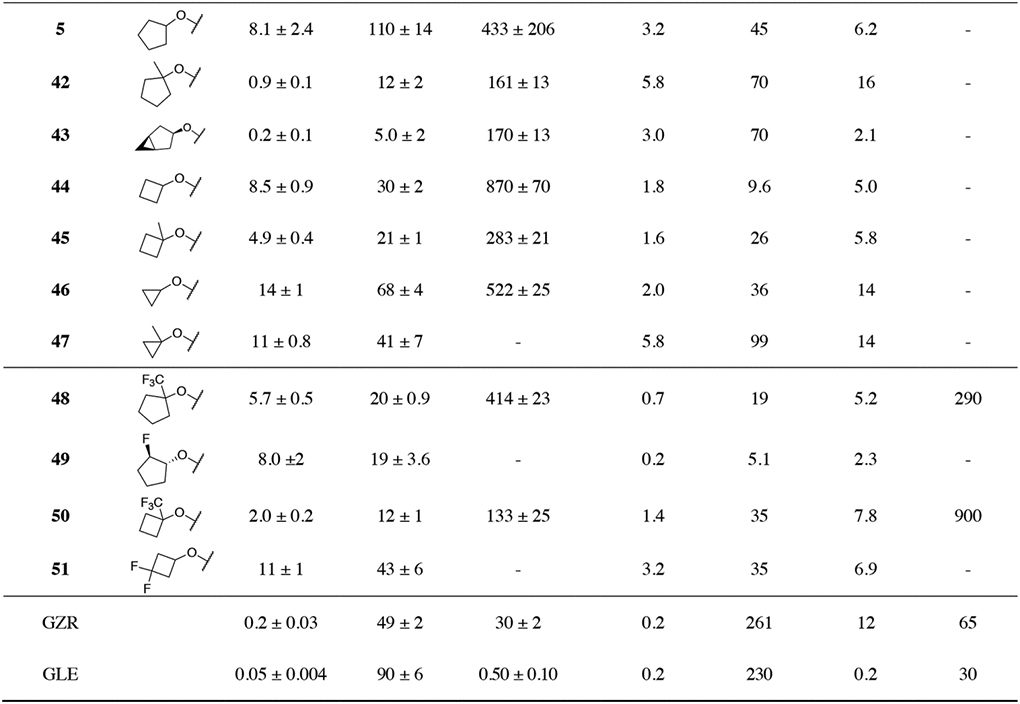
To test this hypothesis, we designed and synthesized two series of PIs containing 3-methyquinoxaline and 3-(trifluoromethyl)-quinoxaline moieties at the P2 position, in combination with a range of diverse P4 capping groups. The potency and resistance profiles of PIs were evaluated using enzyme inhibition and cell-based replicon assays. Inhibition constants (Ki) were determined against the wild-type (WT) GT-1a NS3/4A protease, D168A variant, and GT-3a NS3/4A protease. The antiviral potencies (EC50) were determined against the WT HCV, resistant variants D168A and A156T, and HCV GT-3a (Table 1 and 2). GZR and GLE were used as controls in all assays. Generally, and as previously observed, all PIs were more potent in replicon assays compared to enzyme inhibition assays.35 In replicon assays, the lead compound 2 exhibited improved antiviral activity and resistance profile compared to GZR with EC50 values in the low nM range against the A156T (EC50 = 2.1 nM vs 261 nM for GZR) and D168A (EC50 = 2.2 nM vs 12 nM for GZR) variants. The previously reported compound 3 with a 3-(trifluoromethyl)-quinoxaline at the P2 position also showed excellent antiviral potency and resistance profiles with EC50 values comparable to that of 2 against WT HCV (EC50 = 0.9 nM) and the D168A variant (EC50 = 4.7 nM). Although 3 was 5-fold less potent against the A156T variant (EC50 = 12 nM) relative to 2, both compounds maintained substantially better potency against this variant compared to the approved drugs GZR and GLE (EC50 >200 nM). Thus, the quinoxaline-based P1–P3 macrocyclic analogues 2 and 3 are promising lead compounds for developing robust pan-genotypic NS3/4A PIs with improved resistance profile.
Table 1.
Enzyme and antiviral activity of HCV NS3/4A protease inhibitors 6–27
|
|
Table 2.
Enzyme and antiviral activity of HCV NS3/4A protease inhibitors 28–51

|
|
Exploring the SAR of P4 with 3-Methylquinoxaline at the P2 Position.
The reduced potency of NS3/4A PIs against GT-3a protease and drug-resistant variants mainly results from disruption of the electrostatic network between residues R155, D168, and R123, which causes major structural changes in the active site and destabilizes inhibitor binding.16, 17 In addition, the direct effects of substitutions at positions 168 and 123 result in diminished hydrophobic packing in the S4 pocket. Compound 2 maintains low nM potency against common drug-resistant HCV variants, because the flexible P2 quinoxaline moiety predominantly interacts with the catalytic residues and avoids direct contacts with RAS sites. However, compared to GZR, 2 is less active against the GT-3a protease. Analysis of the cocrystal structures of 2 bound to the WT protease (PDB 5VOJ) and D168A variant (PDB 6UE3) show that the P4 tert-butyl group loses hydrophobic packing in the mutant enzyme.35, 36 Therefore, our goal was to design analogues of 2 that maintain hydrophobic packing in the altered S4 pocket while avoiding direct contacts with A156 to improve antiviral activity against drug-resistant HCV variants and GT-3a.
Acyclic P4 groups.
To enhance hydrophobic packing in the S4 pocket, asymmetric secondary alkyl carbamates were considered to control substituent configuration at P4 and orient specific groups into the S4 subsite. This was evident by the reduced inhibitory potency of the smaller P4 isopropyl compound 6 against the D168A protease. Replacement of the P4 tert-butyl group in 2 with the slightly larger structural isomer 1,2-dimethypropyl, bearing methyl and isopropyl groups at the C1-position, provided compounds 7 and 8 as a pair of P4 stereoisomers. Compounds 7 and 8 were 2-fold more potent against the D168A protease, with no preference observed for either of the stereoisomers (Table 1). This lack of chiral preference was also observed in antiviral data, as both stereoisomers 7 and 8 exhibited similar antiviral activity, with EC50 values against WT HCV and the D168A variant comparable to that of 2. However, these analogues experienced a 3-fold loss in antiviral activity against the A156T variant. Next, we compared isopropyl versus cyclopropyl at the C1-position by preparing compounds 9 and 10 containing the (S)- and (R)-1-cyclopropylethyl P4 groups. These compounds also showed comparable inhibition of the D168A protease. However, in replicon assays 9 and 10 displayed distinct resistance profiles depending on the configuration of the P4 group. The P4 (S)-epimer 9 was significantly less potent across the panel of HCV replicons tested compared to the (R)-epimer 10, which exhibited similar potency as 2, except against the A156T variant. The C1-cyclopropyl group in the (S)-configuration (9) is likely orientated toward residue 156, and the reduced conformational flexibility of cyclopropyl compared to isopropyl (7) leads to lower overall potency. The closely related primary carbamate analogue 11, where the C1-methyl group is moved to the C2-position, displayed similar potency to that of 7 and 8 in both biochemical and replicon assays. These results indicate that asymmetric secondary alkyl carbamates allow control of the binding conformations of P4 substituents to enhance hydrophobic packing in the variable S4 subsite. However, larger P4 groups that bind more deeply into the S4 pocket may also interact with residue A156 increasing inhibitor vulnerability to RASs at this position.
In addition to optimizing hydrophobic packing in the S4 subsite, we explored hydrogen to fluorine substitutions as a general strategy to improve antiviral activity against drug-resistant HCV variants and GT-3a. Due to the high lipophilicity, non-polarizability, and larger vdW radius, fluorine is unique in its ability to influence inhibitor conformation, cell permeability, potency, and pharmacokinetic properties.37–39 The multifaceted properties of fluorine make it an ideal substitution to investigate binding in the dynamic and variable S4 pocket of the NS3/4A protease, as fluorine is able to participate in favorable hydrophobic and electrostatic interactions. Since the S4 subsite is the location of major RASs and polymorphisms between GT-1a and GT-3a, we hypothesized that introducing fluorine atoms at the P4 moiety would allow the inhibitors to adapt to the topology and electrostatic changes in the pocket.
To test this hypothesis, we explored fluorine substitutions in a variety of P4 groups. In compound 2, substitution of a methyl group with trifluoromethyl in P4 tert-butyl provided analogue 12, which displayed excellent antiviral activity against WT HCV and resistant variants A156T and D168A with EC50 values < 5 nM. Compound 12 also showed improved inhibitory activity against the GT-3a protease and was comparable to GZR in GT-3a replicon assays (GT-3a EC50 = 160 nM vs 65 nM for GZR). A recently reported cocrystal structure of the WT NS3/4A protease with BMS-986144 (PDB 7D5L), which contains the same P4 group, shows that the trifluoromethyl group makes favorable interactions with residues D168, R123, and V158 in the S4 pocket.41 The trifluoro tert-butyl group in 12 likely binds in a similar conformation, and the trifluoromethyl group, while avoiding direct contacts with A156, provides enhanced interactions with key variable residues in the S4 pocket that are beneficial to activity against resistant variants and GT-3a.
Next, methyl to trifluoromethyl substitution in the P4 isopropyl analogue 6 was explored. Compounds 13 and 14, prepared as P4 stereoisomers, showed improved biochemical potency against the WT protease relative to both 2 and 12, but only 14 was more potent against the D168A protease. In replicon assays with WT HCV and the resistant variant D168A, both 13 and 14 exhibited EC50 values comparable to that of 2, but lost potency against the A156T variant. Interestingly, while 13 maintained an excellent overall potency profile with EC50 value < 10 nM against the resistant variants, 14 exhibited 2-fold better potency against the D168A variant, in line with the improvement in the biochemical potency. The improved potency of 14 against the D168A variant with concomitant loss in potency against the A156T variant indicates that the trifluoromethyl group is binding close to residues 168 and 156. However, compound 14 showed slightly lower GT-3a potency relative to 12, indicating relatively less favorable interactions in the S4 pocket. Similarly, in compound 11 containing the (1-methylcyclopropyl)methyl carbamate at P4, substitution of the C2-methyl with a trifluoromethyl group to provide analogue 15 also resulted in improvement in antiviral activity compared to the parent compound. Compound 15 exhibited an excellent potency profile with WT EC50 of 0.1 nM and the D168A EC50 of 1.4 nM. However, 15 showed 2-fold lower activity against GT-3a in both enzyme and replicon assays relative to 12. Finally, substitution of the C3-methylene hydrogens of P4 cyclopropyl in 11 with fluorine atoms to generate 16, which was prepared as a mixture of stereoisomers, led to a 5-fold improvement in inhibitory potency against the D168A protease variant; however, in replicon assays 16 showed similar antiviral activity as the parent 11. These results indicate that fluorine substitutions at the P4 capping group generally results in improvements in both biochemical and antiviral potency. Moreover, specific orientation of the fluorinated groups in the S4 subsite is crucial to form favorable intermolecular interactions to improve potency against GT-3a.
Cyclic P4 groups.
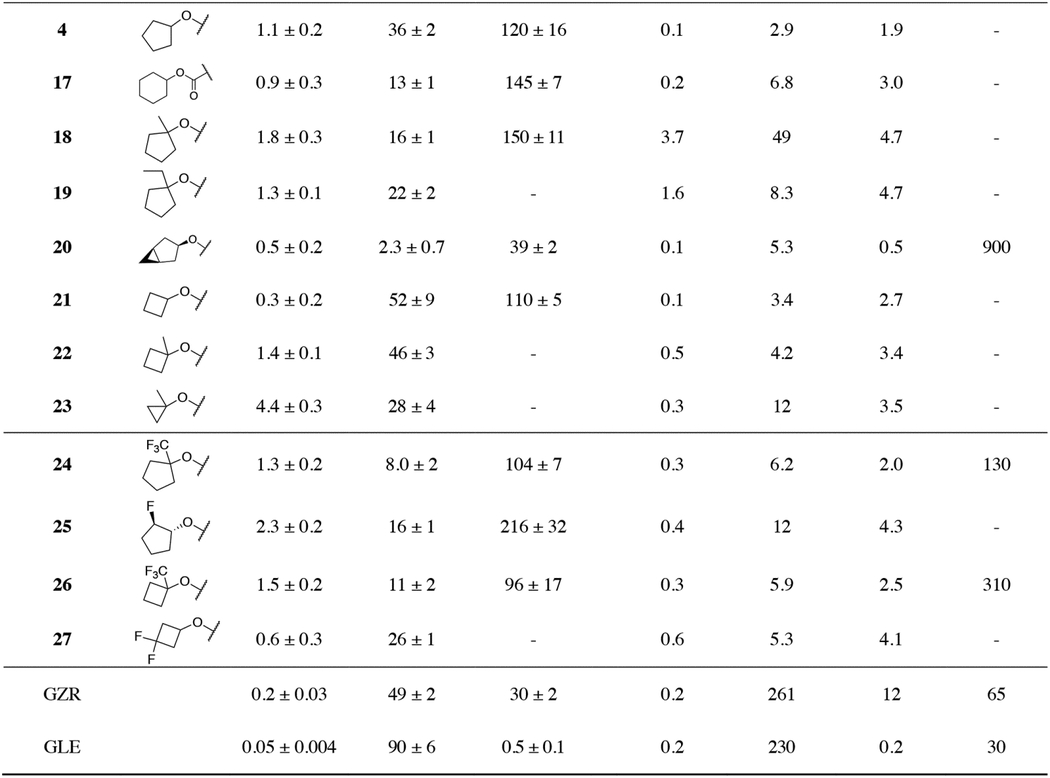
Following the investigation of the skeletal isomers of tert-butyl group in 2, cyclic P4 capping groups of various ring sizes were explored. Our prior SAR effort on the P1–P3 macrocyclic scaffold only showed a slight improvement in potency with the cyclopentyl (4) compared to the tert-butyl (2) P4 capping group.35 Following that work, we undertook a detailed structural study of PIs with cyclic P4 capping groups focusing on elucidating the impact of P4 ring size on the inhibitory activity against the D168A protease variant.36 Detailed structural analysis of compounds 4, 17–20 and 22–23, all designed based on the parent inhibitor 4, emphasized that shape complementarity of the P4 group to the S4 pocket, rather than the ring size, is critical for enhancing hydrophobic interactions to improve inhibitory potency against the D168A protease variant (Figure 3).36 Here these compounds were further evaluated, and inhibitory activity against the GT-3a protease and antiviral activity against our panel of HCV replicons were determined.
Figure 3.
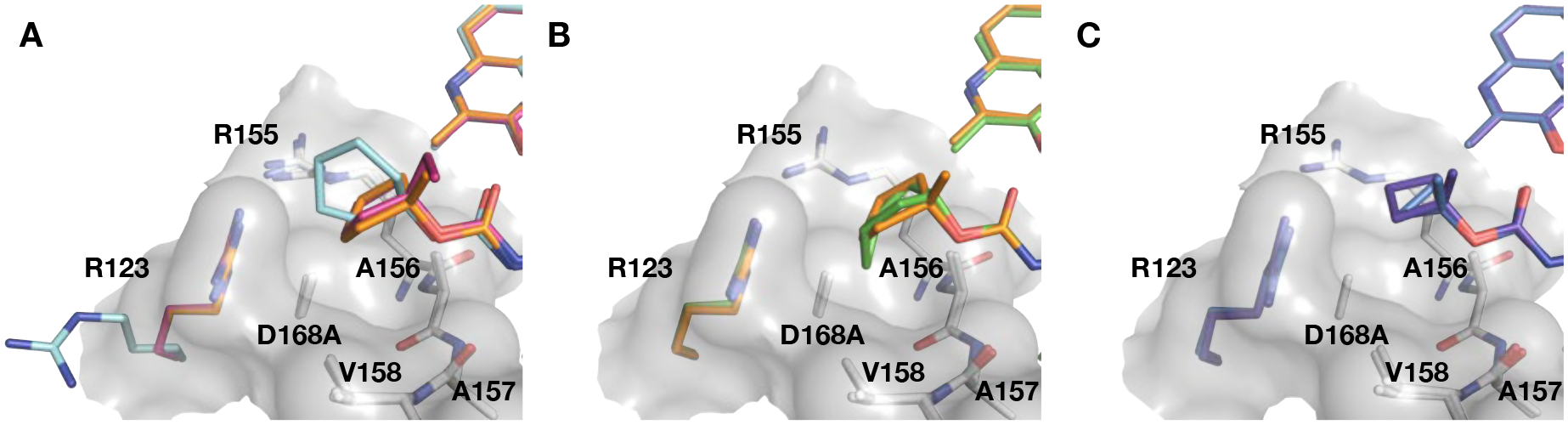
Comparison of the binding conformations of cyclic P4 groups in the S4 subsite. Superposition of the D168A NS3/4A protease complexes with compounds (A) 17 (PDB 6PIW), 18 (PDB 6PJ1), and 19 (PDB 6PJ), (B) 18 (PDB 6PJ1) and 20 (PDB 6PIV), and (C) 22 (PDB 6PIY) and 23 (PDB 6PIZ), focusing on the differences at the P4 group. The residue R123 is colored according to the respective bound inhibitor, while the other residues comprising the S4 pocket are shown as white sticks. The carved surface shows the depth and topology of the S4 pocket.
The P4 cyclopentyl compound 4 has an excellent potency profile with EC50 values similar to that of 2. Compound 17 with a larger cyclohexyl group at the P4 position maintained antiviral activity against the D168A variant but was 2-fold less active against the A156T variant. Structural analysis of 4 and 17 bound the D168A protease variant showed that the cyclohexyl ring adopted a boat conformation with portion of the ring shifted toward solvent. To accommodate the larger cyclohexyl group, the R123 side chain is flipped out of the S4 pocket (Figure 3A). These observations provided an indication to the optimal size of lipophilic group in the S4 pocket and guided the design of additional cyclic P4 analogues. Next, we explored the effect of C1-methyl substitution to orient the cyclopentyl group deeper into the S4 pocket. Compound 18 was less active than the parent 4 in replicon assays, particularly against the A156T variant. The lower antiviral activity observed with 18 is somewhat surprising, as cyclopentyl rings in both compounds adopted similar conformations, with only a slight change in the ring pucker (Figure 3A). To understand the impact of ring pucker on activity, we generated 19, replacing methyl with ethyl at the C1-position of the cyclopentyl group. The 1-ethyl derivative 19 exhibited improved antiviral activity against the A156T variant relative to 18. The cocrystal structures showed that the cyclopentyl ring in 19 adopted a conformation similar to the unsubstituted ring in 4, likely allowing the inhibitor to maintain activity against the A156T variant relative to the 1-methyl analogue 18. Thus, the binding conformation and packing of the cyclopentyl ring is important for maintaining activity against both D168A and A156T variants.
Our previous structural analysis showed that the fused bicyclic P4 moiety of compound 20 contoured the topology, optimally filling the S4 subsite of the NS3/4A protease, thereby avoiding the formation of a frustrated pocket (Figure 3B). The enhanced packing in the S4 pocket led to improved potency in replicon assays. Compound 20 exhibited exceptional antiviral activity with EC50 values of 0.1 and 0.5 nM against WT HCV and the D168A variant, respectively, and maintained low nM activity against the A156T variant. This derivative also showed improved inhibitory activity against the GT-3a protease comparable to that of GZR; however, in GT3a replicon assays 20 was significantly less active than GZR. The P4 cyclobutyl analogue 21 also displayed an excellent antiviral activity profile with EC50 values < 5 nM against the resistant variants tested. The corresponding 1-methylbutyl analogue 22 also showed an excellent antiviral activity profile. The P4 1-methylcyclopropyl compound 23 was equipotent to 22 against WT HCV and the D168A variant but was 3-fold less active against the A156T variant. Overall, our SAR results in the cyclic P4 series indicate that P4 ring size has minimal impact on antiviral activity and resistance profile.
To investigate the effects of fluorine substitutions on potency and resistance profile in cyclic P4 capping groups, we first designed and synthesized 24 and 25, with a trifluoromethyl moiety at the C1-position and a single fluorine substitution at the C2-position of the cyclopentyl group. Converting C1-methyl to C1-trifluoromethyl of the cyclopentyl P4 capping group of 18 provided 24 with a 2-fold improvement in potency against the D168A variant in both biochemical and replicon assays relative to the parent 18. Also, 24 showed an 8-fold improvement in antiviral activity against the A156T variant. The improved potency of 24 against the A156T variant was surprising; the C1-trifluoromethyl substitution is possibly causing cyclopentyl ring to adopt an alternate conformation relative to the C1-methyl derivative. More significantly, 24 showed potent antiviral activity against GT-3a with an EC50 value comparable to that of GZR (EC50 = 130 nM vs 65 nM for GZR). A significant improvement in inhibitory potency against the D168A protease variant was also realized by replacing the 1-methycyclobutyl P4 group in 22 with a 1-(trifluoromethyl)-cyclobutyl (26). Analogue 26 had a 4-fold improvement in potency compared to the non-fluorinated parent inhibitor 22. Introducing fluorine atoms at the C3-methylene position of the cyclobutyl P4 capping group did not affect the overall potency profile of the resulting analogue 27 compared to the parent compound 21. Of the P4 fluorinated PIs, all but one (25) maintained excellent antiviral activity against the resistant variants. Compared to the C1-methyl derivatives, C1-trifluoromethyl substitution provided compounds with improved overall antiviral activity including against GT-3a.
Exploring the SAR of P4 with 3-(Trifluoromethyl)-quinoxaline at the P2 Position.
To investigate how the interplay between the P2 quinoxaline moiety and the P4 capping group might affect antiviral activity and resistance profile, we designed and synthesized analogues of the 3-(trifluoromethyl)-quinoxaline inhibitor 3 with diverse P4 capping groups (Table 2). In our previous SAR studies, compounds with a 3-(trifluoromethyl)-quinoxaline moiety at the P2 position generally exhibited slightly lower antiviral activities compared to the corresponding 3-methylquinoxaline analogues, particularly against the A156T variant.35 At that time, our efforts to co-crystallize the NS3/4A protease complex with a 3-(trifluoromethyl)-quinoxaline analogue were unsuccessful. Hence the reduced potency of these compounds could not be explained by molecular modeling alone, where the P2 moiety was predicted to bind similarly with a slight shift towards the catalytic histidine (H57).35
The results from the current SAR studies in the 3-(trifluoromethyl)-quinoxaline series showed a stronger influence of variations at the P4 capping group on antiviral activity compared to the 3-methylquinoxaline series. Based on the modeling, these potency differences were not expected to arise from subtle differences in the binding conformation of the two quinoxaline moieties. The trifluoromethyl group at the 3-position of the P2 quinoxaline was likely influencing the binding mode of the P4 capping groups, and as a result, significantly impacting antiviral activity against the D168A and A156T variants. To understand the binding of 3-(trifluoromethyl)-quinoxaline compounds, we again attempted, and while this manuscript was in preparation, succeeded in determining cocrystal structures of compound 3 and three new analogues bound to the D168A protease variant. To our surprise, the structures revealed that the P2 quinoxaline moiety has the methoxy group at the 6-position rather than the 7-position. In our previous work, we had described the substitution pattern on the quinoxaline moiety in 3 to be 7-methoxy-3-(trifluoromethyl). This discrepancy resulted from the inadvertent mistake of assuming the 7-methoxy-3-(trifluoromethyl)-quinoxaline, rather than the 6-methoxy-3-(trifluoromethyl)-quinoxaline, as the major regioisomer in the condensation reaction between 4-methoxybenzene-1,2-diamine and ethyl trifluoropyruvate. While the other 3-substituted-quinoxalines (3-methyl, ethyl, isopropyl) we synthesized had the 3susbtituted-7-methoxy-quinoxaline as the major regioisomer.
Comparison of the D168A NS3/4A protease cocrystal structures with 2 and 3 showed overall similar binding conformations, with major structural differences occurring only at the quinoxaline moiety (Figure 4). The 6-methoxy-3-(trifluoromethyl)-quinoxaline moiety in 3 is shifted towards the catalytic histidine (H57) relative to the position of the 7-methoxy-3-methylquinoxaline in 2 (Figure 4B). The lateral shift of the quinoxaline moiety in 3 is also evident when compared to the quinoxaline moiety in GZR bound to the D168A protease variant (Figure 4C). Even with this lateral shift, the quinoxaline moieties in 2 and 3 maintain similar stacking interactions with the catalytic residues. The 3-trifluoromethyl group of 3 occupies a similar position in space as the 3-methyl group of 2, despite the shift of the quinoxaline. Therefore, the trifluoromethyl substituent contributes to greater hydrophobic contacts due to the larger vdW radius of fluorine relative to hydrogen; however, we did not observe fluorine-specific molecular interactions in the cocrystal structure with 3. The increased vdW contacts of the trifluoromethyl group are beneficial for potency against WT and D168A variants, but detrimental against the A156T variant due to a potential steric clash with the threonine. The shift in the position of the quinoxaline also avoids a steric clash between the 6-methoxy substituent and residues V78 and Y56, which are located directly above the catalytic H57, forming part of the hydrophobic binding site for the quinoxaline moiety. This structural analysis also explains the observed potency profiles of the 3-(trifluoromethyl)-quinoxaline compounds (3 and 5) in our previous SAR studies.35 Although the analogues of 2 and 3 with modified P4 groups are not exact matched pairs, the SAR analysis provides valuable insights into the interplay between the P2 and P4 moieties and its impact on potency against resistant variants.
Figure 4.
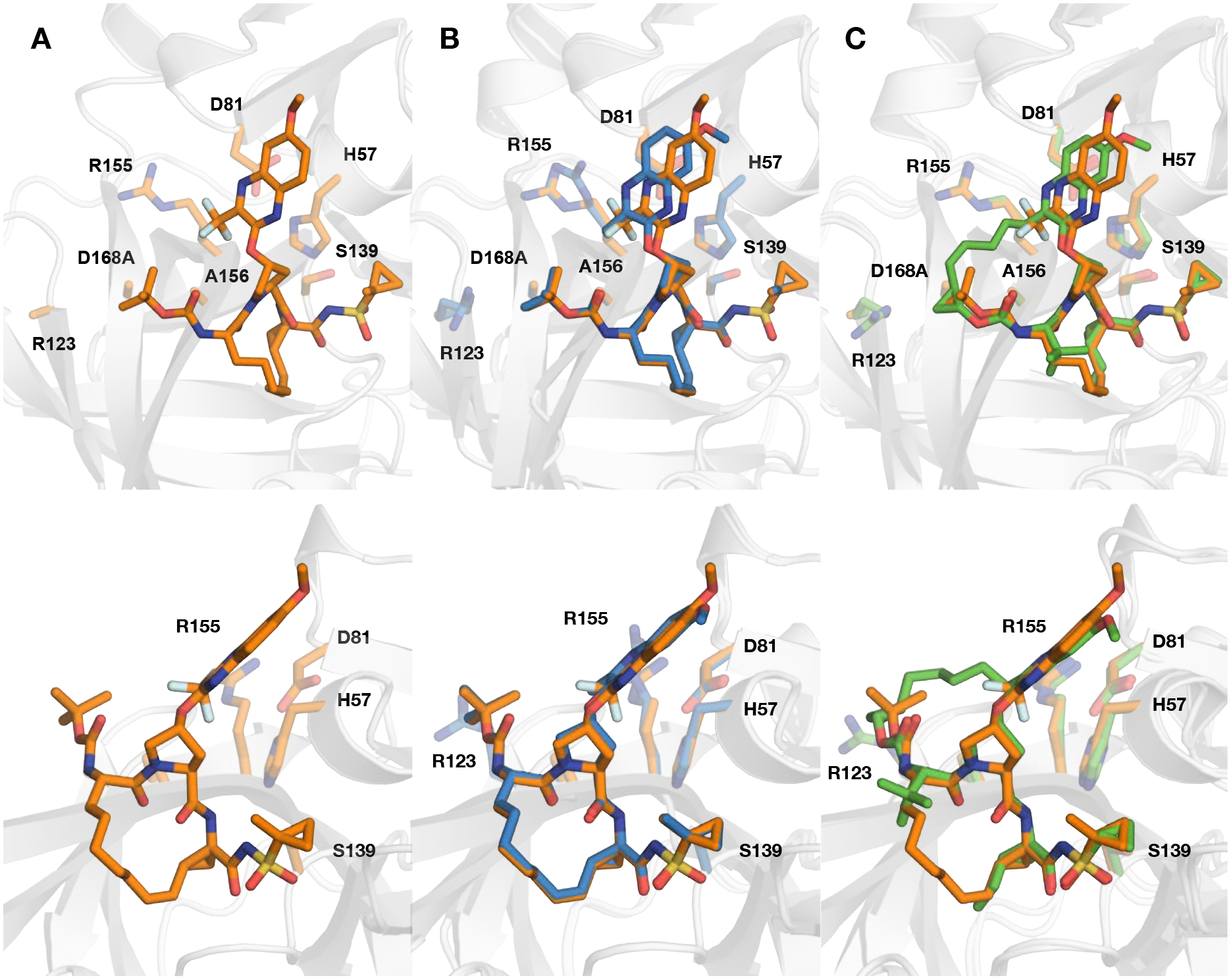
(A) Crystal structure of D168A NS3/4A protease variant in complex with compound 3. (B) Comparison of the binding conformations of compound 3 with (B) 2 (PDB 6UE3) and (C) grazoprevir (PDB 3USF) in the active site of D168A NS3/4A protease, focusing on the differences at P2 quinoxaline. The protease is in ribbon representation with bound inhibitors 3 (orange), 2 (blue), and grazoprevir (green) shown as sticks. The side chains of the catalytic triad (D81, H57, S139) and resistance-associated substitutions and/or natural polymorphism residues (R123, R155, A156, and D168) are shown as sticks.
Acyclic P4 groups.
Replacement of the P4 tert-butyl group in 3 with slightly larger structural derivatives generally resulted in improvement in inhibitory activity against the D168A protease variants but antiviral activity against resistant variants varied depending on the shape of the P4 group. Compound 28 with a slightly larger tert-pentyl carbamate showed improved biochemical potency but 2-fold lower antiviral activity against the D168A variant in replicon assays. However, 28 maintained similar antiviral activity as 3 against the A156T variant. In contrast, the P4 pentan-3-yl compound 29 was 7-fold less potent against the A156T variant compared to both 3 and 28, indicating that the P4 group of 29 may bind too deeply into the S4 pocket, making contact with A156 in the WT protease, hence becoming readily susceptible to RASs at this position.
Next, similar to the 3-methylquinoxaline series, we explored configurational control to orient the hydrophobic substituents into the S4 subsite. Secondary alkyl carbamates bearing asymmetric substituents at the C1 position provided pairs of P4 stereoisomers. The P4 1,2-dimethypropyl derivatives 30 and 31, bearing a methyl and an isopropyl group at the C1-position, exhibited comparable antiviral potencies against WT replicon and D168A variant but differed in loss of potency against the replicon with A156T substitution. The analogue 31 with the (R)-configuration at the P4 group was 2-fold less active compared to the (S)-stereoisomer 30 against both WT and D168A replicons. Additionally, 31 was 5-fold less active against the A156T replicon, indicating that the (R)-1,2-dimethypropyl group is directed towards position 156, while the corresponding (S)-stereoisomer in 30 is likely oriented in the opposite direction. Notably, enzyme inhibition data showed a preference for the (R)-stereoisomer at P4, with 31 showing a 2-fold improvement in Ki values compared to the (S)-epimer 30. Similar trends were observed for compounds 32 and 33, containing the (S)- and (R)-enantiomers of 1-cyclcopropylethyl group at the P4 position, respectively. While both analogues showed similar antiviral potency against the WT and D168A variants, 32, with the (S)-configuration at P4, maintained better potency against the A156T variant than the (R)-epimer 33. Interestingly, compounds in the 3-(trifluoromethyl)-quinoxaline series (30–33) showed different trends in EC50 values against the A156T variant than the corresponding analogues in the 3-methylquinoxaline series (7–10). Comparison of the A156T EC50 data suggests that the trifluoromethyl substitution on the quinoxaline is likely affecting the binding conformation of the P4 capping groups. The 3-methylquinoxaline compounds 7 and 8 showed similar potencies against the resistant variants, but the corresponding 3-(trifluoromethyl)-quinoxaline analogues 30 and 31 diverged from each other. The analogue 31 is 6-fold more susceptible to the A156T RAS compared to 8, indicating that the substituent at the 3-position of quinoxaline plays a role in orienting the P4 capping group. The trifluoromethyl group may form additional hydrophobic interactions with the 1,2-dimethylpropyl P4 group in WT protease above position 156, making 31 susceptible to the A156T RAS, while the methyl substituent in 8 may not form these contacts due to the smaller vdW surface area and lower hydrophobicity.38, 39
In contrast, both series of compounds with a 1-cyclcopropylethyl group at the P4 position showed different antiviral potencies against the A156T variant depending on the configuration of the P4 group. From the reduced potency of 9 against the A156T variant relative to 10, we can conclude that the cyclopropyl ring in the (S)-stereoisomer of the P4 group is packing closer to residue 156. However, the 3-trifluoromethyl substituent on the quinoxaline, which generally increases inhibitor susceptibility to A156T due to increased interaction with the larger 156 side chain, seems to prevent the (S)-stereoisomer (32) from interacting with residue 156. The larger 3-trifluoromethyl group may be forcing the P4 capping group of 32 to adopt an alternate binding conformation relative to the corresponding 3-methylquinoxaline analogue (9) resulting in much lower susceptibility to A156T. Thus, for the 3-(trifluoromethyl)-quinoxaline series, as the size of the P4 groups increases, the potential for steric clash between these two proximal moieties seem to promote an alternate binding for the P4 capping group, which could be exploited to avoid inhibitor susceptibility to A156T.
Reducing the size of the acyclic P4 group also proved to be detrimental to antiviral potency. The P4 isobutyl analogue 34 was less potent against the D168A and A156T variants than 30 and 31, which contain a methyl group at the C1-position. The corresponding neopentyl carbamate 35, with similar antiviral potency against WT HCV and the D168A variant as 34, experienced further loss in potency against the A156T variant. Among the primary alkyl carbamates, the (1-methylcyclopropyl)methyl derivative 36 exhibited better antiviral potency against both the A156T and D168A variants. Together, these data showed that in the 3-(trifluoromethyl)-quinoxaline series, inhibitor potency was more sensitive to minor changes at the P4 capping group compared to the 3-methylquinoxaline series.
Introduction of fluorine substitutions at the P4 position in the 3-(trifluoromethyl)-quinoxaline series resulted in distinct potency profiles depending on the location of the fluorine atoms. The P4 trifluoro tert-butyl analogue 37 exhibited an excellent potency profile in replicon assays with EC50 values similar to that of 3 against the WT HCV (EC50 = 1.0 nM), A156T (EC50 = 10 nM), and D168A (EC50 = 4.3 nM) variants. However, this analogue was less active against the GT-3a relative to its 3-methylquinoxaline counterpart 12. The cocrystal structure of 37 with the D168A protease variant showed a similar binding conformation of the P2 quinoxaline moiety as observed for 3 (Figure 5A). The P4 trifluoro tert-butyl group adopts a binding conformation identical to that of the P4 group in BMS-986155 (PDB 7D5L). One of the P4 geminal methyl groups is oriented toward the trifluoromethyl group of the P2 quinoxaline, while the other points away from the protein surface. The trifluoromethyl group is positioned in the S4 pocket interacting with residues D168, R123, and V158 (Figure 5A). The enhanced interactions in the S4 pocket are likely contributing to the excellent antiviral activity of 37 against drug-resistant HCV variants.
Figure 5.
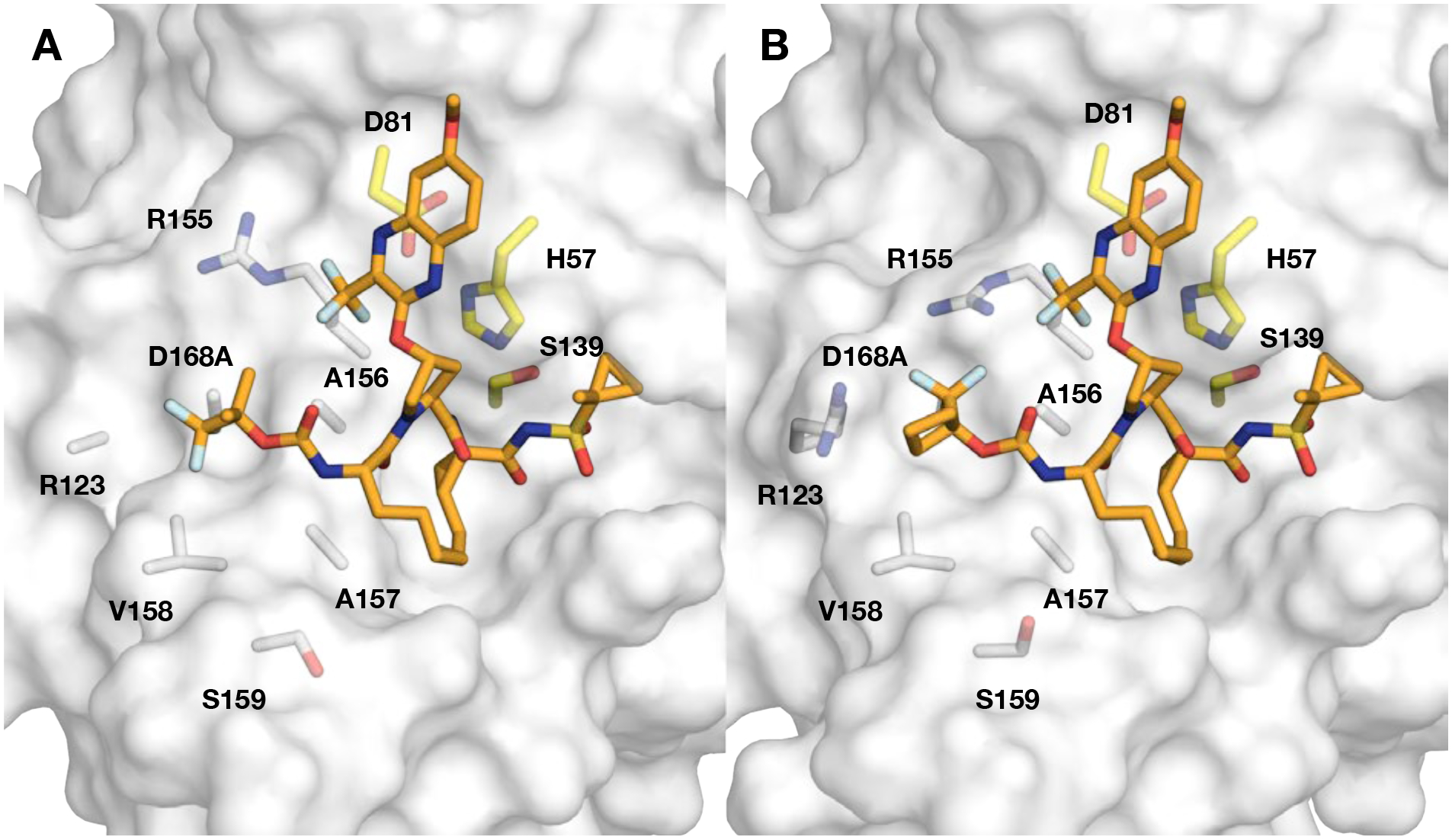
Crystal structures of compounds (A) 37 and (B) 50 in complex with the D168A NS3/4A protease variant. The protease active site is shown as a light grey surface with bound inhibitors 37 and 50 shown as orange sticks. The catalytic triad is highlighted in yellow, and residues comprising the S4 pocket, R123, R155, A156, A157, V158, S159, and DA168A, are shown as white sticks.
The P4 1-(trifluoromethyl)ethyl derivatives 38 and 39 exhibited distinct potency profiles depending on the stereochemistry at the P4 group. Compound 39, with the (R)-configuration at P4, was at least 2-fold more potent in both biochemical and replicon assays than 38, which has the (S)-configuration at P4. Interestingly, in both the 3-methylquinoxaline and 3-(trifluoromethyl)-quinoxaline series, compounds with the (R)-1-(trifluoromethyl)ethyl carbamate at P4 (14 and 39) were more potent against WT HCV and the D168A variant, relative to the epimeric congeners (13 and 38). While the same trend was observed for antiviral potency against the A156T variant for 38 and 39, the preference for (R)-configuration at P4 was reversed in 13 and 14, where the (S)-configuration was preferred instead. The reversed preference for the (R)-stereoisomer in the 3-(trifluoromethyl)-quinoxaline compounds is likely due to repulsive interactions between trifluoromethyl groups at the 3-position of the P2 quinoxaline and the P4 group.
Substitution of the P4 C2-methyl group in 36 with trifluoromethyl provided the (1-(trifluoromethyl)cyclopropyl)methyl compound 40, which was 2-fold more active in replicon assays against WT HCV and the D168A variant, without loss in potency against the A156T variant. Compound 41, which was generated by replacing the C3-methylene hydrogens of the P4 cyclopropyl group in 36 with fluorine atoms, prepared as a P4 racemic mixture, also showed 2-fold improvement in antiviral activity against WT HCV. However, analogue 41, while equipotent to 36 against the D168A variant, lost 3-fold potency against the A156T variant.
Cyclic P4 groups.
After exploring different design strategies with acyclic P4 carbamates, we used a similar approach to identify cyclic carbamate moieties that can be combined with the 3-(trifluoromethyl)-quinoxaline P2 moiety to optimally target the S4 subsite. We have previously shown that relative to 3, the P4 cyclopentyl analogue 5 was less potent against WT HCV and the A156T variant, though it maintained similar potency against the D168A variant. The reduced potency of 5, particularly against the A156T variant, likely results from the larger size of the P4 group. This was further supported by the observed potency profile of the corresponding 1-methylcyclocpentyl analogue 42, which showed further loss in potency across the three HCV replicons tested. In contrast to reduced potency in replicon assays, 42 showed improved potency in biochemical assays relative to 5, even against the GT-3a protease. The fused bicyclic P4 analogue 43 showed a different trend, with comparable antiviral activity against WT and a 3-fold improvement in potency against the D168A variant. However, similar to analogues with a large cyclic P4 group, 43 experienced significant loss in antiviral potency against the A156T variant. The cocrystal structure of 43 bound to the D168A protease variant showed a shift in the position of the P2 quinoxaline compared to the corresponding 3-methylquinoxaline compound 20, but the bicyclic P4 group in both compounds bound in a similar conformation (Figure 6). Similar to 20, compound 43 showed significantly improved Ki values against the WT, D168A, and GT-3a proteases. The improved biochemical and antiviral potency of 43 relative to 5 is likely due to enhanced hydrophobic interactions in the S4 subsite. However, the SAR data show that larger cyclic P4 groups that bind deeply into the S4 pocket are detrimental to antiviral activity against the A156T variant, and the C1-methyl substitution results in a further loss of potency against this variant.
Figure 6.
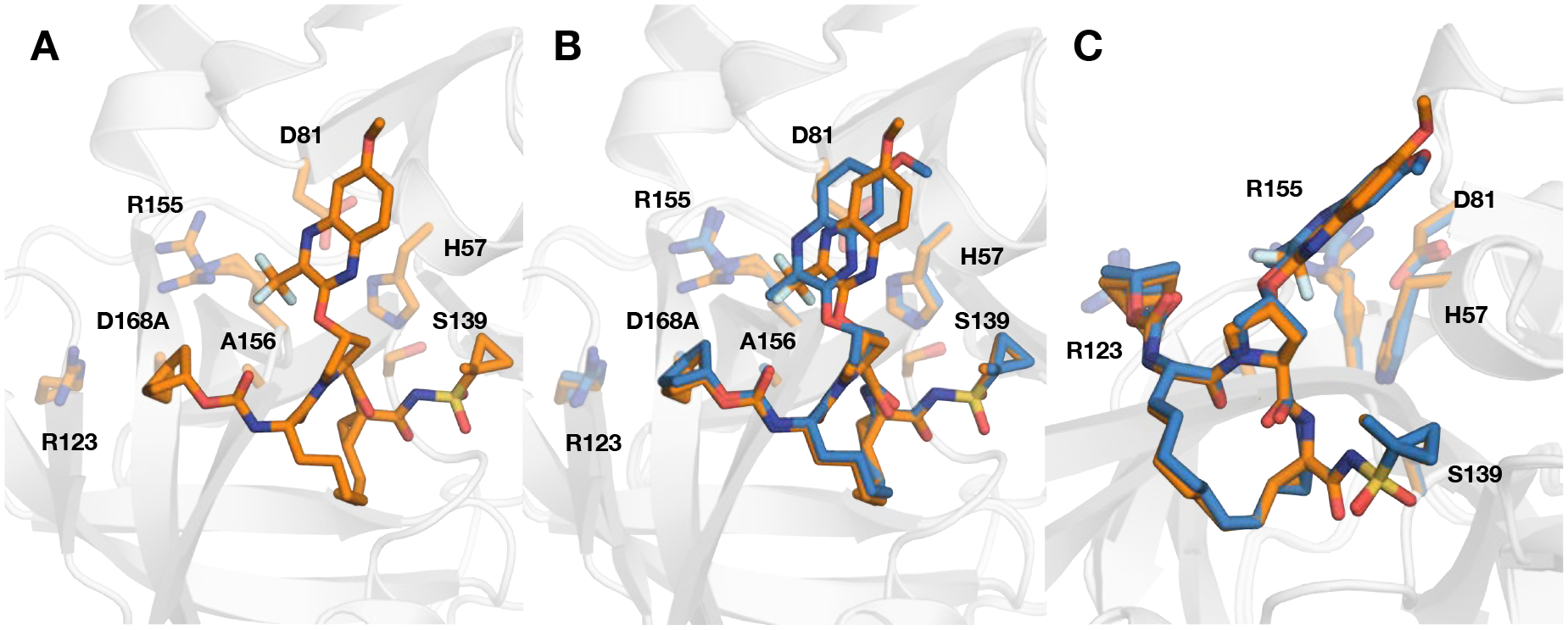
(A) Crystal structure of compound 43 in complex with the D168A NS3/4A protease variant. Comparison of the binding conformations of compound 43 with (B) 20 (PDB 6PJ1) in the active site of D168A NS3/4A protease, focusing on the differences at P2 quinoxaline containing and (C) stacking interactions with the catalytic residues. Both compounds contain the same fused bicyclic P4 group but different P2 quinoxaline moieties. The protease is in ribbon representation with bound inhibitors 43 (orange) and 20 (blue). The side chains of the catalytic triad (D81, H57, S139) and resistance-associated substitutions and/or natural polymorphism residues (R123, R155, A156, and D168) are shown as sticks.
Next, we investigated reducing ring size at the P4 position, and found the cyclobutyl group to be optimal for maintaining antiviral potency against resistant variants. Compound 44 showed an excellent potency profile with EC50 values against WT HCV and the D168A variant similar to that of 5, but 4-fold better potency against the A156T variant. A similar trend was observed for the 1-methylcyclobutyl analogue 45, which was equipotent to 44 against the D168A variant with a 2-fold improvement in potency against the A156T variant relative to 5. Further reducing the ring size at P4 to cyclopropyl, as in 46, proved to be detrimental to biochemical potency. A slight loss in antiviral potency against the D168A variant was also observed. The addition of a methyl group at the C1 position of cyclopropyl to provide analogue 47 did not improve antiviral activity against the D168A variant and led to a further loss in potency against the A156T variant. Together, the SAR data for the cyclic P4 inhibitors indicates that in the 3-methylquinoxaline series increasing lipophilic bulk at the P4 position was mostly beneficial to antiviral potency. However, in the 3-(trifluoromethyl)-quinoxaline series a smaller cyclobutyl ring was preferred to maintain potency against resistance variants. This data provides further evidence for the interplay between the substituent at the 3-position of the P2 quinoxaline and the P4 capping group.
Fluorine substitution in cyclic P4 groups generally maintained or, in some cases, improved potency against WT HCV and resistant variants. Compared to the 1-methylcyclocpentyl compound (42) the 1-(trifluoromethyl)-cyclopentyl analogue 48 was significantly more potent in replicon assays against WT HCV and exhibited 3-fold better potency against the resistant variants D168A and A156T. The improved potency profile of 48 indicates that the trifluoromethyl group at the 3-position of P2 quinoxaline may be causing the P4 moiety to adopt an alternate conformation relative to the P4 group in 42, resulting in enhanced interactions in the S4 pocket while reducing contacts with residue 156. The increased interactions of 48 in the S4 pocket are also likely responsible for the improved potency against GT-3a.
Next, the trans-2-fluorocyclopentyl analogue 49 was designed to further evaluate the interplay between the P2 and P4 moieties. Interestingly, 49 exhibited an exceptional potency profile with EC50 values comparable to that of 2 and 3. Notably, 49 was significantly more potent against the A156T variant relative to the P4 cyclopentyl analogue 5. The 1-(trifluoromethyl)cyclobutyl analogue 50, generated by substitution of the C1-methyl in 45 with trifluoromethyl group, displayed a similar potency profile in replicon assays as the parent compound. Compound 50 was more active against GT-3a protease relative to 48 but exhibited lower antiviral activity. Hydrogen to fluorine substitution at C3-position of P4 cyclobutyl group in 44, leading to 51, did not affect potency in both biochemical and replicon assays. Thus, fluorine substitution in cyclic P4 groups was generally beneficial to potency, particularly against the A156T variant.
We also determined the cocrystal structure of 50 bound the D168A protease variant (Figure 5B). Overall, the binding mode of 50 is similar to that of 37. The P4 1-trifluoromethyl cyclobutyl group is orientated such that both the cyclobutyl moiety and fluorine atoms interact with residues in the S4 pocket. One of the 1-trifluoromethyl fluorine atoms is nested directly above the alanine side chain at position 168, another is orientated towards the guanidinium group of R123, and the third points at the 3-trifluoromethyl substituent on the P2 quinoxaline. Moreover, part of the cyclobutyl moiety forms hydrophobic packing interactions with V158. Essentially, the 1-trifluoromethyl cyclobutyl P4 group is optimally oriented to complement the contour of the S4 pocket. Surprisingly, in the cocrystal structure, the side chain of R155 samples a rotamer not observed in other cocrystal structures with 3-methylquinoxaline containing inhibitors, indicating a potential favorable interaction between the arginine guanidinium group and fluorine.
Overall, compounds in the 3-(trifluoromethyl)-quinoxaline series generally showed lower antiviral activity against the A156T and D168A variants compared to the 3-methylquinoxaline analogues. The larger trifluoromethyl group at the 3-position of P2 quinoxaline has increased interaction with residue 156, causing susceptibility to substitutions at this position. While potency loss against the D168A variant was addressed by enhancing hydrophobic packing in the S4 pocket and fluorine substitutions, maintaining potency against the A156T variant was more challenging. Also, in this series improving potency against GT-3a proved to be challenging. However, fluorine substitutions in the tert-butyl and cyclopentyl P4 groups decreased susceptibility to A156T RAS, providing PIs with improved potency against resistant variants and GT-3a.
CHEMISTRY
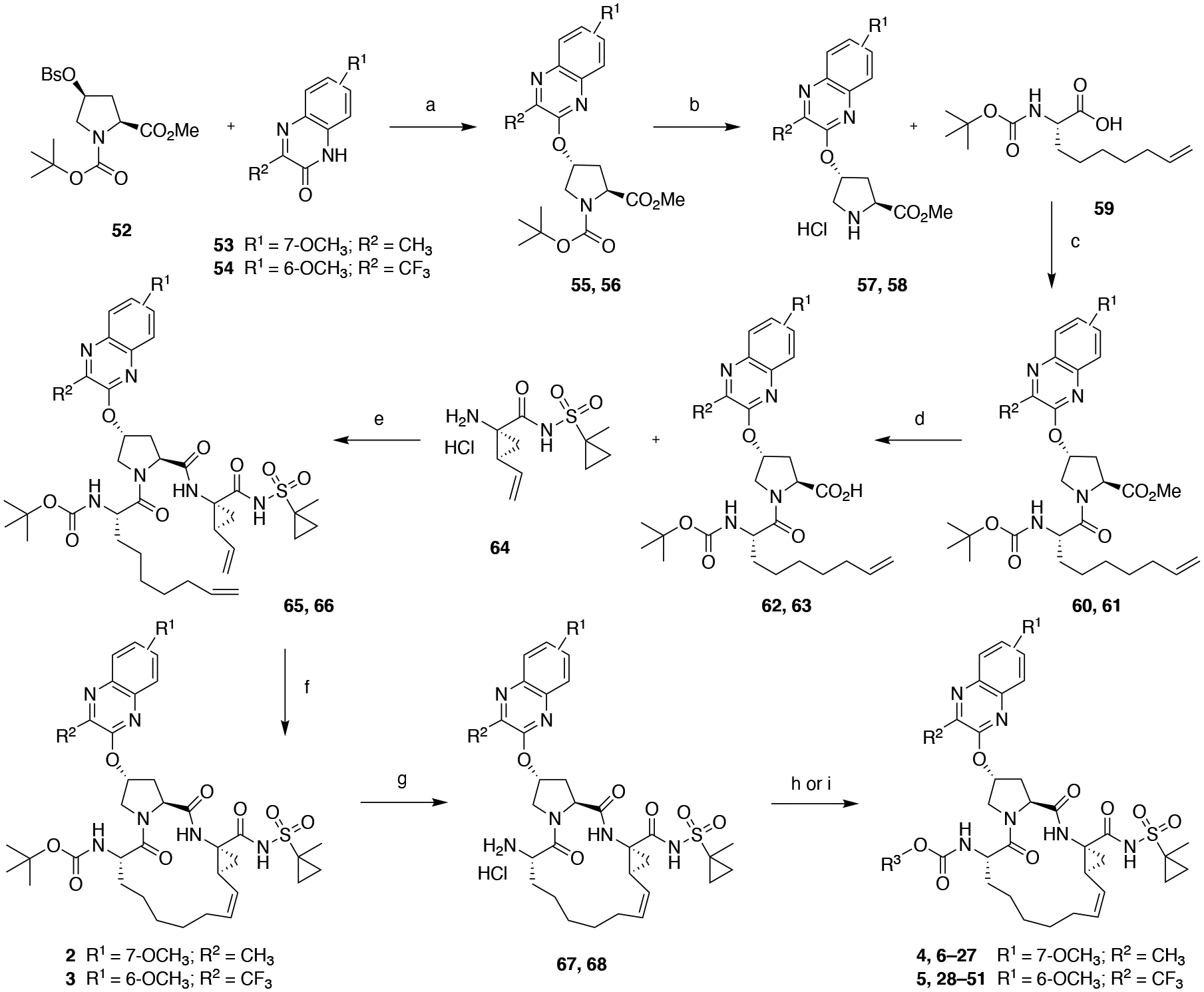
The P1–P3 macrocyclic NS3/4A PIs with 3-methyl- and 3-(trifluoromethyl)- quinoxalines as P2 moieties and diverse P4 capping groups were synthesized using the reaction sequence outlined in Scheme 1. The key Boc-protected P2 intermediates 55 and 56 were prepared from the corresponding 3-substituted quinoxalin-2-ones 53 and 54 by a cesium carbonate-mediated nucleophilic substitution reaction with the activated cis-hydroxyproline derivative 52 as described previously.35 The target P1–P3 macrocyclic inhibitors were assembled from the P2 intermediates using a sequence of deprotection and peptide coupling steps followed by the ring-closing metathesis (RCM) reaction. Briefly, removal of the Boc group in 55 and 56 using 4 N HCl in 1,4-dioxane provided the amine salts 57 and 58, which were coupled with the (S)-2-((tert-butoxycarbonyl)amino)non-8-enoic acid 59 using HATU and diisopropylethylamine (DIEA) in DMF to yield the P2–P3 ester intermediates 60 and 61. These esters were treated with LiOH.H2O in a 1:1 mixture of THF and H2O to afford of carboxylic acids 62 and 63 in quantitative yield. Reaction of the P2–P3 acid intermediates with the P1–P1’ acyl sulfonamide intermediate 64,42, 43 which was synthesized following reported methods, under HATU/DIEA coupling conditions provided the bis-olefin intermediates 65 and 66. The P1–P3 macrocyclization of the bis-olefin intermediates was accomplished using a highly efficient RCM catalyst Zhan Catalyst-1B in anhydrous 1,2-dichloroethane at 70 °C for 6 h and provided the P1–P3 macrocyclic inhibitors 2 and 3 in 70% yield. Finally, deprotection of Boc group and reaction of the resulting amine salts 67 and 68 with chloroformate or carbonates 69a–x in the presence of DIEA in acetonitrile provided the target compounds 6–51. The carbonate intermediates 69a–x were synthesized from the corresponding alcohols by reaction with p-nitrophenyl chloroformate in the presence of pyridine in dichloromethane (see Supporting Information for details).
Scheme 1.
Synthesis of P1–P3 Macrocyclic HCV NS3/4A Protease Inhibitors 6–51
Reagents and Conditions: (a) Cs2CO3, NMP, 55 °C, 6 h; (b) 4 N HCl in dioxane, CH2Cl2, RT, 3 h; (c) HATU, DIEA, DMF, RT, 4 h; (d) LiOH.H2O, THF, H2O, RT, 24 h; (e) HATU, DIEA, DMF, RT, 2 h; (f) Zhan catalyst-1B, 1,2-DCE, 70 °C, 6 h; (g) 4 N HCl in dioxane, RT, 3 h; (h) R3OCOCl, CH2Cl2, RT, 12 h; (i) R3O-(4-nitrophenyl) carbonate 69a–x, DIEA, CH3CN, RT, 36 h.
CONCLUSIONS
The structure-activity relationships of quinoxaline-based P1–P3 macrocyclic NS3/4A PIs were explored to identify compounds with improved resistance profiles and GT-3a potency. The P2 quinoxaline moiety and P4 capping group significantly impacted inhibitor potency and resistance profile, suggesting the interplay between these groups likely influenced the binding conformation of the P4 group. Compounds in the 3-methylquinoxaline series exhibited excellent potency profiles with EC50 values < 5 nM against the D168A variant, and nearly all compounds maintained low nM antiviral potency against the A156T variant, significantly better than the two clinically approved drugs GZR and GLE. In the 3-(trifluoromethyl)-quinoxaline series, the P4 modifications significantly impacted antiviral potency against both the A156T and D168A variants. In both quinoxaline series, the PIs that maintain hydrophobic packing in the S4 pocket exhibited improved antiviral activity against drug-resistant HCV variants and GT-3a. However, larger P4 groups that bind too deeply into the S4 pocket were susceptible to RASs at position 156. Notably, PIs containing the fluorinated P4 groups trifluoro tert-butyl (12 and 37) and 1-(trifluoromethyl)-cyclopentyl (24 and 48) displayed excellent antiviral potency across HCV variants including against GT-3a. Moreover, the improved potency profiles of PIs in both series indicate that these fluorinated P4 groups are unique in their ability to maintain favorable interactions in the S4 pocket. Thus, our strategy of using fluorinated motifs to target the variable S4 pocket has proven effective in improving potency against common resistant variants (D168A, A156T) and GT-3a. Overall, our SAR results demonstrate that optimal P4 modifications can improve potency across drug-resistant HCV variants and GT-3a, providing compounds with potential pan-genotypic potency profiles.
EXPERIMENTAL SECTION
General.
All reactions were performed in oven-dried round-bottom flasks fitted with rubber septa under argon atmosphere unless otherwise noted. All reagents and solvents, including anhydrous solvents, were purchased from commercial sources and used as received. Flash column chromatography was performed on an automated Teledyne ISCO CombiFlash Rf+ system equipped with a UV-vis detector using disposable Redisep Gold high performance silica gel columns or was performed manually using silica gel (230−400 mesh, EMD Millipore). Thin- layer chromatography (TLC) was performed using silica gel (60 F254) coated aluminum plates (EMD Millipore), and spots were visualized by exposure to ultraviolet light (UV), exposure to iodine adsorbed on silica gel, and/or staining with alcohol solutions of phosphomolybdic acid (PMA) and ninhydrin followed by brief heating. 1H NMR and 13C NMR spectra were acquired on a Bruker Avance III HD 500 MHz NMR instrument. Chemical shifts are reported in ppm (δ scale) with the residual solvent signal used as a reference and coupling constant (J) values are reported in hertz (Hz). Data are presented as follows: chemical shift, multiplicity (s = singlet, d = doublet, dd = doublet of doublet, dd = doublet of triplet, t = triplet, m = multiplet, br s = broad singlet), coupling constant in Hz, and integration. High-resolution mass spectra (HRMS) were recorded on a Thermo Scientific Orbitrap Velos Pro mass spectrometer coupled with a Thermo Scientific Accela 1250 UPLC and an autosampler using electrospray ionization (ESI) in the positive mode. The purity of final compounds was determined by analytical HPLC and was found to be ≥95% pure. HPLC was performed on an Agilent 1200 system equipped with a multiple wavelength detector and a manual injector under the following conditions: column, Phenomenex Hypersil-BDS-5u-C18 (5 μm, 4.6 mm × 250 mm, 130 Å); solvent A, H2O containing 0.1% trifluoroacetic acid (TFA); solvent B, CH3CN containing 0.1% TFA; gradient, 20% B to 100% B over 15 min followed by 100% B over 5 min; injection volume, 20 μL; flow rate, 1 mL/min. The wavelengths of detection were 254 nm and 280 nm. Retention times and purity data for each target compound are provided in the Experimental Section.
Isopropyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (6).
A solution of the amine salt 67 (0.10 g, 0.15 mmol) in anhydrous CH2Cl2 (10 mL) was treated with Et3N (0.083 mL, 0.60 mmol) followed by slow addition of isopropyl chloroformate solution (1M in toluene) (0.30 mL, 0.30 mmol). The reaction mixture was stirred at room temperature for 12 h, then concentrated under reduced pressure and dried under high vacuum. The residue was purified by flash column chromatography (RediSep Gold column, 12 g, gradient elution with 50–90% EtOAc/hexanes) to provide the target compound 6 (0.073 g, 66%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.12 (s, 1 H), 7.80 (d, J = 9.5 Hz, 1 H), 7.21−7.15 (m, 2 H), 6.90 (s, 1 H), 5.92 (br s, 1 H), 5.71 (q, J = 8.5 Hz, 1 H), 5.22 (d, J = 7.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.73−4.65 (m, 1 H), 4.62 (t, J = 7.5 Hz, 1 H), 4.43 (d, J = 11.5 Hz, 1 H), 4.31 (t, J = 7.5 Hz, 1 H), 4.06 (dd, J = 11.0, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.75−2.66 (m, 1 H), 2.64−2.47 (m, 5 H), 2.30 (q, J = 8.5 Hz, 1 H), 1.97−1.74 (m, 4 H), 1.62−1.22 (m, 12 H), 1.15−1.07 (m, 6 H), 0.86−0.76 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.21, 173.25, 166.99, 160.42, 155.60, 155.46, 144.80, 141.10, 136.31, 134.45, 129.12, 125.03, 118.94, 106.17, 74.83, 68.78, 59.60, 55.86, 53.21, 52.38, 44.85, 36.60, 34.68, 32.79, 29.87, 27.33, 27.16, 26.19, 22.38, 22.16, 21.07, 19.94, 18.34, 14.64, 12.72 ppm.
(S)-3-Methylbutan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (7).
A solution of the amine salt 67 (0.20 g, 0.29 mmol) in anhydrous CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (S)-3-methylbutan-2-yl (4-nitrophenyl) carbonate 69c (0.081 g, 0.32 mmol). The reaction mixture was stirred at room temperature for 36 h, then concentrated under reduced pressure and dried under high vacuum. The residue was purified by flash column chromatography (RediSep Gold column, 24 g, gradient elution with 50–90% EtOAc/hexanes) to provide the target compound 7 (0.20 g, 90%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.80 (d, J = 10.0 Hz, 1 H), 7.22−7.14 (m, 2 H), 6.93 (s, 1 H), 5.92 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.23 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.62 (t, J = 7.5 Hz, 1 H), 4.44 (d, J = 11.5 Hz, 1 H), 4.40 (t, J = 6.0 Hz, 1 H), 4.32 (t, J = 7.5 Hz, 1 H), 4.05 (dd, J = 11.0, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.74−2.64 (m, 1 H), 2.63−2.48 (m, 5 H), 2.30 (q, J = 9.0 Hz, 1 H), 1.94−1.75 (m, 4 H), 1.73−1.25 (m, 13 H), 1.05 (d, J = 6.0 Hz, 3 H), 0.84−0.75 (m, 7 H), 0.63−0.60 (m, 1 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.29, 173.24, 167.10, 160.45, 155.92, 155.47, 144.72, 141.15, 136.32, 134.26, 128.89, 125.02, 118.99, 106.18, 76.25, 74.89, 59.57, 55.86, 53.21, 52.39, 44.82, 36.60, 34.71, 32.86, 32.77, 29.83, 27.33, 27.17, 26.23, 22.35, 21.04, 19.86, 18.32, 18.06, 17.95, 16.87, 14.63, 12.69 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H53N6O9S, 769.3589; found 769.3553; Anal. HPLC: tR 12.16 min, purity 99.6%.
(R)-3-Methylbutan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (8).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (R)-3-methylbutan-2-yl (4-nitrophenyl) carbonate 69d (0.081 g, 0.32 mmol) to provide the target compound 8 (0.21 g, 94%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.13 (s, 1 H), 7.80 (d, J = 10.0 Hz, 1 H), 7.22−7.16 (m, 2 H), 6.89 (s, 1 H), 5.92 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.27 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.61 (t, J = 7.5 Hz, 1 H), 4.44 (d, J = 11.5 Hz, 1 H), 4.41 (t, J = 6.0 Hz, 1 H), 4.34 (t, J = 7.5 Hz, 1 H), 4.06 (dd, J = 11.0, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.73−2.65 (m, 1 H), 2.63−2.49 (m, 5 H), 2.31 (q, J = 9.0 Hz, 1 H), 1.95−1.76 (m, 4 H), 1.71−1.26 (m, 13 H), 1.03 (d, J = 6.0 Hz, 3 H), 0.88−0.76 (m, 8 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.23, 173.20, 167.03, 160.45, 155.88, 155.46, 144.69, 141.15, 136.34, 134.31, 128.94, 125.03, 118.99, 106.17, 76.34, 74.88, 59.56, 55.87, 53.21, 52.38, 44.84, 38.75, 36.60, 34.70, 32.83, 29.84, 27.31, 27.18, 26.21, 22.41, 21.06, 19.88, 18.33, 18.13, 17.96, 16.74, 14.64, 12.71 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H53N6O9S, 769.3589; found 769.3561; Anal. HPLC: tR 11.96 min, purity 99.6%.
(S)-1-Cyclopropylethyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (9).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (S)-1-cyclopropylethyl (4-nitrophenyl) carbonate 69e (0.08 g, 0.32 mmol) to provide the target compound 9 (0.20 g, 90%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.19 (s, 1 H), 7.79 (d, J = 10.0 Hz, 1 H), 7.22−7.16 (m, 2 H), 7.07 (s, 1 H), 5.90 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.51 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.62 (t, J = 7.5 Hz, 1 H), 4.46 (d, J = 11.0 Hz, 1 H), 4.31 (t, J = 8.0 Hz, 1 H), 4.04 (dd, J = 11.5, 4.0 Hz, 1 H), 3.98−3.95 (m, 1 H), 3.94 (s, 3 H), 2.72−2.63 (m, 1 H), 2.62−2.47 (m, 5 H), 2.32 (q, J = 8.5 Hz, 1 H), 1.93−1.68 (m, 5 H), 1.63−1.54 (m, 1 H), 1.52−1.26 (m, 10 H), 1.18 (d, J = 6.5 Hz, 3 H), 0.87−0.79 (m, 3 H), 0.49−0.33 (m, 2 H), 0.19−0.12 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.39, 173.13, 167.10, 160.42, 155.72, 155.46, 144.70, 141.12, 136.30, 134.33, 128.95, 125.05, 118.94, 106.15, 76.45, 74.91, 59.65, 55.85, 53.15, 52.41, 44.87, 38.75, 36.59, 34.71, 32.72, 29.77, 27.24, 26.26, 22.37, 20.95, 20.15, 19.88, 18.32, 16.58, 14.64, 12.68, 3.68, 2.64 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H51N6O9S, 767.3433; found 767.3408; Anal. HPLC: tR 11.37 min, purity 100%.
(R)-1-Cyclopropylethyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (10).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (R)-1-cyclopropylethyl (4-nitrophenyl) carbonate 69f (0.080 g, 0.32 mmol) to provide the target compound 10 (0.21 g, 94%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.80 (d, J = 9.5 Hz, 1 H), 7.22−7.16 (m, 2 H), 6.97 (s, 1 H), 5.91 (br s, 1 H), 5.71 (q, J = 8.0 Hz, 1 H), 5.37 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.63 (t, J = 7.5 Hz, 1 H), 4.42 (d, J = 11.5 Hz, 1 H), 4.33 (t, J = 7.5 Hz, 1 H), 4.06 (dd, J = 11.0, 3.5 Hz, 1 H), 4.03−3.97 (m, 1 H), 3.94 (s, 3 H), 2.73−2.65 (m, 1 H), 2.63−2.48 (m, 5 H), 2.31 (q, J = 8.5 Hz, 1 H), 1.97−1.77 (m, 4 H), 1.72−1.55 (m, 2 H), 1.52−1.26 (m, 10 H), 1.16 (d, J = 6.0 Hz, 3 H), 0.89−0.79 (m, 3 H), 0.50−0.38 (m, 2 H), 0.30−0.13 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.25, 173.05, 167.05, 160.45, 155.73, 155.46, 144.65, 141.14, 136.31, 134.33, 128.94, 125.04, 119.01, 106.17, 76.37, 74.91, 59.59, 55.87, 53.18, 52.40, 44.83, 38.75, 36.60, 34.73, 32.86, 29.81, 27.35, 27.19, 26.23, 22.38, 21.01, 20.03, 19.90, 18.34, 16.40, 14.64, 12.70, 3.55, 2.60 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H51N6O9S, 767.3433; found 767.3409; Anal. HPLC: tR 11.42 min, purity 100%.
(1-Methylcyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (11).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (1-methylcyclopropyl)methyl (4-nitrophenyl) carbonate 69g (0.08 g, 0.32 mmol) to provide the target compound 11 (0.18 g, 81%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.81 (d, J = 10.0 Hz, 1 H), 7.21−7.16 (m, 2 H), 6.98 (s, 1 H), 5.92 (br s, 1 H), 5.70 (q, J = 9.0 Hz, 1 H), 5.40 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.63 (t, J = 8.0 Hz, 1 H), 4.40 (d, J = 11.5 Hz, 1 H), 4.33 (t, J = 7.0 Hz, 1 H), 4.06 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 3.69 (s, 2 H), 2.72−2.65 (m, 1 H), 2.63−2.49 (m, 5 H), 2.31 (q, J = 8.5 Hz, 1 H), 1.95−1.57 (m, 6 H), 1.53−1.26 (m, 10 H), 1.03 (s, 3 H), 0.86−0.79 (m, 2 H), 0.40−0.33 (m, 2 H), 0.32−0.26 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.26, 173.06, 167.07, 160.56, 156.18, 155.48, 144.60, 141.20, 136.30, 134.26, 128.71, 125.02, 119.16, 106.18, 74.97, 73.12, 59.60, 55.88, 53.21, 52.46, 44.82, 38.75, 36.61, 34.71, 32.74, 29.81, 27.37, 27.18, 26.23, 22.35, 20.96, 19.72, 18.33, 15.42, 14.64, 12.70, 11.42, 11.33 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H51N6O9S, 767.3433; found 767.3405; Anal. HPLC: tR 11.55 min, purity 100%.
1,1,1-Trifluoro-2-methylpropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (12).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.25 g, 0.36 mmol) in CH3CN (12 mL) was treated with DIEA (0.45 mL, 2.58 mmol) and 4-nitrophenyl (1,1,1-trifluoro-2-methylpropan-2-yl) carbonate 69h (0.11 g, 0.37 mmol) to provide the target compound 12 (0.28 g, 96%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.79 (d, J = 10.0 Hz, 1 H), 7.19−7.16 (m, 2 H), 7.03 (s, 1 H), 5.87 (br s, 1 H), 5.72−5.64 (m, 2 H), 4.97 (t, J = 9.5 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.48 (d, J = 11.5 Hz, 1 H), 4.26−4.22 (m, 1 H), 4.02 (dd, J = 11.5, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.66−2.63 (m, 2 H), 2.59−2.50 (m, 4 H), 2.30 (q, J = 9.0 Hz, 1 H), 1.89−1.73 (m, 4 H), 1.60−1.22 (m, 18 H), 0.84−0.80 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.28, 172.28, 167.14, 160.43, 155.40, 153.35, 141.61, 141.10, 136.42, 134.39, 129.01, 125.05 (q, J = 281.0 Hz), 124.97, 118.94, 106.16, 79.78 (q, J = 29.3 Hz), 74.99, 59.70, 55.84, 53.27, 52.28, 44.81, 36.60, 34.82, 32.66, 29.62, 27.27, 27.21, 26.28, 22.20, 20.94, 19.88, 19.49, 19.43, 18.29, 14.64, 12.68 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H48F3N6O9S+, 809.3150; found 809.3128; Anal. HPLC: tR 12.08 min, purity 100%.
(S)-1,1,1-Trifluoropropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (13).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (S)-4-nitrophenyl (1,1,1-trifluoropropan-2-yl) carbonate 69i (0.09 g, 0.32 mmol) to provide the target compound 13 (0.20 g, 87%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.13 (s, 1 H), 7.80 (d, J = 9.5 Hz, 1 H), 7.21−7.16 (m, 2 H), 6.96 (s, 1 H), 5.91 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.61 (d, J = 7.5 Hz, 1 H), 5.02−4.94 (m, 2 H), 4.63 (t, J = 8.0 Hz, 1 H), 4.39−4.31 (m, 2 H), 4.06 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.71−2.60 (m, 2 H), 2.55−2.48 (m, 4 H), 2.29 (q, J = 8.5 Hz, 1 H), 1.97−1.82 (m, 2 H), 1.80−1.58 (m, 4 H), 1.52−1.34 (m, 10 H), 1.29 (d, J = 6.5 Hz, 3 H), 0.86−0.80 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.17, 172.40, 167.08, 160.54, 155.39, 153.83, 144.47, 141.15, 136.34, 134.25, 128.90, 124.93, 124.18 (q, J = 280.5 Hz), 119.14, 106.17, 74.93, 67.76 (q, J = 33.7 Hz), 59.66, 55.87, 53.28, 52.59, 44.76, 38.75, 36.62, 34.78, 32.53, 29.77, 27.43, 27.12, 26.25, 22.18, 20.93, 19.80, 18.30, 14.65, 13.80, 12.68 ppm; 19F NMR (470 MHz, CDCl3) δ −79.01 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C36H46F3N6O9S, 795.2994; found 795.2961; Anal. HPLC: tR 11.17 min, purity 100%.
(R)-1,1,1-Trifluoropropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (14).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (R)-4-nitrophenyl (1,1,1-trifluoropropan-2-yl) carbonate 69j (0.09 g, 0.32 mmol) to provide the target compound 14 (0.20 g, 87%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.15 (s, 1 H), 7.87 (d, J= 9.0 Hz, 1 H), 7.23−7.18 (m, 2 H), 7.08 (s, 1 H), 5.92 (br s, 1 H), 5.76−5.64 (m, 2 H), 5.02−4.93 (m, 2 H), 4.64 (t, J = 8.0 Hz, 1 H), 4.40 (d, J = 11.5 Hz, 1 H), 4.36−4.29 (m, 1 H), 4.05 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.72−2.60 (m, 2 H), 2.58−2.48 (m, 4 H), 2.29 (q, J= 9.0 Hz, 1 H), 1.94−1.73 (m, 5 H), 1.64−1.57 (m, 1 H), 1.52−1.25 (m, 13 H), 0.85−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.16, 172.34, 167.05, 160.68, 155.50, 153.92, 144.57, 141.25, 136.31, 128.52, 125.00, 124.08 (q, J = 278.7 Hz), 119.33, 106.18, 75.09, 67.92 (q, J = 33.7 Hz), 59.63, 55.90, 53.22, 52.59, 44.77, 38.76, 36.61, 34.73, 32.55, 29.67, 27.37, 27.16, 26.23, 22.28, 20.83, 19.41, 18.31, 14.64, 13.80, 12.72 ppm; 19F NMR (470 MHz, CDCl3) δ −78.88 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C36H46F3N6O9S, 795.2994; found 795.2958; Anal. HPLC: tR 11.31 min, purity 100%.
(1-(Trifluoromethyl)cyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (15).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.15 g, 0.22 mmol) in ACN (10 mL) was treated with DIEA (0.27 mL, 1.55 mmol) and 4-nitrophenyl ((1-(trifluoromethyl)cyclopropyl)methyl) carbonate 69k (0.073 g, 0.24 mmol) to provide the target compound 15 (0.14 g, 78%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.15 (s, 1 H), 7.81 (d, J = 9.5 Hz, 1 H), 7.21−7.16 (m, 2 H), 7.01 (s, 1 H), 5.90 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.52 (d, J = 7.5 Hz, 1 H), 4.99 (t, J = 9.0 Hz, 1 H), 4.63 (t, J = 7.5 Hz, 1 H), 4.40 (d, J = 11.0 Hz, 1 H), 4.33−4.27 (m, 1 H), 4.04 (dd, J = 11.5, 4.0 Hz, 1 H), 3.99 (d, J = 10.5 Hz, 2 H), 3.95 (s, 3 H), 2.71−2.59 (m, 2 H), 2.58−2.47 (m, 4 H), 2.30 (q, J = 8.5 Hz, 1 H), 1.96−1.55 (m, 6 H), 1.53−1.24 (m, 10 H), 1.03−0.93 (m, 2 H), 0.86−0.79 (m, 2 H), 0.74−0.62 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.24, 172.77, 167.06, 160.48, 155.47, 144.70, 141.14, 136.31, 134.07, 128.89, 126.48 (q, J = 272.5 Hz), 125.01, 119.04, 106.18, 74.94, 65.52, 59.65, 55.86, 53.23, 52.54, 44.82, 38.75, 36.61, 34.73, 32.63, 29.78, 27.35, 27.15, 26.22, 22.95 (q, J = 33.0 Hz), 22.30, 20.98, 19.83, 18.32, 14.65, 12.70, 8.20 ppm; 19F NMR (470 MHz, CDCl3) δ −69.68 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3120; Anal. HPLC: tR 11.48 min, purity 99.5%.
(2,2-Difluoro-1-methylcyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (16).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.15 g, 0.22 mmol) in CH3CN (10 mL) was treated with DIEA (0.27 mL, 1.55 mmol) and (2,2-difluoro-1-methylcyclopropyl)methyl (4-nitrophenyl) carbonate 69l (0.069 g, 0.24 mmol) to provide the target compound 16 (1:1 mixture of P4 stereoisomers) (0.13 g, 74%) as a white solid. 1H NMR (500 MHz, CDCl3) (mixture of P4 stereoisomers) δ 10.14 (s, 0.5 H), 10.13 (s, 0.5 H), 7.82 (d, J = 9.8 Hz, 1 H), 7.22−7.16 (m, 2 H), 6.98 (s, 0.5 H), 6.95 (s, 0.5 H), 5.91 (br s, 1 H), 5.75−5.66 (m, 1 H), 5.49 (d, J = 7.6 Hz, 0.5 H), 5.45 (d, J = 7.4 Hz, 0.5 H), 5.02−4.95 (m, 1 H), 4.63 (t, J = 7.8 Hz, 1 H), 4.44−4.36 (m, 1 H), 4.32 (td, J = 9.8, 3.4 Hz, 1 H), 4.13−4.03 (m, 2 H), 3.95 (s, 3 H), 3.76 (t, J = 11.8 Hz, 1 H), 2.82−2.59 (m, 2 H), 2.61−2.47 (m, 4 H), 2.30 (q, J = 8.5 Hz, 1 H), 1.97−1.58 (m, 6 H), 1.58−1.20 (m, 11 H), 1.18−1.13 (m, 3 H), 1.09−1.00 (m, 1 H), 0.86−0.78 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) (mixture of P4 stereoisomers) δ 177.24 (177.21), 172.83, 167.05, 160.58 (160.54), 155.63 (155.57), 155.48 (155.46), 144.68 (144.54), 141.19, 136.33 (136.30), 128.77, 125.02 (124.99), 119.20 (119.13), 106.19, 74.97, 66.21, 59.63, 55.88, 53.24, 52.51, 44.82 (44.81), 38.76, 36.61, 34.73, 32.72 (32.68), 29.81 (29.80), 27.40 (27.37), 27.16, 26.23, 25.77−25.37 (m), 22.31 (22.30), 21.23−20.77 (m), 19.72, 18.32, 14.87−14.72 (m), 14.65, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −138.81, −138.87 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H49F2N6O9S, 803.3244; found 803.3206; Anal. HPLC: tR 11.15 (11.25) min, purity 99%.
1-(Trifluoromethyl)cyclopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (24).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.15 g, 0.22 mmol) in CH3CN (10 mL) was treated with DIEA (0.27 mL, 1.55 mmol) and 4-nitrophenyl (1-(trifluoromethyl)cyclopentyl) carbonate 69u (0.076 g, 0.24 mmol) to provide the target compound 24 (0.15 g, 82%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.10 (s, 1 H), 7.80 (d, J = 9.5 Hz, 1 H), 7.22−7.16 (m, 2 H), 6.85 (s, 1 H), 5.88 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.63 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.60 (t, J = 8.0 Hz, 1 H), 4.47 (d, J = 11.5 Hz, 1 H), 4.33−4.26 (m, 1 H), 4.04 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.72−2.61 (m, 2 H), 2.59−2.49 (m, 4 H), 2.32 (q, J = 8.5 Hz, 1 H), 2.00−1.82 (m, 7 H), 1.81−1.73 (m, 2 H), 1.70−1.25 (m, 15 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.18, 172.77, 166.99, 160.46, 155.36, 153.25, 144.50, 141.09, 136.42, 134.45, 129.07, 125.82 (q, J = 281.2 Hz), 124.97, 119.02, 106.16, 89.34 (q, J = 28.7 Hz), 74.98, 59.72, 55.85, 53.39, 52.29, 44.85, 36.61, 34.84, 32.98, 32.90, 32.77, 29.69, 27.29, 27.20, 26.27, 25.69, 25.51, 22.25, 21.02, 19.82, 18.33, 14.66, 12.72 ppm; 19F NMR (470 MHz, CDCl3) δ −80.56 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C39H50F3N6O9S, 835.3307; found 835.3276; Anal. HPLC: tR 12.95 min, purity 100%.
(1R,2R)-2-Fluorocyclopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (25).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and (1R,2R)-2-fluorocyclopentyl (4-nitrophenyl) carbonate 69v (0.086 g, 0.32 mmol) to provide the target compound 25 (0.19 g, 83%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.11 (s, 1 H), 7.88 (d, J = 9.0 Hz, 1 H), 7.22−7.18 (m, 2 H), 6.88 (s, 1 H), 5.93 (br s, 1 H), 5.71 (q, J = 8.0 Hz, 1 H), 5.23 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.93−4.77 (m, 2 H), 4.63 (t, J = 7.5 Hz, 1 H), 4.40 (d, J = 11.5 Hz, 1 H), 4.31 (t, J = 7.5 Hz, 1 H), 4.06 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.75−2.67 (m, 1 H), 2.65−2.49 (m, 5 H), 2.29 (q, J = 8.5 Hz, 1 H), 2.02−1.27 (m, 22 H), 0.86−0.78 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.16, 172.92, 166.94, 160.68, 155.53, 155.01, 144.59, 141.25, 136.30, 128.53, 125.00, 119.33, 106.19, 97.56 (d, J = 176.2 Hz), 79.81 (d, J = 30.0 Hz), 75.02, 59.61, 55.91, 53.18, 52.50, 44.84, 38.32, 36.62, 34.68, 32.66, 30.64 (d, J = 21.2 Hz), 29.97, 29.86, 27.36, 27.13, 26.20, 22.30, 21.26, 21.05, 19.53, 18.33, 14.65, 12.73 ppm; 19F NMR (470 MHz, CDCl3) δ −181.06 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50FN6O9S, 785.3339; found 785.3305; Anal. HPLC: tR 11.14 min, purity 98.7%.
1-(Trifluoromethyl)cyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (26).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.15 g, 0.22 mmol) in CH3CN (10 mL) was treated with DIEA (0.27 mL, 1.55 mmol) and 4-nitrophenyl (1-(trifluoromethyl)cyclobutyl) carbonate 69w (0.073 g, 0.24 mmol) to provide the target compound 26 (0.15 g, 83%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.11 (s, 1 H), 7.79 (d, J = 10.0 Hz, 1 H), 7.22−7.16 (m, 2 H), 6.90 (s, 1 H), 5.89 (br s, 1 H), 5.70 (q, J = 8.0 Hz, 1 H), 5.55 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 7.5 Hz, 1 H), 4.44 (d, J = 11.5 Hz, 1 H), 4.33−4.26 (m, 1 H), 4.05 (dd, J = 11.5, 4.0 Hz, 1 H), 3.95 (s, 3 H), 2.72−2.63 (m, 2 H), 2.58−2.50 (m, 6 H), 2.43−2.33 (m, 2 H), 2.31 (q, J = 8.5 Hz, 1 H), 1.94−1.72 (m, 6 H), 1.64−1.56 (m, 1 H), 1.54−1.25 (m, 11 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.19, 172.64, 167.06, 160.43, 155.38, 153.18, 144.63, 141.08, 136.37, 134.40, 129.01, 125.00 (q, J = 282.0 Hz), 124.99, 118.98, 106.16, 78.83 (q, J = 31.2 Hz), 74.92, 59.69, 55.85, 53.31, 52.29, 44.81, 38.75, 36.61, 34.82, 32.69, 29.72, 28.70, 27.31, 27.18, 26.27, 22.23, 21.00, 19.83, 18.31, 14.65, 13.21, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −82.93 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3112; Anal. HPLC: tR 12.08 min, purity 100%.
3,3-Difluorocyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((7-methoxy-3-methylquinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (27).
The same procedure was used as described above for compound 7. A solution of amine salt 67 (0.20 g, 0.29 mmol) in CH3CN (10 mL) was treated with DIEA (0.35 mL, 2.0 mmol) and 3,3-difluorocyclobutyl (4-nitrophenyl) carbonate 69x (0.087 g, 0.32 mmol) to provide the target compound 27 (0.21 g, 92%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.81 (d, J = 8.5 Hz, 1 H), 7.21−7.16 (m, 2 H), 6.98 (s, 1 H), 5.90 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.37 (d, J = 7.0 Hz, 1 H), 4.98 (t, J = 9.5 Hz, 1 H), 4.71−4.64 (m, 1 H), 4.63 (t, J = 8.0 Hz, 1 H), 4.36 (d, J = 11.0 Hz, 1 H), 4.29 (t, J = 7.5 Hz, 1 H), 4.04 (dd, J = 11.5, 4.5 Hz, 1 H), 3.94 (s, 3 H), 2.95−2.79 (m, 2 H), 2.69−2.44 (m, 8 H), 2.28 (q, J = 8.5 Hz, 1 H), 1.95−1.55 (m, 6 H), 1.52−1.26 (m, 10 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.20, 172.59, 167.08, 160.47, 155.41, 154.79, 144.67, 141.12, 136.28, 134.35, 128.99, 124.98, 119.02, 118.15 (dd, J = 280.0, 268.7 Hz), 106.18, 74.85, 59.63, 59.61 (dd, J = 17.5, 5.0 Hz), 55.85, 53.22, 52.43, 44.77, 43.32 (t, J = 23.0 Hz), 43.07 (t, J = 23.2 Hz), 38.75, 36.62, 34.75, 32.60, 29.84, 27.42, 27.11, 26.22, 22.25, 21.01, 19.90, 18.31, 14.65, 12.68 ppm; 19F NMR (470 MHz, CDCl3) δ −85.03 (d, J = 200 Hz), −96.80 (d, J = 200 Hz) ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H47F2N6O9S, 789.3088; found 789.3054; Anal. HPLC: tR 10.74 min, purity 99.8%.
tert-Pentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (28).
A solution of the amine salt 68 (0.20 g, 0.27 mmol) in anhydrous CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl tert-pentyl carbonate 69a (0.075 g, 0.30 mmol). The reaction mixture was stirred at room temperature for 36 h, then concentrated under reduced pressure and dried under high vacuum. The residue was purified by flash column chromatography (RediSep Gold column, 24 g, gradient elution with 50–90% EtOAc/hexanes) to provide the target compound 28 (0.19 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.13 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.48 (dd, J = 9.5, 3.0 Hz, 1 H), 7.43 (d, J = 3.0 Hz, 1 H), 6.91 (s, 1 H), 5.91 (br s, 1 H), 5.69 (q, J = 9.0 Hz, 1 H), 5.13 (d, J = 8.0 Hz, 1 H), 4.99 (t, J = 9.5 Hz, 1 H), 4.63−4.57 (m, 2 H), 4.25−4.18 (m, 1 H), 4.02 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 2.71−2.52 (m, 3 H), 2.32 (q, J = 8.5 Hz, 1 H), 1.93−1.76 (m, 4 H), 1.58−1.23 (m, 14 H), 1.15 (s, 6 H), 0.86−0.79 (m, 2 H), 0.75 (t, J = 7.5 Hz, 3 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.16, 173.46, 167.02, 159.60, 155.08, 151.94, 138.43, 137.14, 136.38, 134.48 (q, J = 36.8 Hz), 128.15, 125.67, 125.11, 120.70 (d, J = 275.7 Hz), 107.60, 82.31, 75.75, 59.61, 56.02, 52.89, 52.06, 44.95, 36.58, 34.78, 33.58, 33.03, 29.73, 27.21, 27.15, 26.22, 25.65, 25.51, 22.43, 21.11, 18.34, 14.65, 12.71, 8.28 ppm; 19F NMR (470 MHz, CDCl3) δ −67.76 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50F3N6O9S, 823.3307; found 823.3279; Anal. HPLC: tR 13.54 min, purity 98.9%.
Pentan-3-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (29).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl pentan-3-yl carbonate 69b (0.083 g, 0.30 mmol) to provide the target compound 29 (0.19 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 3.0 Hz, 1 H), 7.00 (s, 1 H), 5.94 (br s, 1 H), 5.69 (q, J = 9.0 Hz, 1 H), 5.32 (d, J = 8.5 Hz, 1 H), 4.99 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.56 (d, J = 11.5 Hz, 1 H), 4.29−4.19 (m, 2 H), 4.01 (dd, J = 11.5, 3.5 Hz, 1H), 3.94 (s, 3 H), 2.71−2.51 (m, 3 H), 2.30 (q, J= 9.0 Hz, 1 H), 1.94−1.77 (m, 4 H), 1.58−1.23 (m, 16 H), 0.86−0.77 (m, 5 H), 0.71 (t, J = 7.5 Hz, 3 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.18, 173.25, 167.06, 159.58, 156.07, 151.99, 138.46, 137.17, 136.34, 134.68 (q, J = 35.8 Hz), 128.16, 125.62, 125.14, 120.78 (d, J = 275.4 Hz), 107.58, 77.81, 75.68, 59.57, 56.01, 52.86, 52.33, 44.93, 36.58, 34.67, 32.91, 29.74, 27.16, 26.57, 26.50, 26.17, 22.48, 21.07, 18.33, 14.63, 12.70, 9.52, 9.42 ppm; 19F NMR (470 MHz, CDCl3) δ −67.78 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50F3N6O9S, 823.3307; found 823.3281; Anal. HPLC: tR 13.56 min, purity 99.5%.
(S)-3-Methylbutan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (30).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (S)-3-methylbutan-2-yl (4-nitrophenyl) carbonate 69c (0.075 g, 0.30 mmol) to provide the target compound 30 (0.19 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 2.5 Hz, 1 H), 6.92 (s, 1 H), 5.94 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.19 (d, J = 8.0 Hz, 1 H), 5.01 (t, J = 9.5 Hz, 1 H), 4.62 (t, J = 8.0 Hz, 1 H), 4.57 (d, J = 12.0 Hz, 1 H), 4.26−4.18 (m, 2 H), 3.99 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 2.70−2.52 (m, 3 H), 2.31 (q, J = 8.5 Hz, 1 H), 1.94−1.77 (m, 4 H), 1.61−1.23 (m, 13 H), 1.01 (d, J = 6.5 Hz, 3 H), 0.85−0.79 (m, 2 H), 0.76 (d, J = 7.0 Hz, 3 H), 0.73 (d, J = 6.5 Hz, 3 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.23, 173.24, 167.08, 159.57, 155.83, 152.03, 138.45, 137.17, 136.33, 134.71 (q, J = 36.5 Hz), 128.16, 125.61, 125.15, 120.78 (d, J = 275.2 Hz), 107.57, 76.13, 75.67, 59.60, 55.99, 52.80, 52.32, 44.93, 36.58, 34.65, 32.84, 32.81, 29.78, 27.16, 26.19, 22.45, 21.14, 18.33, 18.11, 17.87, 16.59, 14.63, 12.69 ppm; 19F NMR (470 MHz, CDCl3) δ −67.81 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50F3N6O9S, 823.3307; found 823.3279; Anal. HPLC: tR 13.69 min, purity 98.8%.
(R)-3-Methylbutan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (31).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (R)-3-methylbutan-2-yl (4-nitrophenyl) carbonate 69d (0.075 g, 0.30 mmol) to provide the target compound 31 (0.18 g, 81%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 2.5 Hz, 1 H), 6.96 (s, 1 H), 5.94 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.27 (d, J = 8.0 Hz, 1 H), 5.01 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.56 (d, J = 11.5 Hz, 1 H), 4.28−4.19 (m, 2 H), 4.00 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 2.70−2.52 (m, 3 H), 2.30 (q, J = 9.0 Hz, 1 H), 1.93−1.76 (m, 4 H), 1.65−1.24 (m, 13 H), 0.93 (d, J = 6.0 Hz, 3 H), 0.86−0.78 (m, 8 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.21, 173.19, 167.14, 159.57, 155.80, 151.99, 138.47, 137.17, 136.36, 134.68 (q, J = 35.5 Hz), 128.14, 125.60, 125.13, 120.78 (d, J = 275.5 Hz), 107.59, 76.28, 75.68, 59.55, 56.00, 52.85, 52.29, 44.89, 36.57, 34.67, 32.91, 32.73, 29.72, 27.17, 26.19, 22.49, 21.05, 18.32, 17.99, 17.83, 16.68, 14.62, 12.69 ppm; 19F NMR (470 MHz, CDCl3) δ −67.78 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50F3N6O9S, 823.3307; found 823.3282; Anal. HPLC: tR 13.51 min, purity 99.1%.
(S)-1-Cyclopropylethyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (32).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (S)-1-cyclopropylethyl (4-nitrophenyl) carbonate 69e (0.080 g, 0.30 mmol) to provide the target compound 32 (0.20 g, 90%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.20 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 3.0 Hz, 1 H), 6.93 (s, 1 H), 5.93 (br s, 1 H), 5.70 (q, J = 9.0 Hz, 1 H), 5.60 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.64 (d, J = 11.5 Hz, 1 H), 4.59 (t, J = 8.0 Hz, 1 H), 4.23−4.17 (m, 1 H), 3.98 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H, overlapping), 3.76−3.69 (m, 1 H), 2.71−2.54 (m, 3 H), 2.33 (q, J = 8.5 Hz, 1 H), 1.91−1.77 (m, 4 H), 1.58−1.21 (m, 12 H), 1.16 (d, J = 6.5 Hz, 3 H), 0.86−0.79 (m, 2 H), 0.78−0.72 (m, 1 H), 0.53−0.25 (m, 2 H), 0.11−0.08 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.39, 173.25, 166.97, 159.58, 155.68, 152.05, 138.41, 137.16, 136.33, 134.77 (q, J = 35.4 Hz), 128.14, 125.64, 125.17, 120.77 (d, J = 275.3 Hz), 107.52, 76.46, 75.74, 59.76, 56.01, 52.74, 52.30, 45.08, 36.57, 34.67, 32.63, 29.70, 27.28, 26.95, 26.24, 22.48, 21.02, 19.86, 18.34, 16.62, 14.67, 12.68, 3.72, 2.59 ppm; 19F NMR (470 MHz, CDCl3) δ −67.97 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3124; Anal. HPLC: tR 12.79 min, purity 100%.
(R)-1-Cyclopropylethyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (33).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (R)-1-cyclopropylethyl (4-nitrophenyl) carbonate 69f (0.080 g, 0.30 mmol) to provide the target compound 33 (0.19 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.18 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.41 (d, J = 3.0 Hz, 1 H), 7.02 (s, 1 H), 5.94 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.46 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.62 (t, J = 7.5 Hz, 1 H), 4.55 (d, J = 11.5 Hz, 1 H), 4.23 (t, J = 8.0 Hz, 1 H), 4.00 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 3.85−3.79 (m, 1 H), 2.73−2.51 (m, 3 H), 2.31 (q, J = 9.0 Hz, 1 H), 1.91−1.77 (m, 4 H), 1.58−1.23 (m, 12 H), 1.06 (d, J= 6.5 Hz, 3 H), 0.88−0.77 (m, 3 H), 0.50−0.37 (m, 2 H), 0.30−0.16 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.25, 173.08, 167.06, 159.59, 155.67, 152.00, 138.48, 137.16, 136.31, 134.67 (q, J = 36.2 Hz), 128.14, 125.63, 125.15, 120.77 (d, J = 275.4 Hz), 107.58, 76.50, 75.71, 59.59, 56.02, 52.80, 52.31, 44.94, 36.57, 34.69, 32.84, 29.71, 27.20, 27.15, 26.20, 22.47, 21.00, 19.92, 18.33, 16.23, 14.64, 12.70, 3.31, 2.56 ppm; 19F NMR (470 MHz, CDCl3) δ −67.78 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3130; Anal. HPLC: tR 12.94 min, purity 99.8%.
Isobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (34).
A solution of the amine salt 68 (0.20 g, 0.27 mmol) in anhydrous CH2Cl2 (10 mL) was treated with Et3N (0.24 mL, 1.60 mmol) followed by slow addition of isobutyl chloroformate (0.077 g, 0.56 mmol). The reaction mixture was stirred at room temperature for 12 h, then concentrated under reduced pressure and dried under high vacuum. The residue was purified by flash column chromatography (RediSep Gold column, 12 g, gradient elution with 50–90% EtOAc/hexanes) to provide the target compound 34 (0.15 g, 69%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.12 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 3.0 Hz, 1 H), 6.78 (s, 1 H), 5.96 (br s, 1 H), 5.71 (q, J = 8.5 Hz, 1 H), 5.19 (d, J = 8.0 Hz, 1 H), 5.02 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.52 (d, J = 11.5 Hz, 1 H), 4.27−4.21 (m, 1 H), 4.00 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 3.58−3.51 (m, 2 H), 2.73−2.51 (m, 3 H), 2.31 (q, J = 9.0 Hz, 1 H), 1.97−1.70 (m, 5 H), 1.58−1.23 (m, 12 H), 0.86−0.72 (m, 8 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.20, 173.12, 167.04, 159.61, 156.05, 151.98, 138.49, 137.16, 136.32, 134.68 (q, J = 36.3 Hz), 128.15, 125.65, 125.12, 120.78 (d, J= 274.8 Hz), 107.59, 75.63, 71.29, 59.61, 56.00, 52.87, 52.34, 44.92, 36.58, 34.65, 32.75, 29.76, 27.94, 27.19, 27.16, 26.18, 22.47, 21.11, 19.04, 19.00, 18.33, 14.64, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.80 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H48F3N6O9S, 809.3150; found 809.3121; Anal. HPLC: tR 13.11 min, purity 99.3%.
Neopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (35).
The same procedure was used as described above for compound 34. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH2Cl2 (10 mL) was treated with Et3N (0.24 mL, 1.60 mmol) and neopentyl chloroformate (0.077 g, 0.56 mmol) to provide the target compound 35 (0.12 g, 54%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.42 (d, J = 2.5 Hz, 1 H), 6.87 (s, 1 H), 5.95 (br s, 1 H), 5.70 (q, J = 9.0 Hz, 1 H), 5.23 (d, J = 8.5 Hz, 1 H), 5.01 (t, J = 9.0 Hz, 1 H), 4.62 (t, J = 8.0 Hz, 1 H), 4.52 (d, J = 11.5 Hz, 1 H), 4.28−4.22 (m, 1 H), 4.00 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 3.47−3.42 (m, 2 H), 2.70−2.51 (m, 3 H), 2.32 (q, J = 8.5 Hz, 1 H), 1.95−1.77 (m, 4 H), 1.59−1.24 (m, 12 H), 0.86−0.79 (m, 11 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.20, 173.11, 167.06, 159.61, 156.16, 151.98, 138.49, 137.18, 136.34, 134.69 (q, J = 35.7 Hz), 128.15, 125.65, 125.11, 120.78 (d, J = 275.2 Hz), 107.58, 75.63, 74.49, 59.59, 55.99, 52.87, 52.36, 44.90, 36.58, 34.66, 32.79, 31.40, 29.79, 27.22, 27.14, 26.37, 26.19, 22.46, 21.13, 18.32, 14.64, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.77 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H50F3N6O9S, 823.3307; found 823.3283; Anal. HPLC: tR 13.75 min, purity 97.9%.
(1-Methylcyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (36).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (1-methylcyclopropyl)methyl (4-nitrophenyl) carbonate 69g (0.075 g, 0.30 mmol) to provide the target compound 36 (0.17 g, 77%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.12 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.49 (dd, J = 9.0, 3.0 Hz, 1 H), 7.43 (d, J = 2.5 Hz, 1 H), 6.80 (s, 1 H), 5.96 (br s, 1 H), 5.70 (q, J = 9.5 Hz, 1 H), 5.36 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.60 (t, J = 8.0 Hz, 1 H), 4.54 (d, J = 11.5 Hz, 1 H), 4.27−4.21 (m, 1 H), 4.00 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 3.56 (q, J = 11.0 Hz, 2 H), 2.73−2.52 (m, 3 H), 2.31 (q, J = 9.0 Hz, 1 H), 1.95−1.74 (m, 4 H), 1.60−1.25 (m, 12 H), 1.00 (s, 3 H), 0.86−0.78 (m, 2 H), 0.38−0.22 (m, 4 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.34, 173.02, 167.12, 159.60, 156.12, 151.97, 138.46, 137.16, 136.30, 134.67 (q, J = 36.5 Hz), 128.13, 125.65, 125.12, 120.76 (d, J = 275.3 Hz), 107.56, 75.68, 73.03, 59.66, 56.00, 52.85, 52.32, 44.92, 36.56, 34.66, 32.66, 29.66, 27.23, 27.14, 26.24, 22.43, 20.97, 20.85, 18.31, 15.36, 14.63, 12.67, 11.42, 11.28 ppm; 19F NMR (470 MHz, CDCl3) δ −67.82 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3124; Anal. HPLC: tR 12.94 min, purity 99.7%.
1,1,1-Trifluoro-2-methylpropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (37).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl (1,1,1-trifluoro-2-methylpropan-2-yl) carbonate 69h (0.087 g, 0.30 mmol) to provide the target compound 37 (0.19 g, 82%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.13 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.49 (dd, J = 9.5, 2.5 Hz, 1 H), 7.43 (d, J = 2.5 Hz, 1 H), 6.96 (s, 1 H), 5.91 (br s, 1 H), 5.72−5.62 (m, 2 H), 4.99 (t, J = 9.0 Hz, 1 H), 4.59 (t, J = 8.0 Hz, 1 H), 4.55 (d, J = 11.5 Hz, 1 H), 4.23−4.16 (m, 1 H), 4.01 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 2.66 (dd, J = 8.0, 2.5 Hz, 2 H), 2.63−2.53 (m, 1 H), 2.31 (q, J = 8.5 Hz, 1 H), 1.93−1.75 (m, 4 H), 1.58−1.22 (m, 18 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.13, 172.78, 167.05, 159.67, 153.28, 151.91, 138.42, 137.11, 136.45, 134.56 (q, J = 35.9 Hz), 128.13, 125.77, 125.05, 125.04 (q, J = 282.7 Hz), 120.75 (d, J = 275.2 Hz), 107.55, 79.79 (q, J = 29.4 Hz), 75.80, 59.65, 56.04, 52.96, 52.21, 44.88, 36.58, 34.79, 32.81, 29.56, 27.20, 27.18, 26.27, 22.26, 20.97, 19.44, 19.20, 18.32, 14.64, 12.71 ppm; 19F NMR (470 MHz, CDCl3) δ −67.81, −83.78 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H45F6N6O9S, 863.2867; found 863.2844; Anal. HPLC: tR 13.41 min, purity 99.8%.
(S)-1,1,1-Trifluoropropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (38).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (S)-4-nitrophenyl (1,1,1-trifluoropropan-2-yl) carbonate 69i (0.082 g, 0.30 mmol) to provide the target compound 38 (0.19 g, 83%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.12 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.50 (dd, J = 9.5, 3.0 Hz, 1 H), 7.42 (d, J = 3.0 Hz, 1 H), 6.81 (s, 1 H), 5.95 (br s, 1 H), 5.85 (d, J = 7.0 Hz, 1 H), 5.69 (q, J = 9.5 Hz, 1 H), 4.99 (t, J = 9.0 Hz, 1 H), 4.81−4.75 (m, 1 H), 4.60−4.53 (m, 2 H), 4.23−4.17 (m, 1 H), 3.97 (d, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H, overlaping), 2.66 (dd, J = 8.5, 2.5 Hz, 2 H), 2.63−2.54 (m, 1 H), 2.30 (q, J= 8.5 Hz, 1 H), 1.92−1.77 (m, 4 H), 1.57−1.31 (m, 12 H), 1.29 (d, J = 6.5 Hz, 3 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.13, 172.68, 166.91, 159.76, 153.86, 151.94, 138.55, 137.20, 136.38, 134.62 (q, J = 35.7 Hz), 128.11, 125.87, 125.06, 124.15 (q, J = 280.0 Hz), 120.85 (d, J = 275.2 Hz), 107.43, 75.77, 67.57 (q, J = 33.2 Hz), 59.79, 56.00, 52.89, 52.54, 44.97, 36.58, 34.73, 32.53, 29.62, 27.17, 27.08, 26.22, 22.32, 21.05, 18.32, 14.67, 13.44, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.81, −79.03 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C36H43F6N6O9S, 849.2711; found 849.2684; Anal. HPLC: tR 12.58 min, purity 100%.
(R)-1,1,1-Trifluoropropan-2-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (39).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (R)-4-nitrophenyl (1,1,1-trifluoropropan-2-yl) carbonate 69j (0.082 g, 0.30 mmol) to provide the target compound 39 (0.20 g, 87%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.12 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.49 (dd, J = 9.5, 3.0 Hz, 1 H), 7.44 (d, J = 3.0 Hz, 1 H), 6.94 (s, 1 H), 5.95 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.64 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.87−4.78 (m, 1 H), 4.62 (t, J = 8.0 Hz, 1 H), 4.47 (d, J = 11.5 Hz, 1 H), 4.29−4.23 (m, 1 H), 4.01 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 2.71−2.62 (m, 2 H), 2.59−2.50 (m, 1 H), 2.28 (q, J = 9.0 Hz, 1 H), 1.93−1.75 (m, 4 H), 1.58−1.24 (m, 12 H), 1.21 (d, J = 6.5 Hz, 3 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.16, 172.28, 167.33, 159.63, 153.88, 151.94, 138.49, 137.11, 136.31, 134.76 (q, J = 36.1 Hz), 128.11, 125.66, 125.13 (q, J = 280.4 Hz), 125.10, 120.68 (d, J = 275.2 Hz), 107.52, 75.77, 67.96 (q, J = 33.3 Hz), 59.57, 56.00, 52.93, 52.49, 44.70, 36.56, 34.63, 32.50, 29.43, 27.27, 27.13, 26.20, 22.33, 20.67, 18.27, 14.56, 13.77, 12.69 ppm; 19F NMR (470 MHz, CDCl3) δ −68.04, −79.04 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C36H43F6N6O9S, 849.2711; found 849.2683; Anal. HPLC: tR 12.61 min, purity 99.8%.
(1-(Trifluoromethyl)cyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (40).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl ((1-(trifluoromethyl)cyclopropyl)methyl) carbonate 69k (0.083 g, 0.30 mmol) to provide the target compound 40 (0.20 g, 85%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.17 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.49 (dd, J = 9.0, 3.0 Hz, 1 H), 7.42 (d, J = 2.5 Hz, 1 H), 6.93 (s, 1 H), 5.93 (br s, 1 H), 5.75−5.65 (m, 2 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.60 (t, J = 8.0 Hz, 1 H), 4.56 (d, J = 11.5 Hz, 1 H), 4.23−4.17 (m, 1 H), 3.98 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H, overlaping), 3.87 (q, J = 12.5 Hz, 2 H), 2.74−2.51 (m, 3 H), 2.31 (q, J= 8.5 Hz, 1 H), 1.94−1.77 (m, 4 H), 1.59−1.23 (m, 12 H), 1.02−0.90 (m, 2 H), 0.86−0.78 (m, 2 H), 0.72−0.53 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.26, 172.88, 167.00, 159.66, 155.54, 152.01, 138.43, 137.16, 136.34, 134.71 (q, J = 35.8 Hz), 128.16, 126.78 (q, J = 211.4 Hz), 125.71, 125.09, 120.79 (d, J = 275.2 Hz), 107.51, 75.77, 65.28, 59.72, 56.01, 52.84, 52.44, 44.97, 36.57, 34.64, 32.52, 29.65, 27.19, 27.10, 26.21, 22.91 (q, J= 33.0 Hz), 22.39, 21.04, 18.32, 14.65, 12.69, 8.21, 8.00 ppm; 19F NMR (470 MHz, CDCl3) δ −67.86, −69.67 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H45F6N6O9S, 875.2867; found 875.2839; Anal. HPLC: tR 12.82 min, purity 99.5%.
(2,2-Difluoro-1-methylcyclopropyl)methyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (41).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (2,2-difluoro-1-methylcyclopropyl)methyl (4-nitrophenyl) carbonate 69l (0.085 g, 0.30 mmol) to provide the target compound 41 (1:1 mixture of P4 stereoisomers) (0.20 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) (mixture of P4 stereoisomers) δ 10.16 (s, 0.5 H), 10.15 (s, 0.5 H), 7.83 (d, J = 10.0 Hz, 0.5 H), 7.82 (d, J = 9.9 Hz, 0.5 H), 7.48 (dt, J = 9.2, 2.5 Hz, 1 H), 7.42 (t, J = 2.9 Hz, 1 H), 6.96 (s, 0.5 H), 6.94 (s, 0.5 H), 5.95 (s, 0.5 H), 5.94 (s, 0.5 H), 5.75−5.65 (m, 1 H), 5.59 (d, J = 8.2 Hz, 0.5 H), 5.54 (d, J = 7.9 Hz, 0.5 H), 5.04−4.96 (m, 1 H), 4.61 (td, J = 8.0, 2.8 Hz, 1 H), 4.56−4.48 (m, 1 H), 4.24 (ddd, J = 19.9, 10.9, 2.9 Hz, 1 H), 4.03−3.90 (m, 2 H), 3.94 (s, 3 H), 3.66 (d, J = 11.7 Hz, 0.5 H), 3.63 (d, J = 11.4 Hz, 0.5 H), 2.74−2.50 (m, 3 H), 2.36−2.25 (m, 1 H), 1.94−1.72 (m, 4 H), 1.58−1.23 (m, 13 H), 1.14 (d, J = 14.4 Hz, 3 H), 1.02 (ddd, J = 28.0, 16.8, 7.7 Hz, 1 H), 0.87−0.78 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) (mixture of P4 stereoisomers) δ 177.22 (177.19), 172.89, 167.03, 159.69 (159.64), 155.63 (155.53), 151.99 (151.92), 138.51, 137.18 (137.16), 136.35 (136.31), 134.59 (q, J = 36.2 Hz), 128.14 (128.10), 125.76 (125.71), 125.12 (125.09), 120.80 (d, J = 272.4 Hz) (120.67 (d, J = 273.0 Hz)), 114.02 (d, J = 287.7 Hz), 107.59 (107.55), 75.71 (75.69), 66.13 (d, J = 21.3 Hz) (65.95 (d, J = 21.2 Hz)), 59.68 (59.66), 53.57, 52.97, 52.87, 52.42 (52.39), 44.93 (44.92), 36.58, 34.67 (34.64), 32.76, 32.65, 29.72, 27.18 (27.15), 26.22 (26.19), 25.63 (t, J = 8.1 Hz) (25.48 (t, J = 8.3 Hz)), 22.43, 21.08, 21.02 (20.94), 18.32, 14.71 (d, J = 4.2 Hz), 14.63, 14.57 (d, J = 4.2 Hz), 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.74 (−67.84), −138.67 (d, J = 116 Hz), −138.70 (d, J = 105 Hz) ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H46F5N6O9S, 857.2962; found 857.2922; Anal. HPLC: tR 12.35 (12.46) min, purity 99%.
1-Methylcyclopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (42).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 1-methylcyclopentyl (4-nitrophenyl) carbonate 69n (0.078 g, 0.30 mmol) to provide the target compound 42 (0.20 g, 89%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.5, 3.0 Hz, 1 H), 7.43 (d, J = 2.5 Hz, 1 H), 6.89 (s, 1 H), 5.91 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.14 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.63−4.55 (m, 2 H), 4.26−4.19 (m, 1 H), 4.02 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 2.70−2.52 (m, 3 H), 2.33 (q, J = 9.0 Hz, 1 H), 1.94−1.75 (m, 6 H), 1.58−1.23 (m, 18 H), 1.25 (s, 3 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.19, 173.33, 167.08, 159.59, 155.23, 151.93, 138.43, 137.15, 136.38, 134.54 (q, J = 35.9 Hz), 128.15, 125.66, 125.10, 120.76 (d, J = 275.2 Hz), 107.58, 89.55, 75.72, 59.61, 56.02, 52.87, 52.05, 44.91, 39.39, 39.04, 36.58, 34.77, 33.00, 29.71, 27.22, 27.17, 26.24, 24.53, 23.85, 23.80, 22.40, 21.09, 18.33, 14.63, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.76 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C39H50F3N6O9S, 835.3307; found 835.3282; Anal. HPLC: tR 13.99 min, purity 99.9%.
(1R,3R,5S)-Bicyclo[3.1.0]hexan-3-yl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (43).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (1R,3R,5S)-bicyclo[3.1.0]hexan-3-yl (4-nitrophenyl) carbonate 69p (0.078 g, 0.30 mmol) to provide the target compound 43 (0.19 g, 85%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.14 (s, 1 H), 7.82 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.43 (d, J = 3.0 Hz, 1 H), 6.89 (s, 1 H), 5.94 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.10 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.79 (t, J = 7.0 Hz, 1 H), 4.61 (t, J = 7.5 Hz, 1 H), 4.51 (d, J = 11.5 Hz, 1 H), 4.24−4.17 (m, 1 H), 4.00 (dd, J = 12.0, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.74−2.49 (m, 3 H), 2.30 (q, J = 9.0 Hz, 1 H), 2.08−2.01 (m, 1 H), 2.00−1.74 (m, 6 H), 1.70−1.16 (m, 15 H), 0.86−0.79 (m, 2 H), 0.40 (dd, J = 13.0, 8.0 Hz, 1 H), 0.23 (q, J = 4.0 Hz, 1 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.18, 173.17, 167.02, 159.61, 155.36, 151.99, 138.49, 137.15, 136.34, 134.69 (q, J = 36.3 Hz), 128.15, 125.63, 125.11, 120.77 (d, J = 273.8 Hz), 107.57, 75.63, 59.62, 56.03, 52.83, 52.28, 44.93, 36.58, 35.81, 35.60, 34.64, 32.75, 29.77, 27.17, 26.18, 22.45, 21.15, 18.33, 16.84, 16.75, 14.64, 12.71, 10.53 ppm; 19F NMR (470 MHz, CDCl3) δ −67.76 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C39H48F3N6O9S, 833.3150; found 833.3158; Anal. HPLC: tR 13.39 min, purity 97.1%.
Cyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (44).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and cyclobutyl (4-nitrophenyl) carbonate 69q (0.066 g, 0.30 mmol) to provide the target compound 44 (0.19 g, 87%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.44 (d, J = 2.5 Hz, 1 H), 6.93 (s, 1 H), 5.95 (br s, 1 H), 5.70 (q, J = 9.0 Hz, 1 H), 5.44 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 10.0 Hz, 1 H), 4.63−4.55 (m, 2 H), 4.51 (d, J = 11.5 Hz, 1 H), 4.25−4.18 (m, 1 H), 3.99 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 2.70−2.51 (m, 3 H), 2.32 (q, J = 8.5 Hz, 1 H), 2.20−2.13 (m, 1 H), 2.07−1.98 (m, 1 H), 1.94−1.75 (m, 6 H), 1.73−1.23 (m, 14 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.25, 173.00, 167.01, 159.64, 155.14, 151.96, 138.53, 137.17, 136.31, 134.69 (q, J = 35.0 Hz), 128.16, 125.67, 125.10, 120.78 (d, J = 272.5 Hz), 107.55, 75.61, 69.17, 59.65, 56.03, 52.81, 52.19, 44.96, 36.57, 34.65, 32.70, 30.68, 30.19, 29.74, 29.42, 27.20, 27.15, 26.22, 22.41, 21.05, 18.33, 14.65, 13.24, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.88 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H46F3N6O9S, 807.2994; found 807.2966; Anal. HPLC: tR 12.45 min, purity 99.3%.
1-Methylcyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (45).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 1-methylcyclobutyl (4-nitrophenyl) carbonate 69r (0.074 g, 0.30 mmol) to provide the target compound 45 (0.19 g, 86%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.16 (s, 1 H), 7.82 (d, J = 9.5 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.43 (d, J = 3.0 Hz, 1 H), 6.99 (s, 1 H), 5.92 (br s, 1 H), 5.70 (q, J = 9.0 Hz, 1 H), 5.34 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.60 (t, J = 7.5 Hz, 1 H), 4.52 (d, J = 11.5 Hz, 1 H), 4.28−4.22 (m, 1 H), 4.03 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 2.70−2.52 (m, 3 H), 2.33 (q, J = 9.0 Hz, 1 H), 2.14−2.04 (m, 2 H), 1.93−1.75 (m, 6 H), 1.58−1.31 (m, 14 H), 1.28 (s, 3 H, overlapping), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.18, 173.15, 167.07, 159.62, 154.51, 151.89, 138.44, 137.13, 136.35, 134.52 (q, J = 35.7 Hz), 128.15, 125.69, 125.10, 120.75 (d, J = 276.3 Hz), 107.58, 79.54, 75.66, 59.59, 56.03, 52.89, 52.01, 44.91, 36.58, 35.33, 35.24, 34.75, 32.99, 29.71, 27.23, 27.19, 26.26, 23.39, 22.38, 21.02, 18.33, 14.64, 13.66, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.75 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H48F3N6O9S, 821.3150; found 821.3126; Anal. HPLC: tR 13.04 min, purity 99.8%.
Cyclopropyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (46).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and cyclopropyl (4-nitrophenyl) carbonate 69s (0.066 g, 0.30 mmol) to provide the target compound 46 (0.18 g, 84%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.17 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.5, 3.0 Hz, 1 H), 7.42 (d, J = 2.5 Hz, 1 H), 7.01 (s, 1 H), 5.97 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.43 (d, J = 8.0 Hz, 1 H), 4.98 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.51 (d, J = 11.5 Hz, 1 H), 4.30−4.23 (m, 1 H), 4.02 (dd, J = 11.5, 3.5 Hz, 1 H), 3.94 (s, 3 H), 3.78−3.72 (m, 1 H), 2.70−2.51 (m, 3 H), 2.30 (q, J= 9.0 Hz, 1 H), 1.92−1.77 (m, 4 H), 1.59−1.23 (m, 12 H), 0.87−0.79 (m, 2 H), 0.60−0.47 (m, 4 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.22, 172.87, 167.07, 159.63, 156.21, 151.96, 138.52, 137.15, 136.29, 134.64 (q, J = 35.5 Hz), 128.15, 125.66, 125.08, 120.79 (d, J = 275.0 Hz), 107.58, 75.57, 59.61, 56.02, 52.88, 52.31, 49.41, 44.89, 36.58, 34.67, 32.65, 29.72, 27.21, 27.17, 26.21, 22.38, 20.99, 18.32, 14.63, 12.69, 5.03, 4.98 ppm; 19F NMR (470 MHz, CDCl3) δ −67.75 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C36H44F3N6O9S, 793.2837; found 793.2809; Anal. HPLC: tR 11.45 min, purity 100%.
1-Methylcyclopropyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (47).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 1-methylcyclopropyl (4-nitrophenyl) carbonate 69t (0.078 g, 0.30 mmol) to provide the target compound 47 (0.18 g, 83%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.10 (s, 1 H), 7.83 (d, J = 9.5 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.43 (d, J = 3.0 Hz, 1 H), 6.80 (s, 1 H), 5.95 (br s, 1 H), 5.70 (q, J = 8.5 Hz, 1 H), 5.19 (d, J = 7.5 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.62−4.55 (m, 2 H), 4.27−4.20 (m, 1 H), 4.03 (dd, J = 11.5, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.75−2.52 (m, 3 H), 2.30 (q, J = 9.0 Hz, 1 H), 1.96−1.75 (m, 4 H), 1.58−1.23 (m, 12 H), 1.26 (s, 3 H), 0.86−0.79 (m, 2 H), 0.73−0.63 (m, 2 H), 0.51−0.40 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.10, 173.20, 166.97, 159.61, 155.43, 151.95, 138.48, 137.15, 136.34, 134.57 (q, J = 34.2 Hz), 128.14, 125.66, 125.09, 120.20 (d, J = 274.6 Hz), 107.62, 75.61, 59.60, 56.69, 56.03, 52.87, 52.25, 44.94, 36.59, 34.77, 32.87, 29.73, 27.17, 27.14, 26.18, 22.39, 21.28, 21.00, 18.34, 14.64, 12.91, 12.83, 12.73 ppm; 19F NMR (470 MHz, CDCl3) δ −67.65 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H46F3N6O9S, 807.2994; found 807.2968; Anal. HPLC: tR 12.01 min, purity 99.8%.
1-(Trifluoromethyl)cyclopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (48).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl (1-(trifluoromethyl)cyclopentyl) carbonate 69u (0.094 g, 0.30 mmol) to provide the target compound 48 (0.18 g, 75%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.11 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.49 (dd, J = 9.0, 2.5 Hz, 1 H), 7.43 (d, J = 2.5 Hz, 1 H), 6.86 (s, 1 H), 5.91 (br s, 1 H), 5.69 (q, J = 9.0 Hz, 1 H), 5.64 (d, J = 7.5 Hz, 1 H), 4.99 (t, J = 9.0 Hz, 1 H), 4.60 (t, J = 8.0 Hz, 1 H), 4.53 (d, J = 11.5 Hz, 1 H), 4.27−4.21 (m, 1 H), 4.03 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 2.66 (dd, J = 8.0, 2.5 Hz, 2 H), 2.63−2.52 (m, 1 H), 2.33 (q, J = 8.5 Hz, 1 H), 2.05−1.76 (m, 8 H), 1.73−1.23 (m, 16 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.12, 172.75, 167.02, 159.69, 153.20, 151.85, 138.49, 137.10, 136.45, 134.56 (q, J = 35.9 Hz), 128.15, 125.79, 125.04 (q, J = 282.7 Hz), 125.02, 120.75 (d, J = 275.2 Hz), 107.59, 89.24 (q, J = 29.4 Hz), 75.77, 59.73, 56.04, 53.11, 52.19, 44.88, 36.58, 34.82, 32.88, 32.84, 32.76, 29.54, 27.22, 27.19, 26.30, 25.56, 25.47, 22.26, 21.01, 18.32, 14.64, 12.71 ppm; 19F NMR (470 MHz, CDCl3) δ −67.89, −80.60 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C39H47F6N6O9S, 889.3024; found 889.3030; Anal. HPLC: tR 14.21 min, purity 99.3%.
(1R,2R)-2-Fluorocyclopentyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (49).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and (1R,2R)-2-fluorocyclopentyl (4-nitrophenyl) carbonate 69v (0.080 g, 0.30 mmol) to provide the target compound 49 (0.19 g, 84%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.10 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.0, 2.5 Hz, 1 H), 7.45 (d, J = 2.5 Hz, 1 H), 6.77 (s, 1 H), 5.96 (br s, 1 H), 5.72 (q, J = 8.5 Hz, 1 H), 5.15 (d, J = 8.0 Hz, 1 H), 5.02 (t, J = 9.0 Hz, 1 H), 4.86−4.71 (m, 2 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.46 (d, J = 11.5 Hz, 1 H), 4.29−4.21 (m, 1 H), 4.01 (dd, J = 12.0, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.72−2.60 (m, 2 H), 2.59−2.40 (m, 1 H), 2.29 (q, J= 8.5 Hz, 1 H), 1.96−1.63 (m, 8 H), 1.58−1.23 (m, 14 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.10, 172.82, 167.00, 159.63, 154.83, 151.92, 138.57, 137.13, 136.31, 134.64 (q, J = 35.9 Hz), 128.09, 125.72, 125.08, 120.79 (d, J = 275.1 Hz), 107.66, 97.31 (d, J = 176.3 Hz), 79.58 (d, J = 30.6 Hz), 75.60, 59.60, 56.03, 52.90, 52.36, 44.88, 36.59, 34.66, 32.78, 30.59 (d, J = 21.7 Hz), 29.91, 29.78, 27.26, 27.12, 26.17, 22.39, 21.24, 21.20, 18.32, 14.64, 12.70 ppm; 19F NMR (470 MHz, CDCl3) δ −67.78, −181.12 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H47F4N6O9S, 839.3056; found 839.3025; Anal. HPLC: tR 12.59 min, purity 98.6%.
1-(Trifluoromethyl)cyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (50).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 4-nitrophenyl (1-(trifluoromethyl)cyclobutyl) carbonate 69w (0.090 g, 0.30 mmol) to provide the target compound 50 (0.21 g, 89%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.13 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.48 (dd, J = 9.5, 3.0 Hz, 1 H), 7.42 (d, J = 3.0 Hz, 1 H), 6.96 (s, 1 H), 5.93 (br s, 1 H), 5.69 (q, J = 8.5 Hz, 1 H), 5.58 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.5 Hz, 1 H), 4.60 (t, J = 8.0 Hz, 1 H), 4.50 (d, J = 11.5 Hz, 1 H), 4.28−4.22 (m, 1 H), 4.03 (dd, J = 11.5, 4.0 Hz, 1 H), 3.94 (s, 3 H), 2.65 (dd, J = 8.0, 3.0 Hz, 2 H), 2.60−2.45 (m, 3 H), 2.37−2.29 (m, 3 H), 1.92−1.70 (m, 6 H), 1.58−1.23 (m, 12 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.11, 172.59, 167.14, 159.67, 153.07, 151.85, 138.46, 137.10, 136.40, 134.45 (q, J = 36.1 Hz), 128.12, 125.76, 125.04, 124.95 (q, J = 282.1 Hz), 120.76 (d, J = 275.2 Hz), 107.55, 78.75 (q, J = 31.9 Hz), 75.71, 59.67, 56.03, 53.01, 52.21, 44.82, 36.58, 34.79, 32.77, 29.60, 28.64, 28.51, 27.24, 27.19, 26.28, 22.24, 20.95, 18.31, 14.62, 13.13, 12.69 ppm; 19F NMR (470 MHz, CDCl3) δ −67.85, −83.02 ppm; HRMS (ESI) m/z: [M + H]+ calcd for C38H45F6N6O9S, 875.2867; found 875.2840; Anal. HPLC: tR 13.33 min, purity 100%.
3,3-Difluorocyclobutyl ((2R,6S,13aS,14aR,16aS,Z)-2-((6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl)oxy)-14a-(((1-methylcyclopropyl)sulfonyl)carbamoyl)-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl)carbamate (51).
The same procedure was used as described above for compound 28. A solution of amine salt 68 (0.20 g, 0.27 mmol) in CH3CN (10 mL) was treated with DIEA (0.33 mL, 1.90 mmol) and 3,3-difluorocyclobutyl (4-nitrophenyl) carbonate 69x (0.081 g, 0.30 mmol) to provide the target compound 51 (0.19 g, 83%) as a white solid. 1H NMR (500 MHz, CDCl3) δ 10.11 (s, 1 H), 7.83 (d, J = 9.0 Hz, 1 H), 7.49 (dd, J = 9.0, 2.5 Hz, 1 H), 7.44 (d, J = 2.5 Hz, 1 H), 6.79 (s, 1 H), 5.94 (br s, 1 H), 5.71 (q, J = 9.5 Hz, 1 H), 5.23 (d, J = 8.0 Hz, 1 H), 5.00 (t, J = 9.0 Hz, 1 H), 4.61 (t, J = 8.0 Hz, 1 H), 4.55−4.46 (m, 2 H), 4.20−4.13 (m, 1 H), 3.97 (dd, J = 11.5, 3.5 Hz, 1 H), 3.95 (s, 3 H), 2.95−2.84 (m, 1 H), 2.71−2.61 (m, 3 H), 2.60−2.46 (m, 2 H), 2.45−2.34 (m, 1 H), 2.28 (q, J = 8.5 Hz, 1 H), 1.93 (dd, J = 8.0, 6.0 Hz, 1 H), 1.90−1.76 (m, 3 H), 1.58−1.24 (m, 12 H), 0.86−0.79 (m, 2 H) ppm; 13C NMR (125 MHz, CDCl3) δ 177.19, 172.61, 167.18, 159.67, 154.75, 151.96, 138.50, 137.17, 136.30, 134.67 (q, J = 34.3 Hz), 128.12, 125.75, 125.12, 120.77 (d, J = 270.1 Hz), 118.17 (dd, J = 283.1, 268.3 Hz), 107.48, 75.70, 59.64, 59.43 (dd, J = 19.0, 9.0 Hz), 56.00, 52.84, 52.33, 44.85, 43.35 (t, J = 21.4 Hz), 42.85 (t, J = 22.9 Hz), 36.57, 34.66, 32.61, 29.73, 27.18, 27.12, 26.19, 22.38, 21.08, 18.29, 14.63, 12.64 ppm; 19F NMR (470 MHz, CDCl3) δ −67.77, −85.01 (d, J = 200 Hz), −97.19 (d, J = 200 Hz) ppm; HRMS (ESI) m/z: [M + H]+ calcd for C37H44F5N6O9S, 843.2805; found 843.2773; Anal. HPLC: tR 11.98 min, purity 99.4%.
Enzyme Inhibition Assays.
The enzyme inhibition assays were performed as previously described.27, 35 Briefly, for each assay, 2 nM of NS3/4A protease (WT GT-1a, D168A variant, and GT-3a) was pre-incubated at room temperature for 1 h with increasing concentration of inhibitors in assay buffer (50 mM Tris, 5% glycerol, 10 mM DTT, 0.6 mM LDAO, and 4% DMSO, pH 7.5). Inhibition assays were performed in non-binding surface 96-well black half-area plates (Corning) in a reaction volume of 60 μL. The proteolytic reaction was initiated by the injection of 5 μL of HCV NS3/4A protease substrate, Ac-DE-D(Edans)-EE-Abu-c-[COO]-AS-K(Dabcyl)-NH2 (AnaSpec), to a final concentration of 200 nM and kinetically monitored using a Perkin Elmer EnVision plate reader (excitation at 485 nm, emission at 530 nm). Three independent data sets were collected for each inhibitor with each protease construct. Each inhibitor titration included at least 12 inhibitor concentration points, which were globally fit to the Morrison equation to obtain the Ki value. GZR was used as a control in all assays.
Antiviral Activity Assays in HCV GT-1 Replicons.
Antiviral activity of PIs was assessed using HCV Con1 (GT-1b) reporter replicon containing the H77 (GT-1a) NS3 protease region. Mutations (A156T and D168A) were introduced into the WT H77 NS3 protease region of the Con1 (GT-1b) replicon using the “megaprimer” method of site-directed mutagenesis.44 Huh7 cells, previously cured for optimal HCV replicon replication, were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1 mM non-essential amino acids (DMEM – 10% FBS). Huh7 cells were transfected with replicon RNA transcripts using electroporation and maintained in the absence or presence of serially diluted protease inhibitors. Replicon replication was then assessed by measuring luciferase activity (relative light units) 96 h post electroporation. The drug concentrations required to inhibit replicon replication by 50% (EC50) were calculated directly from the drug inhibition curves.
Antiviral Activity Assays in HCV GT-3a Replicon.
Huh7.5 cells harboring the HCV GT-3a replicon, S52/SG-Feo SHI,45 were seeded into a 96-well plate at the density of 8,000 cells per well. The next day, the cells were exposed to different concentrations of protease inhibitors and incubated for another 72 h. The cells were then lysed with 1X Passive Lysis Buffer (Promega) according to the manufacturer’s recommendations and the luciferase activity was measured with the Luciferase Assay System (Promega) using Varioskan Lux (ThermoFisher Scientific). The drug concentrations required to inhibit replicon replication by 50% (EC50) were calculated directly from the drug inhibition curves.
Crystallization and Structure Determination.
Protein expression and purification were carried out as previously described with slight modifications.16 Briefly, the Histrap purified NS3/4A D168A protease variant was thawed, concentrated to 3 mg/mL, and loaded on a HiLoad Superdex75 16/60 column equilibrated with gel filtration buffer (25 mM MES, 500 mM NaCl, 10% glycerol, and 2 mM DTT, pH 6.5). The protease containing fractions were pooled and concentrated to 25 mg/mL with an Amicon Ultra-15 10 kDa filter unit (Millipore). The concentrated samples were incubated for 1 h with 3:1 or 6:1 molar excess of inhibitor. Diffraction-quality crystals were obtained within 2 days by mixing equal volumes of concentrated protein solution with precipitant solution (20–30% PEG-3350, 0.1 M sodium MES buffer, 1–7% ammonium sulfate, pH 6.5) at RT or 15 °C in 24-well VDX hanging drop trays. Crystals were harvested and data was collected at 100 K in cryogenic conditions containing the precipitant solution supplemented with 15% glycerol or ethylene glycol. Although the inhibitors were in excess, the crystals obtained were solved as apo HCV NS3/4A D168A protease variant structures. To obtain electron density for the inhibitors, the apo crystals were soaked overnight in cryogenic conditions supplemented with 10–20 mM concentration of inhibitor in DMF.
X-ray diffraction data were collected in-house using our Rigaku X-ray system with a Saturn 944 detector. All datasets were processed using HKL-3000.46 Structures were solved by molecular replacement using PHASER.47 Model building and refinement were performed using Coot and PHENIX, respectively.48, 49 The final structures were evaluated with MolProbity50 prior to deposition in the PDB. To limit the possibility of model bias throughout the refinement process, 5% of the data were reserved for the free R-value calculation.51 Structure analysis, superposition and figure generation were done using PyMOL.52 X-ray data collection and crystallographic refinement statistics are presented in Table S1.
Structural Analysis.
Superpositions were performed in PyMol using the Cα atoms of active site residues 137–139 and 154–160 of the NS3 protease. The cocrystal structure of inhibitor 2 in complex with the D168A protease variant (PDB 6UE3) was used as a reference structure for alignments.
Molecular Modeling.
Molecular modeling was carried out using MacroModel (Schrödinger, LLC, New York, NY).53 Briefly, inhibitors were modeled into the active site of WT-1a and D168A proteases using the WT-2 (PDB 5VOJ) and D168A-2 (PDB 6UE3) complex structures.35, 36 Structures were prepared using the Protein Preparation tool in Maestro. Two-dimensional chemical structures were modified with the appropriate changes using the Build tool in Maestro. Once modeled, molecular energy minimizations were performed for each inhibitor−protease complex using the PRCG method with 2500 maximum iterations and 0.05 gradient convergence threshold. PDB files of modeled complexes were generated in Maestro for structural analysis in PyMOL.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01 AI085051) and the National Institute of General Medical Sciences (R01 GM135919) of the NIH. JZ and ANM were also supported by the National Institute of General Medical Sciences of the NIH (F31 GM131635 and F31 GM119345). We thank members of the Schiffer and Miller laboratories for helpful discussions.
Funding Sources
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01 AI085051) and the National Institute of General Medical Sciences (R01 GM135919, F31 GM131635 and F31 GM119345) of the NIH.
ABBREVIATIONS
- Boc
tert-butoxycarbonyl
- DAAs
direct-acting antivirals
- DIEA
N,N-diisopropylethylamine
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- GZR
grazoprevir
- GLE
glecaprevir
- GT
genotype
- HATU
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- FRET
fluorescence resonance energy transfer
- PDB
protein data bank
- PIs
protease inhibitors
- SAR
structure-activity relationship
- RAS
resistance-associated substitutions
- VOX
voxilaprevir
- WT
wild-type
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
Synthesis details and characterization data of intermediates 55–68, inhibitors 2–5, and carbonate intermediates 69a–x; X-ray data collection and refinement statistics (PDF)
Molecular formula strings (CSV)
Accessions Codes
The PDB accession codes for X-ray cocrystal structures of HCV NS3/4A D168A protease with compounds 3, 37, 43, and 50 are 7L7P, 7L7L, 7L7O, and 7L7N, respectively. Authors will release the atomic coordinates and experimental data upon article publication.
Dedication
This manuscript is dedicated to the memory of Dr. Wei Huang whose dedication, energy and cheerfulness we all miss.
The authors declare no competing financial interest. AN, CJP and WH are employees of Monogram Biosciences.
REFERENCES
- 1.World Health Organization (WHO). Hepatitis C, Fact Sheet (Updated July 2020) https://www.who.int/news-room/fact-sheets/detail/hepatitis-c. (February 25, 2021).
- 2.Messina JP; Humphreys I; Flaxman A; Brown A; Cooke GS; Pybus OG; Barnes E Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015, 61, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gower E; Estes C; Blach S; Razavi-Shearer K; Razavi H Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol 2014, 61, S45–S57. [DOI] [PubMed] [Google Scholar]
- 4.EASL recommendations on treatment of hepatitis C: Final update of the series. J. Hepatol 2020, 73, 1170–1218. [DOI] [PubMed] [Google Scholar]
- 5.Hepatitis C Guidance: AASLD-IDSA. Recommendations for testing, managing, and treating hepatitis C https://www.hcvguidelines.org. (February 25, 2021).
- 6.Pawlotsky JM Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 2016, 151, 70–86. [DOI] [PubMed] [Google Scholar]
- 7.Dietz J; Susser S; Vermehren J; Peiffer KH; Grammatikos G; Berger A; Ferenci P; Buti M; Mullhaupt B; Hunyady B; Hinrichsen H; Mauss S; Petersen J; Buggisch P; Felten G; Huppe D; Knecht G; Lutz T; Schott E; Berg C; Spengler U; von Hahn T; Berg T; Zeuzem S; Sarrazin C; European HCVRSG Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology 2018, 154, 976–988. [DOI] [PubMed] [Google Scholar]
- 8.Hepatitis C Guidance: AASLD-IDSA. HCV resistance primer. Recommendations for testing, managing, and treating hepatitis C https://www.hcvguidelines.org/evaluate/resistance. (February 25, 2021).
- 9.de Salazar A; Dietz J; di Maio VC; Vermehren J; Paolucci S; Mullhaupt B; Coppola N; Cabezas J; Stauber RE; Puoti M; Arenas Ruiz Tapiador JI; Graf C; Aragri M; Jimenez M; Callegaro A; Pascasio Acevedo JM; Macias Rodriguez MA; Rosales Zabal JM; Micheli V; Garcia Del Toro M; Tellez F; Garcia F; Sarrazin C; Ceccherini-Silberstein F; Gehep-004 cohort, t. E. H. C. V. R. S. G.; the, H. C. V. V. I. R. N. Prevalence of resistance-associated substitutions and retreatment of patients failing a glecaprevir/pibrentasvir regimen. J. Antimicrob. Chemother 2020, 75, 3349–3358. [DOI] [PubMed] [Google Scholar]
- 10.Sarrazin C Treatment failure with DAA therapy: importance of resistance. J. Hepatol 2021, 10.1016/j.jhep.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Meanwell NA 2015 Philip S. Portoghese Medicinal Chemistry Lectureship. Curing hepatitis C virus infection with direct-acting antiviral agents: the arc of a medicinal chemistry triumph. J. Med. Chem 2016, 59, 7311–7351. [DOI] [PubMed] [Google Scholar]
- 12.McCauley JA; Rudd MT Hepatitis C virus NS3/4A protease inhibitors. Curr. Opin. Pharmacol 2016, 30, 84–92. [DOI] [PubMed] [Google Scholar]
- 13.Sarrazin C The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J. Hepatol 2016, 64, 486–504. [DOI] [PubMed] [Google Scholar]
- 14.Lawitz E; Yang JC; Stamm LM; Taylor JG; Cheng G; Brainard DM; Miller MD; Mo H; Dvory-Sobol H Characterization of HCV resistance from a 3-day monotherapy study of voxilaprevir, a novel pangenotypic NS3/4A protease inhibitor. Antivir. Ther 2018, 23, 325–334. [DOI] [PubMed] [Google Scholar]
- 15.Ng TI; Tripathi R; Reisch T; Lu L; Middleton T; Hopkins TA; Pithawalla R; Irvin M; Dekhtyar T; Krishnan P; Schnell G; Beyer J; McDaniel KF; Ma J; Wang G; Jiang LJ; Or YS; Kempf D; Pilot-Matias T; Collins C In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS3/4A protease inhibitor glecaprevir. Antimicrob. Agents Chemother 2018, 62, e01620–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romano KP; Ali A; Aydin C; Soumana D; Ozen A; Deveau LM; Silver C; Cao H; Newton A; Petropoulos CJ; Huang W; Schiffer CA The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog 2012, 8, e1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soumana DI; Kurt Yilmaz N; Ali A; Prachanronarong KL; Schiffer CA Molecular and dynamic mechanism underlying drug resistance in genotype 3 hepatitis C NS3/4A protease. J. Am. Chem. Soc 2016, 138, 11850–11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timm J; Kosovrasti K; Henes M; Leidner F; Hou S; Ali A; Kurt Yilmaz N; Schiffer CA Molecular and structural mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir. ACS Chem. Biol 2020, 15, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summa V; Ludmerer SW; McCauley JA; Fandozzi C; Burlein C; Claudio G; Coleman PJ; DiMuzio JM; Ferrara M; Di Filippo M; Gates AT; Graham DJ; Harper S; Hazuda DJ; McHale C; Monteagudo E; Pucci V; Rowley M; Rudd MT; Soriano A; Stahlhut MW; Vacca JP; Olsen DB; Liverton NJ; Carroll SS MK-5172, a selective inhibitor of hepatitis C virus NS3/4A protease with broad activity across genotypes and resistant variants. Antimicrob. Agents Chemother 2012, 56, 4161–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen SB; Fahnoe U; Pham LV; Serre SBN; Tang Q; Ghanem L; Pedersen MS; Ramirez S; Humes D; Pihl AF; Filskov J; Solund CS; Dietz J; Fourati S; Pawlotsky JM; Sarrazin C; Weis N; Schonning K; Krarup H; Bukh J; Gottwein JM Evolutionary pathways to persistence of highly fit and resistant hepatitis C virus protease inhibitor escape variants. Hepatology 2019, 70, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthew AN; Leidner F; Newton A; Petropoulos CJ; Huang W; Ali A; KurtYilmaz N; Schiffer CA Molecular mechanism of resistance in a clinically significant double-mutant variant of HCV NS3/4A protease. Structure 2018, 26, 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan P; Pilot-Matias T; Schnell G; Tripathi R; Ng TI; Reisch T; Beyer J; Dekhtyar T; Irvin M; Xie W; Larsen L; Mensa FJ; Collins C Pooled Resistance Analysis in Patients with Hepatitis C Virus Genotype 1 to 6 Infection Treated with Glecaprevir-Pibrentasvir in Phase 2 and 3 Clinical Trials. Antimicrob Agents Chemother 2018, 62, e01249–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han B; Parhy B; Lu J; Hsieh D; Camus G; Martin R; Svarovskaia ES; Mo H; Dvory-Sobol H In vitro susceptibility of hepatitis C virus genotype 1 through 6 clinical isolates to the pangenotypic NS3/4A inhibitor voxilaprevir. J. Clin. Microbiol 2019, 57, e01844–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romano KP; Ali A; Royer WE; Schiffer CA Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthew AN; Leidner F; Lockbaum GJ; Henes M; Zephyr J; Hou S; Rao DN; Timm J; Rusere LN; Ragland DA; Paulsen JL; Prachanronarong K; Soumana DI; Nalivaika EA; Kurt Yilmaz N; Ali A; Schiffer CA Drug design strategies to avoid resistance in direct-acting antivirals and beyond. Chem. Rev 2021, 10.1021/acs.chemrev.0c00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozen A; Prachanronarong K; Matthew AN; Soumana DI; Schiffer CA Resistance outside the substrate envelope: hepatitis C NS3/4A protease inhibitors. Crit. Rev. Biochem. Mol. Biol 2019, 54, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali A; Aydin C; Gildemeister R; Romano KP; Cao H; Özen A; Soumana D; Newton A; Petropoulos CJ; Huang W; Schiffer CA Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chem. Biol 2013, 8, 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soumana DI; Kurt Yilmaz N; Prachanronarong KL; Aydin C; Ali A; Schiffer CA Structural and thermodynamic effects of macrocyclization in HCV NS3/4A inhibitor MK-5172. ACS Chem. Biol 2016, 11, 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun LQ; Mull E; Zheng B; D’Andrea S; Zhao Q; Wang AX; Sin N; Venables BL; Sit SY; Chen Y; Chen J; Cocuzza A; Bilder DM; Mathur A; Rampulla R; Chen BC; Palani T; Ganesan S; Arunachalam PN; Falk P; Levine S; Chen C; Friborg J; Yu F; Hernandez D; Sheaffer AK; Knipe JO; Han YH; Schartman R; Donoso M; Mosure K; Sinz MW; Zvyaga T; Rajamani R; Kish K; Tredup J; Klei HE; Gao Q; Ng A; Mueller L; Grasela DM; Adams S; Loy J; Levesque PC; Sun H; Shi H; Sun L; Warner W; Li D; Zhu J; Wang YK; Fang H; Cockett MI; Meanwell NA; McPhee F; Scola PM Discovery of a potent acyclic, tripeptidic, acyl sulfonamide inhibitor of hepatitis C virus NS3 protease as a back-up to asunaprevir with the potential for once-daily dosing. J. Med. Chem 2016, 59, 8042–8060. [DOI] [PubMed] [Google Scholar]
- 30.Llinas-Brunet M; Bailey MD; Bolger G; Brochu C; Faucher A-M; Ferland JM; Garneau M; Ghiro E; Gorys V; Grand-Maitre C; Halmos T; Lapeyre-Paquette N; Liard F; Poirier M; Rheaume M; Tsantrizos YS; Lamarre D Structure-activity study on a novel series of macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of BILN 2061. J. Med. Chem 2004, 47, 1605–1608. [DOI] [PubMed] [Google Scholar]
- 31.Llinàs-Brunet M; Bailey MD; Goudreau N; Bhardwaj PK; Bordeleau J; Bös M; Bousquet Y; Cordingley MG; Duan J; Forgione P; Garneau M; Ghiro E; Gorys V; Goulet S; Halmos T; Kawai SH; Naud J; Poupart M-A; White PW Discovery of a potent and selective noncovalent linear inhibitor of the hepatitis C virus NS3 protease (BI 201335). J. Med. Chem 2010, 53, 6466–6476. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y; Andrews SW; Condroski KR; Buckman B; Serebryany V; Wenglowsky S; Kennedy AL; Madduru MR; Wang B; Lyon M; Doherty GA; Woodard BT; Lemieux C; Do MG; Zhang H; Ballard J; Vigers G; Brandhuber BJ; Stengel P; Josey JA; Beigelman L; Blatt L; Seiwert SD Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A protease. J. Med. Chem 2013, 57, 1753–1769. [DOI] [PubMed] [Google Scholar]
- 33.Kazmierski WM; Hamatake R; Duan M; Wright LL; Smith GK; Jarvest RL; Ji JJ; Cooper JP; Tallant MD; Crosby RM; Creech K; Wang A; Li X; Zhang S; Zhang YK; Liu Y; Ding CZ; Zhou Y; Plattner JJ; Baker SJ; Bu W; Liu L Discovery of novel urea-based hepatitis C protease inhibitors with high potency against protease-inhibitor-resistant mutants. J. Med. Chem 2012, 55, 3021–3026. [DOI] [PubMed] [Google Scholar]
- 34.Moreau B; O’Meara JA; Bordeleau J; Garneau M; Godbout C; Gorys V; Leblanc M; Villemure E; White PW; Llinàs-Brunet M Discovery of hepatitis C virus NS3–4A protease inhibitors with improved barrier to resistance and favorable liver distribution. J. Med. Chem 2014, 57, 1770–1776. [DOI] [PubMed] [Google Scholar]
- 35.Matthew AN; Zephyr J; Hill CJ; Jahangir M; Newton A; Petropoulos CJ; Huang W; Kurt-Yilmaz N; Schiffer CA; Ali A Hepatitis C virus NS3/4A protease inhibitors incorporating flexible P2 quinoxalines target drug resistant viral variants. J. Med. Chem 2017, 60, 5699–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthew AN; Zephyr J; Nageswara Rao D; Henes M; Kamran W; Kosovrasti K; Hedger AK; Lockbaum GJ; Timm J; Ali A; Kurt Yilmaz N; Schiffer CA Avoiding drug resistance by substrate envelope-guided design: toward potent and robust HCV NS3/4A protease inhibitors. mBio 2020, 11, e00172–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meanwell NA Fluorine and fluorinated motifs in the design and application of bioisosteres for drug design. J. Med. Chem 2018, 61, 5822–5880. [DOI] [PubMed] [Google Scholar]
- 38.Gillis EP; Eastman KJ; Hill MD; Donnelly DJ; Meanwell NA Applications of fluorine in medicinal chemistry. J. Med. Chem 2015, 58, 8315–8359. [DOI] [PubMed] [Google Scholar]
- 39.Hunter L The C-F bond as a conformational tool in organic and biological chemistry. Beilstein J. Org. Chem 2010, 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llinas-Brunet M; Bailey MD; Ghiro E; Gorys V; Halmos T; Poirier M; Rancourt J; Goudreau N A systematic approach to the optimization of substrate-based inhibitors of the hepatitis C virus NS3 protease: discovery of potent and specific tripeptide inhibitors. J. Med. Chem 2004, 47, 6584–6594. [DOI] [PubMed] [Google Scholar]
- 41.Sun LQ; Mull E; D’Andrea S; Zheng B; Hiebert S; Gillis E; Bowsher M; Kandhasamy S; Baratam VR; Puttaswamy S; Pulicharla N; Vishwakrishnan S; Reddy S; Trivedi R; Sinha S; Sivaprasad S; Rao A; Desai S; Ghosh K; Anumula R; Kumar A; Rajamani R; Wang YK; Fang H; Mathur A; Rampulla R; Zvyaga TA; Mosure K; Jenkins S; Falk P; Tagore DM; Chen C; Rendunchintala K; Loy J; Meanwell NA; McPhee F; Scola PM Discovery of BMS-986144, a third-generation, pan-genotype NS3/4A protease inhibitor for the treatment of hepatitis C virus infection. J. Med. Chem 2020, 63, 14740–14760. [DOI] [PubMed] [Google Scholar]
- 42.Wang XA; Sun L-Q; Sit S-Y; Sin N; Scola PM; Hewawasam P; Good A,C; Chen Y; Campbell A Hepatitis C virus inhibitors US Patent 6995174, 2006.
- 43.Rudd MT; Butcher JW; Nguyen KT; McIntyre CJ; Romano JJ; Gilbert KF; Bush KJ; Liverton NJ; Holloway MK; Harper S; Ferrara M; DiFilippo M; Summa V; Swestock J; Fritzen J; Carroll SS; Burlein C; DiMuzio JM; Gates A; Graham DJ; Huang Q; McClain S; McHale C; Stahlhut MW; Black S; Chase R; Soriano A; Fandozzi CM; Taylor A; Trainor N; Olsen DB; Coleman PJ; Ludmerer SW; McCauley JA P2-Quinazolinones and bis-macrocycles as new templates for next-generation hepatitis C virus NS3/4A protease inhibitors: discovery of MK-2748 and MK-6325. ChemMedChem 2015, 10, 727–735. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar G; Sommer SS The “megaprimer” method of site-directed mutagenesis. Biotechniques 1990, 8, 404–407. [PubMed] [Google Scholar]
- 45.Saeed M; Scheel TKH; Gottwein JM; Marukian S; Dustin LB; Bukh J; Rice CM Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob. Agents Chemother 2012, 56, 5365–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z; Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 47.McCoy AJ Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 2007, 63, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P; Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung L-W; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis IW; Leaver-Fay A; Chen VB; Block JN; Kapral GJ; Wang X; Murray LW; Arendall WB; Snoeyink J; Richardson JS; Richardson DC MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl. Acids Res 2007, 35, W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunger AT Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- 52.PyMOL. The PyMOL Molecular Graphics System; Version 2.3; Schrödinger, LLC. New York, NY, United States, 2020. [Google Scholar]
- 53.Schrödinger. Schrödinger Release 2020–1; Schrödinger, LLC, New York, NY, United States, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


