Abstract
Resistant starch (RS) enrichments were made using chemostats inoculated with human feces from two individuals at two dilution rates (D = 0.03 h−1 and D = 0.30 h−1) to select for slow- and fast-growing amylolytic communities. The fermentations were studied by analysis of short-chain fatty acids, amylase and α-glucosidase activities, and viable counts of the predominant culturable populations and the use of 16S rRNA-targeted oligonucleotide probes. Considerable butyrate was produced at D = 0.30 h−1, which corresponded with reduced branched-chain fatty acid formation. At both dilution rates, high levels of extracellular amylase activity were produced, while α-glucosidase was predominantly cell associated. Bacteroides and bifidobacteria predominated at the low dilution rate, whereas saccharolytic clostridia became more important at D = 0.30 h−1. Microscopic examination showed that within 48 h of inoculation, one particular bacterial morphotype predominated in RS enrichments at D = 0.30 h−1. This organism attached apically to RS granules and formed rosette-like structures which, with glycocalyx formation, agglomerated to form biofilm networks in the planktonic phase. Attempts to isolate this bacterium in pure culture were repeatedly unsuccessful, although a single colony was eventually obtained. On the basis of its 16S rDNA sequence, this RS-degrading, butyrate-producing organism was identified as being a previously unidentified group I Clostridium sp. A 16S rRNA-targeted probe was designed using this sequence and used to assess the abundance of the population in the enrichments. At 240 h, its contributions to total rRNA in the chemostats were 5 and 23% at D = 0.03 and 0.30 h−1, respectively. This study indicates that bacterial populations with significant metabolic potential can be overlooked using culture-based methodologies. This may provide a paradigm for explaining the discrepancy between the low numbers of butyrate-producing bacteria that are isolated from fecal samples and the actual production of butyrate.
The principal sources of carbon and energy for bacteria growing in the human large intestine are resistant starches (RS), plant cell wall polysaccharides, and host mucopolysaccharides, together with various proteins, peptides, and other low-molecular-weight carbohydrates that escape digestion and absorption in the small bowel (6, 24). The major types of short-chain fatty acid (SCFA) produced during the breakdown of carbohydrates by intestinal bacteria are acetate, propionate, and butyrate, while branched-chain fatty acids such as isobutyrate, 2-methylbutyrate, and isovalerate are mainly formed by the fermentation of branched-chain amino acids (27).
Since SCFA production is directly related to the supply of nutrients to the colon, shifts occur in the metabolism of microbial populations in response to changes in the diet (15). In vitro studies show that compared to the fermentation of non-starch polysaccharides (dietary fiber) by gut microorganisms, butyrate formation mainly occurs during starch breakdown (10, 11, 25). The physiology of butyrate metabolism in the large intestine has been extensively studied (16, 24, 37). Apart from being an important respiratory fuel for the colonocyte (5), this SCFA regulates gene expression and cell growth, and reversibly alters the in vitro properties of human colon cancer cell lines by prolonging doubling times (39). Low concentrations also reduce DNA synthesis and suppress proliferation in a variety of cell types (16).
While butyrate production in the large bowel accounts for approximately 20% of all SCFAs (5), estimates of intestinal bacterial populations that form this substance as a major end product of metabolism indicate that they account for about 1% of total culturable gut anaerobes (28). Butyrate-producing bacteria in the gut are therefore very metabolically active or are particularly difficult to enumerate and culture using traditional anaerobic techniques.
The human gastrointestinal tract harbors large numbers of bacteria, particularly in the distal colon (12, 28, 32, 33). This microbiota has been shown to harbor several hundred different bacterial strains and species, and it has been estimated using culture-based methodologies, that approximately 40% of the microbiota has been described (52). However, advances in molecular phylogeny based on sequence comparisons of 16S rRNA have been used for culture-independent characterizations of complex microbial ecosystems (35, 43–47), and this approach has been applied to human fecal populations (9, 34, 41; R. Sharp and G. T. Macfarlane, submitted for publication).
The initial aims of this study were to use RS at different dilution rates to select for butyrate-producing communities from fecal material. A combination of rRNA-based quantitative dot blot hybridization and conventional culture methods was used to characterize butyrate-producing and amylolytic fecal communities. As the study developed, the work focused on one particular bacterium that microscopic examination showed dominated the RS enrichments at high dilution rates. The organism was recalcitrant to isolation in pure culture, highly amylolytic, degraded RS, and produced butyrate as the major end product of fermentation.
MATERIALS AND METHODS
Chemostat operation.
Fecal samples were collected from two separate individuals, both healthy males (25 and 30 years old) with no history of antibiotic treatment over the preceeding 6-month period, and a normal varied diet. The chemostats were operated at each dilution rate with feces from each donor. Results presented for each dilution rate are an average for the two individuals. Fecal slurries were prepared from fresh human feces homogenized with anaerobic sodium phosphate buffer (100 mM, pH 7.0) to give 10% (wt/vol) suspensions. These were filtered through a 100-μm (pore-size) metal sieve to remove food residues. The filtrates were used to inoculate glass chemostats (280 ml, working volume) containing a culture medium consisting of the following (in g liter−1): NaCl, 4.5; KCl, 2.5; K2HPO4, 1.5; CaCl2 · 6H2O, 0.15; Mg2SO4 · 7H2O, 0.25; NH4Cl, 1.0; cysteine, 0.8; bile salts no. 3, 1.0; Tween 80, 1.0; hemin, 0.01; vitamin B12, 0.005; menadione, 0.005; yeast extract, 2.5; peptone, 2.0; tryptone, 2.0; and resistant starch (Hylon VII), 5.0. In addition, 1.0 ml of a trace elements solution (2) was added per liter of culture medium. Anaerobic conditions were maintained by sparging the cultures with O2-free N2 gas (5 ml min−1). Temperature (37°C) and pH (6.5) were automatically controlled as described elsewhere (25). After overnight equilibration, the culture vessels were operated at two dilution rates, D = 0.03 or 0.30 h−1, to select for slow-growing and fast-growing species, respectively.
Analysis of starch fermentation and starch hydrolysis by cell-associated and soluble amylases.
SCFAs were determined by gas chromatography by using standard procedures (17). These methods did not separate the branched-chain fatty acids isovalerate and 2-methylbutyrate. Cell-free culture supernatants from the chemostats and bacterial cell extracts were prepared as described by Englyst et al. (10). Amylase activities were determined using the 3,5-dinitrosalicylate reagent to measure liberation of reducing end groups (7). α-Glucosidase activities were measured by monitoring the release of p-nitrophenol from p-nitrophenyl α-d-glucopyranoside. One unit of amylase activity is equivalent to 0.36 mg of maltose produced min−1, and one unit of α-glucosidase activity is equal to one μmol of p-nitrophenol produced min−1.
Culture media.
Anaerobic starch-hydrolyzing bacteria were enumerated using plate count methods. Samples (1 ml) from the chemostats were serially diluted in half-strength Wilkins-Chalgren (WC) broth in an anaerobic chamber (atmosphere of 10% H2, 10% CO2, and 80% N2). Bacterial populations were counted using a variety of selective and nonselective agar plates: species belonging to the Bacteroides fragilis group were isolated using a defined mineral salts based medium, containing vancomycin and nalidixic acid as selective agents (29). Facultative anaerobes were counted on MacConkey agar no. 2, while clostridia were determined using perfringens agar containing antibiotic supplements, as stipulated by the manufacturer (Oxoid), and WC agar, with novobiocin and colistin (13). Lactobacilli and bifidobacteria were enumerated by using Rogosa agar and Beerens agar (3), respectively. Total anaerobes and constituent populations of anaerobic gram-positive cocci were counted using WC agar. Soluble starch (10 g liter−1) was added to all of the culture media. The plates were incubated for up to 5 days, aerobically or anaerobically, as appropriate. Amylolytic isolates were detected by the appearance of halos around colonies following the addition of Lugols iodine. The predominant colony types were maintained on WCA-starch plates. Differential counts were made from the different agars and then, after identification of the predominant morphology types by cell wall-fatty acid analysis, the numerics of each bacterial population were surmised. This technique minimizes the overestimation of bacterial groups that grow on different agar groups. For example, bifidobacteria are readily cultured on Rogosa, Beerens, and WC agars; however, with the putative identification of each of the colony types, any potential overlap is accounted for this way.
For determinations of SCFAs and organic acids, individual bacterial species were grown in 5 ml of peptone-yeast extract-glucose (PYG) broth (17) for 48 h. This facilitated their identification and indicated isolates that produced butyrate as a major end product of metabolism.
Analysis of cellular fatty acids for bacterial identification.
Fatty acid methyl esters (FAME) were obtained from cultures (ca. 40 mg [wet weight] of cells) after overnight growth PYG broth by saponification, methylation, and extraction as described previously (31). FAME were separated using a Model 6890A Microbial Identification System (Microbial ID, Inc., Newark, Del.), which comprised a Hewlett-Packard model 6890 gas chromatograph fitted with a 5% phenyl-methyl silicone capillary column (0.2 mm by 25 m), a flame ionization detector, a Hewlett-Packard Model 7637A Automatic Sampler, and a Hewlett-Packard Vectra XM computer (Hewlett-Packard Co., Palo Alto, Calif.). The gas chromatographic parameters were as follows: carrier gas, ultra-high-purity H2; column head pressure, 60 kPa; injection volume, 2 μl; column split ratio, 100:1; septum purge, 5 ml min−1; column temperature, 170 to 270°C; and injection port temperature, 300°C. Peaks were automatically integrated, and fatty acid names and percentages were calculated. The numerical analyses and predictions for bacterial identification were done using standard MIS Library Generation Software.
Total culture rRNA extractions.
Samples (500 mg) in 2.2-ml screw cap tubes were initially subjected to direct phenol extraction with mechanical disruption (Mini-Bead Beater; Biospec Products, Bartlesville, Okla.) using 300 mg of zirconium beads (44). All glassware was baked to 300°C overnight, and solutions were prepared using diethyl pyrocarbonate-treated double-distilled water. Mechanical disruption was followed by another extraction with phenol saturated with Tris-HCl buffer (100 mM, pH 5.1), and two sequential phenol-chloroform-isoamyl alcohol (24:24:1, pH 5.1) and chloroform-isoamyl alcohol extractions. Total rRNA was precipitated at −20°C for 3 h with ammonium acetate (1 M, final concentration). After two washes in 80% ethanol, pellets were resuspended in 50 μl of double-distilled water. The concentration of nucleic acid was estimated spectrophotometrically on the basis that an optical density value of 1.0 at 260 nm corresponded to an RNA concentration of 40 μg of RNA ml−1. The quality of extracted RNA was evaluated using polyacrylamide gel electrophoresis (Mighty Small II, Slab Gel Electrophoresis Unit; Hoefer Instruments, San Francisco, Calif.).
Probes and labeling.
Table 1 lists the oligonucleotide probes used in this study and the source of the sequence. The probe groups targeted were total bacterial rRNA, genus Bifidobacterium, bacteroides cluster, covering the predominant species that occur in the large intestine, and the unknown RS-degrading organism found in this investigation. Synthetic high-pressure liquid chromatography-purified oligonucleotide probes were 5′ end labeled with 32P using polynucleotide kinase (Gibco BRL) and [γ-32P]ATP (ICN) with a specific activity of >5,000 Ci mmol−1 and a concentration of 10 mCi ml−1, as previously described (35).
TABLE 1.
Oligonucleotide probes used in this study
| Target group | Probe sequence (5′ to 3′) | OPD nomenclaturea | Reference |
|---|---|---|---|
| Eubacterial domain | GCTGCCTCCCGTAGGAGT | S-D-Bact-338-a-A-18 | 20 |
| Bacteroides cluster | GCACTTAAGCCGACACCT | S-G-Bac-1080-a-A-18 | 9 |
| Novel clostridium isolate | TCCAATCCCTTGAATT | S-S--RSD-184-a-A-16 | This study |
| Genus Bifidobacterium | CATCCGGCATTACCACCC | S-G-Bif-164-a-A-18 | 21 |
Alm et al. (1).
Total RNA dot blot hybridizations.
Nucleic acids were denatured and diluted to 1.5 ng μl−1 as described earlier. Samples were applied in triplicate, at 50 μl/slot, to Magna Charge membranes (Micron Separation, Inc., Westboro, Mass.) using a slot blot device (Minifold II; Schleicher & Schuell, Inc., Keene, N.H.) under a slight vacuum to pull the entire sample through the membrane in 1 to 2 min. The membranes were air dried and baked for 2 h at 80°C. The baked membranes were then prewetted in hybridization buffer [0.9 M NaCl, 50 mM sodium phosphate (pH 7.0), 5 mM EDTA, 10× Denhardt solution (40), 0.5% sodium dodecyl sulfate, 0.05 mg of poly(A)ml−1] and placed in hybridization tubes (Robbins Scientific, Sunnyvale, Calif.). Membranes were incubated in a rotating incubator with approximately 10 ml of hybridization buffer for 2 h at 40°C. The first hybridization buffer was discarded, and labeled probe was added by inclusion in 10 ml of hybridization buffer. The incubation was then continued at 40°C for 16 to 20 h. After incubation with the probe, the membranes were washed in the hybridization tubes with 100 ml of 1% SDS–1× SSC (0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) for 2 h at 40°C. Membranes were then removed from hybridization tubes and washed twice for 15 min in 300 ml of 1% SDS–1× SSC at the experimentally determined dissociation temperatures for individual probes (47).
Imaging of hybridization signal.
The hybridization signal on air-dried membranes was quantified using an Instant Imager (Canberra Packard, Pangbourne, Berks, United Kingdom). The time of exposure varied depending on the intensity of the 32P signal. Analysis of the signal was done using ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Standard curves were calculated from reference RNA by linear regression. RNA from the following organisms was used: Bifidobacterium longum (bacterial and Bifidobacterium genus), Bacteroides thetaiotaomicron (bacteroides cluster), and the unknown clostridium for that specific population. The abundances of specific groups of organisms are shown as a percentage of total bacterial small-subunit rRNA in the sample, and quantities of RNA for each probe group are also expressed as total quantities of rRNA ml−1 chemostat fluid.
Development of 16S rRNA-targeted hybridization probe.
DNA was extracted from a pure culture of the unknown chemostat isolate using a method specific for clostridia (38). 16S ribosomal DNA (rDNA) was selectively amplified by PCR using bacterial specific primers and sequenced by the dideoxynucleotide method. This process has been detailed elsewhere (19). An oligonucleotide probe was designed using the sequence obtained and a collection of complete 16S rRNA sequences for cluster I true clostridia, obtained from the Ribosomal Database Project (22). The optimum wash temperature for the probe was determined by a sequential washing procedure. Labeled probe that remained bound to target rRNA was quantitated at increasing temperatures as previously described (35). Dot blot hybridization was used to assess the specificity of probe by blotting rRNA (ca. 50 ng) extracted from 48 pure cultures representing a wide range of intestinal species. Of these, 25 cultures were speciated, and the remaining 23 were bacteria isolated from human feces that belong to the Clostridia-Eubacteria group as identified by cell wall fatty acid analysis and SCFA production.
Light microscopy.
Adherence of amylolytic bacteria to RS granules in the chemostats was studied by phase-contrast microscopy using a Zeiss Axiophot photomicroscope.
Scanning electron microscopy.
Samples were taken from the chemostat operated at D = 0.30 h−1 for scanning electron microscopy and placed in 3% (vol/vol) glutaraldehyde in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (100 mM, pH 7.4). They were then fixed with 4% (wt/vol) aqueous OsO4, dehydrated stepwise in ethanol, with three changes (10 min) in each of 50, 75, 95 and, finally, 100% ethanol. The samples were then dried on a Poleron E 5000 critical-point drier, placed on stubs, and gold coated to a depth of 30 nm. A Phillips XL 30 FEG scanning electron microscope was used to visualize the preparations.
Protein measurements.
To calculate specific amylolytic enzyme activities, protein concentrations in bacterial cell extracts were made using the Lowry method.
Chemicals.
Bacteriologic culture media were obtained from Oxoid, Ltd. (Basingstoke, Hamps, United Kingdom). Hylon VII was gift from H. N. Englyst. Unless stated otherwise, all fine chemicals were purchased from the Sigma Chemical Co. (Poole, Dorset, United Kingdom).
RESULTS
Bacterial starch-degrading enzymes.
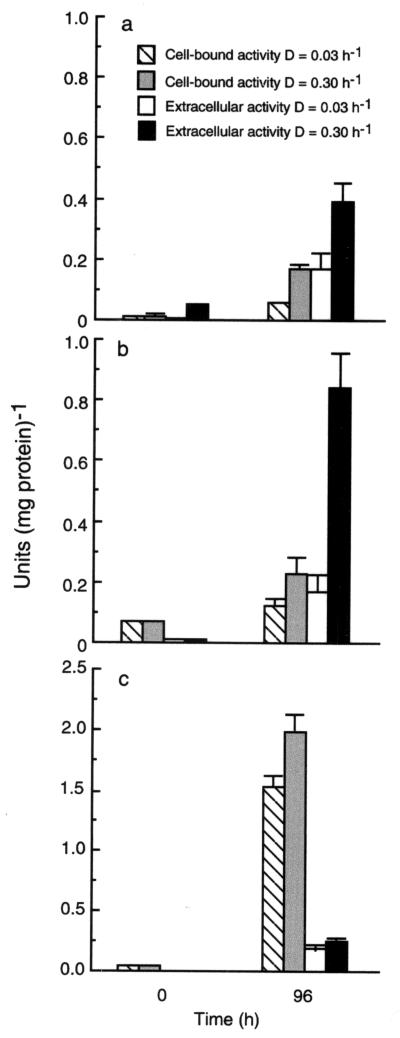
Amylolytic enzyme activities against RS and soluble starch are shown in Fig. 1. Results are given for bacterial cell extracts and cell-free culture supernatants. Significant increases in activity for all enzymes associated with starch degradation were observed after the inoculation of fecal material into the chemostats. Amylase activity was predominantly extracellular, while the majority of α-glucosidase was cell associated. Greater expression of cell-bound and extracellular amylases was seen at D = 0.30 h−1, and bacterial amylases were more active against soluble starch than RS granules.
FIG. 1.
Hydrolysis of resistant starch (a), soluble starch (b), and p-nitrophenyl α-d-glucopyranoside (c) by bacterial cell extracts and cell-free culture supernatants from chemostat enrichments at different dilution rates. Results show zero time and 96-h measurements and are the means of three determinations for two individuals ± the standard deviation.
Starch fermentation.
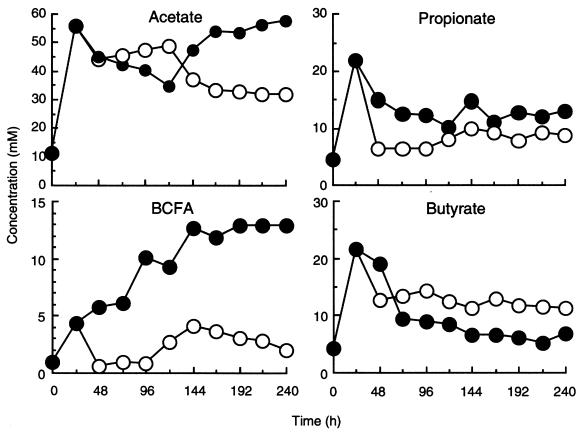
Figure 2 shows acetate, propionate, butyrate, and branched-chain fatty acid (isobutyrate, isovalerate–2-methylbutyrate) production in the chemostats. SCFA profiles were markedly different at the two dilution rates. At the 48 h time point, butyrate formation occurred maximally at the high dilution rate, accounting for 13 and 23% of the total D = 0.03 h−1 and h−1, respectively. Total SCFAs were calculated from the accumulative measurement of acetate, propionate, isobutyrate, butyrate, isovalerate, valerate, and caproate. More acetate and branched-chain fatty acids were produced at D = 0.03 h−1.
FIG. 2.
Effect of dilution rate on fermentation product formation by resistant starch-degrading communities. The BCFA (branched-chain fatty acids) are isobutyrate and isovalerate–2-methylbutyrate. Each value is the mean for two chemostats inoculated with different fecal samples, each analyzed in triplicate.
Bacterial identification.
Table 2 shows the principal groups of intestinal bacteria at different dilution rates in the chemostats at 240 h, as identified on the basis of FAME profiles and fermentation product formation in PYG broth. Variations in cell numbers under both cultural conditions were considered significantly different if they were ≥1 log unit (14). Using this criterion, there were no significant differences found in the major culturable populations between the inocula in the two chemostats (D = 0.30 h−1 and 0.30 h−1) for either of the donors. Subsequently, the main difference in the two enrichments occurred with the clostridia, which constituted on average 0.3% (D = 0.03 h−1) and 5.0% (D = 0.30 h−1) of the total anaerobe counts. However, numbers of bacteria belonging to the Bacteroides fragilis group were sixfold lower at D = 0.30 h−1. The predominant amylolytic isolates in each chemostat are listed in Table 3, where the species indicated in boldface produced butyrate (>5 mM in PYG broth) as a major fermentation end product. These results show that, although there was little difference in amylolytic species diversity at the two dilution rates, the enrichment at D = 0.03 h−1 was mainly characterized by organisms belonging to the B. fragilis group and bifidobacteria, whereas at D = 0.30 h−1 the saccharolytic clostridia were important amylolytic bacteria.
TABLE 2.
Bacterial populations in the chemostats operated at different dilution rates at 240 ha
| Bacterial group | Mean log10 viable counts ml−1 ± SE at dilution rate (h−1):
|
|
|---|---|---|
| 0.03 | 0.30 | |
| Total anaerobes | 9.9 ± 0.3 | 10.2 ± 0.2 |
| Nonsporing anaerobes | 8.8 ± 0.4 | 9.2 ± 0.2 |
| Gram-negative anaerobes | 8.5 ± 0.3 | 9.0 ± 0.3 |
| Bifidobacteria | 9.1 ± 0.3 | 9.0 ± 0.2 |
| Bacteroides | 9.2 ± 0.2 | 8.4 ± 0.3 |
| Clostridia | 7.4 ± 0.3 | 8.9 ± 0.3 |
| Lactobacilli | 7.8 ± 0.2 | 8.2 ± 0.3 |
| Enterococci | 7.8 ± 0.4 | 8.2 ± 0.3 |
| Enterobacteria | 8.9 ± 0.3 | 8.6 ± 0.3 |
Results (log10 viable counts ml−1) are the means of three measurements ± standard error.
TABLE 3.
Predominant amylolytic bacteria in RS chemostat enrichmentsa
| D (h−1) | Predominant bacteria |
|---|---|
| 0.03 | Bacteroides ovatus, Bacteroides uniformis, Bacteroides thetaiotaomicron, Bacteroides caccae, Bifidobacterium angulatum, Bifidobacterium adolescentis, Bifidobacterium sp. strain 1, Bifidobacterium sp. strain 2, Clostridium sporogenesb, Clostridium sordellii |
| 0.30 | Bacteroides uniformis, Bacteroides thetaiotaomicron, Bifidobacterium angulatum, Clostridium sporogenes, Clostridium sordellii, novel Clostridium sp.c, Clostridium perfringens, Clostridium butyricum, Eubacterium limosum, Peptostreptococcus micros, Lactobacillus acidophilus |
Microscopy.
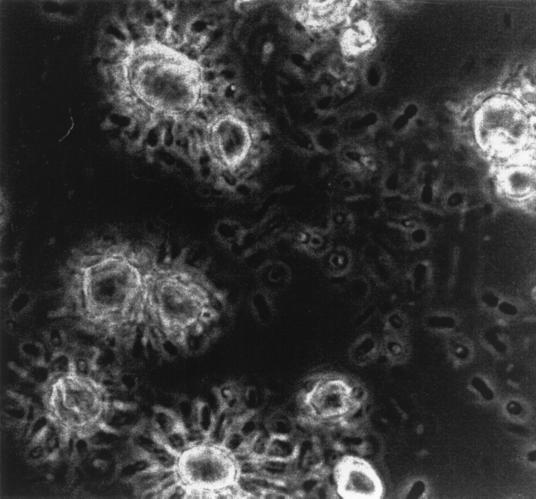
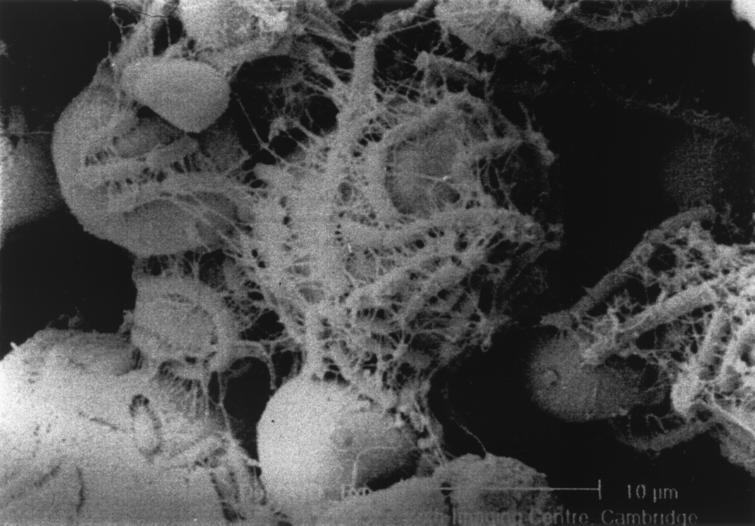
Phase-contrast microscopy of chemostat enrichments showed that at D = 0.30 h−1 the fermentation was dominated by a rod-shaped bacterium that attached apically to the RS granules, forming rosette-like structures (Fig. 3). Extensive mucilage was produced, entrapping other bacteria and forming biofilms in the planktonic phase of the culture. These structures were not formed at D = 0.03 h−1. A scanning electron micrograph of RS granules with these bacteria attached is shown in Fig. 4. The glycocalyx has been dehydrated during sample preparation and appears as condensed fibrillar structures.
FIG. 3.
Phase-contrast light micrograph (×1,600) of bacteria colonizing resistant starch granules in a chemostat operated at D = 0.30 h−1.
FIG. 4.
Scanning electron micrograph of bacteria attached to resistant starch granules during growth in continuous culture at D = 0.30 h−1.
Phylogenetic placement of adherent starch-degrading isolate.
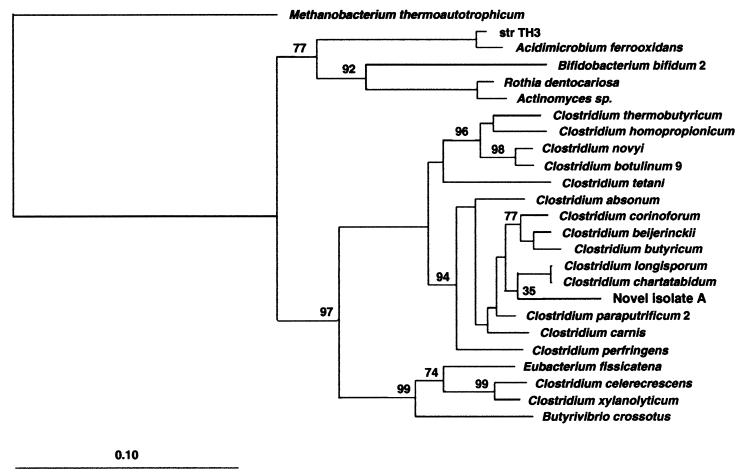
The 16S rRNA sequence for this new species was aligned among several closely related clostridia. The corresponding phylogenetic placement is illustrated in Fig. 5. The sequence has been submitted to the EMBL database under accession no. AJ243511.
FIG. 5.
Phylogenetic tree of the novel starch-degrading clostridium with other representatives of class I clostridia based on SSU rRNA sequences. The tree was constructed using the neighbor-joining method and the correction of Jukes and Cantor (19). The scale bar indicates 0.10 estimated substitutions per nucleotide. The DNABOOT program of PHYLIP (phylogeny inference package) was used to calculate the bootstrap values, placing confidence limits on phylogenies by using parsimony. Methanobacterium thermoautotrophicum was used to root the tree. Bootstrap values are based on the percentages of 100 replications.
Probe design.
Probe design included a search of nontarget group complementarity using the RDP (22). There were no nontarget species with one or two mismatches for the probe. The optimum temperature for the probe was found to be 52°C. The probe was tested against rRNA extracted from 25 different bacteria isolated from human feces, belonging to the genera Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, Peptostreptococcus, Eubacterium, Enterococcus, Oxalobacter, and Escherichia, and no hybridization signal was found.
Total rRNA concentrations.
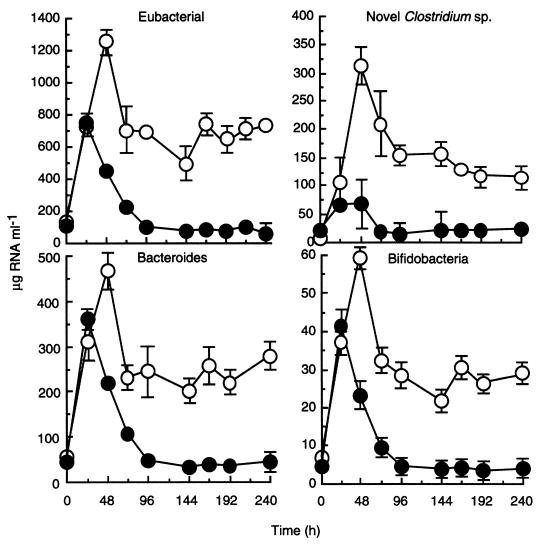
The procedure for nucleic acid extraction was fastidiously quantified throughout the procedure, from sample weights to volumes for analyses. This facilitated the determinations of total SSU rRNA for each population. Total quantities of bacterial rRNA extracted from chemostat samples are shown in Fig. 6. The concentration of rRNA was lowest in the original fecal inoculum (data not included), the time zero values for rRNA concentrations provided are the values taken after overnight equilibration. When the culture medium was fed to the chemostats, differences in nutrient supply resulted in the establishment of characteristic amylolytic communities. Bacterial rRNA concentrations probably reflected nutrient availability and were greater at the high dilution rate (86 and 742 μg ml−1 for D = 0.03 h−1 and 0.30 h−1, respectively) at 240 h.
FIG. 6.
Effect of dilution rate on the abundance of bacterial SSU rRNA (probe S-D-Bact-0338-a-A-18), novel starch-degrading clostridial SSU rRNA (probe S-S-RSD-184-a-A-16), bacteroides SSU rRNA (probe S-D-Bact-0338-a-A-18), and bifidobacterial SSU rRNA (probe S-G-Bif-164-a-A-18) in chemostats inoculated with human feces. Each value is the mean for two chemostats inoculated with different fecal samples, each analyzed in triplicate hybridizations ± the standard deviation.
Abundance of rRNA from the novel clostridial isolate.
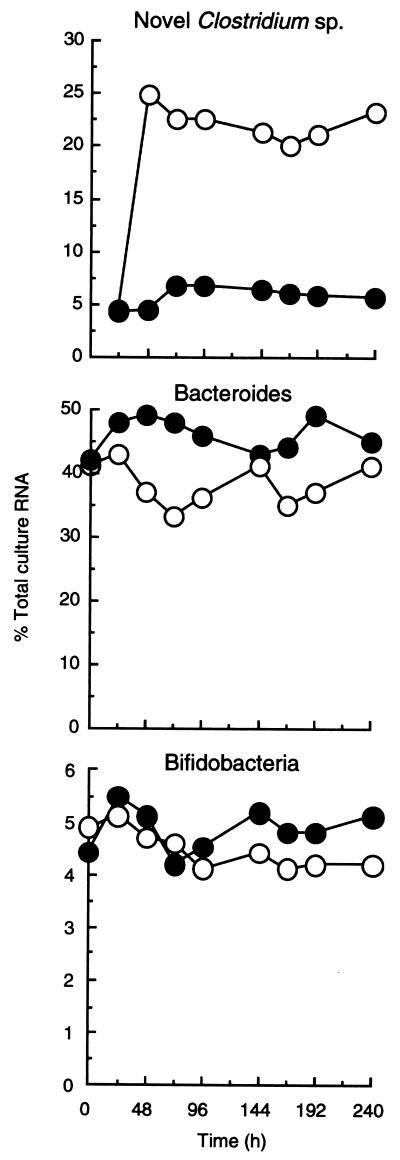
Figure 6 shows the concentration of rRNA in the chemostats relating to the novel clostridium probe. The importance of this population at D = 0.30 h−1, when starch availability was greatest (0.9 g day−1 at D = 0.03 h−1 and 9.0 g day−1 at D = 0.03 h−1), was confirmed. After an initial peak in both chemostats due to the breakdown of accumulated RS, rRNA concentrations stabilized at approximately 20 (D = 0.03 h−1) and 120 (D = 0.30 h−1) μg ml−1. The abundance of rRNA for this population is expressed as a proportion of total bacterial rRNA in Fig. 7, where it can be seen that the clostridia rapidly became predominant members of the community at the high dilution rate, accounting for between 20 and 25% of the culture RNA. In contrast, at D = 0.03 h−1, rRNA from this bacterium constituted approximately 5% of total RNA in the microbiota.
FIG. 7.
Relative abundances (percent total bacterial RNA) of novel starch-degrading clostridium, bacteroides, and bifidobacterial SSU rRNA in chemostats inoculated with human feces at different dilution rates.
Abundance of Bacteroides group rRNA.
Total Bacteroides rRNA was similarly expressed as a concentration and as a proportion of total bacterial rRNA in the chemostats (Fig. 6 and 7). At 240 h, the concentrations of bacteroides group rRNA were 50 and 290 μg ml−1 at D = 0.03 h−1 and 0.30 h−1, respectively, amounting to 40 and 45% of the total community RNA.
Abundance of Bifidobacterium genus rRNA.
The total bifidobacterial rRNA levels at 240 h were 5.9 (D = 0.03 h−1) and 31.2 (D = 0.30 h−1) μg ml−1 (Fig. 6). This represented a low proportion of total ribosomal abundance at 5.1 and 4.2%, respectively (Fig. 7).
DISCUSSION
Cooking and processing of starchy foods results in a portion of the starch becoming resistant to small intestinal hydrolases. This RS can, however, be degraded by amylolytic enzymes formed by colonic bacteria and is one of the most important sources of carbohydrate for these organisms (11, 24).
The same bulk quantity of starch (Hylon VII) was used throughout this study, which contains a consistent mixture of insoluble RS granules and soluble starch. Hylon VII contains 70% amylose, in comparison to Lintners starch, which contains 27% amylose (11). In earlier work, we showed that starch breakdown (irrespective of its physical form) increases butyrate production by intestinal microorganisms (10, 11, 25), and this also occurred in the chemostat enrichments in this investigation (Fig. 2). Saccharolytic clostridia such as Clostridium perfringens and C. butyricum, as well as some Eubacterium spp., are thought to be important in butyrate formation in the gut (51). Members of the phylogenetically incoherent genus Fusobacterium (23) may also be of significance (25). In this study a number of bacterial isolates produced large amounts of butyrate in pure culture, including Eubacterium limosum, C. butyricum, C. sordellii, and C. perfringens. Increased RS supply at D = 0.30 h−1 resulted in high levels of butyrate production and a greater frequency of isolation of butyrate-forming species (Table 3). Overall, these results highlight different strategies in substrate utilization in disparate groups of intestinal bacteria. Saccharolytic clostridia, including the newly identified species, were best adapted to fast growth rates and high substrate concentrations, whereas the bacteroides competed more effectively at low growth rates under conditions of limiting substrate availability. Although there were qualitative variations in species diversity, in terms of overall population size, bifidobacterial growth on RS was less affected than bacteroides or clostridia by environmental conditions in the chemostats. In addition to these populations, the increased production of branched-chain fatty acids at D = 0.03 h−1 was indicative of carbohydrate limitation and increased activities of amino acid-fermenting species (27).
In humans, starch degradation in the large gut is dependent on the activities of bacterial amylases and residual pancreatic amylase (25), whereas in poultry, for example, this is done solely by bacterial enzymes (4). Amylolytic activity in important saccharolytic species such as the bacteroides and bifidobacteria is largely cell associated (8, 24). Although α-glucosidase was principally cell bound in the chemostat enrichments, production of high levels of extracellular amylase was not characteristic of these organisms (Fig. 1). The novel Clostridium sp. that formed rosette-like structures on RS granules and dominated enrichments at D = 0.30 h−1 secreted an extracellular amylase when grown in pure culture (data not shown). Close physical proximity between these bacteria and the starch granule ensured that hydrolytic products were accessible to the organisms and that enzyme secretion was not energetically disadvantageous. However, bacterial attachment to the surface of substrates is often the initial step for degradation by cell-associated bacterial enzymes.
Traditional anaerobic culture methods repeatedly failed to isolate this adherent species. Nonetheless, a single colony was eventually obtained that was identical morphologically to bacteria in the high dilution rate enrichment. This was achievable because these organisms were present in considerable numbers in the chemostat and could be isolated on the same culture medium, using serial dilutions of fermentor contents. In these isolations, the end dilution tube with growth was, on one occasion only, found to contain a pure culture. RS granules were found to be essential for growth, and the tubes had to be constantly agitated. The phenotype was confirmed by its ability to form the same rosettes around RS granules, with subsequent biofilm formation when grown axenically in chemostats. Butyrate (20 mM) and acetate (5 mM) were the predominant SCFAs produced by the pure culture. Since there are at present no reliable means for freezing and regeneration of this species, these experiments provide a paradigm for the use of molecular methods for identification and quantitation of bacteria that are difficult to grow in pure culture.
Population abundance using the 16S rRNA probes is expressed as both total quantities of SSU rRNA per milliliter of chemostat fluid and as a proportion of the total. This was made possible by the meticulous quantification of the procedure used to extract rRNA (44). Expressing the abundance of a specific microbial population as total quantities facilitates a more confident expression of the contribution of a microbial population to the overall bacterial population. Shifts in population abundance measured and expressed as a proportion of the total may represent shifts in the total community structure and ribosomal abundance and not changes in the absolute amounts. Proportional values allow a more direct comparison both between samples of various concentrations and between experiments.
The initial development of 16S rRNA targeted oligonucleotide probes involves (i) comparison of sequences by alignment, (ii) identification of a target sequence which is unique for the species, (iii) synthesis and labeling of complementary nucleic acid probes, and (iv) experimental evaluation of the probe. Differences in the secondary and tertiary structure of the 16S molecule may result in steric hindrance, which prevents effective probe binding, so probe evaluation is important. We used four probes in this study, a Bacteroides group probe (9); a Bifidobacterium genus probe, evaluated for fluorescent whole-cell hybridizations (21); a domain Bacteria probe (20); and a species-specific probe for the novel RS degrading isolate, which was evaluated in this study. Ideally, probe evaluation would be done with database searches, producing no nontarget species with one or two mismatches. However, given the complexity of the gut microbiota, the absence of nontarget species in the databases is not an indication of their absence in intestinal microorganisms. Therefore, precise determination of a Td value specific for this probe was essential. No hybridization signal was obtained using the labeled probe and rRNA from a diverse range of 25 other gut species. The relative rRNA index, which signifies the abundance of the bacteria, was >20% of the total rRNA in chemostats at D = 0.30 h−1 but only 5% at the low dilution rate, at the 240 h end point (Fig. 7), largely confirming microscopic observations.
The importance of this bacterium to RS breakdown in the large gut will depend on its occurrence in different individuals, its relative numbers in the colon, and its metabolic activities in the microbiota. The organisms do not appear to have been anomalies, since they were present in the feces of the two individuals sampled in this study, and two further individuals who provided inocula for subsequent RS enrichments.
Total rRNA concentrations in the chemostats provided a clear measure of the metabolic activity of bacterial populations over the course of the experiment. As expected, marked differences were evident in ribosomal abundance at each dilution rate and were higher at D = 0.30 h−1.
The importance of species belonging to the Bacteroides fragilis group in carbohydrate metabolism in the colon is well established (24, 26); however, the taxonomy of the genus Bacteroides is confused, and several separate genera have been named from within the original grouping (42). Bacteroides nutritional diversity renders most culture media unsuitable for the simultaneous isolation of a number of different species, and currently used selective media contain additives that may lead to underestimations of the total numbers for some species. The Bacteroides-Porphyromonas-Prevotella group probe (9, 30) has been shown to be a useful molecular marker for this phylogenetic cluster and includes many species in the large bowel, such as B. vulgatus, B. fragilis, B. thetaiotaomicron, B. ovatus, B. uniformis, and Prevotella distasonis. Bacteroides RNA accounted for >40% of the total RNA at 240 h. This is consistent with molecular analyses of fecal microflora in a separate study using the same probe sequence (9). Interestingly, viable counts indicated that members of the B. fragilis group constituted only 2% (D = 0.30 h−1) and 20% (D = 0.03 h−1) of the total anaerobes in the chemostats (Table 2). However, caution should be taken in comparing plate counts and molecular data due to the differences inherent in the two techniques.
In contrast to other important genera in the colon, such as Eubacterium and Bacteroides, bifidobacteria form a monophyletic cluster on the basis of 16S rRNA sequencing. This has facilitated the development of genus-specific probes. The bifidobacterium probe was developed for fluorescent-labeling studies in feces (21), and its specificity was determined using dot blot hybridization against a range of gut microorganisms. We determined the precise Td of the probe (62°C) for quantitative hybridization studies with community 16S rRNA.
The abundance of rRNA corresponding to this genus was surprisingly low at 4.8% (D = 0.03 h−1) and 4.1% (D = 0.30 h−1) and did not correlate with relative bifidobacterial abundances of 15.8 and 12.6% using plate counts. Again, while exercising caution in comparing plate counts with molecular data, a potential explanation for this can be found in previous studies which have shown that fecal bifidobacterial numbers can be overestimated by culture-dependent methods. The data suggest that these organisms are very culturable, much more reliably so than many other colonic anaerobes. For example, while fluorescent-probe-based estimations of fecal bifidobacteria equate with culture counts, they indicate that total anaerobes have been underestimated by a factor of 10 using culture-based techniques (21, 51). This may have led to an overestimation of the significance of bifidobacteria in the microbiota when using plate counts.
The similarity between the two dilution rates in terms of bifidobacterial abundance was surprising, since a recent investigation indicated that high-amylose maize starch enhanced the survival of these organisms in the mouse intestinal tract (49). One of the species in that study adhered to the starch granules in a similar way to the novel Clostridium sp. discovered in the present work. However, butyrate is not produced by bifidobacteria, and their ecologic role may be restricted to the initial stages of substrate depolymerization. Butyrate is formed by many saccharolytic clostridia, and C. butyricum is involved in the breakdown of high-amylose starch granules (50). Clostridia are known to degrade starch in the porcine large intestine (36) but are generally considered to be less important in the human colon.
An interesting feature of these results is that the greater proportion of the fecal microbiota remains unaccounted for by the summation of the different group and genus probes. Although it should be acknowledged that circumscribing fecal bacterial diversity using rRNA targeted oligonucleotide probes was not a goal of this experiment, the observation is of interest and consistent with other published material (34). Direct analysis of genes encoding 16S rRNA from fecal bacterial communities has revealed many novel molecular species in the gut (48, 52); indeed, one comprehensive study suggests that 76% of generated rDNA sequences did not correspond to known organisms (48).
In summary, the framework provided by 16S rRNA sequencing provided a useful approach to the study of colonic microbial populations involved in RS breakdown, allowing an essentially unculturable species to be investigated. While only a small part of the RS-degrading community was accessible to the probes used here, this work shows the validity of combining 16S rRNA probes and the chemostat for studying the ecology of intestinal bacteria (41; Sharp and Macfarlane, submitted). The dot blot hybridization base quantitation technique preferably requires a pure culture to provide reference rRNA. However, 16S rDNA transcripts with reverse transcriptase to provide reference rRNA is a possible alternative that would allow quantitation of unculturable bacteria whose presence had been established using 16S rDNA sequencing.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: revaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerens H. Elective and selective isolation medium for Bifidobacterium spp. Lett Appl Microbiol. 1991;11:155–157. [Google Scholar]
- 4.Champ M, Szylit O, Gallant D J. The influence of microflora on the breakdown of maize starch granules in the digestive tract of chickens. Poultry Sci. 1981;60:179–187. doi: 10.3382/ps.0600179. [DOI] [PubMed] [Google Scholar]
- 5.Cummings J H. Short chain fatty acids in the human colon. Gut. 1981;22:773–779. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummings J H, Macfarlane G T. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 7.Dahlqvist A. A methods for the determination of amylase in intestinal contents. Scand J Clin Lab Investig. 1962;14:145–151. doi: 10.3109/00365516209079686. [DOI] [PubMed] [Google Scholar]
- 8.Degnan B A. Transport and catabolism of carbohydrates by anaerobic gut bacteria. Ph.D thesis. Cambridge, England: University of Cambridge; 1993. [Google Scholar]
- 9.Dore J, Sghir A, Hannequart-Gramet G, Corthier G, Pochart P. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst Appl Microbiol. 1998;21:65–71. doi: 10.1016/S0723-2020(98)80009-X. [DOI] [PubMed] [Google Scholar]
- 10.Englyst H N, Hay S, Macfarlane G T. Polysaccharide breakdown by mixed populations of human faecal bacteria. FEMS Microbiol Ecol. 1987;95:163–171. [Google Scholar]
- 11.Englyst H N, Macfarlane G T. Breakdown of resistant starch by human gut bacteria. J Sci Food Agric. 1986;37:699–706. [Google Scholar]
- 12.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous intestinal microflora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 3–31. [Google Scholar]
- 13.Fujisawa, T., K. Namba, K. Hirayama, W. K. Lee, and T. Mitsuoka, T. 1995. New selective media for isolation of clostridia from fecal specimens. J. Appl. Bacteriol. 78:481–486. [DOI] [PubMed]
- 14.Gibson G R, Beatty E R, Wang X, Cummings J H. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;106:975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 15.Goodlad J S, Mathers J C. Large bowel fermentation in rats given diets containing raw peas (Pisum sativum) Br J Nutr. 1990;64:569–587. doi: 10.1079/bjn19900057. [DOI] [PubMed] [Google Scholar]
- 16.Hagopien H K, Riggs M G, Swartz L A, Ingram V M. Effect of n-butyrate on DNA synthesis in chick fibroplast and HeLa cells. Cell. 1977;12:855–860. doi: 10.1016/0092-8674(77)90284-7. [DOI] [PubMed] [Google Scholar]
- 17.Holdeman L V, Cato E P, Moore W E C, editors. Anaerobe laboratory manual. 4th ed. Blacksburg, Va: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 18.Jukes T H, Cantor C P. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 19.Krumholz L R, Sharp R, Fishbain S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene. Appl Environ Microbiol. 1996;61:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D J, Pace B, Olsen B G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphius G R, Wilkinson M H, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application to fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen N, Olsen G J, Maidak B L, McCaughey B L M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The ribosomal RNA database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson P A, Gharbia S E, Shah H N, Clark D R. Recognition of Fusobacterium nucleatum subgroups Fn-1, Fn-2 and Fn-3 ribosomal RNA gene restriction patterns. FEMS Microbiol Lett. 1989;65:41–45. doi: 10.1016/0378-1097(89)90363-7. [DOI] [PubMed] [Google Scholar]
- 24.Macfarlane G T, Cummings J H. The colonic flora, fermentation, and large bowel digestive function. In: Philips S F, Pemberton J H, Shorter R G, editors. The large intestine: physiology, pathophysiology and disease. New York, N.Y: Raven Press; 1991. pp. 51–92. [Google Scholar]
- 25.Macfarlane G T, Englyst H N. Starch utilisation by the human large intestinal microflora. J Appl Bacteriol. 1986;60:195–201. doi: 10.1111/j.1365-2672.1986.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane G T, Gibson G R. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. In: Mackie R I, White B A, editors. Ecology and physiology of gastrointestinal microbes. 1. Gastrointestinal fermentations and ecosystems. New York, N.Y: Chapman & Hall; 1996. pp. 269–318. [Google Scholar]
- 27.Macfarlane G T, Gibson G R, Beatty E, Cummings J H. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol Ecol. 1992;101:81–88. [Google Scholar]
- 28.Macfarlane G T, Gibson G R, Cummings J H. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol. 1992;72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane G T, Hay S, Gibson G R. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989;66:407–417. doi: 10.1111/j.1365-2672.1989.tb05110.x. [DOI] [PubMed] [Google Scholar]
- 30.Manz W, Amann R, Ludwig W, Vancaneyt M, Schleifer K H. Application of a suite of 16S rRNA specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 31.Miller L, Berger T. Bacteria identification by gas chromatography of whole cell fatty acids. Hewlett-Packard application note 228-41. Avondale, Pa: Hewlett-Packard Co.; 1989. [Google Scholar]
- 32.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese Hawaiians. Appl Microbiol. 1974;27:9611–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore W E C, Holdeman L V. Some current concepts in intestinal microbiology. Am J Clin Nutr. 1976;32:164–172. doi: 10.1093/ajcn/31.10.S33. [DOI] [PubMed] [Google Scholar]
- 34.Pochart P, Gramet G, Goderel L, Dore J. Molecular characterization of the human fecal flora using rRNA-targeted hybridisation probes. Gastroenterology. 1998;114:3697. [Google Scholar]
- 35.Raskin L, Stromley J M, Rittman B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid C A, Hillman K, Henderson C, Glass H. Fermentation of native and processed starches by the porcine fecal anaerobe Clostridium butyricum (NCIMB 7423) J Appl Bacteriol. 1996;80:191–198. [Google Scholar]
- 37.Roediger W E W. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rood J I. Isolation and characterization of multiple-antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob Agents Chemother. 1978;13:871–880. doi: 10.1128/aac.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakata T. Stimulatory effect of short chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic effects. Br J Nutr. 1987;58:95–103. doi: 10.1079/bjn19870073. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Manniatis J. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sghir A, Chow J M, Mackie R I. Continuous culture selection of bifidobacteria and lactobacilli from human fecal samples using fructooligosaccharide as selective substrate. J Appl Microbiol. 1998;85:769–777. doi: 10.1111/j.1365-2672.1998.00590.x. [DOI] [PubMed] [Google Scholar]
- 42.Shah H N, Collins M D. Proposal to restrict the genus Bacteroides (Castellani and Chalmers) to Bacteroides fragilis and closely related species. Int J Syst Bacteriol. 1989;39:85–87. [Google Scholar]
- 43.Sharp R, Ziemer C J. Taxonomy, systematics, and beyond: application of molecular techniques to intestinal microbiology. In: Gibson G R, Roberfroid M B, editors. Colonic microbiota, nutrition, and health. London, England: Chapman and Hall; 1999. pp. 235–259. [Google Scholar]
- 44.Sharp R, Zeimer C J, Stern M D, Cotta M A, Whitehead T A, Stahl D A. Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol Ecol. 1998;26:71–78. [Google Scholar]
- 45.Stahl D A. Application of phylogenetically-based hybridization probes to molecular microbial ecology. Mol Ecol. 1995;4:535–542. [Google Scholar]
- 46.Stahl D A, Amann R I. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 205–248. [Google Scholar]
- 47.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suau A, Bonnet R, Sutren M, Godon J J, Gibson G R, Collins M D, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Brown I L, Evans A J, Conway P L. The protective effects of high amylose maize (amylomaize) starch granules on the survival of Bifidobacterium spp. in the mouse intestinal tract. J Appl Microbiol. 1999;87:631–639. doi: 10.1046/j.1365-2672.1999.00836.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Conway P L, Brown I L, Evans A J. In vitro utilization of amylopectin and high-amylose maize (amylomaize) starch granules by human colonic bacteria. Appl Environ Microbiol. 1999;65:4848–4854. doi: 10.1128/aem.65.11.4848-4854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welling G W, Elfferich P, Raangs G C, Wilderboer-Veloo A C M, Jansen G J, Degener J E. 16S rRNA-targeted oligonucleotide probes for monitoring of intestinal tract bacteria. Scand J Gastroenterol. 1997;32:17–19. doi: 10.1080/00365521.1997.11720711. [DOI] [PubMed] [Google Scholar]
- 52.Wilson K H, Ikeda J S, Blitchington R B. Phylogenetic placement of community members of human colonic biota. Clin Infect Dis. 1997;25:S114–S116. doi: 10.1086/516230. [DOI] [PubMed] [Google Scholar]