Abstract
Background and Objectives: Krűppel-like factor 10 (KLF10) participates in the tumorigenesis of several human cancers by binding to the GC-rich region within the promoter regions of specific genes. KLF10 is downregulated in human cancers. However, the role of KLF10 in gastric cancer formation remains unclear. Materials and Methods: In this study, we performed immunohistochemical staining for KLF10 expression in 121 gastric cancer sections. Results: The loss of KLF10 expression was correlated with advanced stages and T status. Kaplan–Meier analysis revealed that patients with higher KLF10 levels had longer overall survival than those with lower KLF10 levels. Univariate analysis revealed that in patients with gastric cancer, advanced stages and low KLF10 levels were associated with survival. Multivariate analysis indicated that age, gender, advanced stages, and KLF10 expression were independent prognostic factors of the survival of patients with gastric cancer. After adjusting for age, gender, and stage, KLF10 expression was also found to be an independent prognostic factor in the survival of patients with gastric cancer. Conclusion: Our results collectively suggested that KLF10 may play a critical role in gastric cancer formation and is an independent prognosis factor of gastric cancer.
Keywords: KLF10, gastric cancer, prognosis
1. Introduction
Krűppel-like factors (KLFs) are highly conserved zinc-finger proteins that regulate cellular transcription machinery [1,2]. KLFs regulate a wide range of cellular functions, including cell proliferation, apoptosis, differentiation, and neoplastic transformation, by binding to GC-rich promoter regions [1,2].
Subramaniam et al. first identified KLF10, which was previously named as tumor growth factor-beta (TGF-beta)-inducible early gene (TIEG1), from a human osteoblast cDNA library [3]. Recently, reports demonstrate that KLF10 play a critical role in human carcinogenesis [4,5,6]. In TGF-beta-resistant pancreatic cancer cells, the ectopic expression of KLF10 increases chemosensitivity to gemcitabine and induces apoptosis [7]. Hsu et al. demonstrated that the overexpression of KLF10 results in the reduction in Bax-inhibitor-1 expression and disrupts intracellular calcium homeostasis, which leads to the apoptosis of estrogen-dependent adenocarcinoma cells [8]. KLF10 recruits HDAC to suppress the expression of epithelial growth factor receptor (EGFR), and as a consequence, represses the EGF signals that are involved in the invasion and metastasis of breast cancer [9]. However, the association between the expression of KLF10 and the clinical outcomes of patients with cancer remains controversial. The cirFUT8, a circular RNA, enhances the expression of KLF10 and subsequently reduces the metastasis of bladder cancer [10]. The downregulation of KLF10 in pancreatic ductal adenocarcinoma stably expressing KRasG12D promotes invasion and metastasis via the activation of the SDF-1/CXCR4 and AP-1 pathways [11]. KLF10 is significantly diminished in multiple myeloma relative to in normal tissues [12]. KLF10 attenuates the nuclear translocation of beta-catenin, inhibits the expression of PTTG1, and then decreases the proliferation of multiple myeloma [12]. Expression of KLF10 was associated with a higher 5-year survival rate and was an independent factor for the overall survival rate [13]. Yang et al. demonstrated that increased KLF10 and then downregulation of EGFR enhanced the sensitivity of gemcitabine-resistant cholangiocarcinoma to photodynamic therapy [14]. In a randomized phase III trial, higher expression of KLF10 was correlated with better overall survival and recurrence-free survival [15]. Downregulation of KLF10 was found in multiple myeloma tissues compared to normal groups, whereas overexpression of KLF10 mitigated the proliferation of multiple myeloma cells by controlling PTTG1 expression [12]. By contrast, KLF10 is highly expressed in prostate cancers [16]. Ivanov et al. detected high KLF10 expression in renal cell carcinoma, colon, lung, and small intestine cancers [17].
Gastric cancer has a high prevalence and is one of the leading causes of cancer-related deaths in the Asian region [18]. In accordance with histopathological analysis, gastric cancer can be divided into three subtypes: intestinal, diffuse, and mixed [18]. Each subtype has a distinct genetic background. For example, the germline mutation of E-cadherin is found in the diffuse type, whereas Helicobacter pylori infection is associated with intestinal-type gastric cancer [19]. Our previous reports have shown that several KLF proteins are correlated with gastric tumorigenesis. KLF5 and KLF8 expression is associated with advanced tumor stages and poor prognosis [20,21]. Although KLF10 is associated with several cancers, its role in gastric cancer remains to be elucidated. In this study, we demonstrated that KLf10 expression is associated with clinical stages and overall survival rate. Our findings provided evidence that KLF10 may function as a tumor suppressor gene in gastric cancer.
2. Materials and Methods
2.1. Sample Collection
A total of 121 gastric cancer patients who underwent radical subtotal gastrectomy from 1999 to 2005 were enrolled in this study. The inclusion criteria were the cases performed by radically subtotal gastrectomy. The exclusion criteria were the cases not performed by radically subtotal gastrectomy and neoadjuvant chemotherapy. Chemotherapy such as adriamycin and 5-fluorouracil was applied to the status of nodal metastasis. The postoperation nutrition was oral nutritional intervention including dietary advice, oral nutritional supplements, or both. Monthly follow-up was recommended. These conditions showed no relevant differences between patients with high and low levels of KLF10 would be sufficient.
Formalin-fixed and paraffin-embedded gastric cancer specimens were obtained from patients who underwent surgical resection at the Department of Surgery, Changhua Christian Hospital, and tissue blocks were obtained from the Department of Surgical Pathology, Changhua Christian Hospital. Stages and grades were classified in accordance with the TNM and World Health Organization classification systems. All samples were arranged in tissue array blocks. Histopathological and clinical data were obtained from the cancer registry of Changhua Christian Hospital. Disease-free survival was measured as the time interval between surgical operation and either the date of death or the end of follow-up. This research was approved by the internal review board of Changhua Christian Hospital.
2.2. Tissue Microarrays
A tissue core section (2 mm in diameter) from cancer and tumor-adjacent normal part tissues in each paraffin block was longitudinally cut and arranged into new paraffin blocks by using a fine steel needle to generate tissue microarrays (TMAs). A 4 µm section was stained with hematoxylin and eosin to confirm the presence of the morphologically representative areas of the original cancers.
2.3. Analysis of KLF10 Expression via Immunohistochemistry (IHC)
IHC analysis was performed by using the streptavidin peroxidase protocol. Briefly, 4 µm sections of the paraffin-embedded cancer tissues and paired noncancerous tissue sections were deparaffinized. After blocking with 3% H2O2, the sections were rehydrated, and the antigen was retrieved by heating at 100 °C for 20 min in 10 mM citrate buffer (pH 6.0). After incubation with the anti-KLF10 antibody (SC-130408, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 20 min at room temperature, the slides were incubated with a horseradish peroxidase/Fab polymer conjugate for an additional 30 min, then washed thoroughly three times with PBS. Color was developed by using 3,3′-diamino-benzidine tetrahydrochloride as a chromogen, and the samples were counterstained with hematoxylin. Appropriate positive and negative controls were also included in IHC staining. After staining, the slides were scored by two pathologists. The paraffin-embedded and fresh frozen sections of the normal colonic epithelium of a homogeneous immunophenotype for the studied antigens were included as the positive controls. Negative controls had the primary antibody omitted and replaced with PBS. The intensity of the cytoplasmic and nuclear staining of the KLF10 protein was scored semiquantitatively in accordance with the percentage of positive cells in individual lesions. Here, 0–10% KLF10 protein staining was defined as negative (−), and greater than 10% KLF10 protein staining was defined as positive (+) as previously described [22]. The staining intensity of the cytoplasm was scored from 0 to 4 as previously reported. Intensity was classified as either weak (<2) or strong (≥2) as previously described [23].
2.4. Statistical Analysis
The relationship between the KLF10 expression level and different clinicopathological parameters was determined through χ2 and Fisher exact tests. The overall survival rate was analyzed via Kaplan–Meier plot and log-rank test. Multivariate analysis based on a Cox proportional hazard regression model was used to evaluate the prognostic significance of the clinical variables. All statistical analyses were performed by using SPSS statistical software (version 15.0; SPSS Inc., Chicago, IL, USA). p < 0.05 was considered to be statistically significant.
3. Results
3.1. Association of KLF10 Expression with the Clinical Parameters of Patients with Gastric Cancer
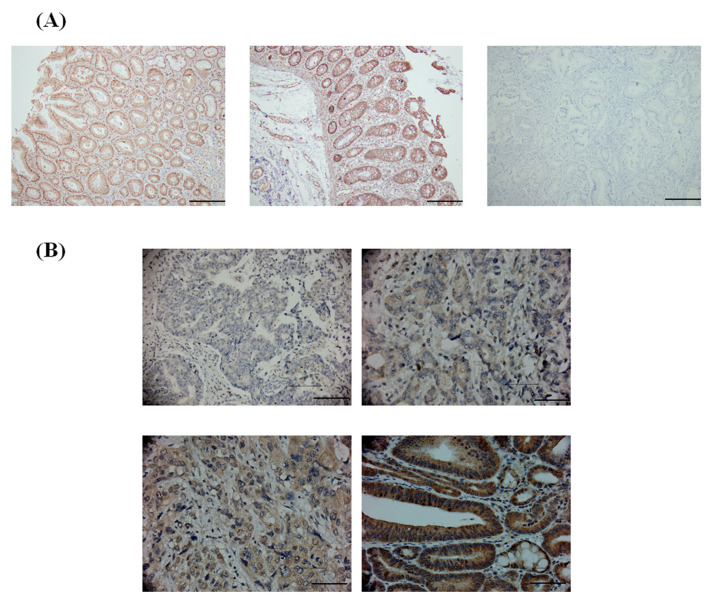
Previous reports have shown that KLF10 may play a critical role in several human cancers [3]. We performed IHC staining by using gastric cancer TMAs to evaluate the relationship between the expression level of KLF10 and the clinical parameters of gastric cancer. The expression level of KLF10 was subdivided into two categories, namely, low and high, on the basis of IHC staining intensity as described in the Section 2 (Figure 1).
Figure 1.
The expression patterns of KLF10 in gastric cancer. (A) Representative Immunohistochemistry (IHC) images of immunohistochemical staining of KLF10 gastric tissues. Left panel: IHC staining of normal gastric tissue. Middle panel: positive IHC staining region in normal gastric tissues. Right: IHC staining without KLF10 antibody in gastric tissues as negative control. All images magnification ×100. Scale bar: 5 µM. (B) Immunohistochemical staining of KLF10 in gastric cancer. KLF10 was positively detected in the nuclear and cytoplasmic regions. Representative IHC intensity of images of negative (upper panel, left), 1+ (upper panel, right), 2+ (lower panel, left) and 3+ (lower panel, right) were shown. All images magnification ×400. Scale bar: 20 µM.
Significantly higher KLF10 expression was found in intestinal and mixed types than in diffused gastric cancer types (p < 0.001). KLF10 expression had an inverse relationship with tumor stages (p = 0.005) as shown in Table 1.
Table 1.
Relationships of KFL10 expression with clinical parameters in 121 gastric cancer patients.
| Parameters | Case Number | KFL10 Expression | p-Value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Age (year) | 69.4 ± 12.0 | 72.4 ± 13.4 | 0.194 | |
| Gender | ||||
| Female | 35 | 20 (57.1) | 15 (42.9) | 0.550 |
| Male | 86 | 44 (51.2) | 42 (48.8) | |
| Type | ||||
| Diffuse | 23 | 21 (91.3) | 2 (8.7) | <0.001 |
| Intestinal | 70 | 32 (45.7) | 38 (54.3) | |
| Mix | 28 | 11 (39.3) | 17 (60.7) | |
| Stage | ||||
| I | 19 | 13 (68.4) | 6 (31.6) | 0.005 |
| II | 31 | 10 (32.3) | 21 (67.7) | |
| III | 49 | 24 (49.0) | 25 (51.0) | |
| IV | 22 | 17 (77.3) | 5 (22.7) | |
| t value | ||||
| 1 | 14 | 8 (57.1) | 6 (42.9) | 0.055 |
| 2 | 16 | 5 (31.3) | 11 (68.8) | |
| 3 | 76 | 39 (51.3) | 37 (48.7) | |
| 4 | 15 | 12 (80.0) | 3 (20.0) | |
| n value | ||||
| 0 | 36 | 21 (58.3) | 15 (41.7) | 0.810 |
| 1 | 29 | 14 (48.3) | 15 (51.7) | |
| 2 | 25 | 12 (48.0) | 13 (52.0) | |
| 3 | 31 | 17 (54.8) | 14 (45.2) | |
| m value | ||||
| 0 | 116 | 61 (52.6) | 55 (47.4) | 0.745 |
| 1 | 5 | 3 (60.0) | 2 (40.0) | |
| Grade | ||||
| Well | 8 | 5 (62.5) | 3 (37.5) | 0.723 |
| Moderate | 45 | 22 (48.9) | 23 (51.1) | |
| Poor | 68 | 37 (54.4) | 31 (45.6) | |
3.2. KLF10 Expression Was Correlated with the Survival of Patients with Gastric Cancer
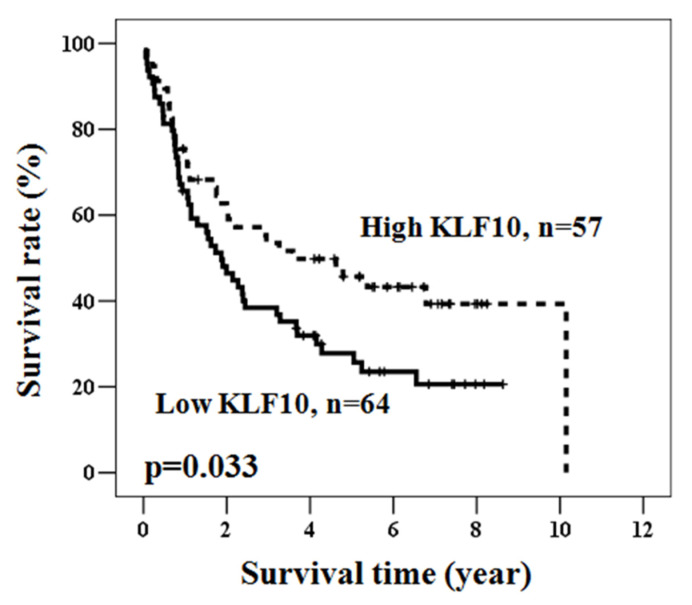
Next, the overall survival rate was analyzed through the Kaplan–Meier method to address whether KLF10 expression level was correlated with patient survival. As shown in Figure 2, overall survival was significantly longer in patients with higher KLF10 expression than in those with low KLF10 expression.
Figure 2.
The prognostic significance of KLF10 in gastric cancer. Kaplan–Meier survival curve for the 121 gastric cancer patients with KLF10 protein expression. Patients with high KLF10 expression had a significantly better overall survival rate compared to patients with low KLF10 expression, as defined by a log-rank test (p = 0.033).
3.3. KLF10 Was an Independent Prognosis Factor of Gastric Cancer
We performed univariate Cox regression analysis to investigate the relationship between possible prognostic factors and patient survival. Advanced stages (hazard ratio = 3.723, 95% confidence interval = 1.615–8.583, p = 0.002) and low KLF10 expression (hazard ratio = 1.623, 95% confidence interval = 1.036–2.544, p = 0.035) were found to be prognostic factors (Table 2). Multivariate Cox regression analysis revealed that age, advanced stages, and low KLF10 expression were independent prognostic factors (Table 3). After adjusting for gender, age, and stage, a high KLF10 expression level was found to be significantly associated with a good overall survival rate of patients with advanced ages (≥71), both genders (female and male), intestinal-type gastric cancer, early stage (I + II), t value, and n value (Table 4).
Table 2.
Univariate analysis of the influence of various parameters on overall survival in 121 gastric cancer patients.
| Parameter | Category | Overall Survival | |||
|---|---|---|---|---|---|
| 5-Year Survival (%) | Hard ratio (HR) | 95% CI | p-Value | ||
| Age (year) | ≥71/<71 | 31.7/41.2 | 1.433 | 0.919–2.233 | 0.112 |
| Gender | Male/female | 31.1/49.3 | 1.613 | 0.971–2.681 | 0.065 |
| Stage | II + III + IV/I | 29.3/72.9 | 3.723 | 1.615–8.583 | 0.002 |
| KFL10 expression | Low/high | 27.8/45.7 | 1.623 | 1.036–2.544 | 0.035 |
Table 3.
Multivariate analysis of the influence of various parameters on overall survival in 121 gastric cancer patients.
| Parameter | Category | Overall Survival | |||
|---|---|---|---|---|---|
| 5-Year Survival (%) | HR | 95% CI | p-Value | ||
| Age (year) | ≥71/<71 | 31.7/41.2 | 2.314 | 1.443–3.712 | <0.001 |
| Gender | Male/female | 31.1/49.3 | 1.561 | 0.935–2.605 | 0.088 |
| Stage | II + III +I V/I | 29.3/72.9 | 5.344 | 2.266–12.599 | <0.001 |
| KFL10 expression | Low/high | 27.8/45.7 | 2.344 | 1.460–3.762 | <0.001 |
Table 4.
Multivariate analysis of the influence of KFL10 expression according to clinical parameters on overall survival in gastric cancer patients.
| Parameter | Overall Survival 1 | |||
|---|---|---|---|---|
| 5-Year Survival (%) | HR | 95% CI | p-Value | |
| All cases | 27.8/45.7 | 2.344 | 1.460–3.762 | <0.001 |
| Age (year) | ||||
| <71 | 32.5/55.7 | 2.117 | 0.984–4.556 | 0.055 |
| ≥71 | 21.3/39.3 | 2.637 | 1.436–4.844 | 0.002 |
| Gender | ||||
| Female | 41.7/59.3 | 3.001 | 1.129–7.977 | 0.028 |
| Male | 22.7/40.7 | 2.085 | 1.218–3.570 | 0.007 |
| Type | ||||
| Diffuse | 19.0/0.0 | 1.373 | 0.286–6.596 | 0.692 |
| Intestinal | 36.9/57.1 | 2.565 | 1.326–4.960 | 0.005 |
| Mix | 16.4/23.2 | 2.199 | 0.700–6.906 | 0.177 |
| Stage | ||||
| I + II | 45.2/66.2 | 5.138 | 2.064–12.793 | <0.001 |
| III + IV | 18.8/25.9 | 1.467 | 0.815–2.642 | 0.201 |
| t value | ||||
| 1 + 2 | 59.3/70.6 | 22.757 | 3.377–153.357 | 0.001 |
| 3 + 4 | 19.8/34.6 | 1.762 | 1.051–2.953 | 0.032 |
| n value | ||||
| 0 | 44.4/73.3 | 4.676 | 1.447–15.111 | 0.010 |
| 1 + 2 + 3 | 20.3/35.5 | 1.898 | 1.117–3.227 | 0.018 |
1 Adjusted for age, gender, and stage.
4. Discussion
Gastric cancer is a high-mortality cancer in the Asian region [18]. KLF family proteins play an important role in several biological functions [5]. KLF10 may also be associated with several diseases [3]. In this study, IHC staining demonstrated that the loss of KLF10 was associated with advanced tumor stages and short survival. Univariate and multivariate analyses also revealed that KLF10 could be a prognostic factor of gastric cancer. Moreover, in the gastric cancer line, KLF10 overexpression altered the Bcl-2/Bax ratio, caused caspase-3 activation, and finally, caused apoptosis.
Decreased KLF10 expression has been found in several human cancers. Gene expression profiling analysis indicated that KLF10 expression is low in metastatic brain tumors [24]. Chang et al. demonstrated that KLF10 expression is downregulated by hypermethylation in pancreatic cancer [25]. The loss of KLF10 expression is correlated with pancreatic cancer stages. Multiple logistic regression analysis also revealed that KLF10 is an independent prognostic factor of prognosis-free survival and overall survival in pancreatic cancer [25]. In mice, KLF10 knockout facilitates skin tumorigenesis and papilloma formation in response to DMBA/TPA treatment [26]. Very recently, Yeh et al. demonstrated that KLF10 is an independent prognosis factor for oral squamous cell carcinoma [13]. In line with these observations, our findings indicated that KLF10 was low in gastric cancers with advanced stages. In addition, KLF10 was an independent factor in the overall survival of patients with gastric cancers. Heo et al. reported lower tumor incidence and proliferation of cell nuclear antigen in the liver of KLF10 null mice exposed to N-nitrosodiethylamine than in wild-type mice [27]. Our findings collectively suggested that KLF10 may be a tumor suppressor in gastric cancer.
Several proteins have been shown to be differentially expressed in the diffuse and intestinal types of gastric cancers. B cell translocation gene 1 is more likely expressed in the intestinal type than in the diffuse type of gastric cancer [28,29]. The mRNA of mitochondrial cytochrome c oxidase I and NADH dehydrogenase 4 is predominantly expressed in diffuse-type gastric cancer [30]. The loss of the expression of the breast cancer type 1 susceptibility protein is associated with the diffuse type of gastric cancer, whereas the loss of mediator of DNA damage checkpoint protein 1 is correlated with the intestinal type of gastric cancer [31]. Moreover, reports from our laboratory indicated that distinct KLF expression patterns are found in different types of gastric cancer. High nuclear KLF8 expression is observed in the diffuse type of gastric cancer [20]. In this study, KLF10 expression was significantly higher in intestinal-type gastric cancer than in other types. After adjusting for gender, age, and stage, KLF10 expression was found to be an independent prognostic factor of intestinal gastric cancer. Tanabe et al. found extensive epithelial-to-mesenchymal transition (EMT) in the diffuse type, but not the intestinal type, of gastric cancer [32]. This phenomenon results in the poor prognosis of diffuse-type gastric cancer [32]. By contrast, silencing KLF10 leads to the suppression of EMT protein expression [33]. The exact role of KLF10 in the development of intestinal-type gastric cancer remains to be investigated. Our results collectively revealed that KLF10 may be involved in the carcinogenesis of gastric cancer, especially intestinal-type gastric cancer. Since the pathogenesis of gastric cancer involves several steps and several factors/co-factors, future studies should focalize on the relationship between KLF10 expression and some of these factors, for example, Helicobacter pylori infection.
5. Conclusions
In conclusion, our findings provided the first piece of evidence supporting the role of KLF10 in gastric carcinogenesis. KLF10 expression was inversely correlated with tumor stages. Furthermore, KLF10 expression was low in the diffuse type of gastric cancer. KLF10 was also an independent prognostic factor of gastric cancer, especially the intestinal type of gastric cancer.
Author Contributions
Conceptualization, Y.-M.L. and K.-T.Y.; data curation, K.-T.Y. and C.-M.Y.; formal analysis, C.-M.Y.; funding acquisition, M.-S.S. and K.-T.Y.; investigation, C.-M.Y. and M.-S.S.; methodology, Y.-M.L. and K.-T.Y.; supervision, L.-S.H.; validation, Y.-M.L. and C.-M.Y.; writing—original draft, L.-S.H.; writing—review and editing, K.-T.Y. and L.-S.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Institutional Review Board and Ethics Committee of the Changhua Christian Hospital, Changhua, Taiwan (IRB no. 131014).
Informed Consent Statement
These tissues were obtained from established tissue bank, the informed consents were waived.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work is supported by Changhua Christian Hospital, 102-CCH-IRP-006.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zakeri S., Aminian H., Sadeghi S., Esmaeilzadeh-Gharehdaghi E., Razmara E. Kruppel-like factors in bone biology. Cell Signal. 2022;93:110308. doi: 10.1016/j.cellsig.2022.110308. [DOI] [PubMed] [Google Scholar]
- 2.Nicoleau S., Fellows A., Wojciak-Stothard B. Role of Kruppel-like factors in pulmonary arterial hypertension. Int. J. Biochem. Cell Biol. 2021;134:105977. doi: 10.1016/j.biocel.2021.105977. [DOI] [PubMed] [Google Scholar]
- 3.Subramaniam M., Hawse J.R., Johnsen S.A., Spelsberg T.C. Role of TIEG1 in biological processes and disease states. J. Cell. Biochem. 2007;102:539–548. doi: 10.1002/jcb.21492. [DOI] [PubMed] [Google Scholar]
- 4.Taracha-Wisniewska A., Kotarba G., Dworkin S., Wilanowski T. Recent Discoveries on the Involvement of Kruppel-Like Factor 4 in the Most Common Cancer Types. Int. J. Mol. Sci. 2020;21:8843. doi: 10.3390/ijms21228843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Li G., Feng L., Lu H., Wang X. Kruppel-like factors in breast cancer: Function, regulation and clinical relevance. Biomed. Pharm. 2020;123:109778. doi: 10.1016/j.biopha.2019.109778. [DOI] [PubMed] [Google Scholar]
- 6.Subramaniam M., Hawse J.R., Rajamannan N.M., Ingle J.N., Spelsberg T.C. Functional role of KLF10 in multiple disease processes. Biofactors. 2010;36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang L., Chen Y., Chan C.Y., Wang X., Lin L., He M.L., Lin M.C., Yew D.T., Sung J.J., Li J.C., et al. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009;274:101–108. doi: 10.1016/j.canlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Hsu C.F., Sui C.L., Wu W.C., Wang J.J., Yang D.H., Chen Y.C., Yu W.C., Chang H.S. Klf10 induces cell apoptosis through modulation of BI-1 expression and Ca2+ homeostasis in estrogen-responding adenocarcinoma cells. Int. J. Biochem. Cell Biol. 2011;43:666–673. doi: 10.1016/j.biocel.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Jin W., Chen B.B., Li J.Y., Zhu H., Huang M., Gu S.M., Wang Q.Q., Chen J.Y., Yu S., Wu J., et al. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol. Cell Biol. 2012;32:50–63. doi: 10.1128/MCB.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Q., Yan D., Dong W., Bi J., Huang L., Yang M., Huang J., Qin H., Lin T. circRNA circFUT8 Upregulates Krupple-like Factor 10 to Inhibit the Metastasis of Bladder Cancer via Sponging miR-570-3p. Mol. Oncolytics. 2020;16:172–187. doi: 10.1016/j.omto.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng C.C., Hawse J.R., Subramaniam M., Chang V.H.S., Yu W.C.Y., Hung W.C., Chen L.T., Cheng K.H. KLF10 loss in the pancreas provokes activation of SDF-1 and induces distant metastases of pancreatic ductal adenocarcinoma in the Kras(G12D) p53(flox/flox) model. Oncogene. 2017;36:5532–5543. doi: 10.1038/onc.2017.155. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M., Chen J., Zhang H., Liu H., Yao H., Wang X., Zhang W., Zhao Y., Yang N. KLF10 inhibits cell growth by regulating PTTG1 in multiple myeloma under the regulation of microRNA-106b-5p. Int. J. Biol. Sci. 2020;16:2063–2071. doi: 10.7150/ijbs.45999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh C.M., Lee Y.J., Ko P.Y., Lin Y.M., Sung W.W. High Expression of KLF10 Is Associated with Favorable Survival in Patients with Oral Squamous Cell Carcinoma. Medicina. 2020;57:17. doi: 10.3390/medicina57010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Li J., Yao L., Wu L. Effect of Photodynamic Therapy on Gemcitabine-Resistant Cholangiocarcinoma in vitro and in vivo Through KLF10 and EGFR. Front. Cell Dev. Biol. 2021;9:710721. doi: 10.3389/fcell.2021.710721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pen S.L., Shan Y.S., Hsiao C.F., Liu T.W., Chen J.S., Ho C.L., Chou W.C., Hsieh R.K., Chen L.T., Ch’ang H.J. High expression of kruppel-like factor 10 or Smad4 predicts clinical benefit of adjuvant chemoradiotherapy in curatively resected pancreatic adenocarcinoma: From a randomized phase III trial. Radiother. Oncol. 2021;158:146–154. doi: 10.1016/j.radonc.2021.02.037. [DOI] [PubMed] [Google Scholar]
- 16.Eid M.A., Kumar M.V., Iczkowski K.A., Bostwick D.G., Tindall D.J. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998;58:2461–2468. [PubMed] [Google Scholar]
- 17.Ivanov S.V., Ivanova A.V., Salnikow K., Timofeeva O., Subramaniam M., Lerman M.I. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem. Biophys. Res. Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivey A., Pratt H., Boone B.A. Molecular pathogenesis and emerging targets of gastric adenocarcinoma. J. Surg. Oncol. 2022;125:1079–1095. doi: 10.1002/jso.26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin X., Zhao Y., Song W.M., Zhang B. Molecular classification and prediction in gastric cancer. Comput. Struct. Biotechnol. J. 2015;13:448–458. doi: 10.1016/j.csbj.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu L.S., Wu P.R., Yeh K.T., Yeh C.M., Shen K.H., Chen C.J., Soon M.S. Positive nuclear expression of KLF8 might be correlated with shorter survival in gastric adenocarcinoma. Ann. Diagn. Pathol. 2014;18:74–77. doi: 10.1016/j.anndiagpath.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Soon M.S., Hsu L.S., Chen C.J., Chu P.Y., Liou J.H., Lin S.H., Hsu J.D., Yeh K.T. Expression of Kruppel-like factor 5 in gastric cancer and its clinical correlation in Taiwan. Virchows. Arch. 2011;459:161–166. doi: 10.1007/s00428-011-1111-0. [DOI] [PubMed] [Google Scholar]
- 22.Hsu H.T., Wu P.R., Chen C.J., Hsu L.S., Yeh C.M., Hsing M.T., Chiang Y.S., Lai M.T., Yeh K.T. High cytoplasmic expression of Kruppel-like factor 4 is an independent prognostic factor of better survival in hepatocellular carcinoma. Int. J. Mol. Sci. 2014;15:9894–9906. doi: 10.3390/ijms15069894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandya A.Y., Talley L.I., Frost A.R., Fitzgerald T.J., Trivedi V., Chakravarthy M., Chhieng D.C., Grizzle W.E., Engler J.A., Krontiras H., et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin. Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.CCR-03-0484. [DOI] [PubMed] [Google Scholar]
- 24.Zohrabian V.M., Nandu H., Gulati N., Khitrov G., Zhao C., Mohan A., Demattia J., Braun A., Das K., Murali R., et al. Gene expression profiling of metastatic brain cancer. Oncol. Rep. 2007;18:321–328. doi: 10.3892/or.18.2.321. [DOI] [PubMed] [Google Scholar]
- 25.Chang V.H., Chu P.Y., Peng S.L., Mao T.L., Shan Y.S., Hsu C.F., Lin C.Y., Tsai K.K., Yu W.C., Ch’ang H.J. Kruppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am. J. Pathol. 2012;181:423–430. doi: 10.1016/j.ajpath.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Song K.D., Kim D.J., Lee J.E., Yun C.H., Lee W.K. KLF10, transforming growth factor-beta-inducible early gene 1, acts as a tumor suppressor. Biochem. Biophys. Res. Commun. 2012;419:388–394. doi: 10.1016/j.bbrc.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Heo S.H., Jeong E.S., Lee K.S., Seo J.H., Lee W.K., Choi Y.K. Kruppel-like factor 10 null mice exhibit lower tumor incidence and suppressed cellular proliferation activity following chemically induced liver tumorigenesis. Oncol. Rep. 2015;33:2037–2044. doi: 10.3892/or.2015.3801. [DOI] [PubMed] [Google Scholar]
- 28.Kanda M., Oya H., Nomoto S., Takami H., Shimizu D., Hashimoto R., Sueoka S., Kobayashi D., Tanaka C., Yamada S., et al. Diversity of clinical implication of B-cell translocation gene 1 expression by histopathologic and anatomic subtypes of gastric cancer. Dig. Dis. Sci. 2015;60:1256–1264. doi: 10.1007/s10620-014-3477-8. [DOI] [PubMed] [Google Scholar]
- 29.Zheng H.C., Li J., Shen D.F., Yang X.F., Zhao S., Wu Y.Z., Takano Y., Sun H.Z., Su R.J., Luo J.S., et al. BTG1 expression correlates with pathogenesis, aggressive behaviors and prognosis of gastric cancer: A potential target for gene therapy. Oncotarget. 2015;6:19685–19705. doi: 10.18632/oncotarget.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J.T., Han C.B., Zhou Y., Zhao J.Z., Jing W., Zou H.W. Altered expression of mitochondrial cytochrome c oxidase I and NADH dehydrogenase 4 transcripts associated with gastric tumorigenesis and tumor dedifferentiation. Mol. Med. Rep. 2012;5:1526–1530. doi: 10.3892/mmr.2012.832. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z.Z., Liu Y.J., Yin X.L., Zhan P., Gu Y., Ni X.Z. Loss of BRCA1 expression leads to worse survival in patients with gastric carcinoma. World J. Gastroenterol. 2013;19:1968–1974. doi: 10.3748/wjg.v19.i12.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanabe S., Aoyagi K., Yokozaki H., Sasaki H. Gene expression signatures for identifying diffuse-type gastric cancer associated with epithelial-mesenchymal transition. Int. J. Oncol. 2014;44:1955–1970. doi: 10.3892/ijo.2014.2387. [DOI] [PubMed] [Google Scholar]
- 33.Venkov C., Plieth D., Ni T., Karmaker A., Bian A., George A.L., Jr., Neilson E.G. Transcriptional networks in epithelial-mesenchymal transition. PLoS ONE. 2011;6:e25354. doi: 10.1371/journal.pone.0025354. [DOI] [PMC free article] [PubMed] [Google Scholar]