Abstract
The extension of ecological tolerance limits may be an important mechanism by which microorganisms adapt to novel environments, but it may come at the evolutionary cost of reduced performance under ancestral conditions. We combined a comparative physiological approach with phylogenetic analyses to study the evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Among the 20 laboratory clones of Synechococcus isolated from collections made along an Oregon hot spring thermal gradient, four different 16S rRNA gene sequences were identified. Phylogenies constructed by using the sequence data indicated that the clones were polyphyletic but that three of the four sequence groups formed a clade. Differences in thermotolerance were observed for clones with different 16S rRNA gene sequences, and comparison of these physiological differences within a phylogenetic framework provided evidence that more thermotolerant lineages of Synechococcus evolved from less thermotolerant ancestors. The extension of the thermal limit in these bacteria was correlated with a reduction in the breadth of the temperature range for growth, which provides evidence that enhanced thermotolerance has come at the evolutionary cost of increased thermal specialization. This study illustrates the utility of using phylogenetic comparative methods to investigate how evolutionary processes have shaped historical patterns of ecological diversification in microorganisms.
Has adaptation to novel environments by the extension of tolerance limits historically been an important mechanism by which the ecological requirements of microorganisms have diverged? While the breadth of physiological diversity exhibited by microorganisms suggests that this may be the case, the relevance of this mechanism is yet to be firmly established. Similarly, little is known about the consequences of this mode of adaptation for fitness in ancestral environments. For example, are there trade-offs or costs of adaptation such that there is a correlated loss in performance under ancestral conditions? Providing answers to the above questions may help microbiologists to better make sense of extant microbial diversity and its distribution on the planet.
The cyanobacteria provide an excellent system for testing hypotheses concerning the evolution of tolerance. During its evolutionary history, this ancient lineage of oxygen-evolving, photoautotrophic bacteria has established itself in diverse aquatic and terrestrial habitats exhibiting wide ranges in temperature, salinity, water potential, pH, and irradiance (36). An example of ecological diversification in the cyanobacteria is the invasion of alkaline hot spring habitats across western North America, Asia, Africa, and possibly Europe by members of the genus Synechococcus (8). Hot spring outflows typically exhibit marked temperature gradients, and microbial communities containing Synechococcus generally develop in these systems at temperatures between ∼45 and 73°C, the thermal maximum for photosynthetic life (3, 4). Peary and Castenholz (26) demonstrated with laboratory isolates that at least four temperature strains of Synechococcus inhabited a single channel at Hunter's Hot Springs in Oregon. It was later hypothesized that more thermotolerant Synechococcus strains evolved from less thermotolerant ancestors growing at lower temperatures (5). It was not possible to test this hypothesis, however, because the data required to establish the branching order of the temperature strains in a phylogeny (i.e., their order of evolutionary divergence) were not available.
Conversely, substantial cyanobacterial molecular diversity has been identified among environmental 16S rRNA and rRNA gene fragment sequences recovered from microbial mat communities in Octopus Spring in Yellowstone National Park (32). Among these, phylogenetically similar sequences which clearly belong to Synechococcus have distribution patterns consistent with the hypothesis that genotypically distinct lines have diverged in temperature range (11, 13). However, while it is possible to gain an understanding of genealogical patterns from these molecular data, the lack of phenotypic data for the organisms limits the testing of evolutionary ecological hypotheses.
The ability to infer the patterns and processes of past ecological diversification therefore depends upon both the availability of comparative phenotypic data for the study organisms and an understanding of their phylogenetic relatedness. We have combined comparative physiology of laboratory isolates with phylogenetic approaches in order to investigate the evolution of thermotolerance in Synechococcus from Hunter's Hot Springs in Oregon. We first reconstructed molecular phylogenies to infer the evolutionary relationships of Synechococcus temperature strains isolated in laboratory culture from Hunter's Hot Springs. We next collected comparative growth rate data for these isolates at a series of temperatures in order to distinguish four groups of strains based on both identity of 16S rRNA gene sequence data and thermotolerance characteristics. From the phylogeny we then determined the order of evolutionary divergence of groups of strains with different thermotolerances to test the hypothesis that more thermotolerant strains of Synechococcus evolved from less thermotolerant ancestors. Finally, we incorporated phylogenetic information into a statistical analysis of the comparative physiological data in order to reveal possible phenotypic trade-offs during the evolution of increased thermotolerance.
MATERIALS AND METHODS
Sample collection, enrichment, and clone isolation.
Hunter's Hot Springs comprises a cluster of several alkaline thermal sources located ∼3 km north of Lakeview, Oreg. Between 1994 and 1998, several collections of microbial mat were taken at four temperatures (45, 55, 65, and 70°C) along one of the outflow channels (Jack's Spring). For each collection, the upper, Synechococcus-containing component of the mat was harvested with a sterile 10-ml syringe, transferred to a sterile 20-ml scintillation vial, and stored at ambient temperature in the dark until it was taken to the University of Oregon.
The general procedure used for enrichment and isolation of Synechococcus clones in laboratory culture was as follows. Each sample was homogenized in the laboratory with a sterile syringe, and the density of Synechococcus cells in the resulting cell suspension was estimated with a hemocytometer. The suspension was diluted serially by successive transfers of 1 ml of suspension into 9 ml of liquid BG11 medium (7) to create a series of suspensions ranging in density from 101 to 107 cells ml−1. Aliquots (1 ml) from each tube in the series were used to inoculate individual liquid enrichment flasks (75 ml) containing D medium and BG11 medium (7). Most of the enrichment flasks were incubated at the collection temperature; the only exceptions were the flasks prepared from 70°C collections, which were incubated at 67°C. This temperature had previously been found to be the upper limit of the optimal temperature range for laboratory cultures of the high-temperature form of Synechococcus cf. lividus (22). To minimize evaporation during enrichment, isolation, and maintenance of Synechococcus cultures at 65 and 67°C, metal-capped Bellco flasks were used. Enrichments grown at 45 and 55°C were incubated in incubators with an irradiance of 120 microeinsteins m−2 s−1 (provided by cool-white fluorescent lamps), while 65 and 67°C enrichments were incubated in water baths with an irradiance of 30 microeinsteins m−2 s−1 (provided by very-high-output, cool-white fluorescent lamps).
Isolation attempts were made with both the most- and least-diluted positive enrichments in a series. Synechococcus clones were isolated from 45 and 55°C enrichments after at least three rounds of picking and restreaking of isolated colonies on 1.5% agar plates containing the appropriate medium. Clones were obtained from 65 and 67°C enrichments by using the filter isolation method of Meeks and Castenholz (22). Laboratory clones were maintained under the same conditions as the enrichments.
The isolation procedure was different for one 55°C clone (OH2). In this case, 1 drop of a homogenized cell suspension made from a collection taken in March 1996 was spread on an agar plate. Individual Synechococcus cells were visualized under a dissecting microscope, transferred to test tubes containing 7 ml of either D or BG11 medium, and incubated as described above. Ten isolations were attempted for each medium. One of the BG11 tubes was successful, and clone OH2 was obtained from this tube after repeated plating as described above.
DNA isolation, amplification, and sequencing.
Genomic DNA of clones were isolated as described by Pitcher et al. (27). Fragments (ca. 950 bp) of the 16S rRNA gene were amplified from these DNA preparations. This gene was chosen because the database for it is the most extensive database among those for cyanobacterial loci, which facilitated adequate taxon sampling and placement of the Synechococcus strains within the context of the cyanobacterial phylogeny. The 50-μl amplification reaction mixtures contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.12 mM MgCl2, 1.25 U of Taq polymerase (Perkin-Elmer), ∼10 ng of genomic DNA, 0.2 μM primer CYA359F (25), and 0.2 μM primer PLG2.3 (5′CTTCA[C/T]G[C/T]AGGCGAGTTGCAGC3′), a modification of PLG2.1 (31). The reaction conditions used were 40 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. Amplification products were purified with a QIAquick-spin PCR purification kit (Qiagen), and cleaned amplification products were directly sequenced with an ABI Prism 377 sequencer at the DNA Sequencing Facility, University of Oregon.
Sequence alignment.
16S rRNA gene sequences were preliminarily aligned with Clustal W1.6 (29). Since the stem-loop structure of the mature 16S rRNA molecule contains information with respect to nucleotide positional homology, secondary structure was then syntactically imposed on each sequence for the fragment spanning Escherichia coli nucleotide positions 360 to 1326 as described by Titus and Frost (30). Previously published secondary-structure models aided identification of stem-encoding nucleotides of cyanobacterial and outgroup sequences (14, 37). A final pairwise alignment was made by using Malign 2.7 (35) with a gap cost to substitution cost of 3. Missing data were treated as unknown nucleotides.
Phylogeny reconstruction.
Phylogenetic trees were constructed by using three optimality criteria. A neighbor-joining tree was built with MEGA 1.01 (20). For calculation of a distance matrix, Kimura's two-parameter model was assumed (19), and nucleotide positions containing gaps or missing data were deleted in a pairwise fashion. The tree was inferred with pairwise deletion of gaps and with 1,000 bootstrap pseudoreplicates. A maximum-parsimony phylogeny was constructed by using PAUP 3.1.1 (28). A heuristic search was performed by using the tree bisection-reconnection branch-swapping algorithm. Starting trees were obtained by stepwise addition of sequences and 10 replications of random sequence addition. The analysis was bootstrap pseudoreplicated 100 times. A maximum-likelihood tree was created with the “dnaml” program in PHYLIP 3.5c (10) by using a transition-to-transversion ratio of 2, sequential addition of sequences, and 100 bootstrap pseudoreplicates. All trees were rooted with Aquifex pyrophilus.
Determination of clone growth temperature ranges.
Exponential growth rates were estimated at intervals along a clone's thermal range for growth by using the following method. Beginning at the clone's maintenance temperature, the growth rate was determined as described below. The experimental temperature was then shifted up, usually by 5°C, and cells were transferred to fresh medium as described below. Near the predicted upper thermal limit for the clone, based on data for temperature strains of Synechococcus from a previous study (26), the magnitude of the shift was decreased in order to resolve differences among clones with respect to thermal maxima. Growth rates were again estimated, and this procedure was repeated until the clone's lethal temperature was reached, as indicated by a lack of growth and cell bleaching. To evaluate clone performance at temperatures below that used for routine maintenance, an analogous procedure was employed with thermal down-shifts accompanying clone transfer.
For clones of 16S rRNA gene sequence group I (Table 1), the experiment was initiated by inoculating duplicate flasks with cells from an early-stationary-phase batch culture grown under standard maintenance conditions. Cells were concentrated by centrifugation and resuspended in fresh medium to an A750 of ∼0.75. Aliquots (1 ml) of this suspension were delivered to duplicate 125-ml Erlenmeyer flasks containing 75 ml of the appropriate medium to obtain an initial experimental A750 of ∼0.01. The flasks were supplemented with 1 ml of 0.5 M NaHCO3, previously found to stimulate growth in several Synechococcus clones (unpublished data), and were incubated at the experimental temperature under 120 microeinsteins of cool-white fluorescent light m−2 s−1. The positions of the flasks in the incubator were randomized every 24 h. The population densities in the flasks were monitored spectrophotometrically by determining the increase in A750. Between four and five generations of exponential population growth were supported under the above conditions. The generation time during exponential growth was estimated by determining log 2/b, where b is the slope of logarithmically transformed A750 data regressed on time (in hours). This value was transformed and reported as number of population doublings per day. The experimental temperature was then shifted up or down as described above. After 4 h, one of the experimental replicates was randomly chosen and transferred to duplicate flasks as described above. The flasks were then used to determine the growth rate at the new temperature.
TABLE 1.
16S rRNA gene sequence groups of Synechococcus strains isolated from Hunter's Hot Springs in Oregon
| Sequence groupa | Clone | Collectionb
|
Isolation
|
|||
|---|---|---|---|---|---|---|
| Temp (°C) | Date | Inoculum (no. of cells) | Medium | Temp (°C) | ||
| I | OH5 | 45 | 1 Oct. 1994 | 10 | BG11 | 45 |
| OH9 | 45 | 1 Oct. 1994 | 10 | D | 45 | |
| OH10 | 45 | 30 May 1995 | 105 | BG11 | 45 | |
| OH13 | 45 | 1 Oct. 1994 | 10 | BG11 | 45 | |
| OH14 | 45 | 30 May 1995 | 10 | BG11 | 45 | |
| OH18 | 45 | 1 Oct. 1994 | 10 | D | 45 | |
| OH12 | 55 | 1 Oct. 1994 | 106 | BG11 | 55 | |
| OH16 | 55 | 1 Oct. 1994 | 106 | BG11 | 55 | |
| OH19 | 55 | 1 Oct. 1994 | 106 | BG11 | 55 | |
| OH26 | 55 | 9 Sept. 1998 | 104 | BG11 | 55 | |
| II | OH4 | 55 | 30 May 1995 | 10 | BG11 | 55 |
| OH20 | 55 | 30 May 1995 | 107 | BG11 | 55 | |
| OH31 | 55 | 9 Sept. 1998 | 104 | D | 55 | |
| OH32 | 55 | 9 Sept. 1998 | 102 | BG11 | 55 | |
| III | OH2 | 55 | 9 Mar. 1996 | 1c | BG11 | 55 |
| IV | OH28 | 70 | 9 Sept. 1998 | 107 | D | 67 |
| OH29 | 70 | 9 Sept. 1998 | 107 | D | 67 | |
| OH30 | 70 | 20 Mar. 1998 | 107 | BG11 | 67 | |
| OH33 | 65 | 9 Sept. 1998 | 104 | D | 65 | |
| OH34 | 65 | 9 Sept. 1998 | 104 | D | 65 | |
Clones belonging to the same sequence group have identical sequences.
Most samples were obtained from Jack's Spring; the only exception was the 55°C sample collected on 1 October 1994, which was collected from an adjacent outflow.
See Materials and Methods.
The procedure used to determine the growth rates of clones belonging to all other 16S rRNA sequence groups (Table 1) was the same as the procedure described above except that no bicarbonate was added to the experimental flasks. This was because bicarbonate had an unexpected deleterious effect on the growth rates of clones isolated from the 70°C collections (sequence group IV) (Table 1). Due to the difference in procedure, the growth rate data obtained for the group I clones were not included in statistical analyses of data obtained for clones belonging to the other 16S rRNA gene sequence groups.
Statistical analyses.
Growth rate data were analyzed with analysis of variance (ANOVA) models, and pairwise comparisons of means were made by using Bonferroni tests with a significance level of α = 0.05. All analyses were done with SPSS Base 8.0 (SPSS Inc.).
Correlations between thermotolerance traits were estimated after the data were transformed by using the method of independent contrasts (9), a phylogenetic comparative method for comparing two or more continuously distributed variables. Phylogenetic comparative methods (16, 21) incorporate information from phylogenies into the analysis of comparative phenotypic data both to ensure that the data meet the assumptions of the statistical methods used to analyze them and to facilitate the investigation of past evolution by using data collected from extant organisms. The method of independent contrasts accounts for the statistical problem of nonindependence of comparative data which arises due to shared evolutionary histories of taxa. It assumes a Brownian motion model of phenotypic evolution, i.e., that the phylogeny branch lengths in units of sequence divergence are proportional to the expected amount of phenotypic divergence. Brownian motion models the pattern of phenotypic change expected for characters evolving under random genetic drift or directional selection (15). To estimate the expected amount of phenotypic change, we used the branch lengths inferred from our phylogeny reconstructions. Correlations between the contrasts were then estimated, and t tests with 2 degrees of freedom were used to test whether estimated correlation coefficients were significantly different from zero at the α = 0.05 level.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence data obtained for the Synechococcus strains used in this study have been deposited in the GenBank database under accession numbers AF285244 to AF285253 (group I), AF285240 to AF285243 (group II), AF285259 (group III), AF285254 to AF285258 (Group IV), and AF285260 (Synechococcus sp. strain SH-94-5).
The GenBank nucleotide sequence accession numbers for the taxa used in phylogenetic analyses are as follows: Aquifex pyrophilus, M83548; Escherichia coli, J01859; Bacillus subtilis, X60646; Synechococcus sp. strain PCC 6307, AF001477; Synechococcus sp. strain PCC 6301, X03538; Leptolyngbya sp. strain PCC 73110, X84810; Microcoleus sp. strain PCC 7420, X70770; “Mastigocladus” sp. strain PCC 7518, X68780; Nostoc sp. strain PCC 7120, X59559; Nostoc sp. strain PCC 73102, AF027655; Prochloron sp., X63141; Pleurocapsa sp. strain PCC 7516, X78681; Synechococcus sp. strain PCC 7002, D88289; Spirulina sp. strain PCC 6313, X75044; Microcystis sp. strain PCC 7941, U40340; Synechococcus sp. strain PCC 6803, D64000; and Synechococcus sp. strain C9, L35481 to L35483). Sequences for Chamaesiphon sp. strain PCC 7430, Chroococcidiopsis sp. strain PCC 7203, and Gloeobacter PCC 7421 were obtained from the Ribosomal Database Project II (http://www.cme.msu.edu/RDP).
RESULTS
Synechococcus clones isolated from Hunter's Hot Springs.
Twenty Synechococcus clones were isolated in two kinds of mineral salts media from samples collected at four temperatures at Hunter's Hot Springs (Table 1). Isolates were obtained from both diluted and undiluted enrichments (Table 1). Sixteen of these clones were obtained from along the temperature gradient of a single outflow channel (Jack's Stream). Clones OH12, OH16, and OH19 were derived from an adjacent site at Hunter's Hot Springs sampled in October 1994. At this time central Oregon was experiencing drought conditions, and Jack's Stream was dry at temperatures above ∼45°C. By the following collection date in May 1995, the source of Jack's Stream was flowing again, and a microbial mat had become reestablished at higher temperatures.
16S rRNA gene sequence groups of Hunter's Hot Springs Synechococcus clones.
All of the clones isolated in this study fall into four groups based on 100% sequence identity of an ∼950-bp fragment of the 16S rRNA gene. All Synechococcus clones isolated from 45°C collections, as well as some clones obtained from 55°C collections, have identical sequences (group I) (Table 1). Clones representing two additional sequence groups, groups II and III, were isolated from 55°C collections, while all isolates from samples collected at 65°C or above have identical sequences (group IV) (Table 1).
While the 16S rRNA gene sequence data for group I Synechococcus isolates exhibit no more than 89.2% identity to the data for all other sequence groups obtained in this study, the latter groups exhibit lower levels of sequence divergence. Groups II and IV differ at 44 (4.7%) of the 944 nucleotide positions used for phylogenetic analysis. The level of dissimilarity between groups II and III is 3.2% (30 of 944 positions), and the level of dissimilarity between groups III and IV is 1.8% (17 of 944 positions).
In addition, none of the sequences of the Hunter's Hot Springs isolates is identical to any environmentally derived cyanobacterial 16S rRNA gene sequence from Octopus Spring in Yellowstone National Park (11, 13, 34), although the group II, III, and IV sequences exhibit a high degree of similarity to these sequences. If comparisons are restricted to a 174-bp fragment (E. coli nucleotide positions 1146 to 1319) for which data are available for all of the Octopus Spring sequences, group II Synechococcus differs from sequence OSB by four nucleotides. Group III Synechococcus differs from OSA by a single nucleotide, whereas group IV Synechococcus differs from OSA′ at two positions.
Phylogenetic reconstructions.
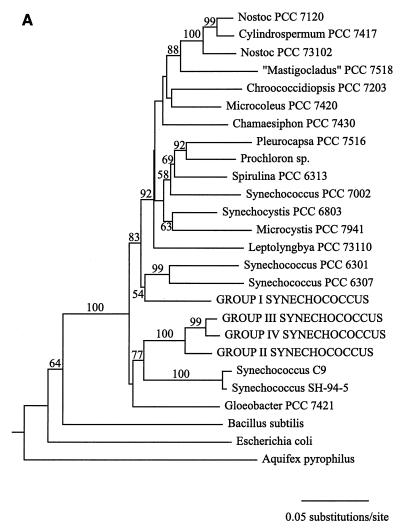
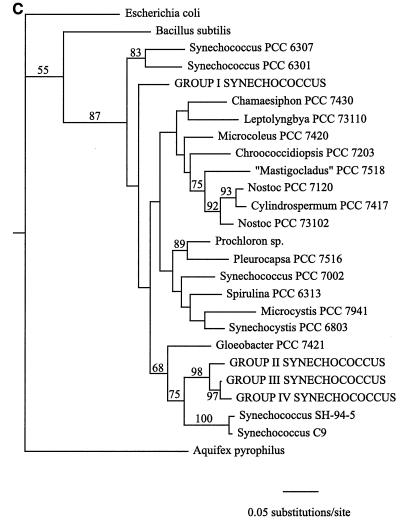
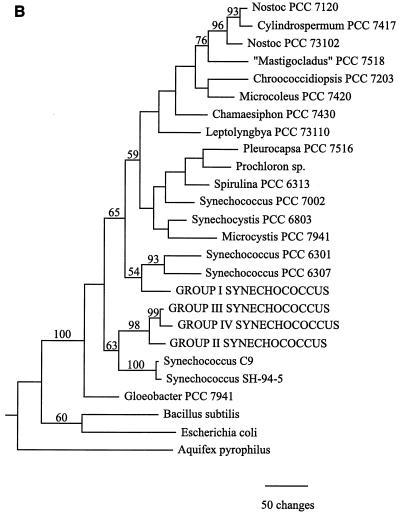
Cyanobacterial 16S rRNA gene sequence phylogenies were inferred by using maximum-parsimony, neighbor-joining, and maximum-likelihood methods (Fig. 1). While there are topological differences among the three phylogenies, they are similar in several important respects. First, Synechococcus clones belonging to sequence groups II, III, and IV, which were all derived from collections obtained at 55°C or above, form a clade in all three trees. The topology of this clade is consistent for all methods and is strongly supported by bootstrap analysis data in all cases (Fig. 1). The maximum-likelihood analysis, which provides error estimates for the estimated evolutionary distances between taxa, further supports the inferred structure of this clade. The estimated evolutionary distance between group II and IV Synechococcus clones is 0.051 substitution per site, with a 95% confidence interval of (0.024,0.079). The estimated evolutionary distance between group II and III Synechococcus clones is 0.038 substitution per site, with a 95% confidence interval of (0.017,0.061), while the distance between group III and IV Synechococcus clones was estimated to be 0.018 substitution per site, with a 95% confidence interval of (0.007,0.031). The fact that none of the confidence intervals overlaps zero provides strong evidence that a statistically significant evolutionary distance separates each pair of taxa and, therefore, that the 16S rRNA gene fragment used in this study contains sufficient genetic variation to resolve the phylogeny of the Hunter's Hot Springs sequence groups.
FIG. 1.
Cyanobacterial phylogenies inferred from ∼950 bp of 16S rRNA gene sequence data by using neighbor-joining (A), maximum-parsimony (B), and maximum-likelihood (C) methods. Synechococcus clones from Hunter's Hot Springs are indicated by capital letters. A value at a node indicates the percentage of the time that the taxa to the right of the node formed a clade for 100 (A and C) or 1,000 (B) bootstrap pseudoreplicates. Only bootstrap values greater than 50% are indicated.
In addition, sequence groups II, III, and IV are part of a larger clade which includes Synechococcus sp. strains SH-94-5 and C9 (Fig. 1). The former strain was isolated from a sample collected at 50°C from South Harney Hot Springs in Oregon, ∼150 km northeast of Hunter's Hot Springs (unpublished data), while the latter strain was isolated from Octopus Spring in Yellowstone National Park (12). Group I Synechococcus clones from Hunter's Hot Springs fall outside this clade in all three phylogenies (Fig. 1). This indicates that Synechococcus clones collected along the Hunter's Hot Springs environmental temperature gradient are not monophyletic (i.e., they do not share a common ancestor to the exclusion of all other taxa) and that Synechococcus clones from samples collected at temperatures greater than 55°C were not derived from an ancestor resembling the group I Synechococcus clones predominant at lower temperatures. The fact that the genus Synechococcus, a morphologically defined taxonomic unit encompassing a diverse assemblage of taxa ranging from the hot spring representatives to open-ocean picoplankton, is not a natural group has long been recognized by cyanobacterial taxonomists, as has the need for revision of the classification scheme of these unicellular organisms to reflect their phylogeny (33).
Temperature range of group I Synechococcus clones.
Growth rate data were collected at temperatures between 30 and 60°C for five group I clones isolated from 45°C collections (Table 2). No differences were found among the clones in terms of growth temperature range; the highest temperature for growth was estimated to be 57°C, and the lower limit was less than 30°C (Table 2). However, there was variation in the mean growth rate among temperatures (Table 3). One-way ANOVAs were performed for each temperature treatment to partition this variation into among-clone and within-clone components. No clone effect was found at temperatures between 40 and 55°C (Table 3). The estimated mean growth rates at these temperatures (Table 3) could therefore be directly compared by using Bonferroni tests. The growth rate estimates increased in the order 55°C < 40°C (=50°C) < 45°C, which indicates that the optimal growth temperature of these group I clones was 45°C. The relative amount of variation in the growth rate (i.e., the coefficient of variation) increased at the temperature range extremes, and differences among clones explained much of this variation, as indicated by the high R2 values (Table 3). The existence of genetic variation in the growth rate among clones at the extremes of the temperature range may indicate that the organisms do not typically experience these temperatures in situ; i.e., this genetic variation is not expressed phenotypically and can therefore accumulate without being subject to natural selection. Otherwise, it would be expected that this genetic diversity would be reduced by natural selection, as appears to be the case at intermediate temperatures, including the optimal temperature (Table 3). An alternative explanation for this pattern is that the variation was generated by mutation during laboratory cultivation. Distinguishing between these two hypotheses will require further experimentation.
TABLE 2.
Exponential growth rates of group I Synechococcus clones at different temperatures
| Temp (°C) | Exponential growth rate (no. of doublings day−1) ofa:
|
||||||
|---|---|---|---|---|---|---|---|
| Clone OH5 | Clone OH9 | Clone OH10 | Clone OH14 | Clone OH18 | Clone OH16 | Clone OH26 | |
| 30 | 0.28 ± 0.036 | 0.27 ± 0.020 | 0.67 ± 0.012 | 0.75 ± 0.012 | 0.76 ± 0.020 | NDb | ND |
| 35 | 1.34 ± 0.124 | 1.05 ± 0.032 | 1.55 ± 0.080 | 1.43 ± 0.080 | 1.81 ± 0.080 | ND | ND |
| 40 | 1.84 ± 0.108 | 1.92 ± 0.064 | 2.10 ± 0.088 | 1.94 ± 0.056 | 2.19 ± 0.112 | ND | ND |
| 45 | 2.24 ± 0.124 | 2.51 ± 0.151 | 2.40 ± 0.148 | 2.42 ± 0.044 | 2.85 ± 0.044 | ND | ND |
| 50 | 2.20 ± 0.036 | 2.20 ± 0.032 | 2.26 ± 0.120 | 2.05 ± 0.036 | 1.93 ± 0.223 | ND | ND |
| 55 | 1.43 ± 0.036 | 2.05 ± 0.183 | 1.46 ± 0.187 | 1.65 ± 0.020 | 1.49 ± 0.144 | 2.18 ± 0.052 | 1.93 ± 0.024 |
| 57 | 0.66 ± 0.000 | 1.24 ± 0.205 | 1.17 ± 0.144 | 1.40 ± 0.104 | 2.00 ± 0.355 | 1.25 ± 0.084 | 1.36 ± 0.064 |
| 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Mean ± standard error.
ND, not determined.
TABLE 3.
Summary statistics for growth rate data and ANOVA models for group I Synechococcus clones grown at different temperatures
| Temp (°C) | No. of doublings day−1a | Coefficient of variationa | ANOVA modelb
|
|
|---|---|---|---|---|
| Fc | R2 | |||
| 30 | 0.55 ± 0.075 | 44.5 ± 9.71 | 149.4*** | 0.992 |
| 35 | 1.44 ± 0.086 | 19.4 ± 4.24 | 17.1** | 0.932 |
| 40 | 2.00 ± 0.052 | 8.5 ± 1.85 | 2.7 | 0 |
| 45 | 2.48 ± 0.078 | 10.1 ± 2.21 | 4.1 | 0 |
| 50 | 2.13 ± 0.056 | 8.5 ± 1.86 | 1.4 | 0 |
| 55 | 1.62 ± 0.091 | 18.2 ± 3.97 | 3.0 | 0 |
| 57 | 1.30 ± 0.157 | 39.4 ± 8.59 | 6.0* | 0.829 |
Mean ± standard error.
One-way ANOVA was used to model the effect of the clone on the observed variation in the growth rate at each temperature. Nonsignificant F values and R2 values of zero indicate no effect.
Significance levels are based on an F[4,5] distribution: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Some growth rate data were also determined for two group I clones isolated from 55°C collections (OH16 and OH26) (Table 2). These clones did not show any significant differences in performance compared with the 45°C clones, as determined by Bonferroni tests comparing growth estimates obtained at 55 and 57°C for these clones with growth estimates obtained at 55 and 57°C for 45°C clones. The demonstration that these clones also could not survive at temperatures above 57°C indicates that although they were collected from samples having a temperature higher than the temperatures of the samples that yielded the other group I clones examined, they do not have an extended thermal limit.
Temperature range diversification in the group II-group III-group IV clade.
The two group II Synechococcus clones which were examined (OH4 and OH20) grew at temperatures between 40 and 61°C (Table 4). ANOVA with clone and temperature as factors indicated that there was no difference between the clones in the exponential growth rate (F[1,16] = 0.02; P = 0.89) and no clone-by-temperature interaction (F[7,16] = 0.77; P = 0.62); i.e., the two clones had the same temperature response profile. Growth data for these clones were therefore pooled, and estimated growth rates at different temperatures were compared with Bonferroni tests. The mean growth rate increased in the order 40°C < 61°C (= 45°C = 50°C = 55°C) < 57°C, which indicates that the optimum temperature for growth of these clones is 57°C.
TABLE 4.
Growth rates of group II, III, and IV Synechococcus clones and inferred sister taxa at different temperatures
| Temp (°C) | Growth rates (no. of doublings day−1)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group II clones
|
Group III clone
|
Group IV clones
|
Sister taxa
|
|||||||
| OH4 | OH20 | Pool | OH2 | OH28 | OH29 | OH30 | Pool | SH-94-5 | C9 | |
| 30 | NDb | ND | ND | ND | ND | ND | ND | ND | 0.37 ± 0.020 | 0.20 ± 0.020 |
| 35 | 0 | 0 | 0 | ND | ND | ND | ND | ND | 0.57 ± 0.008 | 0.60 ± 0.020 |
| 40 | 0.42 ± 0.064 | 0.67 ± 0.040 | 0.55 ± 0.078 | 0 | ND | ND | ND | ND | 1.20 ± 0.016 | 0.94 ± 0.064 |
| 45 | 0.83 ± 0.159 | 0.94 ± 0.108 | 0.88 ± 0.084 | 0.74 ± 0.012 | ND | ND | ND | ND | 1.08 ± 0.359 | 1.43 ± 0.303 |
| 50 | ND | ND | ND | ND | 0 | 0 | 0 | 0 | ND | ND |
| 55 | 1.11 ± 0.124 | 1.09 ± 0.028 | 1.10 ± 0.052 | 1.36 ± 0.175 | 0.26 ± 0.040 | 0.45 ± 0.016 | 0.36 ± 0.096 | 0.36 ± 0.045 | 1.05 ± 0.108 | 0.91 ± 0.088 |
| 57 | 1.73 ± 0.207 | 1.57 ± 0.052 | 1.65 ± 0.099 | 1.60 ± 0.028 | ND | ND | ND | ND | 0 | 0 |
| 61 | 0.99 ± 0.195 | 0.81 ± 0.231 | 0.90 ± 0.134 | 1.36 ± 0.012 | 0.63 ± 0.016 | 0.55 ± 0.060 | 0.41 ± 0.080 | 0.53 ± 0.045 | ND | ND |
| 63 | 0 | 0 | 0 | 0.93 ± 0.056 | ND | ND | ND | ND | ND | ND |
| 65 | ND | ND | ND | 0 | 0.64 ± 0.028 | 0.74 ± 0.072 | 0.77 ± 0.020 | 0.72 ± 0.032 | ND | ND |
| 70 | ND | ND | ND | ND | 0.69 ± 0.060 | 0.54 ± 0.179 | 0.69 ± 0.040 | 0.64 ± 0.059 | ND | ND |
| 73 | ND | ND | ND | ND | 0 | 0 | 0 | 0 | ND | ND |
Mean ± standard error.
ND, not determined.
The single group III Synechococcus clone obtained in culture (OH2) grew at temperatures between 45 and 63°C (Table 4). This clone's upper and lower thermal limits are thus higher than those of the group II clones. However, no detectable difference in optimal temperature or in maximum growth rate was observed between group II and III Synechococcus clones (Table 4).
Group IV Synechococcus clones exhibited the greatest thermotolerance. Growth of the three clones tested occurred at temperatures between 55 and 70°C, and optimal growth occurred at 65°C, although the value was not significantly different from the value obtained at 60 or 70°C (Table 4).
Growth rates were also determined for Synechococcus sp. strains SH-94-5 and C9 (12) (C9 was a gift from David Ward, Montana State University), taxa belonging to the inferred sister group of Synechococcus groups II, III, and IV. These strains, which are ∼1% divergent in the 16S rRNA gene fragment used for phylogenetic analysis, did not exhibit statistically significant differences in their responses to temperature, as indicated by the lack of a clone effect (F[1,22] = 0.15; P = 0.71) or a clone-temperature interaction (F[5,22] = 1.03; P = 0.41) in an ANOVA. Growth was observed at temperatures between 30 and 55°C (Table 4). Both clones died at 57°C and therefore had a lower maximum temperature than the group I clones.
Evolution of enhanced thermotolerance in Synechococcus.
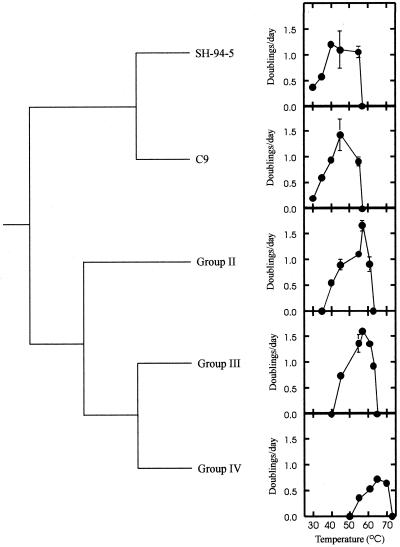
The phylogenetic branching order of lineages which have different phenotypes for a particular trait provides information on the direction of phenotypic change during cladogenesis (21). For the clade comprising group II, III, and IV Synechococcus clones and their inferred sister taxa (Fig. 1), the least thermotolerant lineages are basal, while isolates with greater thermotolerance branch later along the phylogeny (Fig. 2). This pattern of diversification supports the hypothesis that Synechococcus strains capable of growing at increasingly higher temperatures evolved from less thermotolerant ancestors. This is the pattern of diversification expected if there has been an adaptive radiation of Synechococcus temperature strains up the thermal gradient in response to past directional selection along a hot spring outflow channel. The common ancestor of these three groups, however, was not group I-like in terms of its phylogenetic status, and a comparable radiation has not occurred in the group I Synechococcus lineage (Fig. 1; Table 2).
FIG. 2.
Phylogenetic pattern of thermotolerance diversification in the group II-group III-group IV Synechococcus clade.
Evidence for trade-offs.
The observation that more thermophilic lineages of Synechococcus have lost the ability to grow at lower temperatures (Fig. 2) is evidence that there have been evolutionary costs of adapting to high temperatures. We wished to determine whether there may have been additional trade-offs during the evolution of thermotolerance in this clade. This can be assessed by testing whether different aspects of thermal performance are negatively correlated across taxa (18). However, because the study organisms are related to each other to different degrees due to common ancestry, phenotypic data collected from them may be nonindependent and consequently must first be transformed to meet the assumption of correlational analysis that data points are independent. We used Felsenstein's (9) method of independent contrasts (see Materials and Methods) to transform the data prior to estimating correlations.
We estimated correlation coefficients for the following thermal performance traits: optimal temperature, maximum temperature, maximum growth rate, and temperature range. The values for each sequence group (Table 5) were derived from the growth rate data (Table 4). The optimal temperature is the temperature at which the maximum growth rate was estimated, the maximum temperature is the highest observed temperature for growth, the maximum growth rate is the growth rate estimated at the optimal temperature, and the temperature range is the estimated difference between the observed maximum temperature and minimum temperature. Independent contrasts for the above traits were estimated separately by using branch length data from the three cyanobacterial phylogenies (Fig. 1), and correlation coefficients were then estimated for pairs of contrasts (Table 5). In all cases, significant negative correlations were found between an organism's maximum temperature and its temperature range. This indicates that enhanced thermotolerance may have come at the cost of increased thermal specialization. The maximum growth rate was positively correlated with the temperature range in all three analyses and was negatively correlated with the maximum temperature in the neighbor-joining analysis (Table 5). These results suggest that a reduced maximum growth rate may also have been a consequence of evolved thermotolerance.
TABLE 5.
Analysis of correlated character evolution during the diversification of temperature range by Synechococcusa
| Characterb | Group II | Group III | Group IV | Clone SH-94-5 | Clone C9 | Correlation coefficientsc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maximum parsimony
|
Neighbor joining
|
Maximum likelihood
|
||||||||||||
| Tmax | Ratemax | Range | Tmax | Ratemax | Range | Tmax | Ratemax | Range | ||||||
| Topt | 57 | 57 | 65 | 40 | 45 | 0.80 | −0.52 | −0.73 | 0.81 | −0.56 | −0.75 | 0.79 | −0.53 | −0.73 |
| Tmax | 61 | 63 | 70 | 55 | 55 | −0.87 | −0.96* | −0.89* | −0.97* | −0.88 | −0.97* | |||
| Ratemax | 1.65 | 1.60 | 0.72 | 1.20 | 1.43 | 0.96* | 0.96* | 0.96* | ||||||
| Range | 21 | 18 | 15 | 25 | 25 | |||||||||
Phenotypic data for the five sequence groups were transformed into four independent contrasts as described by Felsenstein (9). Branch length data from the three phylogenies in Fig. 1 were used in separate transformations, and correlations were determined for contrast pairs.
Optimal temperature (Topt), maximum temperature (Tmax), and temperature range (Range) are expressed in degrees Centigrade. The units for maximum growth rate (Ratemax) are number of doublings per day. Character values were estimated from Table 4 as described in the text.
An asterisk indicates that the correlation coefficient is significantly different from zero at the α = 0.05 level as determined by a t test with df = 2.
DISCUSSION
A hot spring outflow produces a thermal gradient which directly selects for enhanced performance at higher temperatures. Phylogenetic analyses of ∼950 nucleotides of the 16S rRNA gene were able to resolve a clade of increasingly more thermotolerant lineages of the cyanobacterium Synechococcus which have radiated in response to this directional selection to exploit previously inhospitable thermal niches (Fig. 2). While extension of the maximum temperature was integral to the evolution of thermotolerance in Synechococcus, there was not necessarily a corresponding increase in the optimal temperature, as evidenced by the lack of a detectable difference in this trait between group II clones and group III clone OH2 (Tables 4 and 5). The minimum temperature, however, has clearly shifted to higher values along with the maximum temperature (Fig. 2). Although we cannot infer the underlying mechanism(s) for this apparent trade-off, this is the expected pattern in the case of antagonistic pleiotropy in which there has been evolution of a gene or genes influencing performance at both ends of the thermal tolerance range (17). These genes could include many enzyme-encoding loci whose products are subject to thermodynamic constraints on performance (1). Because performance at lower temperatures decreased faster than performance at higher temperatures increased, the net evolutionary result has been a reduction in the breadth of the temperature range (Table 5), which further indicates that adaptation of Synechococcus to higher temperatures has come at the cost of increased thermal specialization.
The increase in thermal specialization may be related to the biogeographical structure suggested by the lack of identity between the 16S rRNA gene sequences of group II, III, and IV Synechococcus clones and clones from Octopus Spring in Yellowstone National Park. Hot spring Synechococcus isolates have low tolerance for freezing and desiccation, factors likely to be important in dispersal (6, 8). The more rapid loss of tolerance to lower temperatures during adaptation to higher temperatures may have limited the ability of cells to survive dispersal between habitats. Increased ecological specialization may therefore be an agent of geographical isolation.
It is worthwhile to compare the evolutionary trends revealed by this study with results obtained with E. coli lines selected for enhanced thermotolerance. A clone was more likely to extend its thermal limit if it was exposed to a lethal temperature (44°C) than if it was exposed to a high but nonlethal temperature (42°C) (2, 24). However, clones previously propagated at 42°C tended to be more predisposed to evolving an extended thermal limit upon lethal selection than were clones formerly maintained at the ancestral temperature, 37°C (24). An analogous predisposition for more thermotolerant lines to be derived from lines with a previous history of exposure to elevated temperatures has been recorded in the topology of the Synechococcus clade (Fig. 2). On the other hand, in contrast to the pattern suggested by our comparative data (Table 5), Mongold et al. (24) found that extension of the thermal limit in E. coli was not necessarily accompanied by a trade-off with performance at the lower thermal limit.
Directional selection still acts on Synechococcus at the thermal limit in nature, yet there is no evidence of positive population growth at temperatures above 73°C (3, 4). Since many nonphotosynthetic prokaryotes can grow at temperatures higher than 73°C, it has been suggested that the lack of a response to selection may be due to intrinsic limits in the stability of the photosynthetic apparatus (3). However, studies on a Synechococcus isolate from Hunter's Hot Springs with a group IV-like phenotype indicated that carbon reduction was more thermolabile than photosynthetic oxygen evolution (23). Clearly, more information is needed to determine the factor(s) underlying the upper temperature limit for photosynthesis. Having established a phylogenetic framework in which to study thermotolerance in Synechococcus, we are now in a position to trace the mechanistic history of biochemical adaptation in this clade and to investigate the possible evolutionary constraints which have prevented a further response to natural selection.
ACKNOWLEDGMENTS
We thank Emília Martins, Stephen Giovannoni, Doug Gordon, Kevin Vergin, and Tracie-Lynn Nadeau for training, advice, and valuable discussions. We also thank two anonymous reviewers for their comments and suggestions.
This work was supported by National Science Foundation grant IBN-9630674 to R.W.C. and by a National Science Foundation training grant to study the “genetic mechanisms of evolution” to S.R.M.
REFERENCES
- 1.Alexandrov Y Y. Cells, molecules and temperature. New York, N.Y: Springer-Verlag; 1977. [Google Scholar]
- 2.Bennett A F, Lenski R E. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution. 1993;47:1–12. doi: 10.1111/j.1558-5646.1993.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 3.Brock T D. Micro-organisms adapted to high temperatures. Nature. 1967;214:882–885. doi: 10.1038/214882a0. [DOI] [PubMed] [Google Scholar]
- 4.Castenholz R W. Thermophilic blue-green algae and the thermal environment. Bacteriol Rev. 1969;33:476–504. doi: 10.1128/br.33.4.476-504.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castenholz R W. Ecology of blue-green algae in hot springs. In: Carr N G, Whitton B A, editors. The biology of blue-green algae. Oxford, United Kingdom: Blackwell Scientific Publications; 1973. pp. 379–414. [Google Scholar]
- 6.Castenholz R W. The biogeography of hot spring algae through enrichment cultures. Mitt Int Ver Limnol. 1978;21:296–315. [Google Scholar]
- 7.Castenholz R W. Culturing methods for cyanobacteria. Methods Enzymol. 1988;167:68–93. [Google Scholar]
- 8.Castenholz R W. Endemism and biodiversity of thermophilic cyanobacteria. Nova Hedwigia. 1996;112:33–47. [Google Scholar]
- 9.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 10.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 11.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris M J, Ruff-Roberts A L, Kopczynski E D, Bateson M M, Ward D M. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl Environ Microbiol. 1996;62:1045–1050. doi: 10.1128/aem.62.3.1045-1050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acids Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 15.Hansen T F, Martins E P. Translating between microevolutionary process and macroevolutionary patterns: the correlation structure of interspecific data. Evolution. 1996;50:1404–1417. doi: 10.1111/j.1558-5646.1996.tb03914.x. [DOI] [PubMed] [Google Scholar]
- 16.Harvey P H, Pagel M D. The comparative method in evolutionary biology. Oxford, United Kingdom: Oxford University Press; 1991. [Google Scholar]
- 17.Huey R B, Kingsolver J G. Evolution of thermal sensitivity of ectotherm performance. Trends Ecol Evol. 1989;4:131–135. doi: 10.1016/0169-5347(89)90211-5. [DOI] [PubMed] [Google Scholar]
- 18.Huey R B, Kingsolver J G. Evolution of resistance to high temperature in ectotherms. Am Nat. 1993;142:S21–S46. [Google Scholar]
- 19.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. University Park: The Pennsylvania University; 1993. [Google Scholar]
- 21.Martins E P, Hansen T F. Phylogenetic comparative methods: the statistical analysis of interspecific data. In: Martins E P, editor. Phylogenies and the comparative method in animal behavior. Oxford, United Kingdom: Oxford University Press; 1996. pp. 22–75. [Google Scholar]
- 22.Meeks J C, Castenholz R W. Growth and photosynthesis in an extreme thermophile, Synechococcus lividus (Cyanophyta) Arch Mikrobiol. 1971;78:25–41. doi: 10.1007/BF00409086. [DOI] [PubMed] [Google Scholar]
- 23.Meeks J C, Castenholz R W. Photosynthetic properties of the extreme thermophile Synechococcus lividus. II. Stoichiometry between oxygen evolution and CO2 assimilation. J Therm Biol. 1978;3:19–24. [Google Scholar]
- 24.Mongold J A, Bennett A F, Lenski R E. Evolutionary adaptation to temperature. VII. Extension of the upper thermal limit of Escherichia coli. Evolution. 1999;53:386–394. doi: 10.1111/j.1558-5646.1999.tb03774.x. [DOI] [PubMed] [Google Scholar]
- 25.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peary J A, Castenholz R W. Temperature strains of a thermophilic blue-green alga. Nature. 1964;202:720–721. [Google Scholar]
- 27.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 28.Swofford D L. PAUP 3.1.1. Sunderland, Mass: Sinauer Associates; 1996. [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titus T A, Frost D R. Molecular homology assessment and phylogeny in the lizard family Opluridae (Squamata: Iguania) Mol Phylogenet Evol. 1996;6:49–62. doi: 10.1006/mpev.1996.0057. [DOI] [PubMed] [Google Scholar]
- 31.Urbach E, Robertson D, Chisholm S W. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature. 1992;355:267–269. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- 32.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial diversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterbury J B, Rippka R. Subsection I. Order Chroococcales. In: Holt J G, Staley J T, Bryant M P, Pfennig N, editors. Bergey's manual of systematic bacteriology. Vol. 3. Baltimore, Md: The Williams and Wilkins Co.; 1989. pp. 1728–1746. [Google Scholar]
- 34.Weller R, Bateson M M, Heimbuch B K, Kopczynski E D, Ward D M. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992;58:3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler W, Gladstein D. MALIGN: a multiple sequence alignment program. J Hered. 1994;85:417–418. [Google Scholar]
- 36.Whitton B A, Potts M, editors. Ecology of cyanobacteria: their diversity in time and space. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. [Google Scholar]
- 37.Wilmotte A, Van der Auwera G, De Wachter R. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC 7518, and phylogenetic analysis. FEBS Lett. 1993;317:96–100. doi: 10.1016/0014-5793(93)81499-p. [DOI] [PubMed] [Google Scholar]