Abstract
Bacterial atrazine catabolism is initiated by the enzyme atrazine chlorohydrolase (AtzA) in Pseudomonas sp. strain ADP. Other triazine herbicides are metabolized by bacteria, but the enzymological basis of this is unclear. Here we begin to address this by investigating the catalytic activity of AtzA by using substrate analogs. Purified AtzA from Pseudomonas sp. strain ADP catalyzed the hydrolysis of an atrazine analog that was substituted at the chlorine substituent by fluorine. AtzA did not catalyze the hydrolysis of atrazine analogs containing the pseudohalide azido, methoxy, and cyano groups or thiomethyl and amino groups. Atrazine analogs with a chlorine substituent at carbon 2 and N-alkyl groups, ranging in size from methyl to t-butyl, all underwent dechlorination by AtzA. AtzA catalyzed hydrolytic dechlorination when one nitrogen substituent was alkylated and the other was a free amino group. However, when both amino groups were unalkylated, no reaction occurred. Cell extracts were prepared from five strains capable of atrazine dechlorination and known to contain atzA or closely homologous gene sequences: Pseudomonas sp. strain ADP, Rhizobium strain PATR, Alcaligenes strain SG1, Agrobacterium radiobacter J14a, and Ralstonia picketti D. All showed identical substrate specificity to purified AtzA from Pseudomonas sp. strain ADP. Cell extracts from Clavibacter michiganensis ATZ1, which also contains a gene homologous to atzA, were able to transform atrazine analogs containing pseudohalide and thiomethyl groups, in addition to the substrates used by AtzA from Pseudomonas sp. strain ADP. This suggests that either (i) another enzyme(s) is present which confers the broader substrate range or (ii) the AtzA itself has a broader substrate range.
The s-triazine ring is present in many compounds used to make dyes, resins, and herbicides (15, 19, 29). Herbicides are directly applied to soils or plants, and their environmental longevity is largely dependent on bacterial degradation. Many s-triazines were initially thought to be poorly degraded, but more recent studies indicate that they are completely mineralized by a number of bacterial isolates (9, 21, 25). For instance, in 1937, melamine (2,4,6-triamino-1,3,5-s-triazine) was reported to be nonbiodegradable. In 1964, however, bacteria capable of slow degradation were isolated. More recently, melamine was reported to be readily biodegraded (8). In a similar manner, bacteria capable of catabolizing the s-triazine herbicide atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) to carbon dioxide were not reported during the first 30 years of application to agricultural fields. Atrazine is the most heavily used s-triazine herbicide in the United States, with about 68 × 106 to 73 × 106 lb applied to fields each year (17, 20, 35, 36). Identification of numerous atrazine-degrading bacteria in the last decade suggests that bacteria have evolved new degradative abilities due to exposure to atrazine.
Initially, bacteria capable of dealkylation and deamination of atrazine and atrazine analogs were isolated (4, 15, 22–24, 26, 33). A cytochrome P-450 in Rhodococcus sp. strains TE1 and N186/21 was shown to remove the alkyl substituents of atrazine via an oxygenative mechanism (26, 33, 37). Triazine hydrolase (TrzA) from Rhodococcus corallinus NRRL B-15444R catalyzes the deamination of unalkylated aminotriazines, such as melamine or 2-chloro-4,6-diamino-1,3,5-s-triazine (CAAT). TrzA catalyzes a slow dechlorination reaction with desethylsimazine (CEAT) but fails to catalyze either deamination or dechlorination reactions with atrazine (9). Other bacteria and consortia with the ability to dechlorinate less highly substituted triazines like desethylatrazine were also identified; however, these bacteria were unable to dechlorinate atrazine (4, 9).
Prior to 1993, there was no knowledge of a bacterial catabolic pathway for atrazine that proceeded via an initial dechlorination reaction (20, 30, 39). Shortly thereafter, a number of bacteria with atrazine dechlorination activity were isolated (1, 7, 11, 30, 36; K. Boundy-Mills, unpublished data). Pseudomonas sp. strain ADP was one of the first bacteria shown to metabolize atrazine to carbon dioxide, ammonia, and chloride. In this bacterium, the atrazine metabolic pathway proceeds via three consecutive hydrolytic reactions, which remove the chloride, N-ethylamine, and N-isopropylamine substituents, thereby converting atrazine to cyanuric acid. The enzymes that catalyze these reactions are atrazine chlorohydrolase (AtzA), hydroxyatrazine ethylaminohydrolase (AtzB), and isopropylammelide isopropylaminohydrolase (AtzC), respectively (6, 14, 31). All three enzymes were identified as members of the amidohydrolase protein superfamily by computer-based sequence analysis (31).
Other bacteria capable of atrazine dechlorination have been independently isolated from different locations around the world under a variety of isolation conditions: (i) Rhizobium strain PATR (7), (ii) Alcaligenes strain SG1 (Boundy-Mills, unpublished), (iii) Agrobacterium radiobacter J14a (36), (iv) Ralstonia picketti D (M. L. de Souza, N. R. Plechacek, L. P. Wackett, M. J. Sadowsky, and B. L. Hoyle, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. 453, 1998), and (v) Clavibacter michiganensis ATZ1 (1, 11). Data from PCR and Southern hybridization experiments indicate that all of these bacteria contain genes homologous to those that are active in the atrazine degradation pathway of Pseudomonas sp. strain ADP. Moreover, the sequence of an internal 0.5-kb region of the atzA gene from each of the bacteria was more than 99% identical to that of atzA from Pseudomonas sp. strain ADP (13).
Previous studies suggest that strains containing homologs to the Pseudomonas sp. strain ADP atzA have slightly different triazine degradation profiles (7, 11, 14, 36). Purified AtzA from Escherichia coli DH5α(pMD4) containing the atzA gene from Pseudomonas sp. strain ADP was reported to degrade atrazine and simazine but not melamine, terbuthylazine, or CAAT (14). Cell-free lysates from Rhizobium strain PATR were shown by thin-layer chromatography (TLC) to degrade atrazine, simazine, and terbuthylazine but not propazine or melamine (7). Consequently, these investigators concluded that two isopropyl groups prevented catalysis. A. radiobacter J14a was assayed for degradation of atrazine, ametryn, cyanazine, prometon, simazine, and propazine in culture media with whole cells. High-pressure liquid chromatography (HPLC) analysis showed that 62 to 100% of all triazines tested were degraded in 120 h (36). C. michiganensis ATZ1 was shown to degrade ametryn, prometryn, simazine, propazine, simetryn, and atratone in a whole-cell consortium with Pseudomonas strain CN1 (11). Due to different assay conditions and detection methods, the results described in the literature are difficult to compare with the substrate specificity of purified AtzA.
In this study, in vitro investigations were conducted to determine the substrate specificity of enzymes initiating the metabolism of atrazine and analogous compounds. First, purified AtzA from Pseudomonas sp. strain ADP was incubated with atrazine analogs. To expand the study, we synthesized and assayed numerous s-triazine and pyrimidine compounds that are not commercially available. Subsequently, in vitro enzymatic cell extracts prepared from other atrazine-degrading bacteria were investigated for their ability to degrade the atrazine analogs and the degradation was compared to degradation by AtzA from Pseudomonas sp. strain ADP.
MATERIALS AND METHODS
Bacterial strains and protein purification.
Pseudomonas sp. strain ADP, Rhizobium strain PATR, Alcaligenes strain SG1, A. radiobacter J14a, C. michiganensis ATZ1, and R. picketti D were grown in R minimal medium (13) at 30°C with 100 μg of atrazine per ml as the sole nitrogen source. Cells were harvested by centrifugation at 15,000 × g for 2 min and resuspended in 25 mM morpholinepropanesulfonic acid (MOPS) (pH 7). Crude cell extracts were prepared by lysing cell suspensions via three freeze-thaw cycles followed by sonication three times at 80% intensity for 20 s each, using a Biosonik sonicator (Bronwill Scientific, Rochester, N.Y.). Cell debris was removed by centrifugation at 15,000 × g for 15 min.
Purified AtzA protein was isolated from E. coli DH5α(pMD4) (14). Cultures were grown at 37°C overnight in Luria-Bertani medium (32) supplemented with chloramphenicol (30 μg/ml). The cells were lysed and AtzA was purified as previously described (12).
Atrazine analogs.
Atrazine, simazine, propazine, terbuthylazine, ametryn, prometryn, desethylatrazine, desisopropylatrazine, 2-chloro-4-hydroxy-6-(N-isopropylamino)-1,3,5-s-triazine (CIOT), 2-chloro-4-(N-ethylamino)-6-hydroxy-1,3,5-s-triazine (CEOT), N-isopropylammelide, ammeline, and ammelide were graciously provided by Novartis Crop Protection (Greensboro, N.C.). Commercially available CAAT was purchased from Sigma Chemical Co. (St. Louis, Mo.). All other compounds were synthesized specifically for this study as described below (Table 1).
TABLE 1.
Triazines and pyrimidine substrates used in this study.
| Common name | Abbreviation | Mol mass (m/z)a | Chemical name |
|---|---|---|---|
| Atrazine | CIET | 2-Chloro-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine | |
| CMMT | 158, 160, 173, 175 | 2-Chloro-4,6-di(N-methylamino)-1,3,5-s-triazine | |
| Simazine | CEET | 2-Chloro-4,6-di(N-ethylamino)-1,3,5-s-triazine | |
| Propazine | CIIT | 2-Chloro-4,6-di(N-isopropylamino)-1,3,5-s-triazine | |
| Terbuthylazine | CE(tB)T | 2-Chloro-4-(N-ethylamino)-6-(N-t-butylamino)-1,3,5-s-triazine | |
| C(tB)2T | 242, 244, 257, 259 | 2-Chloro-4,6-di(N-t-butylamino)-1,3,5-s-triazine | |
| Fluoroatrazine | FIET | 157, 184, 199 | 2-Fluoro-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine |
| Azidoatrazine | (N3)IET | 179, 181, 196, 222 | 2-Azido-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine |
| Cyanoatrazine | (CN)IET | 163, 191, 206 | 2-Cyano-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine |
| Atratone | (OMe)IET | 154, 169, 196, 211 | 2-Methoxy-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine |
| Prometon | (OMe)IIT | 2-Methoxy-4,6-di(N-isopropylamino)-1,3,5-s-triazine | |
| Ametryn | (SMe)IET | 2-Methylthiol-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine | |
| Prometryn | (SMe)IIT | 2-Methylthiol-4,6-di(N-ethylamino)-1,3,5-s-triazine | |
| Aminoatrazine | (NH2)IET | 154, 181, 196 | 2-Amino-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine |
| Deethylatrazine | CIAT | 2-Chloro-4-amino-6-(N-isopropylamino)-1,3,5-s-triazine | |
| Deisopropylatrazine | CEAT | 2-Chloro-4-amino-6-(N-ethylamino)-1,3,5-s-triazine | |
| CIOT | 2-Chloro-4-hydroxy-6-(N-isopropylamino)-1,3,5-s-triazine | ||
| CEOT | 2-Chloro-4-(N-ethylamino)-6-hydroxy-1,3,5-s-triazine | ||
| CAAT | 2-Chloro-4,6-diamino-1,3,5-s-triazine | ||
| N-isopropylammelide | IOOT | 2-N-Isopropylamino-4,6-dihydroxy-1,3,5-s-triazine | |
| Ammeline | AAOT | 2,4-Diamino-6-hydroxyl-1,3,5-s-triazine | |
| Ammelide | AOOT | 2-Amino-4,6-dihydroxyl-1,3,5-s-triazine | |
| N-Methyl-deisopropylatrazine | C(EM)AT | 172, 174, 187, 189 | 2-Chloro-4-(N-ethyl-N-methylamino)-6-amino-1,3,5-s-triazine |
| N-Methylsimazine | C(MeE)ET | 200, 202, 215, 217 | 2-Chloro-4-(N-ethyl-N-methylamino)-6-(N′-ethylamino)-1,3,5-s-triazine |
| N,N′-Dimethylsimazine | C(MeE)2T | 214, 216, 229, 231 | 2-Chloro-4,6-di(N-ethyl-N-methylamino)-1,3,5-s-triazine |
| CEEP | 185, 187, 200, 202 | 2-Chloro-di(N-ethylamino)-1,3-pyrimidine | |
| ECEP | 185, 187, 200, 202 | 2,6-Di(N-ethylamino)-4-chloro-1,3-pyrimidine | |
| ICIP | 213, 215, 228, 230 | 2,6-Di(N-isopropylamino)-4-chloro-1,3-pyrimidine | |
| CIEP | 199, 201, 214, 216 | 2-Chloro-4-(N-ethylamino)-6-(N-isopropylamino)-1,3-pyrimidine | |
| C(tB)EP | 213, 215, 228, 230 | 2-Chloro-4-(N-t-butylamino)-6-(N-ethylamino)-1,3-pyrimidine | |
| (tB)ECP | 213, 215, 228, 230 | 2-(N-t-Butylamino)-4-chloro-6-(N-ethylamino)-1,3-pyrimidine | |
| E(tB)CP | 213, 215, 228, 230 | 2-(N-Ethylamino)-4-chloro-6-(N-t-butylamino)-1,3-pyrimidine |
Molecular mass was determined by MS for all atrazine analogs synthesized in this study. The compounds were also shown to be homogeneous by chromatographic analysis as described in Materials and Methods.
All triazines and pyrimidines synthesized for this study were analyzed by gas chromatography mass spectrometry (GC-MS) using an HP 6890/5973 instrument (Hewlett-Packard, San Fernando, Calif.) (Table 1). Compounds were purified by crystallization and silica gel chromatography. Purity was confirmed by GC, HPLC, or TLC analyses.
The 2-substituted atrazine analogs were prepared through substitution reactions which replaced the chloride substituent of atrazine. Fluoroatrazine and cyanoatrazine were prepared by heating atrazine with excess potassium fluoride or cyanide in dimethyl sulfoxide at 140°C or in dimethylformamide at 120°C, respectively (16, 27). Products were characterized by GC-MS and 1H and 13C nuclear magnetic resonance spectroscopy. A similar procedure was used to produce azidoatrazine from the sodium azide salt, with a lithium bromide catalyst in methyl ethyl ketone at 80°C. Aminoatrazine was prepared by hydrogenation of azidoatrazine in isopropanol with a 5% palladium-on-calcium-carbonate catalyst under a hydrogen atmosphere. Treatment of atrazine with excess sodium methoxide in methanol at room temperature yielded atratone (methoxyatrazine).
Production of dichloro-mono-N-alkylamino triazine intermediates were essential for the synthesis of various N-alkylamino triazines. The intermediates were formed by reacting cyanuric chloride (2,4,6-trichloro-1,3,5-s-triazine) (Aldrich Chemical Co., Milwaukee, Wis.) with alkyl amines or ammonia in dichloromethane or anhydrous diethyl ether at 0 to 5°C. The reaction of the dichloro intermediates with alkylamine or ammonia in ethanol was used to aminate the triazine ring a second time, thereby producing chloro-di-N,N′-alkylaminotriazines. Alternately, a cold 10% aqueous solution of ethylamine, methylamine, or ammonia was treated with the dichloro intermediate in acetone or tetrahydrofuran to produce the desired chloro-di-N,N′-alkylaminotriazine. By altering the alkylamines used, N-methyldesisopropylatrazine, N-methylsimazine, and N,N′-dimethylsimazine were prepared.
Preparation of the dichloro-mono-N-alkylaminopyrimidine intermediate resembled that of the dichlorotriazine intermediate, using 2,4,6-trichloropyrimidine (Aldrich Chemical Co.) as starting material, except that the reaction mixtures were maintained at room temperature. The 4,6-dichloro-2-alkylaminopyrimidine and 2,6-dichloro-4-alkylaminopyrimidine isomers formed in roughly equal proportions and were separated by crystallization. As with the triazines, a second alkylamino substituent was added to the dichloro intermediate in ethanol. Gentle heating was required for addition of t-butylamine. The compounds 2-chloro-di(N-ethylamino)-1,3-pyrimidine (CEEP), 2,6-di(N-ethylamino)-4-chloro-1,3-pyrimidine (ECEP), 2,6-di(N-isopropylamino)-4-chloro-1,3-pyrimidine (ICIP), 2-chloro-4-(N-ethylamino)-6-(N-isopropylamino)-1,3-pyrimidine (CIEP), 2-chloro-4-(N-t-butylamino)-6-(N-ethylamino)-1,3-pyrimidine [C(tB)EP], 2-(N-t-butyl amino)-4-chloro-6-N-(ethylamino)-1,3-pyrimidine [(tB)ECP], and 2-(N-ethylamino)-4-chloro-6-(N-t-butylamino)-1,3-pyrimidine [E(tB)CP] were prepared in this manner.
Substrate incubation and analysis.
Saturated solutions of the various triazine and pyrimidine compounds listed in Table 1 were prepared in 10 mM phosphate buffer (pH 7.0). Due to the low solubility of most triazines, saturated solutions provided adequate concentrations for detection but prevented the determination of kinetic constants. The triazine and pyrimidine solutions were incubated with 50 μl of cell extracts for 16 and 48 h at 30°C. Enzymatic reactions were stopped by heating for 2 min at 95 to 100°C. Control samples without enzyme were handled in parallel with the enzyme-treated samples. Samples were analyzed by HPLC, using an Hewlett-Packard HP 1100 system equipped with a photodiode array detector interfaced to an HP ChemStation. An Adsorbosphere C18 5μ column (250 by 4.6 mm) (Alltech, Deerfield, Ill.) was used to separate alkylated triazines and pyrimidines with an acetonitrile-water linear gradient as previously described (6). Unalkylated substrates were separated on an Alltech Inertsil 5μ phenyl column (150 by 4.6 mm) with an isocratic aqueous mobile phase consisting of 5 mM sodium octane sulfate in 0.05% H3PO4.
Two colorimetric assays were developed to monitor fluoride production in enzymatic reactions with fluoroatrazine as a potential substrate. The first was modified from the procedure of Bellack and Schouboe (5), using 500-μl samples. In this method, fluoride disruption of a red complex formed by sodium-2-(p-sulfophenylazo)-1,8-sihydronephthalene-3,6-disulfonate (SPANDS) and zirconium(IV) ions results in a bleaching of color. Standard curves had an 8 to 80 μM linear range. The second method was an alizarin-lanthanum assay modified from Sigma Protocols (Sigma Chemical Co.) for 800-μl sample sizes. This method had a 10 to 130 μM linear range.
Two independent procedures were used to monitor ammonia production in reactions, using aminoatrazine as a potential substrate. The first used the Berthelot method as described by Okamura and Kigasawa (28) and Weatherburn (38) with modifications for 50-μl sample sizes. This assay incorporates the nitrogen from ammonia, through a series of chemical reactions, into indophenol, which produces a blue color. Standard curves had a 10 μM to 10 mM linear range. The second method used an enzymatic assay kit (no. 171-A) developed by Sigma Chemical Co. The reductive amination of 2-oxoglutarate to glutamate by glutamate dehydrogenase occurs with the concurrent oxidation of NADPH to NADP+, which was monitored at 340 nm.
RESULTS AND DISCUSSION
Purified AtzA substrate specificity.
Substrate analogs were synthesized to investigate the catalytic activity of purified AtzA from Pseudomonas sp. strain ADP (Table 1). Previously, only atrazine and simazine had been shown to be substrates. In this study, AtzA was shown to displace the fluoride substituent of fluoroatrazine. Fluoride release was also monitored via two spectroscopic methods. However, the acidic conditions required for chromophore formation hydrolyzed fluoroatrazine in the absence of enzyme, thereby preventing an accurate quantification of enzyme-catalyzed fluoride release. The rate of fluoroatrazine conversion to hydroxyatrazine by AtzA was 108% ± 9% (n = 3) of that of enzymatic atrazine dechlorination at equivalent substrate concentrations of 50 μM. The Vmax of AtzA with fluoroatrazine could not be determined due to the low water solubility of the substrate. However, the observation of statistically similar rates with either fluoride or chloride substituents at equimolar concentrations suggests that the carbon-halogen bond energy does not strongly influence the rate of the AtzA-catalyzed reaction. Bromoatrazine was synthesized, but the high rate of spontaneous hydrolysis in water precluded reliable enzyme assays.
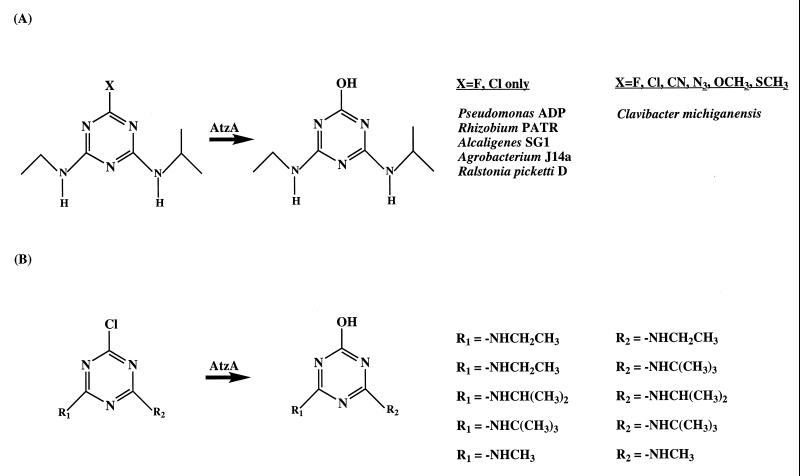
Atrazine analogs containing azido, cyano, methoxy, or thiomethyl groups in place of the chlorine substituent of atrazine were investigated with purified AtzA from Pseudomonas sp. strain ADP. Both analog disappearance and potential formation of hydroxyatrazine were monitored by HPLC. No activity was detected by either criteria. Although AtzA can hydrolytically remove halides, it did not remove pseudohalides (Fig. 1A). Pseudohalides, like azido, methoxy, and cyano groups, have a similar size, electronegativity, and reactivity in substitution reactions to halides, but they are composed of multiple atoms. The thiomethyl group is common to the herbicides ametryn and prometryn. AtzA from Pseudomonas sp. strain ADP catalyzed the dechlorination of atrazine and propazine but did not remove the thiomethyl group of the structurally analogous ametryn and prometryn.
FIG. 1.
Substrate analogs of atrazine. (A) Substituents hydrolyzed by various atrazine-dechlorinating strains. (B) Chlorodialkylamino triazines determined to be dechlorinated by cell extracts of Pseudomonas sp. strain ADP, Rhizobium strain PATR, Alcaligenes strain SG1, A. radiobacter J14a, R. picketti D, and C. michiganensis ATZ1.
AtzA from Pseudomonas sp. strain ADP also failed to remove the free amino group from aminoatrazine. Assays for hydroxyatrazine and ammonia were both negative. Aminoatrazine is of particular interest because AtzA has been identified as being evolutionarily related to enzymes catalyzing deamination reactions (31). Sequence alignments of AtzA with enzymes such as urease and adenosine deaminase have revealed it to be a member of the amidohydrolase protein superfamily (18, 31). It has been known since the 1960s that members of this family are also capable of dechlorinating substrate analogs. Adenosine deaminase, for instance, catalyzes deamination and dechlorination reactions of analogous purine substrates (2, 3, 10). The deamination and dechlorination reactions of s-triazine hydrolase (TrzA), however, are not performed on the same substrates. TrzA catalyzes the deamination of diaminotriazines and pyrimidines and the dechlorination of triazines which have a single alkyl group on one of the two nitrogen substituents, with deamination reactions favored over dechlorination reactions (34). The inability of AtzA to catalyze deamination reactions with aminoatrazine, as well as other aminotriazines and pyrimidines such as melamine and CAAT, places AtzA in a novel dehalogenating subgroup of the amidohydrolase superfamily.
Since numerous members of the amidohydrolase protein superfamily are involved in purine or pyrimidine metabolism, pyrimidine analogs were also synthesized and tested as potential substrates for AtzA. Pyrimidine analogs of atrazine, simazine, terbuthylazine, and propazine [CIEP, CEEP, ECEP, C(tB)ep, (tB)ECP, and E(tB)CP] were prepared with a chlorine substituent in the 2 and 4 positions. None of the pyrimidine substrates tested were substrates for AtzA.
The structural requirements of AtzA with respect to the N-alkyl side chains were also investigated. Mono-N-alkylated triazines with one of the nitrogen side chains alkylated and the other unalkylated (e.g., desethyl and desisopropylatrazine) underwent dechlorination by AtzA. One of the nitrogens on the triazine ring could therefore be unsubstituted, but if both were unsubstituted, as with CAAT, hydrolytic dechlorination was not observed. The requirement of at least one N-alkyl group on the triazine ring is consistent with the alkyl group being significant in substrate positioning and orientation.
The sizes of the alkyl groups on each of the nitrogen substituents were investigated and appeared to be less significant (Fig. 1B). AtzA catalyzed dechlorination with triazine ring substrates containing N-methyl, ethyl, isopropyl, and t-butyl substituents. Compounds with groups larger than t-butyl were too insoluble for analysis. Simazine, propazine, terbuthylazine, 2-chloro-4,6-N,N′-dimethyl-1,3,5-s-triazine, and 2-chloro-4,6-N,N′-ditertbutyl-1,3,5-s-triazine were substrates for AtzA from Pseudomonas sp. strain ADP. Compounds containing tertiary amino side chains like N-methyl-deisopropylatrazine [C(EM)AT], N,N′-dimethylsimazine, and N-methylsimazine were not substrates. These triazines containing tertiary amino substituents were most probably not substrates, either due to size and orientation restrictions or due to the absence of an amino hydrogen. The amino hydrogen has the potential to hydrogen bond in the enzyme active site, thereby assisting in orientation or generating an alternate configuration of electrons in the triazine ring during the dechlorination reaction. The ability of AtzA to dechlorinate compounds with secondary amino side chains which are larger than some of the tertiary amino side chains suggests that electronic factors are potentially more significant than steric factors.
Enzymatic activity of cell extracts from atrazine-degrading strains.
Similar assay conditions were used to compare the substrate degradation profiles of six atrazine-dechlorinating strains. For all of the compounds tested (Table 1), cell extracts of Pseudomonas sp. strain ADP, Rhizobium strain PATR, Alcaligenes strain SG1, A. radiobacter J14a, and R. picketti D had identical substrate degradation profiles to that described for purified AtzA from Pseudomonas sp. strain ADP. Previously, each of those strains were shown by PCR to contain an atzA gene with a sequence nearly identical (>99% identity) to the atzA gene from Pseudomonas sp. strain ADP (13). By contrast, cell extracts from C. michiganensis ATZ1 were able to degrade ametryn, prometryn, atratone, cyanoatrazine, and azidoatrazine in addition to the substrates degraded by the other strains (Fig. 1A).
Pseudomonas sp. strain ADP was reported previously not to degrade terbuthylazine (12). However, in this study, terbuthylazine was shown to be a substrate for AtzA from Pseudomonas sp. strain ADP. A previous report indicates that Rhizobium strain PATR does not degrade propazine (7); however, propazine degradation was observed in this study. The different observations could be because propazine is extremely insoluble in aqueous solution. Therefore, the 1-h incubation time and relative insensitivity of the TLC assay in the previous study may have precluded the observation of propazine degradation. Another apparent difference between the results presented here and those in the literature involves A. radiobacter J14a. In the present study, transformation of triazine-containing methoxy and thiomethyl groups was not observed. Previously, A. radiobacter J14a was reported to degrade ametryn, cyanazine, and prometon. Prometon has a methoxy group similar to atratone. The different observations could be due to methodological differences; the previous study was done with whole cells, while this study was conducted using cell extracts. Although AtzA is soluble and active in cell extracts, membrane-bound enzymes or enzymes that require additional cofactors or cosubstrates might not be active in the soluble cell extracts used here. There is precedence for a cytochrome P-450 being able to carry out atrazine dealkylation (4, 15, 22–24, 26, 33). Dealkylated products of atrazine were detected in whole-cell studies, suggesting that other enzymes might also be involved in atrazine degradation (36). Previously, de Souza et al. examined C. michiganensis ATZ1, in a consortium with Pseudomonas strain CN1, for degradation of s-triazine substrates (11). Whole-cell assays revealed that the consortium degraded methoxy and thiomethyl substrates. However, whether this degradation was due to dealkylation or actual hydrolysis of the substituent in the 2 position of the triazine ring was not determined. In our study, we provide the first evidence that C. michiganensis ATZ1 is capable of this activity and that hydroxyatrazine is the first metabolite produced. C. michiganensis ATZ1 degraded ametryn, prometryn, atratone, cyanoatrazine, and azidoatrazine in addition to the substrates degraded by the other strains. Whether this additional activity is due to activity of an AtzA homolog with broader substrate specificity or to additional enzymes is being investigated.
The specific activity of the atrazine dechlorination reaction for each of the six strains was determined (Table 2). The specific activity of Pseudomonas sp. strain ADP was nearly twice those of other strains. Differences between the strains could either be due to differences in the protein ratio of the AtzA homolog protein to other proteins in the cell or be due to differences in the activity of the enzyme itself. The data presented here do not distinguish between these two cases. C. michiganensis ATZ1 had an extremely low specific activity for atrazine dechlorination (0.05 ± 0.02 nmol/min/mg). The specific activity for the conversion of ametryn to hydroxyatrazine is 0.10 ± 0.03 nmol/min/mg. Although the difference in rates with these two substrates is not large, ametryn was repeatedly degraded more quickly than atrazine. Plates containing concentrations of ametryn above the saturation limit also consistently showed clearing zones before plates containing atrazine. The increased rate of the conversion of ametryn to hydroxyatrazine over the conversion of atrazine to hydroxyatrazine suggests that the strain is better suited for ametryn degradation than for atrazine degradation.
TABLE 2.
Specific activity of cell extracts from various atrazine-dechlorinating bacteria
| Strain | Sp act (nmol/min/mg)a |
|---|---|
| Pseudomonas sp. strain ADP | 7.4 ± 0.1 |
| Rhizobium strain PATR | 4.46 ± 0.08 |
| Alcaligenes strain SG1 | 3.88 ± 0.04 |
| A. radiobacter J14a | 4.79 ± 0.03 |
| R. picketti D | 1.72 ± 0.06 |
| C. michiganensis ATZ1 | 0.05 ± 0.02 |
Mean ± SE of two determinations.
ACKNOWLEDGMENTS
We thank the following researchers for providing strains: David Crowley and coworkers for C. michiganensis ATZ1, T. B. Moorman for A. radiobacter J14a, Kyria Bounty-Mills for Alcaligenes strain SG1, P. Plesiat and C. Bouquard for Rhizobium strain PATR, and Blythe Hoyle and Nathan Pechacek for R. picketti D. Rhodococcus corallinus NRRL B-15444R was obtained from the National Center for Agriculture Utilization Research in Peoria, Ill., with permission of Walter Mulbry. Special acknowledgment of the contributions of Jack Richman should be noted for his initial synthesis of cyanoatrazine and for helpful discussion. We also thank Carol Somody, Janis McFarland, and Andrea Elder of Novartis Crop Protection for providing s-triazine compounds and metabolites.
This research was supported, in part, by a grant from Novartis Crop Protection and by NIH training grant GM08347.
REFERENCES
- 1.Alvey S, Crowley D E. Influence of organic amendments on biodegradation of atrazine as a nitrogen source. J Environ Qual. 1995;24:1156–1162. [Google Scholar]
- 2.Baer H P, Drummond G I, Duncan E L. Formation and deamination of adenosine by cardiac muscle enzymes. Mol Pharmacol. 1966;2:67–76. [PubMed] [Google Scholar]
- 3.Bar H P, Drummond G I. On the mechanism of adenosine deaminase action. Biochem Biophys Res Commun. 1966;24:584–587. doi: 10.1016/0006-291x(66)90361-5. [DOI] [PubMed] [Google Scholar]
- 4.Behki R M, Khan S U. Degradation of atrazine by Pseudomonas: N-dealkylation and dehalogenation of atrazine and its metabolites. J Agric Food Chem. 1986;34:746–749. [Google Scholar]
- 5.Bellack E, Schouboe P J. Rapid photometric determination of fluoride in water. Anal Chem. 1958;30:2032–2034. [Google Scholar]
- 6.Boundy-Mills K, de Souza M L, Mandelbaum R M, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouquard C, Ouazzani J, Prome J-C, Michel-Briand Y, Plesiat P. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl Environ Microbiol. 1997;63:862–866. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook A M, Grossenbacher H, Hutter R. Isolation and cultivation of microbes with biodegradative potential. Experientia. 1983;39:1191–1198. doi: 10.1007/BF01990356. [DOI] [PubMed] [Google Scholar]
- 9.Cook A M, Hutter R. Deethylsimazine: bacterial dechlorination, deamination, and complete degradation. J Agric Food Chem. 1984;32:581–585. [Google Scholar]
- 10.Cory J G, Sunhadolnik R J. Dechlorinase activity of adenosine deaminase. Biochemistry. 1965;4:1733–1735. [Google Scholar]
- 11.de Souza M L, Newcombe D, Alvey S, Crowley D E, Hay A, Sadowsky M J, Wackett L P. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza M L, Seffernick J, Martinez B, Sadowsky S J, Wackett L P. The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol. 1998;180:1951–1954. doi: 10.1128/jb.180.7.1951-1954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Souza M L, Wackett L P, Boundy-Mills K L, Mandelbaum R T, Sadowsky M J. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol. 1995;61:3373–3378. doi: 10.1128/aem.61.9.3373-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson E L, Lee K H. Degradation of atrazine and related s-triazines. Crit Rev Environ Contam. 1989;19:1–13. [Google Scholar]
- 16.Geigy J R, Belg A G. Herbicidal triazines. Swiss Appl. 1962;609:18. [Google Scholar]
- 17.Gianessi L P. Lack of data stymies informed decisions on agricultural pesticides. Resources. 1987;89:1–4. [Google Scholar]
- 18.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 19.LeBaron H. Herbicide resistance in plants. 1982. pp. 9–19. , 99–114. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 20.Mandelbaum R T, Wackett L P, Allan D L. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl Environ Microbiol. 1993;59:1695–1701. doi: 10.1128/aem.59.6.1695-1701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandelbaum R T, Wackett L P, Allan D L. Rapid hydrolysis of atrazine to hydroxyatrazine by soil bacteria. Environ Sci Technol. 1993;27:1943–1946. [Google Scholar]
- 22.Masaphy S, Levanon D, Vaya J, Henis Y. Isolation and characterization of a novel atrazine metabolite produced by the fungus Pleurotus pulmonarius, 2-chloro-4-ethylamino-6-(1-hydroxyisopropyl)amino-1,3,5-triazine. Appl Environ Microbiol. 1993;59:4342–4346. doi: 10.1128/aem.59.12.4342-4346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirgain I, Green G A, Monteil H. Degradation of atrazine in laboratory microcosms: isolation and identification of the biodegrading bacteria. Environ Toxicol Chem. 1993;12:1627–1634. [Google Scholar]
- 24.Mougin C, Laugero C, Asther M, Dubroca J, Frasse P. Biotransformation of the herbicide atrazine by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:705–708. doi: 10.1128/aem.60.2.705-708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulbry W W. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl Environ Microbiol. 1994;60:613–618. doi: 10.1128/aem.60.2.613-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy I, Compernolle F, Ghys K, Vanderleyden J, De Mot R. A single cytochrome P-450 system is involved in degradation of the herbicides EPTC (s-ethyl dipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl Environ Microbiol. 1995;61:2056–2060. doi: 10.1128/aem.61.5.2056-2060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolaeva S V, Kolbin A M, Sapozhnikov Y E, Valitov R B, Ivanov V F. Synthesis of fluoro-substituted symmetrical (di-alkylamino)triazines using phase-transfer catalysis. Khim Geterotsikl Soedin. 1990;10:1370–1372. [Google Scholar]
- 28.Okamura T, Kigasawa K. Re-evaluation of the colorimetric assay for cytidine deaminase activity. Prenatal Diagn. 1994;14:213–218. doi: 10.1002/pd.1970140313. [DOI] [PubMed] [Google Scholar]
- 29.Quirke J M E. 1,3,5-Triazines. In: Katritzky A R, Rees C W, editors. Comprehensive heterocyclic chemistry. Vol. 3. New York, N.Y: Pergamon Press; 1984. pp. 459–529. [Google Scholar]
- 30.Radosevich M, Traina S J, Hao Y, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadowsky M J, Tong Z, de Souza M L, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Shao Z Q, Behki R. Cloning of the genes for degradation of the herbicides EPTC (s-ethyl dipropylthiocarbamate) and atrazine from Rhodococcus sp. strain TE1. Appl Environ Microbiol. 1995;61:2061–2065. doi: 10.1128/aem.61.5.2061-2065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Z Q, Seffens W, Mulbry W, Bahki R M. Cloning and expression of the s-triazine hydrolase gene (trzA) from Rhodococcus corrallinus and development of Rhodococcus recombinant strains capable of dealkylating and dechlorinating the herbicide atrazine. J Bacteriol. 1995;177:5748–5755. doi: 10.1128/jb.177.20.5748-5755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Short P, Colborn T. Pesticide use in the U.S. and policy implications: a focus on herbicides. Toxicol Ind Health. 1999;15:240–275. doi: 10.1191/074823399678846736. [DOI] [PubMed] [Google Scholar]
- 36.Struthers J K, Jayachandran K, Moorman T B. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl Environ Microbiol. 1998;64:3368–3375. doi: 10.1128/aem.64.9.3368-3375.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam A C, Behki R M, Khan S U. Isolation and characterization of an s-ethyl-N,N′-dipropylthiocarbamate-degrading Arthrobacter strain and evidence for plasmid-associated s-ethyl-N,N′-dipropylthiocarbamate degradation. Appl Environ Microbiol. 1987;53:1088–1093. doi: 10.1128/aem.53.5.1088-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weatherburn M W. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- 39.Yanze-Kontchou C, Gschwind N. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl Environ Microbiol. 1994;60:4297–4302. doi: 10.1128/aem.60.12.4297-4302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]