Abstract
The methylotrophic yeast Candida boidinii S2 was found to be able to grow on pectin or polygalacturonate as a carbon source. When cells were grown on 1% (wt/vol) pectin, C. boidinii exhibited induced levels of the pectin-depolymerizing enzymes pectin methylesterase (208 mU/mg of protein), pectin lyase (673 mU/mg), pectate lyase (673 mU/mg), and polygalacturonase (3.45 U/mg) and two methanol-metabolizing peroxisomal enzymes, alcohol oxidase (0.26 U/mg) and dihydroxyacetone synthase (94 mU/mg). The numbers of peroxisomes also increased ca. two- to threefold in cells grown on these pectic compounds (3.34 and 2.76 peroxisomes/cell for cells grown on pectin and polygalacturonate, respectively) compared to the numbers in cells grown on glucose (1.29 peroxisomes/cell). The cell density obtained with pectin increased as the degree of methyl esterification of pectic compounds increased, and it decreased in strains from which genes encoding alcohol oxidase and dihydroxyacetone synthase were deleted and in a peroxisome assembly mutant. Our study showed that methanol metabolism and peroxisome assembly play important roles in the degradation of pectin, especially in the utilization of its methyl ester moieties.
Little is known about the physiological role and environmental significance of methanol-utilizing microorganisms in natural ecological systems. Methanol can be produced through the hydrolysis of pectin, which is the main constituent of primary cell walls and the middle lamellae of higher plant cells, including cells in ripening fruits, germinating seeds, developing pollen, and actively growing and degrading plant tissues. Therefore, pectin is considered to be one of the major sources of methanol in natural environments (12). Hydrolysis of the methyl ester moieties of pectin to methanol and polygalacturonate is catalyzed by pectin methylesterase (PME) (EC 3.1.1.11). Although a methylotroph is assumed to be one of the key organisms in the ecological pectin carbon cycle, no previous reports have dealt with utilization of the methyl ester moieties of pectin by methanol-utilizing organisms. Since many methylotrophic yeasts have been reported to grow on pectin as a carbon source (7), we speculated that methylotrophic yeasts are significantly involved in the ecological pectin carbon cycle. Indeed, the methylotrophic yeast Candida boidinii has been found in pectin-rich sources, including fruits and their products (olives and wine) (1).
In a methylotrophic yeast, the first reaction in methanol metabolism is the oxidation of methanol to formaldehyde catalyzed by alcohol oxidase (AOD) (EC 1.1.3.13) (21), which is localized in peroxisomes (4). The formaldehyde produced by AOD reacts with d-xylulose 5-phosphate and produces dihydroxyacetone and glyceraldehyde 3-phosphate by dihydroxyacetone synthase (DHAS) (EC 2.2.1.3) catalysis in the methanol assimilation pathway (2, 6), and it is dissimilated to CO2 by other enzymes, including glutathione-dependent formaldehyde dehydrogenase (FLD) (EC 1.2.1.1) and formate dehydrogenase (FDH) (EC 1.2.1.2), in the formaldehyde oxidation pathway (18). The physiological significance of these enzymes in methanol metabolism has been revealed through gene disruption analyses using C. boidinii. For example, (i) AOD is an essential enzyme for growth on methanol (8), (ii) the main role of DHAS is fixation of formaldehyde into cell constituents (15), and (iii) the physiological role of FDH has been revealed to be mainly a role in detoxification of formate rather than a role in stimulated energy generation (13). However, the physiological roles of these methanol-metabolic enzymes in pectin degradation are not known.
This study was conducted (i) to reveal the metabolic pathway for pectin degradation in the methylotrophic yeast C. boidinii (ii) and to determine the physiological roles of methanol-metabolizing enzymes and peroxisome assembly in pectin metabolism. Our results revealed that pectin is hydrolyzed to polygalacturonate and methanol by PME. Each of these products is then utilized in an independent pathway, either in conventional methanol metabolism (which requires peroxisome assembly) or in a pectin-degrading enzyme system that includes polygalacturonase (PG) (exo-PG [EC 3.2.1.67] and endo-PG [EC 3.2.1.15]), pectin lyase (PNL) (EC 4.2.2.10), and pectate lyase (PAL) (exo-PAL [EC 4.2.2.9] and endo-PAL [EC 4.2.2.2]).
MATERIALS AND METHODS
Yeast strains, media, and cultivation.
C. boidinii S2 was used as the wild-type strain (22), and the aod1Δ (8), das1Δ (15), fdh1Δ (13), and pex5Δ (17) strains were to test growth on pectic compounds. The C. boidinii GFP-AKL strain producing GFP-PTS1 (green fluorescent protein tagged with an -AKL sequence at the carboxyl terminus that belongs to peroxisome targeting signal type 1 [PTS1]) was used to observe peroxisomal proliferation due to pectic compounds in vivo (17).
Complex yeast extract-peptone-dextrose medium (15) and the mineral synthetic media (MI media) (16) were used for cultivation of the C. boidinii strains. In each experiment the carbon source was one of the following: 2% (wt/vol) glucose, 0.05, 0.1%, 0.15, or 1% (vol/vol) methanol, 1% (wt/vol) pectin, or 1% (wt/vol) polygalacturonate (Sigma Chemical Co., St. Louis, Mo.). The degree of methyl esterification (DE) of pectins was approximately 30, 60, or 90%, and the pectins were from citrus fruit (Sigma Chemical Co.). Pectic compounds were purified by washing them with acidified 60% ethanol (5 ml of concentrated HCl, 100 ml of 60% ethanol) and then with 60 and 90% neutral ethanol (20). The absence of methanol in the pectic compounds was checked by gas chromatography by using a Porapak Q column (0.55 cm [inside diameter] by 2 m) and a Shimadzu GC7A gas chromatograph (Shimadzu, Kyoto, Japan) as previously described (14, 23). The initial pH of the medium was adjusted to 4.0 or 5.0. Cultivation (volume, 50 ml) was performed aerobically at 28°C with rotary shaking at 180 rpm in a 500-ml Erlenmeyer flask, and growth was monitored by measuring the optical density at 660 nm.
Preparation of extracellular and intracellular fractions.
Yeast cells grown to the early log phase on pectic compounds were separated by centrifugation at 12,000 × g for 10 min at 4°C. Supernatants consisting of the culture media were used as extracellular fractions. The yeast cells were resuspended in 50 mM sodium phosphate buffer (pH 7.0) and were broken with a mini bead beater (Biospec Products, Bartlesville, Okla.) by using 30-s pulses with 1-min intermediate cooling periods on ice. The glass beads and cell debris were removed by centrifugation at 12,000 × g for 10 min at 4°C, and the supernatants were used as intracellular fractions for enzyme assays.
Enzyme assays.
PME activity was measured by titration with 0.01 N NaOH. The reaction mixture consisted of 10 ml of 1% pectin (DE, 90%; Sigma Chemical Co.) in 50 mM acetate buffer (pH 5.0) and 1 ml of enzyme solution. Another aliquot of the sample was boiled for 10 min and used as a blank. The reaction mixture was incubated at 30°C for 60 min, and the enzyme reaction was stopped by incubation in boiling water for 10 min. The stopped reaction mixtures were titrated by using 0.01 N NaOH with phenolphthalein, and the amount of 0.01 N NaOH required was determined. One unit was defined as the amount of enzyme that released 1 μmol of carboxy groups per min.
PG activity was assayed by measuring the increase in reducing groups derived from polygalacturonate. The initial reaction mixture consisted of 10 ml of 1% polygalacturonate (DE, 0%; Sigma Chemical Co.) in 50 mM acetate buffer (pH 4.0) and 1 ml of enzyme solution. Another aliquot of the sample was boiled for 10 min and used as a blank. The reaction mixture was incubated at 30°C for 60 min, and the enzyme reaction was stopped by incubation in boiling water for 10 min. Reducing groups derived from polygalacturonate were measured by the method of Somogyi (19) and Nelson (11). One unit was defined as the amount of enzyme that released 1 μmol of reducing groups per min.
PNL and PAL activities were determined by recording the absorbance at 235 nm during incubation with 1% pectin (DE, 90%) or 1% polygalacturonate (DE, 0%). The reaction mixture consisted of 10 ml of 1% substrate in 50 mM acetate buffer (pH 4.0) and 1 ml of enzyme solution. Another aliquot of the sample was boiled for 10 min and used as a blank. The reaction mixture was incubated at 30°C for 60 min, and the enzyme reaction was stopped by incubation in boiling water for 10 min. One unit was defined as an increase of 1.0 unit of absorbance at 235 nm of the reaction mixture per min (5).
AOD (22), DHAS (10, 24), FLD (18), and FDH (18) activities were determined as described previously; each activity was defined as it was defined previously.
Protein was determined by the method of Bradford with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), using bovine serum albumin as the standard.
Fluorescence microscopy.
The C. boidinii GFP-AKL strain was placed on a microscope slide and examined by using the fluorescein isothiocyanate channel of an Axioplan 2 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) equipped with a Plan-NEOFLUAR 100×/1 · 30 (oil) objective and Nomarski attachments and set at the fluorescein isothiocyanate channel (17). Images were acquired with a charge coupled device camera (Carl Zeiss ZVS-47DE) and a CG7 frame grabber (Scion Corp., Frederick, Md.). The number of peroxisomes per cell was determined by examining at least 350 cells in random fields.
RESULTS AND DISCUSSION
C. boidinii S2 was able to grow on pectic compounds.
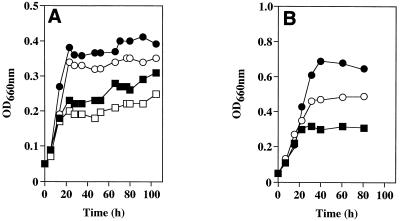
Wild-type C. boidinii strain S2 was able to grow on two pectic compounds, polygalacturonate and pectin. As the DE of pectin increased, the cell density increased (Fig. 1A). The cell density obtained on pectin with a DE of 90% was about twofold greater than that obtained on polygalacturonate (DE, 0%). This difference in growth yield between these two pectic compounds corresponds to the growth yield of C. boidinii on 0.05% methanol, on which 33% of the methyl ester moiety in pectin with a DE of 90% has been calculated to be utilized. These findings show that C. boidinii S2 has the ability to utilize the methyl ester moiety of pectin, as well as the polygalacturonate skeleton, as a sole carbon source. The C. boidinii CCY 27-37-13 strain was reported to be able to grow on a polygalacturonate medium only after adaptation to pectin (20). In contrast, C. boidinii S2 could grow on polygalacturonate medium without adaptation to pectin (Fig. 1A). Thus, the pectin metabolism system of C. boidinii S2 may be different from that of strain CCY 27-37-13, although the ability to grow on pectic compounds seems to be a general feature of this species.
FIG. 1.
(A) Growth of C. boidinii S2 on pectin and polygalacturonate. Symbols: □, 1% (wt/vol) polygalacturonate (DE, 0%); ■, 1% (wt/vol) pectin (DE, 30%); ○, 1% (wt/vol) pectin (DE, 60%); ●, 1% (wt/vol) pectin (DE, 90%) in MI medium. The initial pH and final pH of the medium were 5.7 and 5.5, respectively. (B) Growth on methanol at various concentrations. Symbols: □, 0.05% (vol/vol) methanol (corresponding to the concentration of the methyl ester moiety of 1% pectin with a DE of 33%); ○, 0.1% (vol/vol) methanol (corresponding to a DE of 67%); ●, 0.15% (vol/vol) methanol (corresponding to a DE of 100%). OD660nm, optical density at 660 nm.
PME and pectin-depolymerizing enzyme activities were induced by pectic compounds.
C. boidinii S2 exhibited inducible PME enzyme activity and pectin-depolymerizing enzyme activities (i.e., PEL, PAL, and PG activities). As shown in Table 1, PME activity was not detected in cells grown on glucose medium. In contrast, PME activity was remarkably induced in cells grown on polygalacturonate or pectin. Pectin-grown cells exhibited ca. 11-fold-higher activity than the polygalacturonate-grown cells. In both cases, most PME activity was detected in the extracellular fraction (Table 1). The activities of pectin-depolymerizing enzymes (i.e., PG, PAL and PNL) was also induced in polygalacturonate or pectin medium (Table 2), and most of these activities were detected in the extracellular fraction. Although the growth of C. boidinii S2 on polygalacturonate was weak at the initial pH of the medium, pH 5.0, cells grew well when the initial pH of the medium was adjusted to 4.0 (data not shown). This may have been due to the fact that PG activity was not stable at pH 5.0 (data not shown).
TABLE 1.
Specific activities of PME during growth on glucose, polygalacturonate (DE, 0%), and pectin (DE, 90%)a
| Carbon source | Sp act (mU/mg)
|
|
|---|---|---|
| Intracellular | Extracellular | |
| Glucose | NDb | ND |
| Polygalacturonate | 0.98 ± 0.10 | 19 ± 1.1 |
| Pectin | 1.12 ± 0.09 | 208 ± 5.7 |
Cells were grown on MI medium containing a C source at a concentration of 1%. Intracellular and extracellular fractions were prepared as described in Materials and Methods.
ND, not detected.
TABLE 2.
Specific activities of pectin-depolymerizing enzymes during growth on glucose, polygalacturonate (DE, 0%), and pectin (DE, 90%)a
| Carbon source | Sp act (mU/mg)
|
|||||
|---|---|---|---|---|---|---|
| PEL
|
PAL
|
PG
|
||||
| Intracellular | Extracellular | Intracellular | Extracellular | Intracellular | Extracellular | |
| Glucose | NDb | ND | ND | ND | ND | ND |
| Polygalacturonate | 160 ± 4.0 | 846 ± 31 | 167 ± 3.8 | 615 ± 19 | 0.44 ± 0.02 | 4.48 ± 0.12 |
| Pectin | 181 ± 4.5 | 673 ± 25 | 126 ± 3.0 | 673 ± 22 | 0.33 ± 0.01 | 3.45 ± 0.11 |
Cells were grown on MI medium containing a C source at a concentration of 1% and were disrupted as described in Materials and Methods. Intracellular and extracellular fractions were prepared as described in Materials and Methods.
ND, not detected.
Our results suggest that C. boidinii is able to hydrolyze pectin at the methyl ester moiety by means of PME extracellularly and is able to utilize the polygalacturonate and methanol produced as carbon sources.
Methanol-metabolizing enzymes were induced by pectic compounds.
The DE affected the growth yield of C. boidinii S2 on pectin, and the data suggested that methanol produced from pectin through hydrolysis by PME was utilized by the methanol-metabolic enzymes present in C. boidinii cells. Next, we studied the regulation of the methanol-metabolizing enzymes, AOD, DHAS, FLD, and FDH by pectic compounds. As shown in Table 3, AOD, DHAS, FLD, and FDH activities were induced by pectin, although these activities were lower than those in methanol-grown cells. Also, these enzyme activities in pectin-grown cells were ca. two- to eightfold higher than those in polygalacturonate-grown cells. These results showed that both pectin and polygalacturonate could induce these methanol-metabolizing enzymes.
TABLE 3.
Specific activities of enzymes related to methanol metabolism during growth on methanol, polygalacturonate (DE, 0%), pectin (DE, 90%), and methanola
| Carbon source | Sp act
|
|||
|---|---|---|---|---|
| AOD (U/mg) | DHAS (mU/mg) | FLD (mU/mg) | FDH (mU/mg) | |
| Polygalacturonate | 0.03 ± 0.01 | 71 ± 3.1 | 14 ± 0.1 | 8 ± 0.1 |
| Pectin | 0.26 ± 0.02 | 94 ± 3.4 | 17 ± 0.2 | 50 ± 0.4 |
| Methanol | 1.12 ± 0.09 | 210 ± 7.3 | 201 ± 2.2 | 129 ± 0.9 |
| Glucose | NDb | ND | 11 ± 0.1 | 10 ± 0.1 |
Cells were grown on MI medium containing a C source at a concentration of 1% and were disrupted as described in Materials and Methods.
ND, not detected.
Pectic compounds could induce peroxisome proliferation.
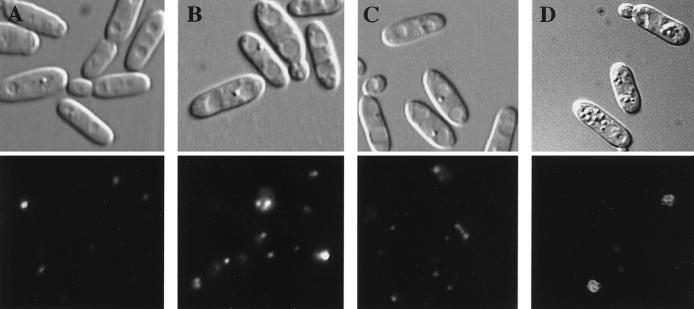
Since both AOD and DHAS are peroxisomal enzymes (3, 4), we examined whether pectin or polygalacturonate could induce peroxisome proliferation. To do this, a morphometric analysis using the C. boidinii GFP-AKL strain producing GFP-PTS1 (green fluorescent protein tagged with an -AKL sequence at the carboxyl terminus) was performed. When the GFP-AKL strain was grown on synthetic glucose medium, there were a few small peroxisomes (1.29 ± 0.13 peroxisomes/cell) (Fig. 2A). The numbers of peroxisomes after growth on pectin and polygalacturonate were 3.34 ± 0.31 and 2.76 ± 0.22 peroxisomes/cell, respectively (Fig. 2B and C). On the other hand, cells grown on methanol had large peroxisomes (4.8 ± 0.41 peroxisomes/cell) (Fig. 2D). The peroxisome inducers in pectin-grown cells seemed to be pectic compounds and not methanol derived from pectin through hydrolysis, based on the following observations: (i) peroxisomes were induced by polygalacturonate, which does not include methyl ester moieties (Fig. 2C), and (ii) the morphology of peroxisomes induced by pectin was different from that in methanol-grown cells (Fig. 2B and D).
FIG. 2.
Fluorescence of GFP-AKL/wt induced by glucose (A), pectin (DE, 90%) (B), polygalacturonate (DE, 0%) (C), and methanol (D). The induced cells were visualized by Nomarski microscopy (upper micrographs) or fluorescence microscopy (lower micrographs).
Disruption of the AOD1, DAS1, FDH1, and PEX5 genes caused a defect in growth on pectin medium but not on polygalacturonate medium.
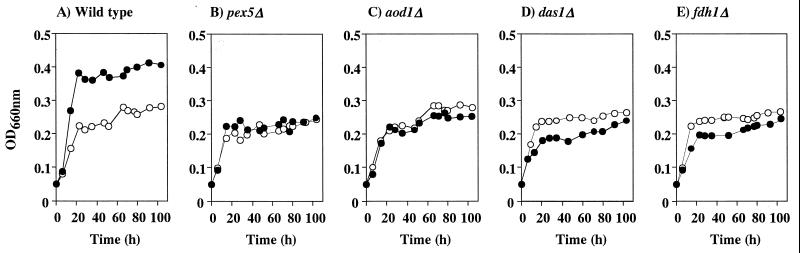
To determine to what extent methanol-metabolic enzymes and peroxisome assembly were directly involved in pectin metabolism, knockout strains depleted of AOD, DHAS, and FDH (i.e., aod1Δ, das1Δ, and fdh1Δ strains), as well as the PTS1 receptor responsible for peroxisome assembly (i.e., a pex5Δ strain), were grown on pectin or polygalacturonate as a carbon source, and the growth of each strain was compared with that of the wild-type strain.
The aod1Δ, das1Δ, fdh1Δ, and pex5Δ strains showed a severe defect in the growth yield when pectin was the sole carbon source compared with the growth yield of the wild-type strain (Fig. 3). These results show that methanol-metabolizing enzymes and peroxisome assembly play significant roles in pectin metabolism.
FIG. 3.
Growth of the wild type (A) and the pex5Δ (B), aod1Δ (C), das1Δ (D), and fdh1Δ (E) strains on polygalacturonate (DE, 0%) (○) and pectin (DE, 90%) (●). The initial pH of the medium was 5.0. OD660nm, optical density at 660 nm.
On the other hand, all of these knockout strains were still able to grow on pectin. Also, the reduced growth yields observed with these knockout strains grown on pectin were not observed when cells were grown on polygalacturonate. This indicated that both methanol-metabolizing enzymes and peroxisome assembly were not directly involved in polygalacturonate metabolism, although they were induced by polygalacturonate. Also, the growth yields of these knockout strains on pectin were almost the same as those on polygalacturonate, suggesting that growth on polygalacturonate was not impaired in these knockout strains.
Mechanism of pectin assimilation in C. boidinii.
In this study, C. boidinii S2 was shown to have the ability to utilize each of two pectic compounds, pectin and polygalacturonate, as a carbon source. This strain produced pectin-depolymerizing enzymes. Furthermore, utilization of methanol derived from hydrolysis of the methyl ester moiety of pectin was found to contribute significantly to the growth yield of C. boidinii on pectin, based on the following observations: (i) the cell density increased as the DE of pectin increased (Fig. 1); (ii) C. boidinii cells grown on pectin had induced levels of PME activity and activities of methanol-metabolizing enzymes (Tables 1 and 3); and (iii) the knockout strains devoid of methanol-metabolizing enzymes (the aod1Δ, das1Δ, and fdh1Δ strains) showed a severe growth defect on pectin (Fig. 3). In addition, peroxisome assembly was also found to be involved in pectin metabolism.
We assume the following scheme for pectin degradation in C. boidinii. (i) Pectin is hydrolyzed by extracellular PME into methanol and polygalacturonate. (ii) Methanol is utilized by conventional methanol-metabolizing enzymes together with peroxisome proliferation. (iii) Polygalacturonate (or pectin) is independently dissimilated by several pectin-depolymerizing enzymes, including PEL, PAL, and polygalacturonase; also, degradation of polygalacturonate depends on neither methanol-metabolizing enzymes nor peroxisome assembly. (iv) Methanol-metabolizing enzymes, pectin-depolymerizing enzymes, and peroxisome assembly are cooperatively induced and regulated during growth on pectin.
This is the first report on an organism which can utilize both the methyl ester moiety and the polygalacturonate skeleton of pectin as carbon sources. It has been reported previously that most methylotrophic yeast strains are able to grow on pectin (7, 20) and that AOD was induced by pectin in Pichia methanolica (9). Based on these facts, the ability to grow on pectin by utilizing the methanol metabolism pathway seems to be a general feature of the methylotrophic yeasts.
ACKNOWLEDGMENTS
We are grateful to Kazuo Komagata, Tokyo University of Agriculture, for valuable suggestions and to Yumiko Uchida and Yu Hosaka for their skillful assistance.
REFERENCES
- 1.Barnett J A, Payne R W, Yarrow D, editors. Yeasts. New York, N.Y: Cambridge University Press; 1983. [Google Scholar]
- 2.Bystrkh L V, Sokolov A P, Trotsenko Y A. Purification and properties of dihydroxyacetone synthase from the methylotrophic yeast, Candida boidinii. FEBS Lett. 1981;132:324–328. [Google Scholar]
- 3.Goodman J M. Dihydroxyacetone synthase is an abundant constituent of the methanol-induced peroxisome of Candida boidinii. J Biol Chem. 1985;260:7108–7113. [PubMed] [Google Scholar]
- 4.Goodman J M, Scott C W, Donahue P N, Atherton J P. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J Biol Chem. 1984;259:8485–8493. [PubMed] [Google Scholar]
- 5.Ishii S, Yokotsuka T. Purification and properties of pectin trans-eliminase from Aspergillus sojae. Agric Biol Chem. 1972;36:146–153. [Google Scholar]
- 6.Kato N, Higuchi T, Sakazawa C, Nishizawa T, Tani Y, Yamada H. Purification and properties of a transketolase responsible for formaldehyde fixation in a methanol-utilizing yeast, Candida boidinii (Kloeckera sp.) No. 2201. Biochim Biophys Acta. 1982;715:143–150. [PubMed] [Google Scholar]
- 7.Lee J D, Komagata K. Taxonomic study of methanol-assimilating yeasts. J Gen Appl Microbiol. 1980;26:133–158. [Google Scholar]
- 8.Nakagawa T, Mukaiyama H, Yurimoto H, Sakai Y, Kato N. Alcohol oxidase hybrid oligomers formed in vivo and in vitro. Yeast. 1999;15:1223–1230. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1223::AID-YEA450>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa T, Uchimura T, Komagata K. Isozymes of methanol oxidase in a methanol-utilizing yeast, Pichia methanolica IAM 12901. J Ferment Bioeng. 1996;81:498–503. [Google Scholar]
- 10.Nash T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J. 1953;55:416–421. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375. [Google Scholar]
- 12.Sakai T, Sakamoto T, Hallaert J, Vandamme E J. Pectin, pectinase and protopectinase: production, properties, and applications. Adv Appl Microbiol. 1993;39:213–294. doi: 10.1016/s0065-2164(08)70597-5. [DOI] [PubMed] [Google Scholar]
- 13.Sakai Y, Murdanoto A P, Konishi T, Iwamatsu A, Kato N. Regulation of the formate dehydrogenase gene, FDH1, in the methylotrophic yeast Candida boidinii and growth characteristics of an FDH1-disrupted strain on methanol, methylamine, and choline. J Bacteriol. 1997;179:4480–4485. doi: 10.1128/jb.179.14.4480-4485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai Y, Murdanoto A P, Sembiring L, Tani Y, Kato N. A novel formaldehyde oxidation pathway in methylotrophic yeasts: methylformate as a possible intermediate. FEMS Microbiol Lett. 1995;127:229–234. doi: 10.1111/j.1574-6968.1995.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 15.Sakai Y, Nakagawa T, Shimase M, Kato N. Regulation and physiological role of the DAS1 gene encoding dihydroxyacetone synthase in the methylotrophic yeast Candida boidinii. J Bacteriol. 1998;180:5885–5890. doi: 10.1128/jb.180.22.5885-5890.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai Y, Rogi T, Takeuchi R, Kato N, Tani Y. Expression of Saccharomyces adenylate kinase gene in Candida boidinii under the regulation of its alcohol oxidase promoter. Appl Microbiol Biotechnol. 1995;42:860–864. doi: 10.1007/BF00191182. [DOI] [PubMed] [Google Scholar]
- 17.Sakai Y, Yurimoto H, Matsuo H, Kato N. Regulation of peroxisomal proteins and organelle proliferation by multiple carbon sources in the methylotrophic yeast, Candida boidinii. Yeast. 1998;14:1175–1187. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1175::AID-YEA319>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Schütte H, Flossdorf J, Sahm H, Kula M-R. Purification and properties of formaldehyde dehydrogenase and formate dehydrogenase from Candida boidinii. Eur J Biochem. 1976;62:151–160. doi: 10.1111/j.1432-1033.1976.tb10108.x. [DOI] [PubMed] [Google Scholar]
- 19.Somogyi M. Notes on sugar determination. J Biol Chem. 1952;159:19–23. [PubMed] [Google Scholar]
- 20.Stratilova E, Breierova E, Vadkertiova R, Machova E, Malovikova A, Slavikova E. The adaptability of the methylotrophic yeast Candida boidinii on media containing pectic substances. Can J Microbiol. 1998;44:116–120. [Google Scholar]
- 21.Tani Y, Kato N, Yamada H. Utilization of methanol by yeasts. Adv Appl Microbiol. 1978;24:165–186. doi: 10.1016/s0065-2164(08)70639-7. [DOI] [PubMed] [Google Scholar]
- 22.Tani Y, Sakai Y, Yamada H. Isolation and characterization of a mutant of a methanol yeast, Candida boidinii S2, with higher formaldehyde productivity. Agric Biol Chem. 1985;49:2699–2706. [Google Scholar]
- 23.Tani Y, Sakai Y, Yamada H. Production of formaldehyde by a mutant of methanol yeast, Candida boidinii S2. J Ferment Technol. 1985;63:443–449. [Google Scholar]
- 24.Yanase H, Okuda M, Kita K, Sato Y, Shibata K, Sakai Y, Kato N. Enzymatic preparation of [1,3-13C]dihydroxyacetone phosphate from [13C]methanol and hydroxypyruvate using the methanol-assimilating system of methylotrophic yeasts. Appl Microbiol Biotechnol. 1995;43:228–234. [Google Scholar]