Abstract
A new multiplexed, bead-based method which utilizes nucleic acid hybridizations on the surface of microscopic polystyrene spheres to identify specific sequences in heterogeneous mixtures of DNA sequences is described. The method consists of three elements: beads (5.6-μm diameter) with oligomer capture probes attached to the surface, three fluorophores for multiplexed detection, and flow cytometry instrumentation. Two fluorophores are impregnated within each bead in varying amounts to create different bead types, each associated with a unique probe. The third fluorophore is a reporter. Following capture of fluorescent cDNA sequences from environmental samples, the beads are analyzed by flow cytometric techniques which yield a signal intensity for each capture probe proportional to the amount of target sequences in the analyte. In this study, a direct hybrid capture assay was developed and evaluated with regard to sequence discrimination and quantitation of abundances. The target sequences (628 to 728 bp in length) were obtained from the 16S/23S intergenic spacer region of microorganisms collected from polluted groundwater at the nuclear waste site in Hanford, Wash. A fluorescence standard consisting of beads with a known number of fluorescent DNA molecules on the surface was developed, and the resolution, sensitivity, and lower detection limit for measuring abundances were determined. The results were compared with those of a DNA microarray using the same sequences. The bead method exhibited far superior sequence discrimination and possesses features which facilitate accurate quantitation.
In environmental microbiology, there is a need to develop more rapid, sensitive, and accurate methods for detecting specific DNA sequences in complex, heterogeneous mixtures of DNA. This would enable the profiling of microbial communities and quantification of changes in soil, sediment, and water microbiota as indicators of bioremediation. The issues in evaluating a method include the ability to discriminate sequences, the quantitative precision and accuracy in measuring abundances of specific sequences, the ability to measure low concentrations of sequences, the ease and rapidity of preparation and detection of samples, the degree of multiplexing (i.e., the number of sequences that can be detected in a single run), and cost.
A variety of molecular methods have been used for microbial profiling (13). One of these methods, oligonucleotide microarrays, can handle a high degree of multiplexing, but at this time, the slides and the equipment for printing the arrays and detecting the signals are expensive and inconvenient to use on a routine basis. An even greater concern is that, despite efforts to construct a useful array for environmental samples, there still is no product which meets the needs for sequence discrimination and quantitation of abundances. Previous oligonucleotide microarrays for environmental samples have targeted genes for monitoring ecotoxicity (3) or rRNA or 16S ribosomal DNA (rDNA) of nitrifying bacteria (7), Escherichia coli and Vibrio proteolyticus (4), and Geobacter chapelleii (D. P. Chandler, D. A. Stahl, and J. F. Gaillard, DOE-NABIR PI Workshop, abstract, 2000). These arrays used samples obtained from culturable microorganisms. Although there are many technical challenges to overcome, the multiplexing capabilities of microarray technology are a powerful tool for dealing with the enormous diversity of natural populations, and for that reason, improvements in this and related technologies are being explored.
In the present study, we develop a new multiplex method, related to oligonucleotide microarrays but based on microscopic polystyrene spheres and flow cytometry, that can identify individual sequences in mixtures of DNA sequences. The “bead method” involves the following elements: microspheres bearing carboxyl groups on the surface, three fluorophores for multiplexed detection, and flow cytometry instrumentation. Two classification fluorophores (in this study, red and orange dyes) are impregnated throughout the volume of the beads in varying discrete amounts, thereby creating distinct populations of “bead types” distinguishable by their red and orange intensities. The DNA assay is conducted on the surface of the beads, and a third fluorophore (in this case, green dye) is used as the reporter. In flow cytometry, the beads are directed single file into a thin fluid column where they are interrogated one at a time by a laser.
An oligonucleotide (“capture probe”), designed to be complementary to a particular target sequence, is attached to the surface of a unique bead type. The analyte consists of a mixture of green-labeled DNA sequences, of which some are targeted by the capture probes. By mixing different bead types in a single tube and exposing them to the same analyte, direct hybrid capture occurs between matching capture probes and sequences. Using flow cytometry instrumentation, multiplexed detection is accomplished through measurements of the red, orange, and green emission intensities and the forward and side scatter. Detection times are typically a few seconds to a few minutes per tube.
Several DNA assays on bead surfaces have been reported. The Luminex Corporation (Austin, Tex.; personal communication) suggests that the best capture probe lengths for sequence discrimination are 18 to 20 bases and that larger signals can be achieved with 22- to 24-mers. Longer capture probes range from 45 to 564 bases (8, 9, 10, 22). For multiplexed bead formats, a competitive assay, a ligation assay, and capture of biotinylated sequences followed by indirect labeling using fluorescent conjugates of streptavidin (5, 8, 20) have been demonstrated with eukaryotic and viral systems. Target sequences were 150 to 462 bp (8) or ∼200 bp (Luminex, personal communication).
In this paper, we describe the design and performance of the bead method for identifying and quantifying prokaryotic sequences obtained from an environmental sample. As in previous studies (5, 8, 20) using a multiplexed bead approach, we worked with a Becton Dickinson flow cytometer, two-color beads, and a similar hybridization buffer. This work differs in other ways. We developed another hybridization procedure consisting of direct hybrid capture of single-stranded (ss), fluorescent target DNA because of the need for high-quality sequence discrimination and accurate quantitation of concentrations. We also used a different fluorescence label and optimized the detection buffer. Longer target sequences, more realistic for environmental studies, were tested. As a step toward absolute quantitation of abundances, a bead standard was prepared with a known number of fluorescent oligonucleotides on the surface. A more refined statistical analysis procedure was adopted to characterize the performance of the bead method. A more fundamental understanding of factors for optimizing detection was achieved.
Where possible, comparative results between the bead method and an oligonucleotide microarray are shown. This work provides a basis for developing quantitative, multiplexed bead-based assays for any set of sequences.
MATERIALS AND METHODS
Capture probe design and oligonucleotide synthesis.
Target sequences were aligned using the PileUp program from the Wisconsin Package (Genetics Computer Group). Oligonucleotides (see Table 1) were designed with OligoTech (Oligos Etc.) to avoid stem-loops and homodimers. The sequences for the beads are substrings of the ones used for the microarray.
TABLE 1.
Oligonucleotide sequencesa
| Oligonucleotide name | Description | Sequence 5′ to 3′ |
|---|---|---|
| 7102 | 204 array capture probe | SS-CT AgC gAA AAg CgA TTC CCT TTC g |
| 7103 | 204 array detection probe | ACC CgA gAg AgA-FL |
| 7105 | 1404 array capture probe | SS-TT CTA TAg gCT TgT gTT CAC AAg TC |
| 7106 | 1404 array detection probe | AAg CgT TCA CAC-FL |
| 7108 | 401 array capture probe | SS-Ag AAA TCA ACA TTC CAC AgC CAC gC |
| 7109 | 401 array detection probe | TgC AAA TgC TCA-FL |
| 7111 | 102 array capture probe | SS-gT TgT ggg CTT ggg TAg Agg gAC TC |
| 7112 | 102 array detection probe | CTC CCT gTg TCC-FL |
| R2 | PCR primer, forward | ggg WgA AgT CgT AAC AAg |
| R5FS | PCR primer, reverse | FL-T∧T ∧A∧g∧C ACg TCC TTC ATC gCC |
| 1234 | Standard | UL-AT ggT CTT CTg gTT gCC CCC-FL |
| 2000 | 102 bead capture probe | UL-gg gCT Tgg gTA gAg ggA CTC |
| 2001 | 1404 bead capture probe | UL-TT CTA gAg gCT TgT gTT CAC |
| 2002 | 204 bead capture probe | UL-CT AgC gAA AAg CgA TTg ggT |
Capture probes and PCR primer sequences for the bead method and the DNA microarray. Additional oligonucleotides, not shown here, were synthesized for testing purposes. SS, disulfide; UL, unilinker; FL, fluorescein; ∧, phosphorothioate linkage.
The capture probes for the beads were synthesized with a 5′-amino modification called “unilinker” (Operon Technologies, Inc., or Oligos Etc.). Oligonucleotide 1234, used on the bead standard, corresponds to human locus 20 and has a 5′-unilinker, 3′-fluorescein modification. These oligonucleotides were reconstituted in water to a concentration of 200 μM.
The PCR primers, R2 (forward) and R5FS (reverse), are located within the 16S and 23S rDNA regions, respectively, flanking the intergenic spacer region (ISR). Primer R5FS (Operon Technologies, Inc.) was modified at the 5′ end with “fluorescein-ON” and four phosphorothioate linkages between the five terminal bases; a phosphodiester bond exists between the fluorescein and the 5′-terminal base.
PCR amplification and quantitation of the amplicon concentration.
The template DNA for the PCR amplifications was plasmid DNA (pDNA) containing inserts corresponding to ISR sequences 102 (728 bp), 204 (647 bp), 401 (591 bp), and 1404 (628 bp). These sequences were obtained by cloning the ISR of microorganisms (6) filtered from polluted groundwater at the U.S. Department of Energy site at Hanford, Wash. (D. Brown and F. Robb, unpublished data). The pDNA was purified using the Qiagen Spin Miniprep kit and resuspended in water.
PCRs were carried out in a total volume of 29.5 μl containing 2.5 U of Amplitaq DNA polymerase (Perkin-Elmer), 1× GeneAmp PCR buffer supplied with the polymerase, final concentrations of 0.68 μM (each) primer and 55 μM (each) deoxynucleoside triphosphate, and 0.5 μl of pDNA. The thermal cycling protocol consisted of a 3-min denaturation step at 94°C, followed by addition of Amplitaq (hot start) and 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 50°C), and extension (1 min 30 s at 72°C), followed by a final extension for 5 min at 72°C. The PCR products were then stored at 4°C for up to several weeks. The concentrations of the PCR products were determined from a 1% agarose gel containing ethidium bromide and a titration of Low DNA Mass Ladder (Life Technologies catalog no. 10068-013). Typical concentrations were about 40 to 70 fmol of DNA per μl.
Preparation of full-length, ss-PCR products.
Based on reference 14, ss-PCR products were prepared by adding 10 to 20 U of T7 gene 6 exonuclease (U.S. Biochemical Corp., Cleveland, Ohio) to a mix of PCR product, water (optional), and 1× exonuclease buffer supplied with the enzyme (5× buffer contains 200 mM Tris-HCl [pH 7.5], 100 mM MgCl2, and 250 mM NaCl). Incubation occurred for 45 min at 37°C, after which no band was visible on a gel. The 5′-to-3′ hydrolytic activity of exonuclease is inhibited by the four phosphorothioate bonds. For the microarray experiments, the exonuclease reactions were stopped with 1 mM EDTA, pH 8. For the bead method, no stopping was used.
Attachment of capture probes to beads.
The beads (2.5 × 106 in number) were initially washed with water, by centrifuging them at 10,000 rpm (5,600 × g) for 1 min and removing the supernatant. The beads were resuspended by vortexing them in 50 μl of 0.1 M MES [2-(N-morpholino)ethanesulfonic acid; Sigma], pH 4.5, containing 1 nmol of the capture probe. EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimidehydrochloride; Pierce] was diluted in water to a concentration of 10 mg/ml; 2.5 μl of the solution was added to the beads. The mix was vortexed and incubated at room temperature in the dark for 30 min. Addition of EDC was repeated. The beads were then washed with 1 ml of 0.02% Tween 20, followed by two washes in 500 μl of 0.1% sodium dodecyl sulfate (SDS). After removal of the supernatant, the beads were resuspended in 0.1 M MES (pH 4.5) to a final concentration of 5 × 104 beads/μl and stored in the refrigerator for several months. The bead concentration was checked with a hemacytometer.
Beads.
Typically, four kinds of carboxylated beads of the same size (5.6 ± 0.06 μm in diameter) were used: plain (i.e., undyed) beads (Bangs Laboratories) and three types of colored beads, labeled 8047, 8058, and 8069 (Luminex Corp.). These colored beads contain equal amounts of orange dye (no. 80) and varying amounts of red dye (no. 47, 58, and 69). A unique capture probe was attached to each bead type. The bead-probes were designated plain/102, 8047/204, 8058/102, and 8069/1404, where 8047/204 means that the capture probe complementary to the 204 PCR product is attached to bead type 8047.
Direct hybrid capture of PCR products on bead surfaces (Fig. 1b).
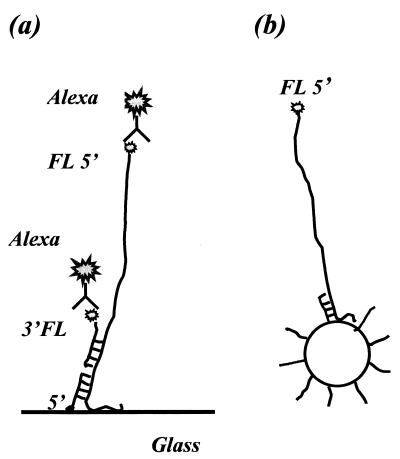
FIG. 1.
Hybridization procedures. (a) Modified sandwich assay on the DNA microarray. Capture probes were attached at the 5′ end to a glass slide. A mixture of 5′-fluoresceinated ssDNA and 3′-fluoresceinated detection probes was pipetted over the microarray. The detection probe was designed to be adjacent to the capture probe. Amplification of the fluorescence signal was achieved with an antifluorescein antibody, Alexa 488. (b) Direct hybrid capture on beads. Capture probes were attached at the 5′ end to the bead surface. The beads were then combined with a mixture of 5′-fluoresceinated ssDNA sequences.
A 1.5× TMAC buffer was prepared according to the directions of Luminex Corp. (personal communication) to contain 3 M tetramethylammonium chloride (Sigma), 0.1% SDS, 50 mM Tris-HCl (pH 8.0) (Amresco), and 4 mM EDTA (pH 8.0). A bead-probe mix was prepared by suspending 25,000 beads of each type in 1.5× TMAC buffer for a total volume of 34 μl and prewarmed at the hybridization temperature of 46°C. The desired volume of ss-PCR product was brought up to 17 μl with water, heated at 95°C for 10 min, combined with the bead mix, and incubated at 46°C for 1 h. The beads were washed one or two times in prewarmed 1× TMAC buffer, resuspended in 200 μl of 1× high-pH TMAC buffer (pH 9.8; see below), and kept at room temperature in the dark for up to 24 h before detection.
Flow cytometry measurements.
The beads were analyzed with a Becton Dickinson FACScan flow cytometer using Ar laser excitation at 488 nm and blood bank saline (0.90% [wt/vol] NaCl [pH 6.0 to 7.5] at 25°C; Fisher) for the sheath fluid. The core stream was 1× high-pH TMAC buffer (3 M TMAC, 0.1% SDS, 50 mM Tris buffer, 4 mM EDTA [pH 8.0]). Forward and side light scattering enabled the separation of monomeric beads signals from other signals based on size discrimination. Green-orange compensation was used. Typically, 100 to 2,000 events per bead type were collected with a total acquisition time of 15 to 180 s per tube. Cytometry data were analyzed with CellQuest software.
With each set of samples, two types of bead standards were run. The response function was determined with the LinearFlow Green Flow Cytometry Low Intensity Calibration Kit (Molecular Probes). Surface-labeled beads (see Results) were used to convert intensity values into the number of fluorescent nucleic acid sequences (FNAS) per bead.
Statistical analysis of flow cytometry data.
The following quantities were used to analyze the data: number (N) of beads of a certain type; mean signal value (ns) for beads with DNA; mean background signal (nb) for beads with no DNA hybridized to the surface; signal (= ns − n b) due to DNA; standard deviation (S); coefficient of variation (CV); standard error in the mean (ς = S/N1/2); standard error in the difference of means ςd = (ςs2 + ςb2)1/2, and probability (p) for the confidence interval. (Subscripts s and b refer to signal and background, respectively). The lowest detectable DNA signal was determined by nmin = nb + kςd, where ςd was calculated from the lowest mean signal with the condition that ns − nb > kςd. In this paper, p = 68% and k = 1 for N ≥ 30. The resolution in the signal is given by δns ≈ kςs√2.
Fluorescence imaging microscopy.
Individual beads were imaged with an upright episcopic fluorescence microscope (Nikon Labophot-2) equipped with a cooled charge-coupled device (CCD) camera (First Magnitude SpecIm-3) and an Ar laser (Laser Physics 150M-006-00B). A linearly polarized laser beam (488 nm) was sent into the microscope and compressed by the microscope objective lens onto the sample plane. The fluorescence emission was selected by two glass cutting filters and a band-pass interference filter (530 nm). The beads were resuspended in high-pH TMAC buffer and placed between two quartz coverslips.
Absorption spectrum measurements.
The concentration and spectroscopic properties of dye-labeled oligonucleotides were tested at room temperature using a Spectronic Genesys-5 spectrophotometer.
DNA microarrays.
The alignments, PCR amplifications, quantitation of amplicons, and preparation of ss-target sequences are described above.
(i) Oligonucleotides (Table 1).
Oligonucleotides (12-, 24-, and 25-mers) were synthesized by Molecular Tool, Inc. (now called Orchid Biocomputer), according to the procedure described in reference 16. Printing of seven-by-seven arrays involved deposition of capture probes onto the surface of mercaptosilane-coated glass slides (Molecular Tool, Inc.). The printing and surface chemistry of the attachment process are described in reference 16.
(ii) Hybridization.
The hybridization procedure is depicted in Fig. 1a, in which a fluoresceinated detection probe is first hybridized to an ss-fluoresceinated target sequence, followed by capture onto the microarray. The capture and detection probes are adjacent to each other. The purposes of the detection probe are to reduce secondary structures near the vicinity of capture, to create a helical twist to the DNA which would promote hybridization, and to provide an additional fluorescent center for detection.
The analyte consisted of 1× GBA salts (1 mM cetyltrimethylammonium chloride, 1.5 M NaCl, 10 mM EDTA), the desired ss-PCR product volume, 1 μM (each) detection probe, and water for a total volume of 40 μl. The microarrays were prewarmed in a moist environment for 30 to 40 min at 37°C. The analyte was initially heated to 95°C for 10 min, held at 37°C for 5 to 10 min, pipetted onto the warm arrays, and incubated in a moist environment for 3 h at 37°C. The slides were washed three times in 1× TNTw (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.05% Tween 20), dried on the back, hydrated with 1× TNTw on the front, and scanned.
(iii) Fluorescence amplification.
To amplify the emission signal from the arrays, a 1:500 dilution of Alexa Fluor 488 conjugate of antifluorescein rabbit polyclonal immunoglobulin G (Molecular Probes) was prepared using a buffer containing 1× TNTw and 1% (wt/vol) fraction V bovine serum albumin and pipetted onto the arrays. Incubation occurred for at least 30 min at room temperature. The slides were washed three times in 1× TNTw, dried on the back, hydrated on the front, and scanned. There were 3.3 Alexa molecules per antibody molecule.
(iv) Detection and analysis.
Each slide was scanned with a Molecular Dynamics 575 FluorImager using an argon laser light source (488 nm) and a 530-nm emission filter. The images were analyzed using mostly ImageQuant software (Molecular Dynamics). Additional images were acquired using a cooled CCD camera with a Fujinon TV lens HF35A-2, 488-nm laser excitation, two barrier glass filters, and a band-pass interference filter for selecting the green fluorescence (530 nm) of the microarray.
The statistical analysis was similar to that for the beads (see above). Small sample statistics were used in which N = 6 was the number of spots of each type.
RESULTS
Spectroscopy, detection buffer, and bead standard.
To find a buffer suitable for the core fluid, we measured the absorption spectra of oligonucleotide 1234 (3′-fluorescein) and a water-soluble fluorescein salt (Aldrich 16,630-8) in water, blood bank saline, TNTw (pH 7.4), MES (pH 4.5), and TMAC buffer (pH 8.5 and 9.8). The results were compared with a fluorescein solution in 0.01 M NaOH which has known spectroscopic characteristics and with literature data for different prototropic forms of fluorescein molecules (11). Oligonucleotide 1234 exhibited the dianion form of fluorescein in TMAC (pH 9.8), the monoanion form in water and in saline, and the mixture of these two forms in the other liquids. In TMAC and water, primer R5FS (5′-fluorescein) had the same absorption spectrum as 1234. Since the dianion form has the highest fluorescence efficiency (11, 19), TMAC buffer (pH 9.8) was used as the reference liquid in all of the calibration procedures and as the core fluid for the flow cytometry measurements.
The percentage of oligonucleotide 1234 labeled with fluorescein was determined to be 70% based upon extinction coefficients and optical densities at 260 and 498 nm, assuming no free fluorescein molecules.
A bead standard was prepared by attaching oligonucleotide 1234 to plain beads. Using fluorescence microscopy, the image intensities of the beads and a solution of oligonucleotide 1234 in high-pH TMAC buffer were compared. The mean intensity value per bead was found to be equivalent to 3.25 × 105 FNAS, corresponding to 4.6 × 105 oligonucleotides attached to the bead surface. From flow cytometry measurements, this bead standard in TMAC buffer has an emission signal 13.6-fold higher than that in water, 15-fold higher than that in saline, and 2-fold higher than that in TNTw due to activation of the dianion form of fluorescein.
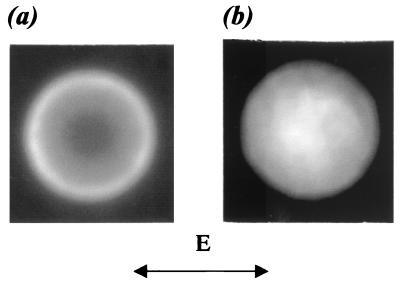
Surface distribution and spatial orientation of capture probes.
Figure 2 shows images of the surface-labeled bead standard (Fig. 2a) and a bead with green dye molecules impregnated in the volume (Fig. 2b). The increase in intensity from the center to the perimeter of the bead in Fig. 2a corresponds to the increase in the emitting surface projected onto a two-dimensional plane. In Fig. 2b, the intensity is greatest in the center, corresponding to the largest emitting volume projected onto the image plane. For Fig. 2a, there is an intensity variation of 15% along the perimeter, possibly corresponding to slight inhomogeneities in the surface attachment. Image analyses also show that fluorescein molecules conjugated to oligonucleotides have a nearly random orientation on the surface of the beads. This is because, with linearly polarized excitation, a difference of intensity between the orthogonal sections of the bead perimeter is expected if transient dipoles in dye molecules have a predominant orientation relative to the bead surface. In fact, we observed random spatial variations in the intensities in Fig. 2a which cannot be attributed to orientational phenomena. From additional polarization measurements, we determined that there is partial immobilization of labeled oligonucleotides on the bead surface during the 4-ns fluorescence lifetime of the dianions (11, 19), which mostly have isotropic orientation.
FIG. 2.
Fluorescence microscopy of individual beads using linearly polarized laser irradiation with electric field vector E. The spatial resolution on the sample plane was 0.4 μm. The photon density of the excitation was 2 × 1017 photons/cm2 · s, and the CCD integration time was 1 s. (a) Bead standard (5.6-μm diameter) with 3′-fluoresceinated oligonucleotides on the surface. (b) Molecular Probes calibration bead (6-μm diameter).
Hybridization procedure.
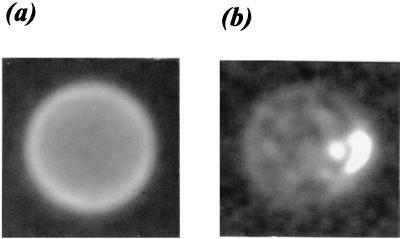
We compared the behavior of ss-PCR and double-stranded (ds)-PCR products in the hybridization procedure. Beads were combined with 500 fmol of ds-PCR product in one tube or 500 fmol of ss-PCR product in another tube. After hybridization, the beads were analyzed by fluorescence microscopy and flow cytometry. As shown in Fig. 3a, uniform fluorescence was observed on the bead surface for the ss-sample. In contrast, the ds-sample showed random patches of hybridization of varying size and shape (Fig. 3b). From flow cytometry measurements, these beads had a mean intensity corresponding to 8.3 × 102 FNAS with CV ≈ 40% and 5.5 × 103 FNAS with CV ≈ 15% in the ds- and ss-samples, respectively. Based on these data, we believe that, for quantitative analyses, hybridizations using ss-PCR products are advantageous.
FIG. 3.
Enhanced fluorescence microscopy images of beads with 102 target DNA hybridized to capture probes. (a) ss target DNA prepared with exonuclease. (b) ds target DNA.
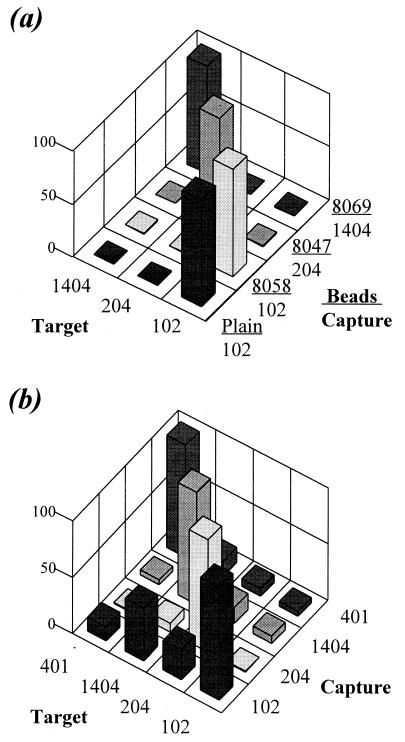
Direct and cross hybridization.
Sequence discrimination was evaluated for the bead method and the DNA microarray using the same environmental target sequences (102, 204, and 1404) and similar capture probe sequences. For the beads, each tube contained four types of beads and 400 fmol of a single target sequence, an amount corresponding to near-saturation conditions for direct hybridization. In Fig. 4a, the direct hybridization signals from matching capture probes and target sequences were normalized to 100%. The cross-hybridization level was determined for each sequence relative to the direct-hybridization level. The lowest cross-hybridization level (0.07%) was observed between the 1404 capture probe and sequence 204; the highest level (0.98%) appeared between the 102 capture probe and sequence 204. Similar results were obtained with 70, 100, 150, and 310 fmol of target sequences in each tube. In all cases, the cross hybridization was not more than 1%.
FIG. 4.
Direct and cross hybridization. The analyte consisted of a single target sequence. The data for matching target and capture probes were normalized to 100%. (a) Bead method. The 102 capture probe was attached to two bead types, plain and 8058. (b) Oligonucleotide microarray. Amplification of fluorescence with Alexa 488 was used.
Results for the DNA microarray are shown in Fig. 4b. Three assays were tested: fluoresceinated target DNA only, fluoresceinated DNA with fluoresceinated detection probe (“no-Alexa”), and fluoresceinated DNA with fluoresceinated detection probe amplified with Alexa 488. Overall, the signals from fluoresceinated DNA were low and considered insufficient for analysis. The Alexa samples, shown in Fig. 4b, had higher signals and lower cross-hybridization levels than the no-Alexa samples. Nevertheless, the cross-hybridization level for microarrays was significantly higher than that for beads. The saturation for direct hybridization was approximately ≥900 fmol of DNA per array. At 890 fmol, the lowest cross-hybridization level was 0.34% between the 204 capture probe and 102 sequence (compared with 0.1% for the bead method). The highest cross-hybridization level was 46% between the 102 capture probe and the 1404 sequence (compared with 0.55 to 0.62% for beads).
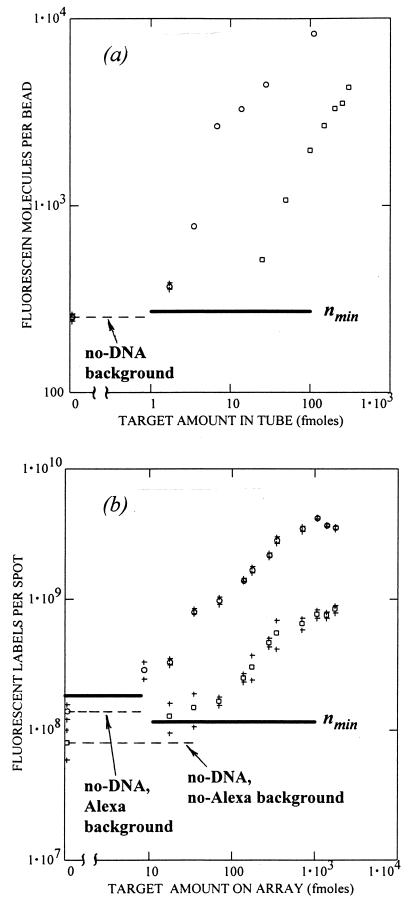
Lower detection limit and resolution of the concentration of DNA.
The minimum detectable amount of DNA in the analyte was determined from an analysis of the concentration curves. Two bead assays were tested. The first had a total reaction volume of 6 μl and 12,000 beads (i.e., 3,000 beads of each type). The second had a total volume of 51 μl and 100,000 beads (i.e., 25,000 beads of each type). In Fig. 5a, ns is plotted as a function of amount C of target sequence 102 hybridized to plain/102 beads. For all points, the relative uncertainties ςs/ns ranged from 0.8 to 4.4%. The background nb of plain beads corresponded to 252 FNAS/bead (ςb = 11 FNAS). Using ςs = 14 FNAS for the lowest target concentration in the 6-μl assay, the minimum detectable signal corresponded to nmin ≈ 270 FNAS/bead. That is, 18 DNA molecules can be detected on each bead above background with 68% probability. The minimum detectable amount of DNA in the tube (Cmin) was determined by extrapolating the concentration curve to nmin to obtain 1.3 fmol for the 6-μl assay and 13 fmol for the 51-μl assay.
FIG. 5.
Dependence of hybridization on target amount (sequence 102) for the bead method and the DNA microarray. (a) Two bead experiments using a 6-μl (○) and a 51-μl (□) total volume in each tube. (b) Results from two microarray experiments, one using no fluorescence amplification (no-Alexa) (□) and the other using Alexa (○). For both graphs, the mean background value (horizontal dashed line) and the lower limit of detection nmin are shown. The 68% confidence intervals are indicated by plus signs; in panel a, only the largest intervals are shown.
We define the sensitivity Sc in the concentrations as the derivative of the signal mean with respect to the concentration at a given point on the concentration curve: Sc = (dns/dC). The resolution in DNA amount in a tube (δC) is given by δC = δns/Sc. For the middle of the curve, we then obtain Sc ≈ 52 FNAS/bead · fmol and δC ≈ 1 fmol for the 6-μl assay and Sc ≈ 13 FNAS/bead · fmol and δC ≈ 7 fmol for the 51-μl assay.
The bead method and the DNA microarray were compared. Quantitation of the microarray was accomplished in terms of the number of fluorophore molecules per square micrometer on the glass slide according to reference 1, in which the fluorescence intensities of the microarray spots were compared with a thin solution layer of the same fluorescence label of known concentration. The microarray results are shown in Fig. 5b, in which the vertical axis represents the number of fluorescent labels per spot and the horizontal axis represents the amount of DNA pipetted over each array. The background signals and uncertainties were different for the no-Alexa (ςs/ns ∼ 7 to 30%) and Alexa (ςs/ns ∼4 to 15%) experiments. The nmin value increased from 1.1 × 108 to 1.8 × 108 fluorescent labels per spot. For the no-Alexa experiment, Sc was ∼1.38 × 106 fluorescent labels/spot · fmol, δC was ∼60 fmol (at the middle), and Cmin was ∼15 fmol. For the Alexa samples, Sc was ∼6.6 × 106 fluorescent labels/spot · fmol, δC was ∼17 fmol, and Cmin was 6 to 10 fmol.
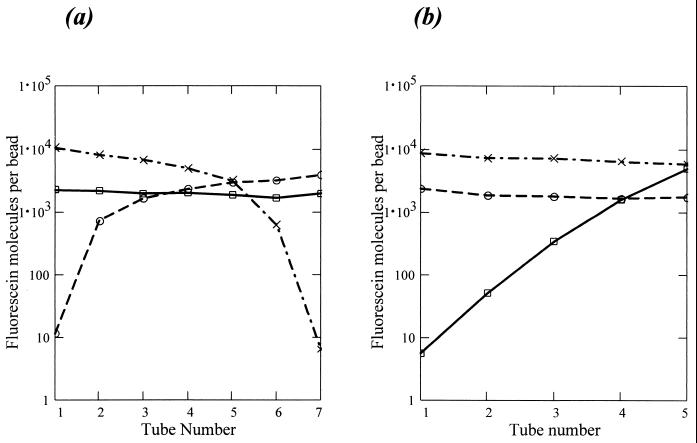
Competition effect in mixtures.
Since the goal of the bead method is to study mixtures of environmental DNA sequences, we evaluated the resolution in measuring concentrations for known mixtures of three DNA sequences, 102, 204, and 1404, using two tests. The first (Fig. 6a) consisted of seven samples (tubes) where the concentrations of 102 and 1404 were varied, keeping 204 constant, and the total concentration was constant at 400 fmol. The second (Fig. 6b) consisted of five samples where the concentration of 204 was increased, holding 102 and 1404 constant. The total reaction volume in each tube was 51 μl with 75,000 beads (25,000 of each type). The intersample variation in the hybridization levels was obtained for the sequences held at a constant concentration and was found to be different for the two tests. In Fig. 6a, the CV was 9% for sequence 204. However, in Fig. 6b, the mean intensity decreased monotonically for 102 and 1404. The CV was no more than 15% across all five tubes. There appears to be a competition effect in mixtures in which the measured signal depends upon whatever else is in the sample. The value of 15% represents the intersample resolution in concentrations if competition effects are disregarded.
FIG. 6.
Dependence of hybridization on the composition of the analyte. The target sequences were 102 (○), 204 (□), and 1404 (×). (a) All tubes contained 400 fmol of DNA in total. The concentration of sequence 204 was kept constant at 100 fmol for tubes 1 to 7. The amount of 102 increased from 0 to 300 fmol, while the amount of 1404 decreased from 300 to 0 fmol in decrements of 50 fmol. (b) The total number of DNA molecules was varied from tube to tube. The amounts of 102 and 1404 were kept constant at 100 and 200 fmol, respectively. The concentration of 204 was 0 fmol in tube 1, 25 fmol in tube 2, 50 fmol in tube 3, 100 fmol in tube 4, and 200 fmol in tube 5.
DISCUSSION
Low levels of cross hybridization are important for quantifying abundances of microorganisms in environmental samples containing many unknown sequences. For the bead method, the cross-hybridization level never exceeded 1% of the direct-hybridization signal. This corresponds to 10 to 30 fluoresceinated DNA molecules per bead, a value comparable to the minimum number of sequences that could be detected in our experiments. Thus, one can expect the actual cross-hybridization level to be even less. These results are far superior to the performance of the oligonucleotide microarray.
Prior studies using a multiplexed bead assay for DNA also demonstrate excellent sequence discrimination. These studies include an HLA tissue typing assay (5); a viral load assay in which six sequences associated with human immunodeficiency virus, hepatitis C virus, and herpes simplex virus were detected at the femtomole level (20); and single nucleotide polymorphism detection of nine single nucleotide polymorphism markers located near the ApoE locus on chromosome 19 (8). Based on these results and ours, we believe that the bead method should be able to distinguish other sets of sequences useful for environmental studies, including 16S rDNA sequences and biodegradation alleles. Our preliminary experiments using 16S rDNA sequences indicate excellent cross-hybridization results but a weaker signal in comparison with ISR sequences.
There are several possible reasons for the high-quality sequence discrimination. All of the hybridizations were conducted in TMAC, which roughly equalizes the stability of the A:T and G:C base pairs, thereby reducing hybridization bias based on sequence variations (21). In addition, thermal tests of the beads with capture probes indicated that the beads are stable at 95°C for short time intervals, and so the hybridization procedure involved 95 and 46°C steps, which are higher than what could be used with the microarray. We did not conduct hybridization tests using more conventional hybridization buffers; it is possible that if the target sequences are different enough, then these buffers may be adequate (8).
The quantitation in the bead experiments was accomplished in terms of FNAS units. MESF units (the number of molecules of equivalent soluble fluorochromes), widely used in flow cytometry, do not indicate the actual number of sequences but only how intense the dye-labeled sample (bead or biological cell) is relative to the same dye solution containing free fluorophore molecules (17, 18). The problem is that, when dealing with labeled DNA, such commercial standards are of limited use because the fluorescent properties of dyes can be significantly changed upon conjugation with biopolymers and are sensitive to pH (2, 11, 12, 15, 19). In this work, a bead standard was prepared with a known number of fluoresceinated oligonucleotides on the surface; this is a spectrally matched standard, responsive to the surrounding fluid, that enabled us to make a transition from relative fluorescence intensities to FNAS units. We believe this is a convenient and comprehensive procedure to directly compare and evaluate the results of different bead experiments and different types of fluorescence detection.
Analysis of Fig. 5a and 6 indicates that <0.1% of the total amount of DNA in the analyte was actually used for fluorescence detection. This occurred even when the bead surface was not fully occupied by target molecules and suggests that the sensitivity and lower detection limit of the bead method can be improved by optimizing the hybridization conditions. However, even at the current level, in which a minimum of several femtomoles of target DNA is needed in the tube, we are still able to detect DNA without additional fluorescence amplification (e.g., multiply labeled target molecules and fluorescent antibodies). This is an advantage for accurate quantitation, where it is necessary to know the uncertainty associated with each preparation step.
Further study of intersample statistics with DNA mixtures is necessary. There may be uncontrolled variability in the preparation conditions (temperature, volume, number of beads, etc.). However, at best, since the intrasample uncertainty was 1 to 4%, we believe this is the minimum CV value for intersample statistics. We speculate that the competition effect which changed the intersample statistics may be due to a decrease in the diffusion rate at higher concentrations complicated by intermolecular interactions.
There have been many technical improvements with DNA microarrays, and further improvements in both the bead method and microarrays will continue to be made. Thus, an unequivocal judgment on the relative performance of these two methods is difficult to make. However, given our data, with regard to the lower detection limit, we did not notice a significant difference between the bead method and the microarray. With regard to other considerations, including surface chemistry, hybridization protocols, detection instrument, quantitation, ease of developing a useful assay, and cost, we believe the bead method has advantages for environmental applications. At this time, the surface chemistry on beads appears to be more stable, enabling higher temperatures and simpler hybridization protocols to be used while still achieving excellent sequence discrimination. The hybridization reactions on beads are faster than those on microarrays; this is most likely due to the three-dimensional nature of the bead system, which allows mixing and close proximity between the beads and target molecules. For developing useful assays, the bead method is far more convenient; modifying the set of sequences to be studied is easily accomplished by attaching new oligonucleotides to the beads and pipetting different beads together. Arrays, however, need to be reprinted, requiring specialized equipment and more time.
The bead method also has several features making it preferable to microarrays for quantitation. First, because hundreds or thousands of beads of each type are examined, good statistics are easy to achieve; on arrays, comparable statistics have to be accomplished by either many replicate spots on high-density arrays or very large spots. Second, for beads, our data show excellent spatial homogeneity in the attachment of the capture probes to the bead surface and also in the hybridization of target sequences to the capture probes. This is due to the spherical symmetry of the beads, which enables uniform conditions for the surrounding DNA. This level of homogeneity cannot be surpassed by microarrays. In fact, for large spots, Bogdanov et al. (1) showed that strong attachment in homogeneities are induced by hydrodynamic redistribution of the DNA concentration inside the solution drop on hydrophobic glass surfaces. Third, the random orientation of the fluorophores on the bead surface produces adequate conditions for quantitative comparison with solutions. In contrast, the fluorophores on microarrays may have a predominant orientation (1), making the arrays sensitive to the geometry of the optical excitation and its polarization.
With regard to cost and the ability to set up similar bead experiments in any lab, there are several considerations. While this paper concentrates on three sequences, we do not anticipate difficulties in dealing with a larger number of capture probes. Currently, 64 red-orange bead types that are suitable for flow cytometers using an argon laser light source are commercially available (Luminex Corp.). Custom beads can also be ordered. Flow cytometers, of the type used in this study, are quite common; thus, it may not be necessary for the researcher to buy specialized detection instrumentation. However, existing software for quantitative analysis of the data is not optimized for real-time multiplexed detection but is effective for slow, careful postprocessing.
Finally, we wish to note that beads can be used to prepare sequences for subsequent steps. By using a cell sorter, a search for new genes in environmental samples may be expedited.
ACKNOWLEDGMENTS
We acknowledge Frank Robb at COMB and the staff at Molecular Tool, Inc. (Yu-Hui Rogers, Miriam Donaldson, Valery L. Bogdanov, and Michael Boyce-Jacino), for the target sequences, DNA microarray work, and hosting M.L.; the staff at Luminex Corporation for sharing their protocols and experience; and Gina Hamlin Green and Mark Soloski at the Johns Hopkins University for use of the flow cytometer.
We acknowledge DOE NABIR grant DE-FG02-99ER62868. Preliminary work was partially funded by DOE DE-FG07-96ER62320 and NSF CTS-9253633.
REFERENCES
- 1.Bogdanov V L, Rogers Y-H, Boyce-Jacino M. Fluorescent imaging and quantitation of solid support bound nucleic acids. Proc SPIE. 1997;2985:129–137. [Google Scholar]
- 2.Der-Ballan G P, Kameda N, Rowley G L. Fluorescein labeling of Fab′ while preserving single thiol. Anal Biochem. 1988;173:59–63. doi: 10.1016/0003-2697(88)90159-5. [DOI] [PubMed] [Google Scholar]
- 3.Doktycz M J, Beattie K L. Genosensors and model hybridization studies. In: Beugelsdiik A J, editor. Automation technologies for genome characterization. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 205–225. [Google Scholar]
- 4.Eggers M D, Balch W J, Mendoza L G, Gangadharan R, Mallik A K, McMahon M G, Hogan M E, Xaio D, Powdrill T R, Iverson B, Fox G E, Willson R C, Maillard K I, Siefert J L, Singh N. Proceedings of the 27th International Conference on Environmental Systems. SAE Technical Paper Series 972422. Warrendale, Pa: Society of Automotive Engineers, Inc.; 1997. Advanced approach to simultaneous monitoring of multiple bacteria in space. [Google Scholar]
- 5.Fulton R J, McDade R L, Smith P L, Kienker L J, Kettman J R. Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 6.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23s rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 7.Guschin D Y, Mobarry B K, Proudnikov D, Stahl D A, Rittmann B E, Mirzabekov A D. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol. 1997;63:2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iannone M A, Taylor J D, Chen J, Li M-S, Rivers P, Slentz-Kesler K A, Weiner M P. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry. 2000;39:131–140. [PubMed] [Google Scholar]
- 9.Imai T, Sumi Y, Hatakeyama M, Fujimoto K, Kawaguchi H, Hayashida N, Shiozaki K, Terada K, Yajima H, Handa H. Selective isolation of DNA or RNA using single-stranded DNA affinity latex particles. J Colloid Interface Sci. 1996;177:245–249. doi: 10.1006/jcis.1996.0027. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen C S. Microscale detection of specific bacterial DNA in soil with a magnetic capture-hybridization and PCR amplification assay. Appl Environ Microbiol. 1995;61:3347–3352. doi: 10.1128/aem.61.9.3347-3352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klonis N, Sawyer W H. Spectral properties of the prototropic forms of fluorescein in aqueous solution. J Fluorescence. 1996;6:147–157. doi: 10.1007/BF00732054. [DOI] [PubMed] [Google Scholar]
- 12.Kumke M U, Li G, McGown L B. Hybridization of fluorescein-labeled DNA oligomers detected by fluorescence anisotropy with protein binding enhancement. Anal Chem. 1995;67:3945–3951. doi: 10.1021/ac00117a020. [DOI] [PubMed] [Google Scholar]
- 13.Madsen E L. Epistemology of environmental microbiology. Environ Sci Technol. 1998;32:429–439. [Google Scholar]
- 14.Nikiforov T T, Rendle R B, Kotewicz M L, Rogers Y-H. The use of phosphorothioate primers and exonuclease hydrolysis for the preparation of single-stranded PCR products and their detection by solid-phase hybridization. PCR Methods Appl. 1994;3:285–291. doi: 10.1101/gr.3.5.285. [DOI] [PubMed] [Google Scholar]
- 15.Nunnally B K, He H, Li L-C, Tucker S A, McGown L B. Characterization of visible dyes for four-decay fluorescence detection in DNA sequencing. Anal Chem. 1997;69:2392–2396. doi: 10.1021/ac961281p. [DOI] [PubMed] [Google Scholar]
- 16.Rogers Y-H, Jiang-Baucom P, Huang Z-J, Bogdanov V, Anderson S, Boyce-Jacino M T. Immobilization of oligonucleotides onto a glass support via disulfide bonds: a method for preparation of DNA microarrays. Anal Biochem. 1999;266:23–30. doi: 10.1006/abio.1998.2857. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz A, Fernandez-Repollet E. Development of clinical standards for flow cytometry. Ann N Y Acad Sci. 1993;677:28–39. doi: 10.1111/j.1749-6632.1993.tb38760.x. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz A, Marti G E, Gratama J W, Fernandez-Repollet E. Standardizing flow cytometry: a classification system of fluorescence standards used for flow cytometry. Flow Cytometry. 1998;33:106–114. [PubMed] [Google Scholar]
- 19.Sjoback R, Nygren J, Kubista M. Absorption and fluorescence properties of fluorescein. Spectrochim Acta. 1995;A51:L7–L21. [Google Scholar]
- 20.Smith P L, WalkerPeach C R, Fulton R J, DuBois D B. A rapid, sensitive, multiplexed assay for detection of viral nucleic acids using the FlowMetrix system. Clin Chem. 1998;44:2054–2056. [PubMed] [Google Scholar]
- 21.Wood I W, Gitschier J, Lasky L A, Lawn R M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci USA. 1985;82:1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zammatteo N, Alexandre I, Ernest I, Le L, Brancart F, Remacle J. Comparison between microwell and bead supports for the detection of human cytomegalovirus amplicons by sandwich hybridization. Anal Biochem. 1997;253:180–189. doi: 10.1006/abio.1997.2352. [DOI] [PubMed] [Google Scholar]