Abstract
PCR techniques have significantly improved the detection and identification of bacterial pathogens. Countless adaptations and applications have been described, including quantitative PCR and the latest innovation, real-time PCR. In real-time PCR, e.g., the 5′-nuclease chemistry renders the automated and direct detection and quantification of PCR products possible (P. M. Holland et al., Proc. Natl. Acad. Sci. USA 88:7276–7280, 1991). We present an assay for the quantitative detection of Listeria monocytogenes based on the 5′-nuclease PCR using a 113-bp amplicon from the listeriolysin O gene (hlyA) as the target. The assay was positive for all isolates of L. monocytogenes tested (65 isolates including the type strain) and negative for all other Listeria strains (16 isolates from five species tested) and several other bacteria (18 species tested). The application of 5′-nuclease PCR in diagnostics requires a quantitative sample preparation step. Several magnetic bead-based strategies were evaluated, since these systems are simple and relatively easy to automate. The combination of nonspecific binding of bacteria to paramagnetic beads, with subsequent DNA purification by use of the same beads, gave the most satisfactory result. The detection limit was approximately 6 to 60 CFU, quantification was linear over at least 7 log units, and the method could be completed within 3 h. In conclusion, a complete quantitative method for L. monocytogenes in water and in skimmed and raw milk was developed.
The listeriae are widely distributed in nature and can be found on decaying vegetation and in soils, animal feces, sewage, silage, and water. Of the listerial species, Listeria monocytogenes is the pathogen of concern for humans. The bacteria can grow over the temperature range of about 1 to 45°C and at a pH of 4.1 to ca. 9.6. It is well established that any fresh food product of animal or plant origin may contain L. monocytogenes. In general, the organism has been found in raw milk, soft cheeses, fresh and frozen meat, poultry, seafood, fruits, and vegetable products. L. monocytogenes has been shown to survive in foods for long periods (11). Some countries have established legal limits on the number of L. monocytogenes organisms that are permissible in foods, especially ready-to-eat products, whereas others have suggested guidelines or criteria that do not have legal standing. The U.S. government has the most rigid policy. Any ready-to-eat food that contains L. monocytogenes can be considered adulterated and thus be subject to recall and/or seizure (11). The European Community directive on milk and milk-based products specifies zero tolerance for soft cheeses and the absence of the organism in 1 g of other products (11). However, these kinds of directives must not hinder research, which can better evaluate the infectious dosages of pathogens and thus perhaps justify a more attainable limit of L. monocytogenes in foods. New risk assessments based on quantitative methods may have an impact on future legislation. Great Britain's provisional guidelines for some ready-to-eat foods establishes four quality groups based on the numbers of L. monocytogenes: not detected in 25 g is satisfactory; <102/25 g is fairly satisfactory; 102 to 103 is unsatisfactory; and numbers of >103 make the food product unacceptable (6). The International Commission on Microbiological Specification for Foods has come to the conclusion that if this organism does not exceed 100 organisms/g of food at the point of consumption, the food is considered acceptable for individuals who are not at risk (11). This means that a quantitative detection method for L. monocytogenes would be useful to control foods before consumption. It will also be of value for determining the infectious dosage of L. monocytogenes during food-poisoning outbreaks as well as for research purposes generally.
DNA-based methods such as the PCR have been increasingly used for rapid, sensitive, and specific nonquantitative detection of L. monocytogenes (4, 16). Among the various PCR strategies available, those based on monitoring the amplification reaction in real time are probably the most promising (9). The approach uses dually labeled fluorogenic hybridization probes incorporated into PCR (7, 9) (Perkin-Elmer Research News [Perkin-Elmer, Norwalk, Conn.], 1995) and exploits of the 5′-3′ nuclease activity of the Taq DNA polymerase to hydrolyze this probe during the DNA polymerization step. The probe is labeled with a reporter dye and a quencher dye, and when both dyes are attached to the same molecule the quencher dye will absorb the “excited state” energy of the reporter dye. If hybridization occurs, the probe is cleaved by the 5′-nuclease activity of the DNA polymerase during the extension of the primer. This separates the reporter dye from the quencher dye, generating an increase in the reporter dye's fluorescence intensity. The ABI Prism 7700 Sequence Detection System (PE Biosystems, Foster City, Calif.) measures the increase in the reporter dye's fluorescence during the thermal cycling of the PCR.
The 5′-nuclease assay demonstrates several advantages over other quantitative PCR approaches. The fluorogenic assay is a convenient, self-contained process. The only necessary steps are the reaction setup and the tube sealing. Unlike other quantitative PCR methods, real-time PCR does not require post-PCR sample handling, thus preventing potential PCR product carryover contamination and resulting in much faster and higher-throughput assays. The real-time PCR method has a very large dynamic range of starting target molecule determination (at least 5 orders of magnitude). Real-time quantitative PCR is extremely accurate and less labor-intensive than current quantitative PCR methods (7).
Our goal was to develop a high-throughput, specific, sensitive, and accurate quantification assay for L. monocytogenes. The most significant virulence factor associated with L. monocytogenes is listeriolysin O (11) encoded by the hlyA gene (14). We describe here the development and evaluation of a primer and probe system toward the hlyA gene, which can be used in quantification of L. monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk using the 5′-nuclease chemistry and the ABI Prism 7700 Sequence Detection System by PE Biosystems. In combination with a magnetic bead-based cell concentration and DNA purification procedure, the 5′-nuclease PCR can be used in the quantification of L. monocytogenes from a number of different environments.
MATERIALS AND METHODS
Bacterial strains, media, and cultures.
A total of 65 isolates of L. monocytogenes including the L. monocytogenes type strain, DSMZ 20600T (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), were used to control the specificity of the primers and the probe. The isolates were mostly of serotypes 1 and 4 from different sources (e.g., poultry, fish, and environmental samples) collected in Norway in the period from 1987 to 1997.
The following bacterial strains were used as negative controls (strains not assigned to any known culture collection came from private collections): Listeria innocua, five strains including the type strain (DSMZ 20649T), Listeria ivanovii, five strains, Listeria seeligeri, three strains, Listeria welshimeri, two strains, Listeria grayi, Bacillus subtilis ATCC 6633 (American Type Culture Collection, Rockville, Md.), Brochothrix thermosphacta ATCC 11509T (type strain), Campylobacter jejuni subsp. jejuni DSMZ 4688T (type strain), Carnobacterium divergens DSMZ 20623T (type strain), Carnobacterium gallinarum NCFB 2766T (National Collection of Food Bacteria, Reading, England) (type strain), Enterobacter aerogenes, Enterococcus faecalis NCDO 2602 (National Collection of Dairy Organisms, Reading, England) (type strain), Escherichia coli ATCC 8739, E. coli O157:H7, Lactobacillus acidophilus (type strain), Lactobacillus casei ATCC 393, Lactobacillus curvatus DSMZ 20019T (type strain), Lactobacillus plantarum DSMZ 20174T (type strain), Lactobacillus sake subsp. sake DSMZ 20017T (type strain), Lactococcus lactis subsp. cremoris, Salmonella enterica serovar Enteritidis, Salmonella enterica serovar Kentucky, Staphylococcus aureus ATCC 25923, and Yersinia enterocolitica.
All strains were plated on blood agar. Listeria spp. were grown in beef heart infusion media (BHI) or Listeria enrichment broth at 30°C. Bacillus, Enterobacter, Enterococcus, Escherichia, Salmonella, Staphylococcus, and Yersinia spp. were grown on BHI at 30°C. Brochothrix was grown in BHI at 25°C. Lactobacillus and Lactococcus spp. were grown in MRS broth at 25°C, and Carnobacterium spp. were grown in Trypticase soy yeast medium at 30°C. Campylobacter was grown microaerobically in BHI at 37°C. All agar and media were from Oxoid, Ltd., Basingstoke, Hampshire, England. Cultures were serially diluted in peptone-water. CFU were enumerated by plating 0.1 ml of each dilution onto Trypticase soy agar (Oxoid, Ltd.), followed by incubation at 30°C for 2 days.
Water, skim milk, and unpasteurized whole milk were artificially contaminated with an overnight culture of L. monocytogenes by adding 10-fold dilutions in defined amounts of water or milk. Aliquots were then subjected to DNA isolation. The bacterial number in the culture was estimated by standard plate counts. Prior to artificial contamination, the CFU from the background flora of the milk were enumerated by plating. The experiment was done twice.
DNA isolation from pure cultures.
Samples (0.5 ml) of overnight cultures were centrifuged at 6,000 × g for 7 min at 4°C, and the supernatants were discarded. The cell pellets were stored at −80°C. For DNA isolation, cell pellets were resuspended in 1× TE buffer (pH 8.0; 10 mM Tris, 1 mM EDTA). Dynabead DNA Direct I (Dynal AS, Oslo, Norway), 200 μl, were then added to the suspension of bacteria, which was incubated at 65°C for 15 min, followed by incubation at room temperature for 2 min. DNA bound to magnetic beads was then drawn to the wall of the microcentrifuge tube by a magnet (MPC-E; Dynal AS) for 2 min. The supernatant containing salts, detergent, and cell debris was carefully removed without disrupting the Dynabead-DNA complex. The beads were washed twice with a washing buffer (buffer 2 from the kit). Finally, the DNA was removed from the beads by resuspension in 20 or 40 μl of 10 mM Tris HCl (pH 8.0; buffer 3 from the kit) and incubation at 65°C for 5 min. The beads, now released from the DNA, were collected with the magnet, and the DNA-containing supernatant was transferred to a fresh tube and used directly in the PCR. The experiment was repeated several times.
DNA isolation from water, skim milk, and unpasteurized whole milk. (i) Dynabead DNA Direct I method.
This technique was identical to isolation of the DNA from pure cultures, except that sample volumes were 1.6 ml and cell pellets were resuspended directly in the Dynabead DNA Direct I bead solution. The experiment was not repeated due to technical difficulties.
(ii) Dynabead modified method.
The Dynabeads (Dynal AS) were used with a modified procedure and buffers (e.g., 4 M guanidine thiocyanate [GTC]–1% Sarkosyl-ethanol) according to the method of Rudi et al. (20). Cell pellets from 1.6 ml of artificially contaminated water, skim milk, or unpasteurized whole milk were subjected to DNA isolation, and the DNA-bead complex was used directly in the PCR. The experiment was not repeated due to technical difficulties.
(iii) Bacterium-binding (BB) bead method.
A total of 1.4 ml of artificially contaminated water, skim milk, or unpasteurized whole milk was added to 5 μl of a 50-mg/ml concentration of BB beads (Genpoint AS, Oslo, Norway) and mixed gently for 15 min at room temperature. Bacteria bound to magnetic beads were then drawn to the wall of the microcentrifuge tube by a magnet (MPC-E; Dynal AS). The supernatant was carefully removed without disrupting the bacterium-bead complex. Then, 50 μl of lysis buffer from the kit was added before incubation at 65°C for 5 min. Next, 100 μl of 96% ethanol was added, and the sample was incubated further for another 5 min. After removing the supernatant, the DNA-bead complex was washed twice with 1 ml of 70% ethanol. The DNA-bead sample was resuspended in 50 μl of H2O and incubated at 65°C for 5 to 10 min to evaporate the residues of ethanol and then used directly in the PCR. The experiment was done once with samples from the first series of artificially contaminated water and milk and four times with samples from the second series of artificially contaminated water and milk.
(iv) Dynabead anti-Listeria beads.
A total of 1.4 ml of artificially contaminated water, skim milk, or unpasteurized whole milk was added to 20 μl of Dynabead anti-Listeria beads (Dynal AS), mixed gently, and incubated for 30 min at room temperature. Bacteria bound to the magnetic beads were then drawn to the wall of the microcentrifuge tube by a magnet (MPC-E; Dynal AS) for 3 min. The supernatant was carefully removed without disrupting the bacterium-bead complex. Then, 50 μl of 4 M GTC–1% Sarkosyl was added, and the suspension was transferred to a new tube before incubated for 10 min at 65°C. Next, 100 μl of 96% ethanol was added, and the sample was incubated for 5 min at room temperature. After removal of the supernatant, the DNA-bead complex was washed twice with 500 μl of 70% ethanol. The DNA-bead sample was resuspended in 50 μl of H2O and transferred to a new tube, and the residual ethanol was removed by evaporation at 65°C for 10 min. The DNA-bead complex was then used directly in the PCR. The experiment was done twice; one set of samples from each series of artificially contaminated water and milk was analyzed.
TaqMan probe and primer design.
GenBank was searched for sequences of the gene encoding listeriolysin O (hlyA). The published sequences were aligned using CLUSTAL W (version 1.7) multiple sequence alignment (23) from the Genetic Computer Group (GCG) program package. An alignment of the gene encoding listeriolysin O (M24199) with the gene encoding streptolysin O from Streptococcus pyogenes (A28468), the gene encoding pneumolysin from Streptococcus pneumoniae (M17717), and the hlyA genes from E. coli O157 (X79839) and Vibrio cholerae (M36855) was carried out. The primers and the probe were selected from a region with 100% homology between the reported L. monocytogenes sequences and with little homology to the sequences reported from the other species. The Primer Express (version 1.0) and guidelines (Perkin-Elmer Research News [Perkin-Elmer, Norwalk, Conn.], 1995) were used for the primer-probe design, together with guidelines from PE Biosystems. The GCG program FastA (17) was used to search for sequence similarities between the selected DNA fragment from the listeriolysin O gene and the sequences reported in GenBank.
TaqMan-based PCR assay.
Amplification reactions (50 μl) contained a DNA sample (1 to 5 μl); 1× TaqMan Buffer A; 5 mM MgCl2; 200 μM dATP, dCTP, and dGTP; 400 μM dUTP; 0.1 μM L. monocytogenes-specific probe; 0.3 μM L. monocytogenes-specific primers (each), and 2.5 U of AmpliTaq Gold DNA polymerase. PCR samples and controls were prepared in triplicates. The reaction tubes were MicroAmp Optical tubes, and the tube caps were MicroAmp optical caps. All consumables were supplied by PE Biosystems.
Before amplification, the PCR mixture was heated to 95°C for 10 min to denature the template DNA. The amplification profile was as follows: 40 cycles of 95°C for 20 s and 60°C for 1 min. Reactions were performed in the ABI Prism 7700 Sequence Detection System (PE Biosystems). Reaction conditions were programmed and data were analyzed on a power Macintosh 4400/20 (Apple Computer, Santa Clara, Calif.) linked directly to the ABI Prism 7700 Sequence Detection System using the SDS 1.6 application software (PE Biosystems) as described by the manufacturer. PCR products were detected directly by monitoring the increase in fluorescence from the dye-labeled L. monocytogenes-specific DNA probe. The TaqMan probe consisted of an oligonucleotide with a 5′-reporter dye and a 3′-quencher dye. The reporter dye, FAM (carboxyfluorescein) was covalently linked to the 5′ end of the oligonucleotide. The fluorescence of the reporter was quenched by TAMRA (6-carboxy-N,N,N′,N′-tetramethylrhodamine), located at the 3′ end. When the probe was intact, the proximity of the reporter dye to the quencher dye resulted in suppression of the reporter fluorescence. If the probe was cleaved, the reporter and quencher dyes were separated, causing the reporter dye fluorescence to increase. The amplification was plotted as ΔRn, which was the normalized reporter signal (reporter signal minus background) against the number of cycles. One then chose a threshold signal, which intersected the amplification curves in the linear region of the semilog plot. This gave threshold cycles (CT), which are defined as PCR cycles in which an increase in fluorescence first occurred, for each amplification plot. Different amplifications could then be compared by their respective threshold cycles. The CT values were plotted against log input and gave standard curves for quantification of unknown samples and possibilities to estimate the amplification efficiency in the reaction (7; Perkin-Elmer Applied Biosystems User Bulletin 2 [ABI PRISM 7700 Sequence Detection System], 1997).
The size of the PCR product was verified with ethidium bromide-stained 2% agarose gels (SeaPlaque GTG Agarose; FMC Bioproducts, Rockland, Maine). Agarose gel electrophoresis was performed essentially as described by Sambrook et al. (21).
RESULTS
PCR fragment specificity.
Specific PCR primers and probe directed against the listeriolysin O gene (hlyA) were designed for L. monocytogenes (Table 1). The probe region was chosen to optimize specificity and amplification efficiency. The 113-bp DNA fragment (positions 1627 to 1740 in GenBank under accession no. M24199) was subjected to a homology search in FastA, which revealed no other identical sequences than those reported for the hlyA gene from L. monocytogenes.
TABLE 1.
Primers and fluorogenic probe for specific detection of L. monocytogenes
| Probe or primer | Sequence (5′–3′) | Denaturation temp (°C)a |
|---|---|---|
| Primers | ||
| Forward | TGC AAG TCC TAA GAC GCC A | 60.3 |
| Reverse | CAC TGC ATC TCC GTG GTA TAC TAA | 60.3 |
| Probe | CGA TTT CAT CCG CGT GTT TCT TTT CG | 70.2 |
Calculated by nearest-neighbor algorithm by the Primer Express program (primer concentrations, 300 nM; probe concentration, 100 nM; salt concentration, 55 nM).
DNA from a total of 65 L. monocytogenes isolates, including the type strain, were analyzed by the 5′-nuclease PCR assay and found to be specific (a signal was achieved using real-time PCR) for the chosen primers and probe. The specificity of the primers and probe were controlled against DNA from 16 Listeria strains of 5 other species and DNA from 18 other species belonging to other genera of phylogenetically related or common foodborne organisms and pathogens (see Materials and Methods), all of which gave no signal during real-time PCR.
PCR amplicons were analyzed on a 2% agarose gel by standard horizontal gel electrophoresis. The experiments showed a fragment of the expected length of 113 bp for L. monocytogenes, while nonspecific PCR products were not detected (results not shown).
5′-Nuclease PCR reaction optimization.
Optimization of PCR was performed to choose the appropriate magnesium concentration yielding the highest intensity of reporter fluorescent signal without sacrificing specificity. The PCR gave identical results at between 3 and 6 mM magnesium (results not shown), and a concentration of 5 mM magnesium was chosen. Different annealing-extension temperatures were also examined. The PCR worked well at all temperatures between 60 and 65°C, but at 65°C a decline in efficiency of the reaction was observed (results not shown). The reaction worked well with smaller reaction volumes (25 μl), since the slopes (−3.49 in 25-μl reactions versus −3.50 in 50-μl reactions) and square regression coefficient (R2 = 0.996 in 25-μl reactions versus 0.993 in 50-μl reactions) of the standard curves were identical.
Using 5 μl of the DNA-BB bead complex in the PCR gave a lower square regression coefficient (R2 = 0.956) than with 1 μl of DNA-BB beads per reaction, while the amplification efficiency showed only small changes (slope of −3.27 versus −3.49).
Analyses of L. monocytogenes in pure cultures.
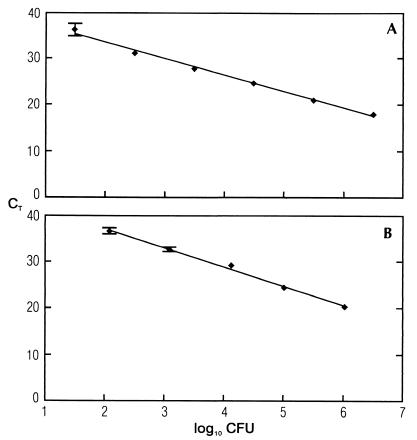
Serial dilutions of purified DNA or cells were made, and two types of standard curves were constructed: DNA standard curves and cell standard curves. For DNA standard curves, DNA isolated from approximately (1.3 ± 0.3) × 108 CFU of L. monocytogenes was diluted 10-fold serially in 1× TE buffer and subjected to PCR. The standard curve based on dilutions of DNA showed a linear relationship between log input DNA and threshold cycles (Fig. 1A). The slope of the curve was −3.56, and the square regression coefficient (R2) after the linear regression was 0.993. When the same DNA was used in a separate PCR experiment, the standard deviations between the two experiments were identical or less than in a triplicate reaction. When DNA from a separate isolation was used, the variations in the amplification efficiency in the standard curves were small (slope of −3.50 versus −3.56), and the square regression coefficient remained constant. For cell standard curves (5.0 ± 0.3) × 107 CFU were diluted into 10-fold dilutions. DNA was isolated from each dilution and subjected to PCR. The standard curve based on five 10-fold dilutions of CFU showed a linear relationship between log input CFU and the threshold cycles (Fig. 1B). The slope of the curve was −4.12, and the square regression coefficient after the linear regression was 0.995. When different DNA isolations were used, the variation in the square regression coefficient in the standard curves was small (R2 = 0.993 versus 0.991), while there was a little more variation in the amplification efficiency (slope of −4.12 versus −3.75).
FIG. 1.
(A) 5′-Nuclease PCR analysis of serial 10-fold dilutions of L. monocytogenes DNA. CT values are plotted against the calculated CFU (i.e., 10-fold dilutions of the bacterial DNA from 3.125 × 106 CFU/μl). The straight line, which is calculated by linear regression (y = −3.56x [CFU] + 40.7) shows a square regression coefficient of R2 = 0.993. The standard deviations based on three PCR reactions are indicated. (B) 5′-Nuclease PCR analysis of serial 10-fold dilutions of L. monocytogenes cells. CT values plotted against the number of CFU of L. monocytogenes. Template DNA was extracted from samples of cells containing serial 10-fold dilutions of approximately (5.0 ± 0.3) × 107 CFU of L. monocytogenes. The straight line, which is calculated by linear regression (y = −4.12x [CFU] + 45.5) shows a square regression coefficient of R2 = 0.995. The standard deviations based on three PCR reactions are indicated.
The detection limit of the PCR assay was approximately 15 ± 10 CFU per PCR (Fig. 1A) using Dynabead DNA Direct I. The detection limit of the DNA isolation was investigated, and DNA was occasionally detected when DNA was purified from a total of 125 CFU (Fig. 1B). However, purification from less than 500 CFU sometimes resulted in false-negative results.
Analyses of L. monocytogenes in water.
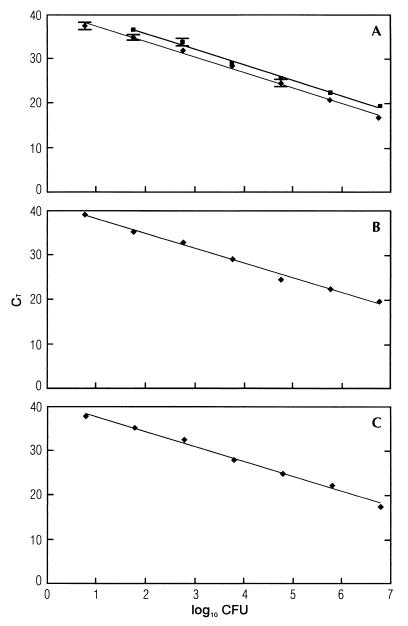
Both BB beads and Dynabead anti-Listeria beads were used successfully to obtain DNA from L. monocytogenes in water. Using both methods, the cell standard curve based on seven 10-fold dilutions showed a linear relationship between log input CFU and the threshold cycles (Fig. 2A). The reproducibility was good (Table 2), but two of the experiments with BB beads showed shorter quantification areas (6 logs) and lower square regression coefficients. When BB beads were used, a detection limit in water of 60 CFU per PCR was observed. However, in some of the experiments 6 CFU per PCR could be detected. This corresponds to a detection limit for the DNA purification method between 200 and 2,000 CFU per ml. The Dynabead anti-Listeria beads gave a detection limit of 60 CFU per PCR in water.
FIG. 2.
5′-Nuclease PCR analysis of serial 10-fold dilutions of L. monocytogenes cells in water (A), skim milk (B), and unpasteurized whole milk (C) purified by BB beads (Genpoint AS) and Dynabead anti-Listeria (Dynal AS). CT values are plotted against the CFU of L. monocytogenes. Template DNA was extracted from samples containing serial 10-fold dilutions of approximately (2.9 ± 0.3) × 108 CFU of L. monocytogenes. The straight lines calculated by linear regression in Dynabead anti-Listeria beads (y = −3.55x [CFU] + 42.8 [■]) (A) and in BB beads (y = −3.33x [CFU] + 44.4 [A], y = −3.49x [CFU] + 43.9 [B], and y = −3.40x [CFU] + 44.1 [⧫] [C]) show square regression coefficients for BB beads of R2 = 0.994 (A) and for Dynabead anti-Listeria beads of R2 = 0.995 (A), R2 = 0.995 (B), and R2 = 0.993 (C). The standard deviations based on three PCRs are indicated.
TABLE 2.
Regression analysis of PCR results of L. monocytogenes isolated from water and milk
| Bead type | Expt no. | Watera
|
Skim milk
|
Unpasteurized whole milk
|
|||
|---|---|---|---|---|---|---|---|
| Slope | R2 | Slope | R2 | Slope | R2 | ||
| BB beads | 1 | −3.33* | 0.994 | −3.49 | 0.991 | −3.40 | 0.993 |
| 2 | −3.03 | 0.991 | −3.28 | 0.991 | −3.14 | 0.984 | |
| 3 | −3.59 | 0.960† | −3.49 | 0.995 | −3.19 | 0.995 | |
| 4 | −3.47 | 0.967† | −3.55 | 0.991 | −3.58 | 0.996 | |
| Dynabead anti- Listeria beads | 1 | −3.55 | 0.995 | NDb | ND | ND | ND |
| 2 | −3.61 | 0.988 | |||||
Symbols: *, values calculated from seven 10-fold dilutions; †, values calculated from six 10-fold dilutions.
ND, no data.
Attempts to isolate DNA from L. monocytogenes in water using the Dynabead DNA Direct I method and the Dynabead modified method were abandoned due to the incapability of the two methods to isolate DNA from L. monocytogenes in milk.
Analyses of L. monocytogenes from skim milk and unpasteurized whole milk.
The BB beads were employed successfully in isolating DNA from L. monocytogenes from both skim milk and unpasteurized whole milk. The cell standard curve based on seven 10-fold dilutions of L. monocytogenes cells showed a linear relationship between log input CFU and the threshold cycles (Fig. 2B and C). The isolation of L. monocytogenes from milk was reproducible (Table 2). In skim milk and unpasteurized whole milk, both detection limits using BB bead DNA isolation were approximately 6 CFU per PCR.
Dynabead anti-Listeria beads could isolate L. monocytogenes from both types of milk samples. However, no cell standard curves could be made because of low linear correlation between the 10-fold dilutions (results not shown). The detection limit was 6,000 CFU per PCR in skim milk and 60 CFU per PCR in unpasteurized whole milk.
The Dynabead DNA Direct I method and the Dynabead modified method were not successful during the attempt to purify bacterial DNA from skim milk or unpasteurized whole milk spiked with L. monocytogenes. Experiments with Dynabead DNA Direct I gave a positive PCR signal for the most concentrated sample of skim milk only and no signal at all from the L. monocytogenes contaminated unpasteurized whole milk. The Dynabead modified method caused extensive binding of the beads to the ingredients in the milk, and the beads could not even be separated from the milk.
DISCUSSION
Specific detection and quantification of L. monocytogenes.
There is a requirement for rapid, quantitative, and accurate measurements of target organisms responsible for food poisoning. In the present study a 5′-nuclease system was constructed and applied to specifically detect and quantify L. monocytogenes. The preferable targets for detecting pathogens are pathogen determinants. The listeriolysin O gene (hlyA) is common in virulent and/or pathogenic strains of L. monocytogenes (11). The 5′-nuclease PCR assay was therefore directed against a 113-bp fragment in this gene. The specificity of the primers and probe was confirmed both by homology searches in nucleotide databases and by screening a number of L. monocytogenes strains, five other Listeria species, and several species of common foodborne organisms and pathogens.
The square regression coefficients after the linear regressions indicated a good correlation between the amount of template (log input CFU) and the amount of product (represented by the CT values) in the standard curves from pure cultures. The slopes in both standard curves from pure cultures showed that the amplification efficiencies were slightly lower than 1 (18;) Perkin-Elmer Applied Biosystems User Bulletin 2 [ABI PRISM 7700 Sequence Detection System], 1997). The linearity of the standard curves and the observation that the PCR operates with a constant efficiency confirm that the assay is well suited for quantitative measurements (7). The detection limit for the PCR was estimated to be approximately 15 ± 10 CFU/PCR. Our reported limit of detection is similar to those of other reports using fluorogenic 5′-nuclease PCR assay for endpoint detection. Bassler et al. (1) obtained a detection level of approximately 50 CFU of L. monocytogenes/PCR, while Chen et al. (2) showed a detection limit of as low as 2 CFU/PCR from a pure culture of Salmonella enterica serovar Typhimurium.
Quantitative DNA purification from pure cultures was carried out using the Dynabead DNA Direct I system, both because of the reproducibility and because of the simplicity of the protocol (19). As for most other DNA purification strategies, the Dynabead DNA Direct purification strategy is designed for DNA isolation from cell cultures and tissues (where the amount of material is not limited), and the manufacturer recommends the use of 2 × 107 to 2 × 108 bacteria for isolation. The Dynabead DNA Direct strategy is based on the coaggregation of beads and DNA (19). This aggregate may not form properly at low cell concentrations and may be the reason for the poor DNA recovery when isolating DNA from fewer than 500 cells. In the study of Rudi et al. (20), this was overcome by utilizing a guanidine thiocyanate and Sarkosyl lysis buffer and by ethanol precipitation in the Dynabead isolation of cyanobacteria, and a detection limit of less than 50 cells was obtained.
We therefore tried various approaches for DNA isolation from the artificially contaminated material using different kinds of magnetic beads and different methods. The most promising approach was to employ the recently developed BB beads. L. monocytogenes isolated from water with BB beads, resulted in cell standard curves with amplification efficiencies very close to 1. However, the reproducibility is not optimal and can probably be improved by modifying the buffer system to decrease the tension between the beads and the water. L. monocytogenes isolated from water by Dynabead anti-Listeria beads gave an amplification efficiency similar to that of the DNA standard curve when isolating L. monocytogenes from pure cultures with Dynabead DNA Direct I. The BB beads were most satisfying when it came to the quantitative detection of L. monocytogenes in milk, the cell standard curves amplification efficiencies being close to 1, and the reproducibility was very high. The square regression coefficients confirmed that the assay was well suited for quantification. However, the DNA-bead samples isolated from unpasteurized milk were difficult to handle due to strong tension between the DNA-bead complex and the plastics in the tubes and tips. This may be possible to improve, e.g., by modifying buffers. The detection limit of the BB beads was approximately 200 CFU per ml in skim milk and unpasteurized whole milk. This is a good recovery compared to, e.g., standard methods such as extractions with organic solvents (21). In addition, by isolating bacteria from larger volumes of the matrix and perhaps by manipulating these, along with the amounts of beads used and the incubation period, the detection limit may be improved further.
Dynabead anti-Listeria could be used to isolate L. monocytogenes from milk samples. Dynabead anti-Listeria beads could isolate DNA from 2,000 cells per ml in unpasteurized milk, while skim milk showed a detection limit of 2 × 105 CFU per ml. However, the cell standard curves from milk samples were not good enough for the quantification of L. monocytogenes.
The Dynabead DNA Direct I and Dynabead modified methods failed both in the isolation of L. monocytogenes from skim milk and in the isolation of L. monocytogenes from unpasteurized whole milk due to technical difficulties with the methods and therefore could not give quantitative measurements of cell concentrations in the artificial contaminated milk samples. These methods were therefore abandoned, and the experiments were not completed since the purpose was to develop a method to be used on several matrixes.
Dynabeads are monosized polystyrene spheres with superparamagnetic properties. The Dynabead anti-Listeria beads are precoated with high-affinity antibodies against surface markers of live Listeria. The Dynabead DNA Direct I relies on the adsorption of released DNA to the surface of the Dynabeads during a brief incubation. BB beads are magnetic polymer beads based on polyvinyl alcohol. The surface is modified and claimed to be unique for the adsorption of bacteria and subsequent DNA purification. To a certain extent, raw milk is an oil-in-water emulsion, but the fat globules are more complicated than emulsion droplets, and milk is a dilute aqueous solution and so behaves accordingly. About four-fifths of the protein consists of casein present as caseinate, which means that it binds cations, primarily calcium and magnesium. Lactose is the distinctive carbohydrate in milk. The mineral substances are primarily K, Na, Ca, Mg, Cl, and phosphate, and 87.1% of the average content in milk is water (24).
Milk components will probably influence the isolation and separation of bacteria, but the interactions need to be elucidated. Both beads and milk are hydrophilic, but since the binding properties of the various beads are not known, the impact of the various components in milk are difficult to evaluate.
Concerns connected to DNA-based methods.
There are several concerns regarding the quantification of bacteria using DNA-based methods. Different DNA extraction methods are likely to affect the yield of DNA and therefore the detection limit of the method. In principle, PCR can detect one DNA gene copy if the DNA preparation is pure and the amplification conditions are optimized. Here we have noted a detection limit of 15 ± 10 CFU/PCR in pure cultures, which is estimated from the uncertainty in the numbers of colony-forming cells determined based on standard plating methods. Estimating such a detection limit is based on the assumption that the number of genome copies is equal to the number of CFU. For microorganisms, the number of genome copies per cell depends on the growth rate. In these particular experiments, the cultures were stationary (109 CFU/ml), and the probability of more than one genome per cell was low. The numbers of dead cells were considered insignificant, since the single-colony culture never was older than 24 h before use. We therefore assume that the DNA purification methods used give relatively good recovery and that the standard deviations for the variation in DNA recovery will be small compared to that of the cell counts. In principle, rapid growth of L. monocytogenes in foods may give several genome copies per cell and blur the exact number of organisms in the sample, but normally, when it comes to the detection of pathogens in foods, the growth rate of bacteria is usually limited, and the errors obtained by 5′-nuclease PCR will probably be lower than those obtained via traditional methods.
Another general aspect using DNA-based techniques is how to differentiate between living and dead cells in a sample (8, 13). This could be a matter of concern, especially in ready-to-eat products and other foods subjected to previous heat treatment. It is therefore important to know the history of the food before interpreting the results and to consider additional or complementary tests if required. One method to consider is the mRNA technique (22), which can be done in a similar 5′-nuclease reverse transcription-PCR (RT-PCR) (5). An earlier study presented a 5′-nuclease RT-PCR targeting the hlyA transcript as a marker for living cells (15). However, due to difficulties in cataloging the nature and physiological history of the cells, Norton and Batt (15) concluded that to make a quantitative assay of viability based upon RNA would be difficult. When it comes to the identification of the contamination source and the determination of infectious dosage, e.g., in water and unpasteurized milk, the aspect of living versus dead cells is not very troublesome, since one would assume that the contaminating L. monocytogenes organisms are alive. As mentioned above, the problem with L. monocytogenes is usually growth in food during storage, even at low temperatures, which means that the problem with detecting dead cells is of minor concern, if one assumes that the growth will result in living cells greatly outnumbering the dead cells. Moreover, an initial screening is preferable, since this will tend to over-report rather than under-report the detection of L. monocytogenes. The consequences of having false-positive results are that the samples are either not used or are analyzed further. The consequences of having false-negative results, on the other hand, are that potentially infectious material will be used, e.g., for human consumption. Work is currently in progress to reduce the background arising from dead cells in similar analyses.
Future developments.
We present here a method for quantifying L. monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk, based on magnetic bead separation and DNA purification and 5′-nuclease PCR analysis. The detection limits and quantitative aspects demonstrated here are within the limits of what is practical for monitoring L. monocytogenes in contaminated foods. Adapting 5′-nuclease technology for the quantification of L. monocytogenes in other foods should be feasible. Since paramagnetic beads are easy to manipulate in automated systems (A. Holmberg, A. Deggerdal, and F. Larsen, AMS '95: 3rd Int. Conf. Automation Mapping DNA Sequencing, abstr. A10, 1995), integrated cell concentration and DNA purification methods should be suited for high-throughput assays. Recently, there have also been efforts in the miniaturization of 5′-nuclease systems (10) and the integration of different processing steps (3, 25). Because of the microscopic size of the beads and the possibilities of performing 5′-nuclease PCR at the nanoliter scale (12), paramagnetic beads and real-time PCR may also be valuable tools in future miniaturized systems.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Ingvild Rosshaug, Anne Gulliksen, and Tove Maugesten. We are very grateful to Liv-Marit Rørvik, Department of Pharmacology, Microbiology and Food Hygiene, Norwegian College of Veterinary Medicine, who provided 60 isolates of L. monocytogenes and some of the other Listeria strains. We appreciated the advice of Lars Melin, PE Biosystems Sweden, when designing the primers and the probe of L. monocytogenes.
This work was financed by the Research Levy on certain agricultural products.
REFERENCES
- 1.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S, Yee A, Griffiths M, Larkin C, Yamashiro C T, Behari R, Paszko-Kolva C, Rahn K, De Grandis S A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol. 1997;35:239–250. doi: 10.1016/s0168-1605(97)01241-5. [DOI] [PubMed] [Google Scholar]
- 3.Cheng J, Sheldon E L, Wu L, Uribe A, Gerrue L O, Carrino J, Heller M J, O'Connell J P. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 4.Deener H G, Boychuk I. Species-specific detection of Listeria monocytogenes by DNA amplification. Appl Environ Microbiol. 1991;57:606–609. doi: 10.1128/aem.57.2.606-609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert R J. Provisional microbiological guidelines for some ready-to-eat foods sampled at point of sale: notes for PHLS Food Examiners. PHLS Microbiol Digest. 1992;9:98–99. [Google Scholar]
- 7.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 8.Herman L. Detection of viable and dead Listeria monocytogenes by PCR. Food Microbiol. 1997;14:103–110. [Google Scholar]
- 9.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim M S, Lofts R S, Jahrling P B, Henchal E A, Weedn V W, Northrup M A, Belgrader P. Real-time microchip PCR for detecting single-base differences in viral and human DNA. Anal Chem. 1998;70:2013–2017. doi: 10.1021/ac971091u. [DOI] [PubMed] [Google Scholar]
- 11.Jay J M. Foodborne listeriosis. In: Heldman D R, editor. Modern food microbiology. 5th ed. New York, N.Y: Chapman & Hall; 1996. pp. 478–506. [Google Scholar]
- 12.Kalinina O, Lebedeva I, Brown J, Silver J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997;25:1999–2004. doi: 10.1093/nar/25.10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKillip J L, Jaykus L A, Drake M. Nucleic acid persistence in heat-killed Escherichia coli O157:H7 from contaminated skim milk. J Food Prot. 1999;62:839–844. doi: 10.4315/0362-028x-62.8.839. [DOI] [PubMed] [Google Scholar]
- 14.Mengaud J, Vicente M-F, Chenevert J, Pereira J M, Geoffroy C, Gicquel-Sanzey B, Baquero F, Perez-Diazand J-C, Cossart P. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect Immun. 1988;56:766–772. doi: 10.1128/iai.56.4.766-772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norton D-M, Batt C A. Detection of viable Listeria monocytogenes with a 5′ nuclease PCR assay. Appl Environ Microbiol. 1999;65:2122–2127. doi: 10.1128/aem.65.5.2122-2127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen J E, Aabo S, Hill W, Notermans S, Wernars K, Granum P E, Popovic T, Rasmussen H N, Olsvik O. Probes and polymerase chain reaction of food-borne bacterial pathogens. Int J Food Microbiol. 1995;28:1–78. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 17.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raeymaekers L. Quantitative PCR: theoretical considerations with practical implications. Anal Biochem. 1993;214:582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- 19.Rudi K, Kroken M, Dahlberg O J, Deggerdal A, Jakobsen K S, Larsen F. Rapid, universal method to isolate PCR-ready DNA using magnetic beads. BioTechniques. 1997;22:506–511. doi: 10.2144/97223rr01. [DOI] [PubMed] [Google Scholar]
- 20.Rudi K, Larsen F, Jakobsen K S. Detection of toxin-producing cyanobacteria by use of paramagnetic beads for cell concentration and DNA purification. Appl Environ Microbiol. 1998;64:34–37. doi: 10.1128/aem.64.1.34-37.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 22.Sheridan G E, Masters C I, Shallcross J A, MacKey B M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walstra P, Geurts T J, Noomen A, Jellema A, von Boekel M A J S. Dairy technology: principles of milk properties and processes. New York, N.Y: Marcel Dekker; 1999. Composition, structure, and properties; pp. 3–26. [Google Scholar]
- 25.Waters L C, Jacobson S C, Kroutchinina N, Khandurina J, Foote R S, Ramsey J M. Microchip device for cell lysis, multiplex PCR amplification, and electrophoretic sizing. Anal Chem. 1998;70:158–162. doi: 10.1021/ac970642d. [DOI] [PubMed] [Google Scholar]