Abstract
A 81-kDa protein from Mycobacterium sp. strain PYR-1 was expressed in response to exposure of the strain to the polycyclic aromatic hydrocarbon pyrene and recovered by two-dimensional gel electrophoresis. The N-terminal sequence of the protein indicated that it was similar to catalase-peroxidase. An oligonucleotide probe designed from this sequence was used to screen a genomic library of Mycobacterium sp. strain PYR-1, and a positive clone, containing a part of the gene encoding the 81-kDa protein, was isolated. A gene-walking technique was used to sequence the entire gene, which was identified as katG for catalase-peroxidase. The deduced KatG protein sequence showed significant homology to KatGII of Mycobacterium fortuitum and clustered with catalase-peroxidase proteins from other Mycobacterium species in a phylogenetic tree. The katG gene was expressed in Escherichia coli to produce a protein with catalase-peroxidase activity. Since the induction of this catalase-peroxidase occurred in pyrene-induced cultures and the exposure of these cultures to hydrogen peroxide reduced pyrene metabolism, our data suggest that this enzyme plays a role in polycyclic aromatic hydrocarbon metabolism by strain PYR-1.
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants, some of which are highly carcinogenic, genotoxic, and a threat to public health (5). Many PAHs, including phenanthrene, benzofluoranthene, pyrene, benz[a]anthracene, benzo[a]pyrene, and chrysene, are therefore listed on the U.S. Environmental Protection Agency's priority-pollutant list (13). Degradation of PAHs by indigenous microorganisms seems to play an important role in the removal of contaminants from the environment, particularly from subsurface material and groundwater (4, 23, 25). Due to the high cost associated with trapping, incinerating, and physically removing toxic chemicals from the environment, bioremediation technologies are being developed to clean up PAH-contaminated environments (19). Especially, nocardioform actinomycetes, including Mycobacterium spp., seem to be involved in the degradation of high-molecular-weight PAHs in soil and sediments (2, 6, 8).
Studies of the molecular basis of PAH degradation mechanisms in Mycobacterium species are lacking but extremely important for better understanding and application of these organisms for bioremediation. In previous studies, Mycobacterium sp. strain PYR-1 has been shown to mineralize anthracene, fluoranthene, pyrene, 1-nitropyrene, phenanthrene, and benzo[a]pyrene (8–10, 14, 15, 20, 24). Mycobacterium sp. strain PYR-1 is known to have an inducible system for PAH degradation (8). Our initial approach to identifying genes involved in PAH degradation in strain PYR-1 was to recover proteins produced upon exposure of the strain to pyrene during pyrene metabolism. In this article, we report the cloning, expression, and characterization of a PAH-inducible catalase-peroxidase gene, katG, from Mycobacterium sp. strain PYR-1.
MATERIALS AND METHODS
Strains, chemicals, and culture media.
All bacterial strains, vectors, and plasmids used in this study are listed in Table 1. The culture media were prepared according to procedures described previously (8, 14). Bacteriological media and reagents were purchased from Becton Dickinson, Co., Franklin Lakes, N.J. All solvents and other chemicals used were of the highest purity. Pyrene was obtained from Aldrich Chemical Company (Milwaukee, Wis.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Mycobacterium sp. strain PYR-1 | Mineralizes high-molecular-weight PAHs such as fluoranthene, pyrene, and phenanthrene | 8 |
| Escherichia coli XL1-Blue MRF′ | lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Escherichia coli XLOLR | lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] Su− (nonsuppressing) λr | Stratagene |
| Escherichia coli TOP10 | One Shot competent cells, F′ mcrA Δ(mmr-hsdRMS-mcrBC) Φ80lacZΔM15 Δ(ara-leu)7697 | Invitrogen |
| Plasmid or vector | ||
| ExAssist helper phage | For in vivo excision of the pBK-CMV phagemid from the ZAP Express vector with E. coli XLOLR | Stratagene |
| Zap Express vector | Lambda vector, prokaryotic and eukaryotic expression, in vivo excision of the pBK-CMV vector | Stratagene |
| pBK-CMV vector | Neor Kanr, ColE1 origin, lacZ, cytomegalovirus promoter | Stratagene |
| Myco-pBK-CMV phagemid | pBK-CMV phagemid with 1.8-kb insert from Mycobacterium sp. strain PYR-1 | This study |
| pBAD/Thio-TOPO vector | Ampr pMB1 origin, six-His-Thioredoxin open reading frame, arabinose induced | Invitrogen |
| katG-pBAD/Thio plasmid | pBAD/ThioFusion with the katG gene of Mycobacterium strain PYR-1 | This study |
PAH induction of the Mycobacterium sp. cultures and two-dimensional (2D) gel electrophoretic analysis of recovered proteins.
Mycobacterium sp. strain PYR-1 was grown in 30 ml of minimal basal salts with nutrient broth (8) in 125-ml Erlenmeyer flasks at 30°C for 48 h with shaking at 150 rpm. The culture was used as a source of inocula (15 ml each) for 2-liter Erlenmeyer flasks containing 800 ml of the same medium and incubated as described above. Pyrene (50 mg/ml) was dissolved in N,N-dimethylformamide, and 80 μl was added to one of the flasks at 24, 48, 72, and 96 h. The control culture had similar incubation conditions except that pyrene was not added. After 120 h of incubation, the cells were harvested by centrifugation and the pellets were washed three times with 10 mM Tris-HCl, pH 7.4, and resuspended in 5 to 8 ml of Tris-HCl buffer. The cells were disrupted by sonication for 15 min at 30-s intervals with the intensity set at 60 (Sonics and Materials, Inc., Newton, Conn.). Tergitol NP-40 (U.S. Biochemicals-Amersham, Cleveland, Ohio) was then added at a concentration of 1%. The lysate was centrifuged at 12,000 × g for 30 min, and the supernatant was centrifuged at 50,000 × g for 90 min. The protein concentration in the supernatant was determined with a protein assay kit (Bio-Rad, Hercules, Calif.) based on the Bradford dye-binding procedure (3).
2D polyacrylamide gel electrophoresis (PAGE) was used to separate and recover proteins from PAH-induced and uninduced cultures (11). About 300 μg of total protein was used for 2D PAGE (Bio-Rad PROTEAN II xi 2-D Cell and Slab Cell) according to the instructions in Bio-Rad Bulletin 1144. The polyacrylamide gel concentrations used were 3.5% in the isoelectric-focusing gel and 7.5% in the second-dimension slab gel. Proteins were visualized by staining with 0.025% Coomassie blue R-250 and quantified by scanning densitometry (NEC model 466; Scanalytics). The proteins on the 2D gel from the PAH-induced culture sample were transferred to a polyvinylidene difluoride membrane and sent to Midwest Analytical Inc. (St. Louis, Mo.) for N-terminal amino acid sequence analysis.
Construction of a genomic library.
Genomic DNA of Mycobacterium sp. strain PYR-1 was digested with BamHI and then separated by 1% agarose gel electrophoresis. The digested 0.7- to 10.0-kb DNA fragments were cut out from the agarose gel and purified with a gel extraction kit (Qiagen, Inc., Valencia, Calif.). The calf intestinal alkaline phosphatase-treated and BamHI-predigested ZAP Express vector (Stratagene Cloning Systems, La Jolla, Calif.) was ligated with these digested DNA fragments. The ligated DNA was packaged in bacteriophage λ, using Gigapack III Gold Packaging Extract according to the instructions of the manufacturer (Stratagene).
Design of an oligonucleotide probe and genomic library screening.
The oligonucleotide probe (P81) was designed from the N-terminal amino acid sequence of the PAH-induced (81-kDa) protein, labeled with a digoxigenin (DIG) oligonucleotide 3′-end labeling kit according to the instructions of the manufacturer (Boehringer Mannheim Co., Indianapolis, Ind.), and used to screen the genomic library. Escherichia coli strain XL1-Blue MRF′ was used to plate the phage library. A plaque lift hybridization procedure (Boehringer Mannheim Co.) was used to screen for the positive clone. DIG Easy Hyb was used as the hybridization solution. The prehybridization time was 2 h at 42°C, and the hybridization time was 16 h at 42°C. Positive plaques were directly picked with a toothpick and plated for purification and confirmation at a low plaque density. After the second screening, the purified positive plaques were plated again for confluent growth, so that a high-titer (approximately 1010 phage/ml) stock of the clone could be obtained and stored at −80°C. In vivo excision protocols were used to produce phagemid clones by using ExAssist Helper Phage with the XLOLR strain (Stratagene Cloning Systems). A Qiaprep spin miniprep kit (Qiagen) was used for phagemid or plasmid preparation. Myco-pBK-CMV phagemid DNA (1 μl, 0.5 μg/μl) was applied to a nylon membrane for dot blot hybridization with the DIG-labeled P81 probe for confirmation. The hybridization conditions were the same as those of the plaque lift hybridization procedure.
DNA sequencing.
The Myco-pBK-CMV phagemid was sequenced first by using the primers Zap-F (5′-CACAGGAAACAGCTATGACC) and Zap-R (5′-CCGCTCTAGAAGTACTCTCG), which were located on the phagemid vector upstream and downstream, respectively, of the insert. An ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.) was used for sequencing according to the manufacturer's instructions. An automatic ABI Prism 310 sequencer was used for electrophoresis. After the insert sequence information from both ends was obtained, the complete insert sequence was further deduced by primer walking. Oligonucleotide primers and probes were purchased from Universal DNA, Inc. (Tigard, Oreg.).
A gene-walking method (7) was modified and used in order to sequence the entire gene. It consists of a DNA-pooling step and a PCR step. For the DNA-pooling step, 1 μg of pUC19 plasmid DNA was first digested with 8 to 12 U each of the following enzymes: BamHI, EcoRI, HindIII, HincII, KpnI, PstI, SacI, SalI, SphI, and XmaI (Promega, Madison, Wis.) in a 10-μl volume at 37°C for 2 h. The reaction mixture was then treated with 1 μl of calf intestinal alkaline phosphatase (20 U/μl; Promega) at 37°C for 30 min. At the same time, 2 μg of genomic DNA of Mycobacterium sp. strain PYR-1 was also digested with the restriction enzymes used to digest the pUC19, in a total volume of 30 μl. The digestions were stopped by heating the reaction mixture at 70°C for 5 min. The digested vector and Mycobacterium DNA were combined and loaded on a 1% agarose gel for electrophoresis. The 1.0- to 9.0-kb DNA fragments containing pUC19 and Mycobacterium DNA were cut out from the agarose gel and purified with a gel extraction kit (Qiagen). The purified DNA fragments were dried by a Speed-Vac concentrator (Savant, Farmingdale, N.Y.) and then resuspended in a reaction mix tube containing 1 μl of T4 DNA ligase (3 U/μl; Promega), 1 μl of 10× ligase buffer (Promega), and 8 μl of water and ligated at 4°C overnight. For the PCR step in gene walking, 2 μl of each DNA pool was added to 23 μl of a PCR mixture as described previously (24). Three primers specific for Mycobacterium strain PYR-1 were used in this study for three rounds of gene walking, and their locations are DNA sequence positions 1678 to 1697 (primer 1), positions 2685 to 2703 (primer 2), and positions 3088 to 3106 (primer 3). Two vector-directed primers, 5′-GGTTTTCCCAGTCAGACG (M13-F) and 5′-CACAGGAAACAGCTATGACC (M13-R), were also used for gene walking. The amplification conditions were one cycle of 95°C for 2 min and then 35 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 2 min, followed by one cycle of 72°C for 8 min, and then the reaction mixture was cooled to 4°C. The PCR products from specific gene walking were directly cloned into a pBAD/Thio-TOPO cloning vector by following the instructions of the manufacturer (Invitrogen, Carlsbad, Calif.), and the clones were used for sequencing in the ABI Prism 310 sequencer.
Phylogenetic analysis.
DNA sequence analysis, translation, and alignment with related genes and proteins were carried out using the computer programs Lasergene (DNASTAR, Inc., Madison, Wis.) and Align Plus (Scientific Educational Software, State Line, Pa.). The GenBank program BLAST (1) was used to find similar genes or proteins. The computer program Megalign (DNASTAR, Inc.) was used to construct a phylogenetic tree by comparison of closely related protein sequences.
Construction of plasmids for overexpression of the gene.
After the gene sequence was determined, the full gene was amplified by PCR from the clone containing the 1.8-kb DNA insert. The PCR product was cloned into the pBAD/Thio-TOPO vector (Invitrogen) according to the manufacturer's instructions. The colonies with recombinant pBAD/ThioFusion plasmids were selected by plating the transformants on ampicillin (50 μg/ml)-Luria broth plates. Positive clones containing a katG-pBAD/Thio plasmid with katG in the correct orientation were screened by PCR using a forward primer, Trx (5′-TTCCTCGACGCTAACCTG-3′), in the insert and a pBad reverse primer (5′-GATTTAATCTGTATCAGG-3′) in the vector (pBAD reverse primer, provided in the Invitrogen kit).
Purification of six-His-tagged recombinant protein.
The pBAD/ThioFusion clones were inoculated into Luria broth-ampicillin (50 μg/ml) medium and cultured to an optical density at 600 nm of ∼0.5. Arabinose was added to a final concentration of 0.02%. After 4 or 16 h of incubation, the cells were collected by centrifugation. The pellet was suspended in buffer B (8 M urea, 0.1 M Na-phosphate, 0.01 M Tris [pH 8.0]), followed by three freeze-thaw cycles. The cleared supernatant (0.6 ml) was loaded on the Ni-nitrilotriacetic acid (NTA) column (Qiagen). The column was centrifuged for 2 min at 700 × g and then washed three times with 0.6 ml of buffer C (same as buffer B but pH 6.3). The six-His-tagged recombinant protein was eluted from the column twice with 0.2 ml of buffer E (same as buffer B but pH 4.5).
Catalase-peroxidase activity test.
To determine the activity of the KatG enzyme, a native gel was used instead of a sodium dodecyl sulfate (SDS)-polyacrylamide gel. After electrophoresis, the gel was washed once with phosphate-buffered saline solution and then stained in a solution containing 30 ml of phosphate-buffered saline (PBS), 30 mg of 3,3′-diaminobenzidine tetrahydrochloride (DAB), and 30 μl of H2O2 for more than 30 min at room temperature. The reacted DAB causes a brown color to appear at the site of the catalase-peroxidase (21).
Response to oxygen stress.
The ability of hydrogen peroxide to induce resistance to oxygen stress was determined as described by Sherman et al. (21). Briefly, Mycobacterium sp. strain PYR-1 was exposed to 100 μM hydrogen peroxide for 3 to 5 h before the addition of a lethal dose (5 mM). Viability was determined by dilution and plating at 1 h and at 3 h after the lethal dose.
Measurement of pyrene metabolism.
The ability of Mycobacterium sp. strain PYR-1 to remove pyrene from the culture medium was monitored spectrophotometrically (22). Complete solubilization of PAHs was accomplished by mixing 2 volumes of culture aliquot with 1 volume of dimethyl sulfoxide before centrifugation at 16,000 × g for 10 min. The absorbance of the supernatant fluids obtained from duplicate cultures was measured at 335 nm for pyrene. Supernatants from cultures receiving equivalent volumes of dimethyl sulfoxide and a methanol carrier were used as blanks. Abiotic removal of pyrene was checked by using boiled cell suspensions incubated similarly. Spectrophotometric results were confirmed by quantifying the amount of pyrene by reversed-phase high-performance liquid chromatography using a C18 column (3.9 by 300 mm). A linear gradient of 50 to 95% methanol in water was developed over 40 min at 1 ml/min. Pyrene was identified by comparing characteristic absorption spectra (from 200 to 400 nm) and retention times to authentic pyrene, using a Waters 910 photodiode array detector with data display, and by analysis using Waters version 2.10 Millennium software. Pyrene was quantified by comparison with the amount of an internal standard of naphthyl myristate (10 μg; 28 nmol) added immediately before threefold extraction of the complete aliquot with ethyl acetate.
Nucleotide sequence accession number.
The katG gene and protein sequences were deposited in the GenBank database under accession number AF207899.
RESULTS AND DISCUSSION
Protein profiles of Mycobacterium sp. strain PYR-1 grown with and without pyrene.
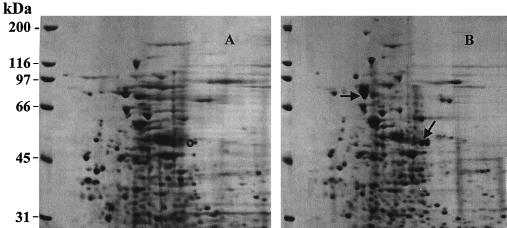
A time course analysis for the expression of pyrene-metabolizing proteins revealed induction 2 h after incubation with pyrene and was followed by complete metabolism of pyrene in 8 h (data not shown). Comparisons concerning the presence and absence of proteins when the organism was grown with and without pyrene were made by 2D SDS-PAGE analysis and revealed two major and several minor overexpressed proteins (Fig. 1). One of the overexpressed proteins had a molecular mass of 81 kDa. The other major protein, which had a molecular mass of 50 kDa and which was identified as a dioxygenase, will be reported in another paper. The protein at 70 kDa was used as a standard of comparison between the pyrene-induced and uninduced cultures because it showed only minor variation when it was quantitated by scanning densitometry. This result suggests that the increases seen in the proteins are absolute changes in expression.
FIG. 1.
2D gel of uninduced and pyrene-induced proteins of Mycobacterium sp. strain PYR-1. (A) proteins of uninduced Mycobacterium sp. strain PYR-1; (B) proteins of pyrene-induced Mycobacterium sp. strain PYR-1. The arrows indicate the PAH-induced proteins, and the circles indicate the same positions in the uninduced sample.
Identification of PAH-induced proteins.
The identity of an 81-kDa protein, produced upon exposure of the strain to pyrene, was studied by transferring the proteins separated by 2D PAGE to membranes and by N-terminal protein sequencing. The N-terminal sequence of the P81 protein resulted in the identification of 24 amino acids: PEATEHPPIGEAQTEPAQSGCPMV. A GenBank homology search revealed that the 24-amino-acid sequence had significant homology to the N terminus of catalase-peroxidase of Mycobacterium fortuitum.
Screening of the genomic library, DNA sequencing, and gene walking.
A genomic library of Mycobacterium strain PYR-1 was prepared and had a titer of 6 × 106 PFU/ml. Almost 99.5% of the library consisted of recombinant phages. An oligonucleotide probe (P81) was designed from the N-terminal sequence of the 81-kDa PAH-induced protein. The probe (P81), which has the sequence CCNGCNCARAGYGGNTGYCCNATGGT (the N is either G, A, T, or C; the R is A or G; and the Y is T or C), represents the codons for PAQSGCPMV (amino acids 16 to 24). The P81 probe was labeled with DIG and used for screening the genomic library. Several positive clones were obtained. Phagemids were prepared of each clone, and the expected sequence was reconfirmed by dot blot hybridization, using the DIG-labeled P81 probe (data not shown). One clone, containing a DNA insert of 1.8 kb, was further analyzed. The nucleotide sequence analysis of the clone confirmed the presence of the 26 bases of the oligonucleotide probe (P81) and the 72 bases of the codons of the 24 amino acids. However, only 155 bases from the 5′ end of the P81 gene were present at the 3′ terminus of the inserted DNA sequence. According to the molecular weight of the induced protein, the P81 gene should be at least 2.2 kb. To clone the full gene for this 81-kDa PAH-induced protein, we used three rounds of gene walking as described in Materials and Methods. A 3,974-bp DNA sequence, including the 1,829-bp DNA insert was obtained from the positive phage clone by gene walking.
The GenBank similarity search showed that the induced 81-kDa protein was similar to catalase-peroxidase of Mycobacterium fortuitum and other mycobacterial enzymes and that in each case the gene was katG. The calculated molecular mass for the deduced KatG protein was 80.84 kDa, which was close to the 2D gel position of the protein at 81 kDa. KatG of Mycobacterium sp. strain PYR-1 is closer to KatGII, but not to KatGI, of Mycobacterium fortuitum (17). However, it clustered with the catalase-peroxidase proteins from other Mycobacterium species (26).
Expression, purification, and enzyme activities of the recombinant proteins.
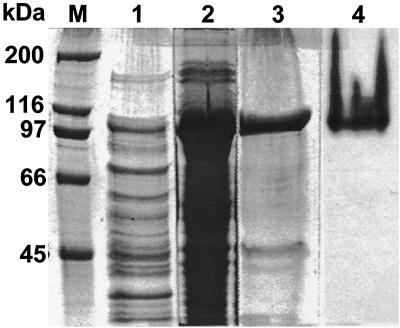
The katG of Mycobacterium sp. strain PYR-1 was subcloned in E. coli. A clone containing the katG-pBAD/Thio plasmid with katG in the correct orientation was obtained and used to produce the recombinant protein for the catalase-peroxidase of Mycobacterium sp. strain PYR-1 (Fig. 2). Most of the recombinant protein was in the pellet, but some was in the supernatant (lanes 1 and 2 of Fig. 2). The six-His-tagged recombinant protein was purified with a Ni-NTA spin column (lanes 3 and 4 of Fig. 2).
FIG. 2.
Expression or recombinant KatG protein for catalase-peroxidase of Mycobacterium sp. strain PYR-1 in E. coli. Lane M, protein size markers; lanes 1 to 3, SDS-polyacrylamide gel stained with Coomassie blue to show the total proteins; lane 1, sample from the supernatant; lane 2, sample from the pellet; lane 3, partially purified recombinant protein from the pellet; lane 4, purified recombinant protein from the supernatant stained with DAB to show catalase-peroxidase enzyme activity.
The pBAD/Thio-TOPO system produces a recombinant protein with horseradish peroxide-thioredoxin (13 kDa) as an N-terminal fusion partner of the cloned gene product in the center and a six-His tag (3 kDa) as a C-terminal fusion partner. Thus, the recombinant protein should have the molecular mass of the gene product plus 13 and 3 kDa. The recombinant protein for Mycobacterium strain PYR-1 KatG had a molecular mass of 97 kDa (Fig. 2); that is, 81 kDa for KatG, 13 kDa for hydrogen peroxide-thioredoxin, and 3 kDa for the tag. Purification of the recombinant protein with Ni-NTA resin confirmed that the recombinant protein contained the six-His tag and that the determined sequence was correct in the open reading frame. The purified recombinant protein from the supernatant of the broken cells was stained with DAB in a native gel and gave a brown color reaction, indicating that the recombinant protein had catalase-peroxidase activity (lane 4 of Fig. 2).
The expression of the 81-kDa catalase-peroxidase found upon exposure of the strain to pyrene was unexpected. To gain insight into the physiological relevance of the KatG protein, uninduced and pyrene-induced cells were exposed to hydrogen peroxide to determine any effect of the protein on resistance to oxidative stress as well as to determine the effect of oxidative stress on the metabolism of the PAH. The conditions of exposure of uninduced cells to hydrogen peroxide followed those employed by Sherman et al. (21) in their study of oxidative stress in mycobacteria. Exposure to 100 μM hydrogen peroxide for 3 h overexpressed the 81-kDa protein by twofold, as was detected and quantified by densitometric scanning of 2D gels. This exposure also resulted in complete protection against a challenge dose of 5 mM hydrogen peroxide that resulted in a 4-log-unit reduction in CFU (106 to 102 CFU) of uninduced cells, also measured after 3 h. Our results are analogous to those obtained by Sherman et al. (21) for Mycobacterium smegmatis, indicating that Mycobacterium sp. strain PYR-1 manifests an OxyR-like response to oxidative stress, coincident with overexpression of the 81-kDa KatG homologue. However, the expression of this protein upon exposure to pyrene resulted in an intermediate level of protection (106 to 104 CFU) from oxygen stress compared to that obtained by exposure to 100 μM hydrogen peroxide. These results indicate that the OxyR-mediated response of this organism to hydrogen peroxide involves other proteins, which are not induced by the PAH in addition to the 81-kDa homologue.
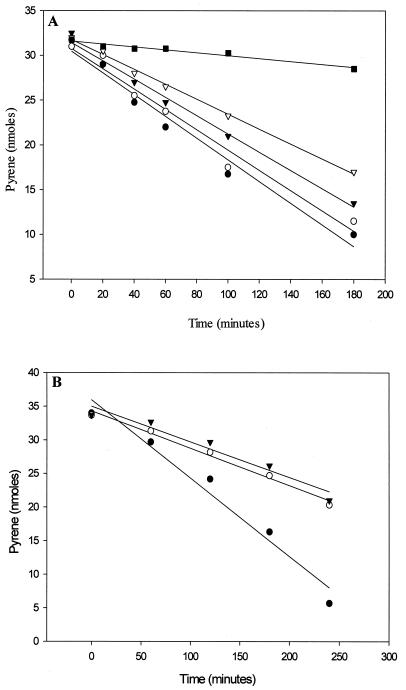
Exposure of pyrene-induced cells to hydrogen peroxide at concentrations up to 2 mM did not affect cell viability as measured by CFU. Doses of hydrogen peroxide from 50 μM to 2 mM added to pyrene-induced cells were employed to determine the effect of oxidative conditions on the metabolism of the PAH (Fig. 3). As shown in Fig. 3A, addition of 100 to 200 μM hydrogen peroxide decreased the metabolism of pyrene over a 3-h period, suggesting that PAH metabolism by this organism is moderately sensitive to redox conditions. Moreover, a 10-fold increase in hydrogen peroxide (1 and 2 mM) reduced the metabolism of pyrene by more than 50% over a 4-h period (Fig. 3B) but did not abolish its metabolism completely. These results indicate that metabolism of the PAH by this organism can withstand highly oxidizing conditions.
FIG. 3.
Effect of hydrogen peroxide on pyrene metabolism by Mycobacterium sp. strain PYR-1. Each point is the average of duplicate determinations. (A) ●, control pyrene incubation (32 nmol); ○, pyrene plus 0.05 mM H2O2; ▾, pyrene plus 0.1 mM H2O2; , pyrene plus 0.2 mM H2O2; ■, heat-inactivated cells plus pyrene. (B) ●, control pyrene inculation (34 nmol); ○, pyrene plus 1 mM H2O2; ▾, pyrene plus 2 mM H2O2.
The 81-kDa KatG homologue may have either one or both of two roles in preserving optimal PAH metabolism and cell viability. First, the enzyme may protect the dioxygenase from oxidative inactivation by exogenous oxidant because our results with whole cells are analogous to the inhibition of purified naphthalene dioxygenase from Pseudomonas sp. (16) and mammalian cytochrome P450 monooxygenase (12) by hydrogen peroxide. Second, the enzyme may be induced to remove endogenous hydrogen peroxide generated as an intermediate in PAH metabolism. Indeed, it has been described that the uncoupling of highly purified naphthalene dioxygenase in vitro leads to hydrogen peroxide production (16). Further, the essential removal of H2O2 has been shown for Brevibacterium fuscum by the production of a protein that has both dioxygenase and catalase activities (18). Along with the results on overexpression of this protein upon exposure to hydrogen peroxide, our results are consistent with the production of H2O2 during dioxygenase activity and show that the removal may be essential for optimal enzyme function in PAH-metabolizing cells.
ACKNOWLEDGMENTS
We thank E. B. Russel for background induction data, Saeed A. Khan and John B. Sutherland for valuable discussion, and Pat Fleischer for clerical assistance.
This work was supported by U.S. Environmental Protection Agency cooperative agreement CR820773.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacteriumsp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Curr Opin Biotechnol. 1993;4:331–338. [Google Scholar]
- 5.Environmental Protection Agency. Evaluation and estimation of potential carcinogenic risks of polynuclear aromatic hydrocarbons. Washington, D.C.: Environmental Protection Agency; 1985. [Google Scholar]
- 6.Grosser R J, Warshawsky D, Vestal J R. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl Environ Microbiol. 1991;57:3462–3469. doi: 10.1128/aem.57.12.3462-3469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison R W, Miller J C, D'Souza M J, Kampo G. Easy gene walking. BioTechniques. 1997;22:650–653. doi: 10.2144/97224bm17. [DOI] [PubMed] [Google Scholar]
- 8.Heitkamp M A, Franklin W, Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol. 1988;54:2549–2555. doi: 10.1128/aem.54.10.2549-2555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitkamp M A, Freeman J P, Miller D W, Cerniglia C E. Pyrene degradation by a Mycobacteriumsp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988;54:2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heitkamp M A, Cerniglia C E. Polycyclic aromatic hydrocarbon degradation by a Mycobacteriumsp. in microcosms containing sediment and water from a pristine ecosystem. Appl Environ Microbiol. 1989;55:1968–1973. doi: 10.1128/aem.55.8.1968-1973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochstrasser D F, Harrington M G, Hochstrasser A C, Miller M J, Merril C R. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 12.Karuzina I I, Archakov A I. Hydrogen peroxide-mediated inactivation of microsomal cytochrome P450 during monoxygenase reactions. Free Radic Biol Med. 1994;17:557–567. doi: 10.1016/0891-5849(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 13.Keith L H, Telliard W A. Priority pollutants I—a perspective view. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 14.Kelley I, Cerniglia C E. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacteriumstrain, PYR-1. J Soil Contam. 1995;4:77–91. [Google Scholar]
- 15.Kelley I, Freeman J P, Evans F E, Cerniglia C E. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacteriumsp. Appl Environ Microbiol. 1991;57:636–641. doi: 10.1128/aem.57.3.636-641.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K. Benzene-induced uncoupling of naphthalene dioxygenase activity and enzyme inactivation by production of hydrogen peroxide. J Bacteriol. 1999;181:2719–2725. doi: 10.1128/jb.181.9.2719-2725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menéndez M C, Ainsa J A, Martin C, Garcia M J. katGI and katGII encode two different catalases-peroxidases in Mycobacterium fortuitum. J Bacteriol. 1997;179:6880–6886. doi: 10.1128/jb.179.22.6880-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller M A, Lipscomb J D. Homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum: a dioxygenase with catalase activity. J Biol Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 19.Mueller J G, Cerniglia C E, Pritchard P H. Bioremediation of environments contaminated by polycyclic aromatic hydrocarbons. In: Crawford R L, Crawford D L, editors. Bioremediation: principles and applications. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 125–194. [Google Scholar]
- 20.Rafii F, Selby A L, Newton R K, Cerniglia C E. Reduction and mutagenic activation of nitroaromatic compounds by a Mycobacteriumsp. Appl Environ Microbiol. 1994;60:4263–4267. doi: 10.1128/aem.60.12.4263-4267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry III C E, Stover C K. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuttleworth K L, Cerniglia C E. Practical methods for the isolation of polycyclic aromatic hydrocarbon (PAH)-degrading microorganisms and the determination of PAH mineralization and biodegradation intermediates. In: Crawford R L, editor. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1996. pp. 766–775. [Google Scholar]
- 23.Sutherland J B, Rafii F, Khan A A, Cerniglia C E. Mechanisms of polycyclic aromatic hydrocarbon degradation. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: Wiley-Liss; 1995. pp. 269–306. [Google Scholar]
- 24.Wang R-F, Luneau A, Cao W-W, Cerniglia C E. PCR detection of polycyclic hydrocarbon-degrading mycobacteria. Environ Sci Technol. 1996;30:307–311. [Google Scholar]
- 25.Wilson S C, Jones K C. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ Pollut. 1993;81:229–249. doi: 10.1016/0269-7491(93)90206-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]